Abstract
Hybrid zones are natural experiments that expose the forces maintaining species differences. But for cases where a trait of one of the hybridizing pair appears shifted into the range of the other, the underlying mechanism can be difficult to infer. For example, hybridization between hermit warbler (Dendroica occidentalis) and Townsend's warbler (Dendroica townsendi) is restricted to narrow hybrid zones in Washington and Oregon, yet hermit mtDNA can be found in phenotypically pure Townsend's populations up to 2000 km north along the Pacific coast. This could reflect introgression of selectively favoured hermit mitochondria north across the hybrid zones, or a neutral genetic wake left behind following southern zone movement. Hermit mitochondrial haplotypes in populations of coastal Townsend's exhibit relatively high genetic diversity and significant divergence from those found in populations of hermit warblers. This contradicts the predictions of selective introgression, but is consistent with a northern population of hermits diverging in a glacial refugium before being replaced by Townsend's via aggressive hybridization. Previous field studies showing Townsend's males to be competitively superior to hermit males support this scenario, and suggest that the extreme hybrid zone movement evidenced by the hermit mitochondrial wake represents an extinction in progress.
Keywords: hybrid zone, selective sweep, genetic wake, glacial refugia
1. Introduction
Hybrid zones, regions of range overlap and interbreeding between differentiated populations, have been valued as ‘natural laboratories’ for the study of speciation (Hewitt 1988; Harrison 1990). Patterns of gene flow across hybrid zones have been used to identify traits contributing to reproductive isolation—phenotypic and genetic markers exhibiting abrupt clines across zones are inferred to be important in maintaining species boundaries, while those exhibiting broad clines are not (Barton & Hewitt 1985). Less clear is the significance of traits whose clines are shifted far into the geographical range of one of the hybridizing pair. Such shifts may reflect selective introgression of adaptive traits across stationary hybrid zones (Parsons et al. 1993; Payseur et al. 2004), or genetic wakes of neutral markers left behind following hybrid zone movement (Gyllensten & Wilson 1987; Marchant et al. 1988; Rohwer et al. 2001). For zones not known to be moving, there has been no formal attempt to test these competing hypotheses. Because zone movement is expected to be transitory (Barton & Hewitt 1985), having a method to detect past zone movement in zones that have ceased to move or are moving too slowly to measure could offer valuable insight into the evolutionary impacts of hybrid zone movement (Buggs 2007).
Dramatic examples of non-coincidence have been observed between mitochondrial and nuclear genetic clines, with numerous cases of mitochondrial genetic clines centred hundreds to thousands of kilometres away from hybrid zones (Ferris et al. 1983; Martinsen et al. 2001; Rohwer et al. 2001; McGuire et al. 2007). Mitochondrial genetic wakes are expected when hybrid zone movement is driven by interspecific differences in male competitive ability resulting in males of the more aggressive species mating with females of the less competitive species, leaving the maternally inherited mtDNA of the less aggressive species behind as the range of the competitively superior species expands (Wirtz 1999). This interpretation assumes selective neutrality of the mtDNA left behind. However, introgression of selectively favoured interspecific mtDNA across a stationary hybrid zone can also explain large-scale shifts of mtDNA relative to nuclear clines (Dasmahapatra et al. 2002). Both genetic wake and selective introgression scenarios have been proposed to explain the 2000 km distance between the centres of mitochondrial genetic and phenotypic clines in Townsend's and hermit warblers (Rohwer et al. 2001; Dasmahapatra et al. 2002).
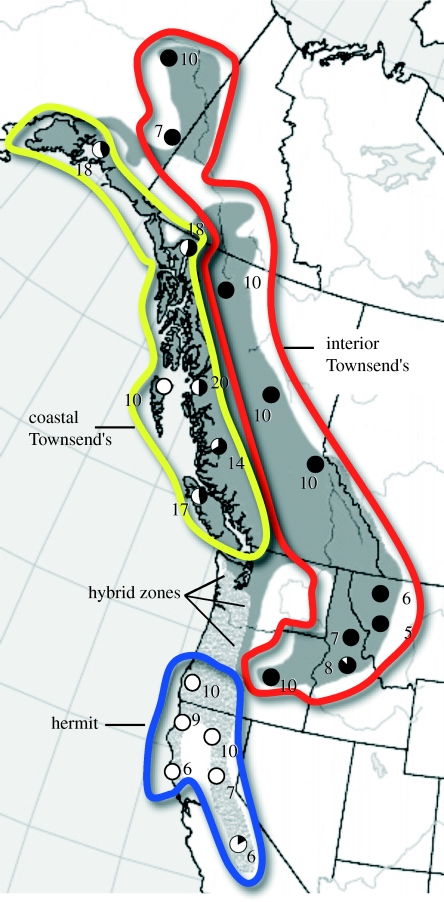
Townsend's and hermit warblers are recently diverged sister species that meet to form three geographically isolated hybrid zones in Washington and Oregon, USA (Bermingham et al. 1992; Rohwer & Wood 1998; Lovette et al. 1999; Rabosky & Lovette 2008). Hermit warblers south of the hybrid zones and Townsend's warblers east of the coastal mountain ranges are fixed for distinct mtDNA restriction fragment length polymorphism (RFLP) clades (Lovette et al. 1999; Rohwer et al. 2001), while coastal populations of Townsend's warbler north of the hybrid zones carry both sets of haplotypes (figure 1; Rohwer et al. 2001). Several features of the system suggested to earlier authors that this pattern reflects a mitochondrial genetic wake generated by southward movement of the zone. First, asymmetric male plumage clines across the hybrid zones suggest that Townsend's males disperse into these zones more successfully than hermit males (Rohwer & Wood 1998; Rohwer et al. 2001). Experimental studies support this contention: Townsend's males have higher testosterone levels and superior fighting ability compared with hermit males (Pearson & Rohwer 2000; Owen-Ashley & Butler 2004). Also, the presence of fixed mitochondrial RFLP haplotypes between hermit and interior Townsend's populations and the estimated date of divergence (ca 400 000 years) of the two clades suggested speciation following isolation in separate coastal and interior glacial refugia, respectively (Lovette et al. 1999; Rohwer et al. 2001). The postglacial range expansion routes of coastal and interior fir-dominated forests in which the two would have diverged suggests that secondary contact between hermit and Townsend's warblers would have occurred ca 5000 years ago, far north of the current hybrid zones (Pielou 1991; Rohwer et al. 2001).
Figure 1.
Sampling localities and sample sizes for 226 hermit and Townsend's warblers. Hermit breeding range is shaded light grey, Townsend's dark grey. Coloured lines encircle the three populations under analysis: coastal Townsend's in yellow, interior Townsend's in red and hermit in blue. Pie diagrams reflect the frequency of ND2 mitochondrial haplotype clades within populations: white pie slices correspond to clade A and black to clade B.
Two distinct mtDNA clades are found in these species (Bermingham et al. 1992; Lovette et al. 1999), and both clades are found in Townsend's warblers (Rohwer et al. 2001). Hermit populations far from the hybrid zones carry only clade A haplotypes and interior Townsend's populations carry only clade B haplotypes, but both clades are found in phenotypically pure coastal Townsend's populations north of the hybrid zones (figure 1; Rohwer et al. 2001). If the clade A haplotypes found in coastal Townsend's populations were derived from selective introgression of hermit mtDNA across a stationary hybrid zone, they should bear the genetic signature of such an event. Selective sweeps accompany a rapid rise in frequency of a mutation under strong positive selection: genetic variation near the locus under selection is erased, as closely linked loci also move to high frequency via genetic hitchhiking, ‘sweeping’ away variation around the locus for the population receiving the adaptive mutation (Maynard Smith & Haigh 1974; Kaplan et al. 1989). Because the mitochondrial genome does not recombine, selective introgression of hermit mtDNA into coastal Townsend's populations across a stationary hybrid zone would result in clade A haplotypes in coastal Townsend's exhibiting low levels of haplotype and nucleotide diversity and few unique mutations, relative to clade A haplotypes found in hermits (Maynard Smith & Haigh 1974; Kaplan et al. 1989).
However, if the clade A haplotypes north of the hybrid zone represent a genetic wake of hermit mtDNA incorporated into Townsend's warblers as the hybrid zone moved south, the resulting genetic pattern should depend on the population history of the replaced hermit warblers. If hermits rapidly expanded north from a southern coastal refugium before being replaced by Townsend's warblers, then we might find a pattern virtually indistinguishable from that produced by a selective sweep. Rapid population expansion from restrictive glacial refugia often results in a pattern of low genetic diversity (Hewitt 1996, 2000), similar to that produced by a selective sweep. However, a growing body of evidence suggests that forested glacial refugia existed north of the hybrid zones on the islands of British Columbia and southeast Alaska (Warner et al. 1982; Zink & Dittmann 1993; Fleming & Cook 2002; Burg et al. 2005). If hermit warblers were present in these northern coastal refugia or if they expanded slowly northwards before being replaced by Townsend's warblers, clade A haplotypes found in coastal Townsend's should not exhibit the lack of diversity expected in the case of a selective sweep (Hewitt 1996; Ibrahim et al. 1996). Indeed, had hermits persisted in northern refugia, then clade A haplotypes from the north might exhibit unique haplotypes and divergence from clade A haplotypes found in pure hermit populations south of the hybrid zone (Hewitt 1996). Thus, relatively high levels of genetic variation and divergence in the clade A haplotypes found in coastal Townsend's populations would be strong evidence against selective introgression as the underlying mechanism for the non-coincidence between the mitochondrial and phenotypic clines.
2. Material and methods
(a) Sampling and DNA sequencing
The complete NADH dehydrogenase subunit 2 gene (ND2) was sequenced from 226 hermit and Townsend's warblers from 22 locations (table S1 in the electronic supplementary material) comprising two transects: (i) a coastal transect from central California to Valdez, Alaska and (ii) an interior transect east of the coastal mountain ranges and mostly west of the Rocky Mountains, from eastern Oregon to Fairbanks, Alaska (figure 1). The hybrid zones themselves were excluded from the study, and no hybrids were found in any of the populations sampled beyond the zones. From each coastal Townsend's locality, we attempted to sequence 10 individuals with clade A haplotypes and 10 with clade B haplotypes (Rohwer et al. 2001), though in several cases available sample sizes could not accommodate this design (figure 1 and table S1 in the electronic supplementary material).
Whole genomic DNA was isolated from frozen muscle tissue using the Qiagen DNAeasy Tissue kit (Valencia, CA, USA). ND2 was amplified using primers L5216 (5′-GGCCCATACCCCGRAAATG-3′) and H6313 (5′-ACTCTTRTTTAAGGCTTTGAAGGC-3′; Sorenson et al. 1999). PCR was run at a total volume of 25 μl using standard protocols and a negative control included with all reactions. PCR products were prepared for sequencing using Exonuclease I and Shrimp Alkaline Phosphatase, then cycle sequenced using each amplification primer and two internal primers, L5758-W (5′-CTAGGYTGAATRGCWATYATAAT-3′) and H5766-W (5′-GAGGAAGATGGTTGCGGT-3′), modified from Sorenson et al. (1999), and the ABI BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). Sequence products were purified using standard ethanol precipitation, and then run on an ABI 3100 (Applied Biosystems) automated sequencer. Sequences were aligned automatically using Sequencher 3.0 (Gene Codes, Ann Arbor, MI, USA) and alignment confirmed by eye. GenBank accession numbers are FJ373895-FJ374120.
(b) Analysis
A haplotype network was constructed to investigate the evolutionary relationships among inter- and intraspecific ND2 haplotypes. Haplotype networks provide the most appropriate representation of the evolutionary relationships of haplotypes within species because, unlike strictly bifurcating trees, population genealogies are often multifurcated and reticulated, and descendant haplotypes coexist with persistent ancestral haplotypes (Posada & Crandall 2001). Haplotype networks are depicted as circles representing unique haplotypes, where the size of a circle reflects the frequency of that haplotype and lines between circles represent single mutations between haplotypes. The sister-species status of hermit and Townsend's warblers (Lovette et al. 1999; Rabosky & Lovette 2008) and the sharing of haplotypes among hermits and coastal Townsend's populations (Rohwer et al. 2001) suggested that a network approach would be most appropriate for analysis of haplotype relationships within and between these species. Haplotype networks were constructed via the median-joining method implemented in Network v. 4.2 (Bandelt et al. 1999).
We estimated coalescent-based divergence times and migration rates using Mdiv (Nielsen & Wakeley 2001). Mdiv uses a Bayesian approach to simultaneously approximate the posterior distribution of three parameters: the population parameter theta (θ=2Neμ); divergence time between populations (T= tdiv/2Ne); and the migration rate between populations since divergence (M=2Nem). We constrained parameters within ranges of θ (0, ∞), M (0, 30) and T (0, 20), and used a Markov chain of 5 000 000 steps preceded by a burn-in of 1 000 000 steps for each population comparison. We ran Mdiv three times for each population pair using different random number seeds to assess the stability of the results. After viewing the results of initial runs, maximum M was raised to 100 and chain length to 10 000 000 steps for clade B haplotypes found in interior versus coastal Townsend's populations. Estimates of T were converted to real time using a range of mtDNA mutation rate estimates for birds (1.0–2.5×10−8 substitutions per site per year; Arbogast et al. 2002; Lovette 2004; Weir & Schluter 2008).
We used the software package Arlequin v. 2.0 (Schneider et al. 2000) to make pairwise Fst comparisons (using 1000 permutations to evaluate significance), to estimate haplotype and nucleotide diversity (Nei 1987) and its 95% CI (1000 replicates), and to calculate Tajima's D (1989a,b) and evaluate its significance using coalescent simulations. A skew in the distribution of polymorphic sites towards rare haplotypes, as indicated by a large negative value of Tajima's D (1989a,b), is a fundamental expectation of selective sweeps (Braverman et al. 1995). However, negative values of Tajima's D are also expected for populations that have undergone recent expansions (Tajima 1989a,b), as expected for populations occupying previously glaciated regions. To better estimate the relative sizes of demographic expansions among groups we used the software package Fluctuate v. 1.3 (Kuhner et al. 1998) to generate coalescent-based maximum-likelihood estimates of population growth (g). Estimates of g>3SD(g) are considered to indicate population expansion (Lessa et al. 2003). We ran Fluctuate separately for each of the four groups, using a transition/transversion ratio of 15, which we estimated using Modeltest v. 3.0.6 (Posada & Crandall 1998). We started each run using Watterson's estimate of θ (Watterson 1975) and ran 20 short chains of 20 000 steps and two long chains of 200 000 steps. We ran each group a minimum of three times using independent random number seeds to guarantee convergence of chains.
3. Results
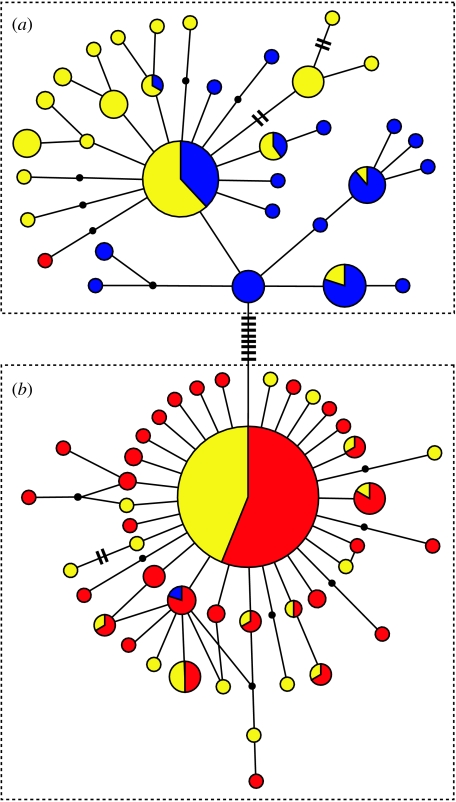
The ND2 haplotype network consists of two mtDNA clades (figure 2), separated by 0.77 per cent sequence divergence. With the exception of one individual (from Placerville, CA, USA; table S1 in the electronic supplementary material), all hermit warblers sampled carry haplotypes from clade A (figures 1 and 2). Interior samples of Townsend's warblers, also with the exception of one individual (from Lowman, ID, USA; table S1 in the electronic supplementary material), carry only haplotypes from clade B (figures 1 and 2). Neither of the two individuals that violated this pattern was a hybrid, but their relatively close proximity to the hybrid zones suggests that they most probably reflect recent hybridization and dispersal events. With the exception of the Queen Charlotte Island population (which carries only clade A haplotypes), all coastal populations of Townsend's warblers included in this study carry both clade A and B haplotypes (figures 1 and 2).
Figure 2.
ND2 haplotype network, divided into (a) clade A and (b) clade B. Haplotypes carried by hermit warblers are coloured blue, coastal Townsend's warblers yellow and interior Townsend's warblers red. The size of each circle reflects the number of individuals sharing that haplotype, a line between two haplotypes represents a single mutational difference, and hash marks reflect the number of mutational differences between haplotypes separated by more than one mutation. Small black dots indicate inferred haplotypes that were not sampled.
Visual inspection of the haplotype network reveals clade A to have dramatically more structure than clade B (figure 2), which exhibits the star-shaped pattern suggestive of rapid population expansion following a population bottleneck (Slatkin & Hudson 1991). Clade A haplotypes found in coastal populations of Townsend's warblers exhibit relatively complex structure and an abundance of unique, non-singleton haplotypes (figure 2).
Pairwise Fst comparisons show significant divergence between clade A haplotypes found in hermit warblers and coastal Townsend's populations, but show no divergence between clade B haplotypes in interior and coastal Townsend's (table 1). Maximum-likelihood estimates of the time of divergence between clade A haplotypes found in coastal Townsend's and hermit warblers are 54 755–136 878 years (table 2). Clade B haplotypes found in coastal and interior Townsend's populations, however, are estimated to have experienced high migration and virtually no divergence (table 2).
Table 1.
Population pairwise Fst values. Values of Fst may vary from 0 (absence of differentiation between populations) to 1 (complete differentiation). *p<0.05 following 1000 permutations of haplotypes among populations.
| clade A | clade B | |||
|---|---|---|---|---|
| hermit | coastal Townsend's | interior Townsend's | coastal Townsend's | |
| clade A | ||||
| hermit | 0.000 | — | — | — |
| coastal Townsend's | 0.188* | 0.000 | — | — |
| clade B | ||||
| interior Townsend's | 0.849* | 0.844* | 0.000 | — |
| coastal Townsend's | 0.844* | 0.835* | −0.005 | 0.000 |
Table 2.
Divergence time and migration rate estimates. A coalescence model was used to estimate maximum-likelihood values of θ (2Neμ), M (2Nem), and T (tdiv/2Ne) (Nielsen & Wakeley 2001), and divergence time estimates in years were based on a range of mtDNA mutation rate estimates for avian taxa (0.01–0.025 substitutions per site per million years). Parenthetical values correspond to standard deviations calculated from the mean of three independent Markov Chain Monte Carlo runs.
| θ (s.d.) | M (s.d.) | T (s.d.) | 0.01 (ss−1Myr−1) | 0.025 (ss−1Myr−1) | |
|---|---|---|---|---|---|
| clade A versus clade B | 14.54 (0.08) | 0.04 (0.01) | 0.81 (0.06) | 565 677 | 226 271 |
| clade A: hermit versus coastal Townsend's | 5.70 (0.11) | 1.17 (0.04) | 0.50 (0.05) | 136 878 | 54 755 |
| clade B: interior versus coastal Townsend's | 8.50 (0.27) | 48.07 (5.66) | 0.001 (0.00) | 408 | 163 |
Summary statistics show that clade A haplotypes in coastal Townsend's populations feature levels of haplotype and nucleotide diversity similar to those found in populations of pure hermits south of the Washington hybrid zones (table 3). Clade B haplotypes found in interior populations of Townsend's feature higher haplotype diversity than found in Clade B haplotypes from coastal Townsend's (table 3).
Table 3.
Genetic diversity and population expansion indices for ND2 haplotypes. N is sample size, Nh is number of haplotypes, h is haplotype diversity, π is nucleotide diversity and its 95% CI, D is Tajima's D with *p<0.05, and g is the growth parameter (with s.d.) from the coalescent model generated with Fluctuate (Kuhner et al. 1998).
| N | Nh | h | π | D | g | |
|---|---|---|---|---|---|---|
| clade A | 99 | 31 | 0.922 | 0.00278±0.00163 | −1.96* | |
| hermit | 46 | 17 | 0.897 | 0.00222±0.00138 | −1.49* | 3655 (898) |
| coastal Townsend's | 52 | 18 | 0.896 | 0.00270±0.00161 | −1.60* | 1479 (318) |
| interior Townsend's | 1 | 1 | n.a. | n.a. | n.a. | n.a. |
| clade B | 127 | 42 | 0.790 | 0.00145±0.00098 | −2.48* | |
| interior Townsend's | 81 | 31 | 0.838 | 0.00149±0.00100 | −2.42* | 5165 (320) |
| coastal Townsend's | 45 | 19 | 0.694 | 0.00140±0.00097 | −2.33* | ≥10 000 (0) |
| hermit | 1 | 1 | n.a. | n.a. | n.a. | n.a. |
Negative values of Tajima's D were seen across all groups (table 3). Tajima's D values were similar for clade A haplotypes found in coastal Townsend's and hermit populations, but negative values of Tajima's D in clade B were twice as large as those found in clade A. Clade B haplotypes in coastal and interior Townsend's had significantly higher g values than clade A haplotypes in hermits or coastal Townsend's (table 3). Clade A haplotypes found in coastal Townsend's were estimated to have a rate of growth approximately half that found in pure hermit populations, and an order of magnitude smaller than the rate found in clade B haplotypes in coastal Townsend's (table 3).
4. Discussion
Our results suggest very different demographic histories for clade A and B. Clade B haplotypes bear strong signals of rapid demographic expansion following a population bottleneck: a star-shaped haplotype network; low haplotype and nucleotide diversity; and a large negative value of Tajima's D. By contrast, clade A haplotypes suggest a more stable population over time, with a relatively complex haplotype network, higher levels of haplotype and nucleotide diversity, and a negative Tajima's D value that is half as large as that for clade B. These differences are consistent with the hypothesis that Townsend's were isolated in the relatively smaller and lower quality glacial refugia available east of the Rockies, while hermit warblers were isolated in the relatively larger and higher quality refugium along the Pacific coast (Rohwer et al. 2001).
This history of isolation is further supported by marked differences in levels of diversity and divergence exhibited by clade A and B haplotypes across populations. Mdiv and Fst analyses reveal no divergence between clade B haplotypes in interior and coastal Townsend's, and a high migration rate between the two. Further, diversity of clade B haplotypes is lower in coastal than interior Townsend's and the estimate of population growth for coastal haplotypes is twice as high as it is for interior Townsend's. These results are consistent with a postglacial expansion of Townsend's from the interior to the coast, followed by a dramatic coastal expansion. By contrast, clade A haplotypes in coastal Townsend's feature numerous unique, non-singleton haplotypes (figure 2). Mdiv and Fst analyses confirm that clade A haplotypes in coastal Townsend's are significantly divergent from those in hermits. Mdiv estimates the age of this divergence to be 54 755–136 878 years (table 2), corresponding to early Wisconsin glaciation (Dawson 1992). This is consistent with an isolated population of hermits diverging in a northern glacial refugium before being replaced by Townsend's populations expanding onto the coast from the interior.
Had hermit warblers rapidly expanded north into coastal British Columbia and Alaska from a southern glacial refugium before being replaced by Townsend's warblers, the expected pattern of low genetic diversity and divergence would be indistinguishable from a selective sweep accompanying selective introgression of clade A haplotypes from hermit warblers into costal populations of Townsend's warblers across a stationary hybrid zone. However, the high levels of genetic diversity and divergence that characterize clade A haplotypes in coastal Townsend's soundly reject a selective sweep as the source of these haplotypes. These results also reject other alternative explanations, such as asymmetric dispersal of hermit females or neutral diffusion of hermit haplotypes north across a stationary hybrid zone: neither could have produced the diversity and divergence of clade A haplotypes in coastal Townsend's warblers.
Yet, there is one alternative explanation that deserves more careful treatment. We have assumed that hermit and Townsend's were reciprocally monophyletic before introgression introduced clade A haplotypes from hermit populations into coastal populations of Townsend's warblers. However, haplotype sharing between populations may also reflect retention of ancestral polymorphism (Funk & Omland 2003). If this were true in this system, then coastal Townsend's warblers would have retained both ancestral haplotype lineages, while interior Townsend's sorted for clade B and hermits sorted for clade A. Our results offer a compelling argument against this scenario. First, ancestral polymorphism retention by Townsend's warblers should not be limited to coastal populations, yet extensive sampling of interior Townsend's populations reveals only a single clade A haplotype in interior Townsend's populations (figure 1). Second, had only coastal populations of Townsend's retained both clades, one should expect clade A and B haplotypes in coastal Townsend's populations to reflect similar demographic histories. Yet, our results clearly point to different histories for the two clades in coastal Townsend's, implying that they were present in different ancestral populations. The lack of divergence between interior and coastal clade B haplotypes, and the congruence of their genetic diversity and growth indices, all suggest that the two sets of clade B haplotypes were carried in the same ancestral population. Our new evidence suggests this population was Townsend's warblers that expanded from an interior glacial refugium, as proposed by Rohwer et al. (2001). Retention of ancestral polymorphism is unlikely to have produced such dramatic demographic contrasts between clade A and B haplotypes in coastal Townsend's warblers.
Our novel yet simple approach has yielded strong support for a genetic wake, offering, to our knowledge, the first rigorous evidence for historical hybrid zone movement in a zone not directly shown to be moving. These results provide valuable insight into the evolutionary impacts of hybrid zone movement. The distance of movement implied by our results is far beyond that seen in any other system (Buggs 2007), partly because measurement was not limited to empirical observation. Also, while many empirically observed cases of hybrid zone movement seem to be driven at least in part by anthropogenic habitat change or global warming (Buggs 2007), the hybrid zone between hermit and Townsend's has been moving south, in direct contrast to the northern movement predicted by postglacial climate amelioration or more recent global warming. Both the direction and degree of zone movement are consistent with it being driven by the competitive superiority of Townsend's.
But what exactly has been moving? Coastal Townsend's do not appear to be hybrids: they are morphologically indistinguishable from interior Townsend's. Yet, our results reveal that they are the product of past hybridization. The frequency of hermit mitochondrial haplotypes in coastal Townsend's is over 50 per cent (Rohwer et al. 2001), suggesting that the alleles underlying Townsend's superiority have moved south while neutral hermit alleles are being left behind. The mechanism of male aggressive superiority and the dramatic extent of zone movement between the two species suggest that we are witnessing an extinction in progress. In another 5000 years, only one warbler will probably remain in the present range of these two species: Townsend's in appearance and behaviour, but with coastal populations that bear the neutral genetic footprint of the extinct hermit warbler.
Acknowledgments
This study was approved by the University of Washington Institutional Animal Care and Use Committee.
Thanks to P. Grant, J. Tewksbury, three anonymous reviewers, and the Rohwer and Bradshaw laboratory groups and Evolution journal club at the University of Washington for their comments on the manuscript; and to J. Felsenstein, M. Kuhner, K. Omland and many others for their helpful discussion. Tissues were supplied by the University of Washington Burke Museum and the University of Alaska, Anchorage Avian Collection; many thanks to the Burke Museum Ornithology Collection staff for their wonderful assistance and to Sergei Drovetski for additional samples. W. Hitchner offered valuable field assistance. Mdiv analyses were conducted using the Computational Biology Computing Service at Cornell University. M.K. was supported by fellowships from NSF, ARCS and the Burke Museum Eddy endowment. Field and laboratory work were funded by the Ornithology Endowment of the Burke Museum.
Supplementary Material
Table 1. Sample localities, sizes, and museum numbers. *Samples from the University of Alaska, Anchorage Avian Collection
References
- Arbogast B.S., Edwards S.V., Wakeley J., Beerli P., Slowinski J.B. Estimating divergence times from molecular data on phylogenetic or population genetic timescales. Annu. Rev. Ecol. Syst. 2002;33:707–825. doi:10.1146/annurev.ecolsys.33.010802.150500 [Google Scholar]
- Bandelt H.J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barton N.H., Hewitt G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–118. doi:10.1146/annurev.es.16.110185.000553 [Google Scholar]
- Bermingham E., Rohwer S., Freeman S., Wood C. Vicariance biogeography in the Pleistocene and speciation in North American wood warblers: a test of Mengel's model. Proc. Natl Acad. Sci. USA. 1992;89:6624–6628. doi: 10.1073/pnas.89.14.6624. doi:10.1073/pnas.89.14.6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman J.M., Hudson R.R., Kaplan N.L., Langley C.H., Stephan W. The hitchhiking effect on the site frequency spectrum of DNA polymorphism. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg T.M., Gaston A.J., Winker K., Friesen V.L. Rapid divergence and postglacial colonization in western North American Steller's Jays. Mol. Ecol. 2005;14:3744–3745. doi: 10.1111/j.1365-294X.2005.02710.x. doi:10.1111/j.1365-294X.2005.02710.x [DOI] [PubMed] [Google Scholar]
- Buggs R.J.A. Empirical study of hybrid zone movement. Heredity. 2007;99:301–312. doi: 10.1038/sj.hdy.6800997. doi:10.1038/sj.hdy.6800997 [DOI] [PubMed] [Google Scholar]
- Dasmahapatra K.K., Blum M.J., Aiello A., Hackwell S., Davies N., Bermingham E.P., Mallet J. Inferences from a rapidly moving hybrid zone. Evolution. 2002;56:741–753. doi: 10.1111/j.0014-3820.2002.tb01385.x. doi:10.1554/0014-3820(2002)056[0741:IFARMH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Dawson A.G. Routledge; New York, NY: 1992. Ice age earth: late quaternary geology and climate. [Google Scholar]
- Ferris S.D., Sage R.D., Huang C.M., Nielsen J.T., Ritte U., Wilson A.C. Flow of mitochondrial DNA across a species boundary. Proc. Natl Acad. Sci. USA. 1983;80:2290–2294. doi: 10.1073/pnas.80.8.2290. doi:10.1073/pnas.80.8.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming M.A., Cook J.A. Phylogeography of endemic ermine (Mustela erminea) in southeast Alaska. Mol. Ecol. 2002;11:795–807. doi: 10.1046/j.1365-294x.2002.01472.x. doi:10.1046/j.1365-294X.2002.01472.x [DOI] [PubMed] [Google Scholar]
- Funk D.J., Omland K.E. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Syst. 2003;34:397–423. doi:10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- Gyllensten U.B., Wilson A.C. Interspecific mitochondrial DNA transfer and the colonization of Scandinavia by mice. Genet. Res. 1987;49:25–29. doi: 10.1017/s0016672300026690. [DOI] [PubMed] [Google Scholar]
- Harrison R.G. Hybrid zones: windows on evolutionary processes. Oxf. Surv. Evol. Biol. 1990;7:129–156. [Google Scholar]
- Hewitt G.M. Hybrid zones: natural laboratories for evolutionary studies. Trends Ecol. Evol. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. doi:10.1016/0169-5347(88)90033-X [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996;58:247–276. doi:10.1111/j.1095-8312.1996.tb01434.x [Google Scholar]
- Hewitt G.M. The genetic legacy of quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. doi:10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- Ibrahim K.M., Nichols R.A., Hewitt G.M. Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity. 1996;77:282–291. doi:10.1038/hdy.1996.142 [Google Scholar]
- Kaplan N.L., Hudson R.R., Langley C.H. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner M.K., Yamato J., Felsenstein J. Maximum likelihood of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa E.P., Cook J.A., Patton J.L. Genetic footprints of demographic expansion in North America, but not Amazonia, during the late Quaternary. Proc. Natl Acad. Sci. USA. 2003;100:10 331–10 334. doi: 10.1073/pnas.1730921100. doi:10.1073/pnas.1730921100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovette I.J. Mitochondrial dating and mixed support for the ‘2%’ rule in birds. Auk. 2004;121:1–6. doi:10.1642/0004-8038(2004)121[0001:MDAMSF]2.0.CO;2 [Google Scholar]
- Lovette I.J., Bermingham E., Rohwer S., Wood C. Mitochondrial RFLP and sequence variation among closely related avian species and the genetic characterization of hybrid Dendroica warblers. Mol. Ecol. 1999;8:1431–1441. doi: 10.1046/j.1365-294x.1999.00706.x. doi:10.1046/j.1365-294x.1999.00706.x [DOI] [PubMed] [Google Scholar]
- Marchant A.D., Arnold M.L., Wilkinson P. Gene flow across a chromosomal tension zone 1. Relicts of ancient hybridization. Heredity. 1988;61:321–328. doi:10.1038/hdy.1988.122 [Google Scholar]
- Martinsen G.D., Whitham T.G., Turek R.J., Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x. doi:10.1111/j.0014-3820.2001.tb00655.x [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J. The hitchhiking effect of a favorable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McGuire J.A., Linkem C.W., Koo M.S., Hutchison D.W., Lappin A.K., Orange D.I., Lemos-Espinal J., Riddle B.R., Jaeger J.R. Mitochondrial introgression and incomplete lineage sorting through space and time: phylogenetics of crotaphytic lizards. Evolution. 2007;61:2879–2897. doi: 10.1111/j.1558-5646.2007.00239.x. doi:10.1111/j.1558-5646.2007.00239.x [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York, NY: 1987. Molecular evolutionary genetics. [Google Scholar]
- Nielsen R., Wakeley J. Distinguishing migration form isolation: a Markov Chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Ashley N.H., Butler L.K. Androgens, interspecific competition and species replacement in hybridizing warblers. Proc. R. Soc. B. 2004;271:S498–S500. doi: 10.1098/rsbl.2004.0230. doi:10.1098/rsbl.2004.0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons T.J., Olson S.L., Braun M.J. Unidirectional spread of secondary sexual plumage traits across an avian hybrid zone. Science. 1993;260:1643–1646. doi: 10.1126/science.260.5114.1643. doi:10.1126/science.260.5114.1643 [DOI] [PubMed] [Google Scholar]
- Payseur B.A., Krenz J.G., Nachman M.W. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mouse. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. doi:10.1554/03-738 [DOI] [PubMed] [Google Scholar]
- Pearson S.F., Rohwer S. Asymmetries in male aggression across an avian hybrid zone. Behav. Ecol. 2000;11:93–101. doi:10.1093/beheco/11.1.93 [Google Scholar]
- Pielou E.C. University of Chicago Press; Chicago, IL: 1991. After the ice age. [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. doi:10.1016/S0169-5347(00)02026-7 [DOI] [PubMed] [Google Scholar]
- Rabosky D.L., Lovette I.J. Density-dependent diversification in North American wood warblers. Proc. R. Soc. B. 2008;275:2363–2371. doi: 10.1098/rspb.2008.0630. doi:10.1098/rspb.2008.0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer S., Wood C. Three hybrid zones between hermit and Townsend's warblers in Washington and Oregon. Auk. 1998;115:284–310. [Google Scholar]
- Rohwer S., Bermingham E.P., Wood C. Plumage and mitochondrial DNA haplotype variation across a moving hybrid zone. Evolution. 2001;55:405–422. doi: 10.1111/j.0014-3820.2001.tb01303.x. doi:10.1554/0014-3820(2001)055[0405:PAMDHV]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Schneider S., Roessli D., Excoffier L. Genetics and Biometry Laboratory, University of Geneva; Switzerland: 2000. Arlequin v. 2.000: a software for population genetics data analysis. [Google Scholar]
- Slatkin M., Hudson R.R. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially expanding populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson M.D., Ast J.C., Dimcheff D.E., Yuri T., Mindell D.P. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol. Phylogenet. Evol. 1999;12:105–114. doi: 10.1006/mpev.1998.0602. doi:10.1006/mpev.1998.0602 [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989a;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989b;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner B.G., Mathewes R.W., Clague J.J. Ice-free conditions on the Queen Charlotte Islands. British Columbia, at the height of late Wisconsonian glaciation. Science. 1982;218:675–677. doi: 10.1126/science.218.4573.675. doi:10.1126/science.218.4573.675 [DOI] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. doi:10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Weir J.T., Schluter D. Calibrating the avian molecular clock. Mol. Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. doi:10.1111/j.1365-294X.2008.03742.x [DOI] [PubMed] [Google Scholar]
- Wirtz P. Mother species–father species: unidirectional hybridization in animals with female choice. Anim. Behav. 1999;58:1–12. doi: 10.1006/anbe.1999.1144. doi:10.1006/anbe.1999.1144 [DOI] [PubMed] [Google Scholar]
- Zink R.M., Dittmann D.L. Gene flow, refugia, and evolution of geographic variation in the song sparrow (Melospiza melodia) Evolution. 1993;47:717–729. doi: 10.1111/j.1558-5646.1993.tb01228.x. doi:10.2307/2410178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Sample localities, sizes, and museum numbers. *Samples from the University of Alaska, Anchorage Avian Collection