Abstract
A fundamental observation across eukaryotic taxa is that mitochondrial genomes have a higher load of deleterious mutations than nuclear genomes. Identifying the evolutionary forces that drive this difference is important to understanding the rates and patterns of sequence evolution, the efficacy of natural selection, the maintenance of sex and recombination and the mechanisms underlying human ageing and many diseases. Recent studies have implicated the presumed asexuality of mitochondrial genomes as responsible for their high mutational load. We review the current body of knowledge on mitochondrial mutation accumulation and recombination, and conclude that asexuality, per se, may not be the primary determinant of the high mutation load in mitochondrial DNA (mtDNA). Very little recombination is required to counter mutation accumulation, and recent evidence suggests that mitochondrial genomes do experience occasional recombination. Instead, a high rate of accumulation of mildly deleterious mutations in mtDNA may result from the small effective population size associated with effectively haploid inheritance. This type of transmission is nearly ubiquitous among mitochondrial genomes. We also describe an experimental framework using variation in mating system between closely related species to disentangle the root causes of mutation accumulation in mitochondrial genomes.
Keywords: recombination, mutation, mitochondrial DNA, Muller's ratchet, Hill–Robertson effect, sex
1. Introduction
The primary defining feature of eukaryotes is the existence of cytoplasmic organelles that have separate genomes and often experience very different modes of inheritance. These mechanisms of inheritance are expected to have profound effects on the evolutionary forces shaping the different genomes (Birky et al. 1983). The nuclear genome, for example, is generally inherited biparentally with regular recombination. In fact, persistently asexual nuclear genomes are notably rare (Bell 1982). Since recombination is required for an effective clearance of deleterious mutations (Charlesworth et al. 1993), one interpretation of this pattern is that lineages with asexually transmitted nuclear genomes are vulnerable to extinction via the accumulation of deleterious mutations (Muller 1964). In contrast to the nuclear genome, mitochondrial genomes are generally assumed to undergo little, if any, recombination among genetically distinct partners. This leads to the expectation of high mutation loads in mitochondrial DNA (mtDNA; e.g. Gabriel et al. 1993; Howell 1996).
Accordingly, comparative surveys of mutation accumulation in plants, animals and fungal mitochondrial and nuclear genomes consistently find that mitochondrial genomes (Lynch 1997; Lynch & Blanchard 1998) and the nuclear genomes of asexuals (Normark & Moran 2000) and selfers (Weinreich & Rand 2000; Bustamante et al. 2002; Glémin et al. 2006) accumulate deleterious mutations at a higher rate than nuclear genomes in outcrossing sexuals (reviewed in Charlesworth & Wright 2001). A similar pattern is seen in portions of the nuclear genome lacking recombination compared with the regions of recombination (e.g. Navarro-Sabaté et al. 2003; Haddrill et al. 2007). Bazin et al. (2006) also showed that mitochondrial genomes are different from nuclear genomes in that polymorphism and rates of evolution are relatively insensitive to variation in census population size in nature. Their interpretation was that for non-recombining mitochondrial genomes, efficient selection in large populations reduces the efficacy of selection at linked sites such that these sites experience a reduced Ne, perhaps similar to that experienced by small census populations (sensu ‘genetic draft’; Gillespie 2000, 2001). There is some discussion in the literature about whether the findings of Bazin et al. reflect positive selection versus purifying selection (reviewed in Meiklejohn et al. 2007), but the central role of asexuality as the driving force in mitochondrial mutation accumulation is assumed throughout.
Recombination can influence mutation accumulation in two ways. First, it can directly reverse the accumulation of deleterious mutations present at low frequency (Muller's ratchet, Muller 1964) by generating offspring genomes that have fewer deleterious mutations than their parent(s). Second, recombination can affect mutation accumulation via its effect on the effective population size (Ne). Absent or very infrequent recombination reduces Ne, and thus the efficacy of selection against deleterious mutations (Ohta & Kimura 1971), by increasing selective interference from linked loci (Hill & Robertson 1966; Birky & Walsh 1988; McVean & Charlesworth 2000; Marais & Charlesworth 2003; e.g. hitchhiking, background selection). The implication of the inverse relationship between the efficacy of selection against deleterious mutations and Ne is that any factor that reduces effective population size (including but not limited to the consequences of absent or very infrequent recombination) will similarly affect mutation accumulation. Thus, the effects of restricted recombination versus other mechanisms of mutation accumulation may be difficult to tease apart. It is generally assumed that the higher mutation load in the mitochondrial genome and in the asexual nuclear genomes results from a lack of recombination (e.g. Bell 1988; Jansen & de Boer 1998; Stewart et al. 2008). However, the fact is that we do not know which of the factors that can reduce Ne are causal.
We review and synthesize several lines of evidence suggesting that relative to the other aspects of mitochondrial transmission, absent/infrequent recombination may be of less central importance for mutation accumulation than that is often assumed. First, it takes very little recombination to counteract mutation accumulation (Pamilo et al. 1987; Charlesworth et al. 1993; Green & Noakes 1995; Haddrill et al. 2007), and there is mounting evidence for mitochondrial recombination in a variety of taxa (Piganeau et al. 2004; Tsaousis et al. 2005; reviewed in Barr et al. 2005; White et al. 2008). Second, high mutational loads have been documented in the mitochondrial genomes of taxa that have biparental inheritance of mitochondria and/or direct evidence for mitochondrial recombination (Lynch & Blanchard 1998). Third, bottlenecking of mitochondrial genomes during transmission is widespread (Rand 2001), and genetic drift during the bottleneck may be an important factor affecting the mitochondrial genomes that have very different patterns of transmission and recombination (Lynch & Blanchard 1998). What is more, the haploid uniparental inheritance usually associated with the bottleneck renders recombination, to the extent that it occurs between identical genomes, irrelevant to the process of mutational clearance.
Identifying the primary determinants of mutation accumulation in mtDNA is important for several reasons. For one, greater understanding of the extent to which the recombination counters mutation accumulation will help to inform the debate surrounding the selective value of genetic recombination and sexual reproduction. Many argue that recombination and sex are advantageous mainly in clearing deleterious mutations (e.g. Kondrashov 2001). However, others focus on the ability of recombination to facilitate the spread of beneficial mutations (Colegrave 2002), or believe that multiple mechanisms are more likely to maintain sex than any mechanism operating alone (West et al. 1999). Determining whether deleterious mutation accumulation is due to the lack of recombination is of direct relevance to this controversy.
More broadly, both somatic and germ-line mitochondrial mutations are often implicated in human disease and ageing (Linnane et al. 1989; Chinnery & Turnbull 2000; Kujoth et al. 2007). Germ-line mutations in mtDNA are what constitutes ‘mutational load’ and are of particular interest because they are transmitted between generations. It is now clear that differences in the number and effect of germ-line mutations are related to the severity of the disease phenotype and patient lifespan. Germ-line mutations can also exacerbate the deleterious effects of somatic mitochondrial mutations, resulting in premature ageing (e.g. Ozawa 1999). Thus, understanding the factors that lead to the accumulation of germ-line mutations in mtDNA can inform research into disease and ageing.
2. The selective sieve
Lynch (1996, 1997) studied mutation accumulation in mitochondrial and nuclear transfer RNA (tRNA) genes in various animals, plants and fungi, and found that mitochondrial tRNAs generally retained a greater number of mildly deleterious mutations than their nuclear counterparts. Next, Lynch & Blanchard (1998) estimated the ratio of non-synonymous substitutions (dN) to synonymous substitutions (dS) in protein-coding genes from the nuclear and mitochondrial genomes of plants, animals and fungal taxa. dN/dS can be taken as a measure of the efficacy of selection because synonymous mutations are assumed to be largely invisible to selection and to accumulate at a rate that approximates the mutation rate, while the usually deleterious non-synonymous mutations are subject to removal by natural selection (Li et al. 1985). Lynch and Blanchard dubbed dN/dS the ‘selective sieve’, and found that mitochondrial genes had wider selective sieves (i.e. accumulated deleterious mutations at a higher rate relative to the underlying rate of mutation) than nuclear genes in plants, animals and fungal taxa.
Lynch & Blanchard (1998) then used their selective sieve estimates to solve for the selection coefficients against mitochondrial and nuclear mutations, and found that the absolute strength of selection was similar in the two genomes (reviewed in Lynch 2007). This result is consistent with the mitochondrial sequence data showing that mitochondrial mutations are, on average, mildly deleterious (Hasegawa et al. 1998; Lynch & Blanchard 1998; Nachman 1998; Elson et al. 2004). Nevertheless, it is notoriously difficult to directly estimate the distribution of mutational effects, and hence separate the intensity of selection from the efficacy of selection in driving sequence evolution (Lynch 2007).
The data presented in Lynch & Blanchard (1998) are consistently cited as some of the best empirical support for the contention that the absence of recombination will inevitably lead to severe fitness loss due to mutation accumulation in mitochondrial genomes (e.g. Johnson & Seger 2001; Gemmell et al. 2004; Rand et al. 2004; Loewe 2006; also see Rokas et al. 2003). However, Lynch and Blanchard used a post hoc evaluation of their data to tentatively attribute this result to reductions in Ne linked to the uniparental transmission of mitochondrial genomes rather than the absence of recombination (also see Blanchard & Lynch 2000). Specifically, they used the standard diffusion approximation for the fixation probability of a mildly deleterious mutation (Crow & Kimura 1970) to calculate the expected dN/dS in nuclear versus organellar genomes, and showed that the increased dN/dS in organelles could be explained entirely by their different mode of inheritance. Uniparental transmission, when combined with the bottlenecking that characterizes mitochondrial transmission and propagation, will usually render mitochondrial genomes ‘effectively’ haploid (Birky et al. 1983). Since the multiple copies of the mitochondrial genome present within each cell will nearly always be identical (Birky et al. 1983; reviewed in Barr et al. 2005, but see White et al. 2008), recombination will not alleviate mutation accumulation. In this case, it is haploidy rather than a lack of physical recombination that results in effective asexuality for mtDNA, and if physical recombination does occur, mitochondria might be more appropriately viewed as selfers rather than as asexuals (sensu, Charlesworth & Wright 2001).
3. How much recombination does it take to counter mutation accumulation?
If compensatory mutation is rare (Wagner & Gabriel 1990), a complete lack of recombination will ultimately lead to extinction (Charlesworth et al. 1993; Lynch et al. 1993). However, simulation-based studies have found that very little recombination is required to achieve most of its evolutionary benefits (Pamilo et al. 1987; Charlesworth et al. 1993; Green & Noakes 1995; also see Bell 1988).
Charlesworth et al. (1993) simulated recombination along a chromosome of 1000 loci to estimate the amounts of recombination required to halt Muller's ratchet and the drift-catalysed fixation of deleterious mutations. They found that for a population size of (N) <100, a recombination rate equivalent to one crossover per chromosome per 100 generations (10−5/locus/generation) effectively countered Muller's ratchet. This is much lower than the minimum of one crossover per chromosome arm per generation that is thought to occur in sexual taxa (Pardo-Manuel de Villena & Sapienza 2001). A higher, but still very low, recombination rate of approximately 10−4 can impede the selective interference that would otherwise enhance the fixation of deleterious mutations due to genetic drift.
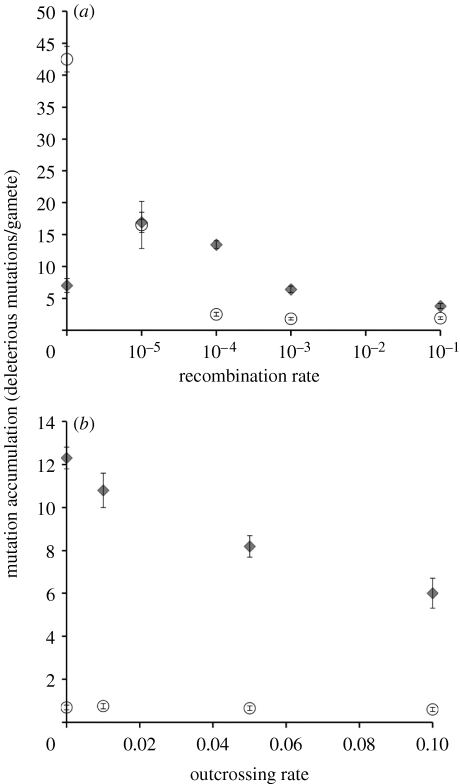
To clarify the difference between genetic drift and Muller's ratchet, we define the process of mutation accumulation as the repeated loss of the class of individuals with the fewest deleterious mutations. In an asexual population, this process is irreversible except by back mutation (Wagner & Gabriel 1990). The loss of the most mutation-free class can occur by two processes: the fixation of mutations at individual loci (which we refer to as drift) and the accumulation of low-frequency mutations at many loci (which we refer to as Muller's ratchet). Recombination can retard both of these processes by increasing the effective population size and the efficacy of selection against the mutations. Under Muller's ratchet, recombination is of added importance because it can always regenerate the least-loaded class. By contrast, fixation due to drift is only affected by recombination during the interval when the mutation is segregating in the population. In large populations, Muller's ratchet and fixation via genetic drift occur more slowly, and even lower rates of recombination will effectively arrest mutation accumulation. Charlesworth et al.'s (1993) results, and our interpretations, are summarized in figure 1.
Figure 1.
Results from the simulation study of Charlesworth et al. (1993) investigating the effect of recombination and mating system on mutation accumulation. The accumulation of low-frequency (polymorphic mutations, circles) alleles by Muller's ratchet is particularly sensitive to (a) recombination rate (probability of recombination among adjacent loci/generation), but not (b) mating system (outcrossing rate). The fixation of deleterious alleles by drift (diamonds) is particularly sensitive to mating system. Therefore, testing whether mutation accumulation is more sensitive to mating system or to recombination, per se, gives insight into the relative importance of these two forces. See tables 1 and 4 in Charlesworth et al. (1993), where N=25 and u=0.1.
The theoretical prediction that very little recombination is needed to retard mutation accumulation finds empirical support from a recent study of patterns of sequence evolution in portions of the Drosophila genome (Haddrill et al. 2007). Haddrill et al. (2007) found that genomic regions where recombination is absent (or so low as to be undetected among marker loci along the chromosomes) were characterized by a pattern of sequence evolution consistent with the higher load of deleterious mutations expected under ‘greatly enhanced’ effects of Hill–Robertson selective interference. However, the pattern of molecular evolution in genomic regions of very low recombination rates of approximately 10−8/bp/generation was indistinguishable from the portions of the Drosophila genome with the highest frequency of recombination. Assuming that current regions of high and low recombination reflect historical patterns, the similar patterns of molecular evolution observed across a wide range of recombination rates suggest that extremely ‘low’ levels of recombination are high enough to counter mutation accumulation. Such levels of recombination may nevertheless be too low to be detected using the sequence data (Posada & Crandall 2001), as we discuss below.
(a) How much recombination occurs in mitochondrial DNA?
The fact that even very rare recombination might have profound effects on mutation accumulation begs the question of how much recombination actually occurs in mitochondrial genomes. A wide range of taxa have been surveyed for the presence of mitochondrial recombination. Most researchers now agree that plant and fungal mitochondrial genomes undergo occasional recombination involving genetically distinct partners (reviewed in Barr et al. 2005). In particular, yeast mtDNA experiences frequent recombination.
In animal mitochondria, there are clear indications that key components of the recombination machinery are present (e.g. Kajander et al. 2001; Rokas et al. 2003; reviewed in Howell 1997). Direct evidence for recombination has now been documented in several taxa (e.g. Lunt & Hyman 1997; Ladoukakis & Zouros 2001; Burzyński et al. 2003; Kraytsberg et al. 2004), and there is a growing body of indirect evidence for mtDNA recombination in a variety of animal taxa (Piganeau et al. 2004; Tsaousis et al. 2005; reviewed in Rokas et al. 2003; Barr et al. 2005; White et al. 2008), although there are other possible interpretations for these patterns (e.g. mutational hot spots; Innan & Nordborg 2002).
(b) Detecting infrequent recombination
There are significant obstacles to obtaining direct estimates of infrequent mitochondrial recombination (White et al. 2008). Indirect estimates are available, which detect the statistical and genealogical effects of recombination in gene trees, but even these tests perform poorly when recombination is very rare (Posada & Crandall 2001). This raises the possibility that even if there is enough mitochondrial recombination to counter mutation accumulation, this level of recombination may be difficult or impossible to detect (Barr et al. 2005).
To illustrate this, consider Posada & Crandall's (2001) estimates of the threshold level of recombination that can be detected from the sequence data. Posada and Crandall used simulations to study the efficacy of 14 different tests for recombination. They defined a population-wide recombination rate occurring at a single locus, ρ, as 4Nrl, where N is the population size; r is the rate of recombination per site per generation; and l is the sequence length. They found that the most powerful methods detected recombination only 50 per cent of the time when ρ=1, which corresponded to three recombination events in the genealogical history of the sequences. To detect recombination more frequently, say, 80 per cent of the time, ρ needs to exceed four (12 recombination events).
Now consider the rate of recombination among loci, which opposes Muller's ratchet (modelled in Charlesworth et al. 1993). Charlesworth et al.'s (1993) model used a chromosome with 1000 loci represented by points along a line. For a population where N=100, a recombination rate of 10−5 between adjacent loci per generation is sufficient to counter Muller's ratchet (Charlesworth et al. 1993). If the genealogy has a neutral coalescent time of 2N (=200) generations (Hudson 1990), then a probability of recombination between adjacent loci of approximately 2−3 (one crossover per 500 loci over the history of the genealogy) opposes Muller's ratchet. If one were to embark on a sequencing study of 1000 bp, located between any pair of loci, there would need to be three recombination events within that stretch of nucleotides over the history of the genealogy to have a 50 per cent chance of detecting recombination (ρ=1; Posada & Crandall 2001). Therefore, a level of recombination that is detectable among contiguous nucleotides will be orders of magnitude higher than the level of recombination required to suppress Muller's ratchet among distant loci.
Sequence data often perform poorly in detecting relevant levels of recombination because recombination across large physical distances can be relevant to mutation accumulation and other evolutionary processes, but will often translate into undetectable recombination rates among adjacent nucleotides. It is therefore critically important to consider the scale of physical distances involved for the question at hand. For example, if we are studying mutation accumulation along an entire chromosome, it would be best to estimate recombination rates using the sequence data from the entire genome, or from non-contiguous nucleotides that span the physical distances over which recombination is more likely to occur. The latter is essentially what is being done in studies that estimate nuclear recombination rates from the marker data (e.g. Haddrill et al. 2007). Alternatively, one might be interested in, say, the possibility that Muller's ratchet operates within a single gene. In this case, Charlesworth et al.'s (1993) threshold rate of one recombination event per 500 loci (or in this instance of a single gene, nucleotides) approaches the detectable level of recombination from the sequence data at this scale. In the present context, the question is whether Muller's ratchet contributes to mutation accumulation in mitochondrial genomes, so that estimates of recombination should use whole-genome sequence data (e.g. Gantenbein et al. 2005; Guo et al. 2006; Ujvari et al. 2007).
The ability to detect relevant rates of recombination also depends upon effective population size. In large populations, the ability to detect rare recombination improves because recombination events are more likely to occur as Ne increases (Posada & Crandall 2001). However, the level of recombination necessary to oppose mutation accumulation decreases with increasing Ne (Charlesworth et al. 1993). Thus, although infrequent recombination becomes easier to detect in larger populations, the threshold level of recombination necessary to counter mutation accumulation is lower. Small populations pose a different problem for detecting recombination. In mtDNA, the relevant effective population size depends on the degree of bottlenecking during mitochondrial transmission, with greater bottlenecking leading to a smaller Ne (Birky et al. 1983; Roze et al. 2005). If the bottleneck during mitochondrial transmission is severe (as is generally the case), low levels of recombination will be especially difficult to detect because the recombining sequences will usually not be divergent enough to identify recombinant progeny. As a logical extension of this last point, recombination becomes not only undetectable, but also irrelevant, if the bottleneck is so severe that the probability of recombination between genetically distinct genomes approaches zero.
In summary, a failure to detect recombination could mean anything from no physical recombination, or no meaningful recombination, to recombination that is sufficient to counter mutation accumulation. Thus, there is really no firm evidence that low recombination is problematic for mitochondrial genomes, even if those genomes appear to be asexual. More rigorous tests of the extent to which mtDNA experiences recombination should involve nucleotides distributed throughout the genome in order to maximize the chance of detecting infrequent recombination.
4. Mutation accumulation and the mitochondrial bottleneck
While mitochondrial and nuclear genomes differ in several important respects, an obvious factor reducing Ne in mitochondrial genomes is their haploid, uniparental transmission (Lynch & Blanchard 1998) and, more specifically, the sharp reduction in mtDNA number that characterizes mitochondrial transmission across a wide variety of taxa (Hauswirth & Laipis 1982; Rand 2001). When and where the bottleneck occurs is the subject of recent debate (Cao et al. 2008; Cree et al. 2008; Wai et al. 2008); the present evidence suggests that both a physical bottlenecking of mitochondrial genomes in primordial germ cells and a genetic bottleneck during post-natal folliculogenesis are responsible (Wai et al. 2008). This bottleneck process culminates in a sharp reduction of the effective number of segregating units of mitochondrial genomes inhabiting a cell (Bendall et al. 1996; Jenuth et al. 1996; Jansen & de Boer 1998; Roze et al. 2005; Wai et al. 2008), rendering the mitochondrial genome effectively haploid (Birky et al. 1983; Jansen & de Boer 1998; Jansen 2000; e.g. Marchington et al. 1998) and reducing the Ne experienced by the mitochondria (Roze et al. 2005).
The expectation that the mitochondrial bottleneck will generate low effective population size has implicated genetic drift during the bottleneck as a causal factor in the high mutation load in mtDNA. For example, Chinnery et al. (2000) highlighted genetic drift due to bottlenecking as a primary explanation for the transmission of deleterious mtDNA mutations in humans (also see Jenuth et al. 1996; Marchington et al. 1998; Brown et al. 2000). Stewart et al. (1996) suggested that bottlenecking might underlie the high rate of substitution in male mitotypes relative to female mitotypes in bivalve species with doubly uniparental inheritance (also see Ort & Pogson 2007) because males experience a more severe bottleneck in mitochondrial number during sperm formation.
By contrast, others have suggested that the mitochondrial bottleneck acts to oppose mutation accumulation by increasing the variance between cells within organisms, or between organisms within populations, thus increasing the efficacy of selection against deleterious mutations (Hauswirth & Laipis 1982; Takahata & Slatkin 1983; Bergstrom & Pritchard 1998; Jansen & de Boer 1998; Krakauer & Mira 1999; Rispe & Moran 2000; Roze et al. 2005; Rand 2008; Stewart et al. 2008; White et al. 2008). There is recent empirical evidence that this type of mechanism can promote the selective elimination of deleterious mitochondrial mutations during oocyte maturation (Fan et al. 2008; Shoubridge & Wai 2008; Stewart et al. 2008).
How the mitochondrial bottleneck ultimately affects the efficacy of selection is of fundamental importance for understanding the mutation accumulation in mitochondrial genomes. If the net effect of the bottleneck is to reduce the efficacy of selection (and increase dN/dS) relative to the nucleus, then it may contribute to the higher mutation load observed in mitochondrial genomes. Instead, if the net effect of the mitochondrial bottleneck is to increase the efficacy of selection in mitochondrial genomes, then the observed patterns of molecular evolution must be due to some other mechanism, and occur despite the net ‘positive’ effects of the bottleneck during transmission.
There are several reasons to believe that this bottleneck is ultimately a contributor to the high mutation load in mtDNA. For one, there is evidence that many phenotypically important mitochondrial mutations escape the bottleneck to segregate at the organismal level (Taylor & Turnbull 2005; Fan et al. 2008; Shoubridge & Wai 2008; Wai et al. 2008). In addition, simulation models indicate that bottlenecking does not increase the efficacy of selection once bottlenecks exceed approximately 20 segregating units (see fig. 5 in Bergstrom & Pritchard 1998), which is smaller than many estimates of bottleneck size (Rand & Harrison 1986; Bendall et al. 1996; Jenuth et al. 1996; Gocke et al. 1998; Cree et al. 2008). Another simulation study used a multilevel selection approach to determine how the selection and drift of mitochondrial mutants are affected by bottlenecks that simultaneously reduce Ne and increase opportunities for selection within and between cells (Roze et al. 2005). They found that the Ne-reducing effect of the bottleneck more than offset the increased efficacy of selection, such that the net effect of the bottleneck was to increase the rate of fixation of deleterious alleles and decreased the rate of fixation of advantageous alleles (Roze et al. 2005).
The data from taxa that experience frequent mitochondrial recombination also point to the mitochondrial bottleneck, rather than uniparental transmission or the absence of recombination, as the root cause of high mutation load in mitochondrial genomes. For example, the mitochondrial genome in the yeast Saccharomyces cerevisiae recombines freely and is biparentally inherited, but nevertheless has a high mutation load relative to the nuclear genome (Lynch 1997; Lynch & Blanchard 1998). One possible explanation for this observation could be the mating system: Saccharomyces yeasts are predominantly selfing (Field & Wills 1998; Johnson et al. 2004), which is expected to increase genomic mutational load (Charlesworth & Wright 2001) relative to biparental inheritance from unrelated individuals (Charlesworth 2003; Glémin 2007; see below). For yeast, however, selfing should increase the selective sieve for both the mitochondrial and the nuclear genomes, since both genomes have the capability for regular biparental transmission. More likely, the example of yeast points to the importance of the mitochondrial bottleneck and vegetative segregation during the rounds of cell division. These combine to result in effectively haploid mitochondrial transmission, even though physically it is biparentally inherited (reviewed in Birky 2001).
In summary, the current body of theory and the data suggest that the mitochondrial bottleneck exacerbates the fixation of deleterious mutations via drift. The high mutational load in yeast illustrates specifically why this bottleneck reduces effective population size relative to the nucleus. The nuclear genome of eukaryotes is transmitted in just two copies in each cell generation, and into each zygote, but mitosis and meiosis ensure that one copy is dutifully retained from each parent. Thus, the nuclear genome is diploid. The mitochondrial bottleneck is superficially less severe in the sense that more genome copies are transmitted from one generation to the next, but without a fair meiosis/mitosis, selection or drift among those genome copies within cell lineages results in haploid inheritance (Birky 2001). Additional empirical research following the dynamics of mitochondrial mutations within and between cells (e.g. Taylor et al. 2002; Roze et al. 2005; Rand 2008; Shoubridge & Wai 2008; Stewart et al. 2008) may clarify the distinct genetic and evolutionary processes that influence mitochondrial genomes.
5. Mating system, effective population size and mutation accumulation
A promising way to investigate what forces affect Ne and drive mutation accumulation is to consider how the Ne experienced by nuclear genomes varies among taxa with different mating systems (Birky et al. 1983; Pollak 1987; Birky & Walsh 1988; Nordborg 2000; Charlesworth & Wright 2001; Butlin 2002; Glémin et al. 2006; Glémin 2007). It would be especially informative to identify the patterns of nucleotide substitution in nuclear genomes under circumstances where they should accumulate mutations differently than their mitochondrial counterparts, and compare these patterns to situations where the nuclear genome should experience the same Ne, or recombination rate, as the mitochondrial genome.
In outcrossing hermaphrodites, a twofold difference in Ne is expected between mitochondrial and nuclear genomes (fourfold for outcrossing dioecious species; Birky et al. 1983). In highly selfing species, nuclear recombination occurs, but the difference in Ne between nuclear and mitochondrial genomes is expected to be sharply reduced. This is, in large part, due to the decrease in efficacy of recombination caused by increased homozygosity in the nuclear genome (Nordborg 2000; Charlesworth & Wright 2001). By contrast, the Ne experienced by a uniparentally inherited mitochondrial genome should be less affected by selfing unless hitch-hiking effects are extreme (Charlesworth 2003; Glémin 2007). The nuclear genome of asexual species should also experience reduced Ne relative to the nuclear genome in sexual species owing to the increased likelihood of hitch-hiking and background selection when recombination is absent (Hill & Robertson 1966).
Although theory indicates that the increase in homozygosity that accompanies selfing can promote mutation accumulation via the reduced efficacy of recombination (Heller & Maynard Smith 1979), simulations demonstrate that it only takes a small amount of outcrossing to counter mutation accumulation in largely selfing populations (Pamilo et al. 1987). Moreover, other simulations show that the sharp reductions in fitness due to mutation accumulation in small selfing populations are more a consequence of restricted outcrossing than limits upon recombination (Charlesworth et al. 1993). We take these results to mean that mutation accumulation in highly selfing populations should be linked to reductions in Ne due more to uniparental reproduction than to restricted effects of recombination. In this context, it is not the presumed asexuality of mitochondrial genomes that is responsible for their high mutation load, but the mechanism of inheritance that essentially turns mitochondria, to the extent that they are ‘sexual’, into selfers.
Following this logic, the width of the selective sieve in the mitochondrial and nuclear genomes should become more similar in selfing species if reduction in Ne, unrelated to recombination, is the crucial determinant of mutation accumulation for both genomes (also see Charlesworth 2003). By contrast, if the nuclear and mitochondrial selective sieves are more similar in asexual species than in selfing and outcrossing species, the lack of recombination must play a central role in mutation clearance.
Evaluating these alternatives requires nuclear and mitochondrial genomic data from closely related taxa that vary in mating system. This type of mating system variation is common in some taxa, such as freshwater snails (Jarne & Städler 1995) and many groups of angiosperms. Although these data are not currently available (Charlesworth & Wright 2001; Charlesworth 2003; Glémin 2007), several studies suggest that processes of molecular evolution (including mutation accumulation) in mitochondrial and nuclear genomes may be more similar in selfing than outcrossing taxa (Weinreich & Rand 2000; Graustein et al. 2002), and in recombining versus non-recombining sections of the nuclear genome (Comeron et al. 1999; Munte et al. 2001; Navarro-Sabaté et al. 2003; Haddrill et al. 2007). Moreover, when closely related sexual and asexual taxa are compared, asexuals (‘effectively asexual’ in the case of the endosymbionts studied in Moran 1996; Woolfit & Bromham 2003) suffer increased retention of deleterious nuclear (Normark & Moran 2000) and mitochondrial (Moran 1996; Woolfit & Bromham 2003, 2005; Paland & Lynch 2006) mutations relative to their sexual counterparts.
Taken together, these studies provide preliminary support for the hypothesis that both the mating system and the presence of recombination are important determinants of mutation accumulation, and that such effects can be parsed out with the appropriate data. This would provide an important test of how reduced recombination and uniparental/haploid transmission combine to explain differences in dN/dS in nuclear versus mitochondrial genomes. Such a test would clarify the fundamental evolutionary mechanisms responsible for mutation accumulation in eukaryotic genomes, and how those mechanisms are affected by the inheritance and recombination of those genomes. Understanding the factors underlying the high mutation load in mtDNA also has applied significance, in the light of the links between mitochondrial mutation, human disease and ageing.
Acknowledgements
We thank A.F. Agrawal, C. Barr, J. Busch, A. Caballero, S. Keller, E. (B.) Neiman, D. Schoen, D. Sloan, D. Sowell, M. Whitlock and two anonymous reviewers for their comments on and critical reviews of an earlier version of the manuscript.
References
- Barr C.M., Neiman M., Taylor D.R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. doi:10.1111/j.1469-8137.2005.01492.x [DOI] [PubMed] [Google Scholar]
- Bazin E., Glémin S., Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. doi:10.1126/science.1122033 [DOI] [PubMed] [Google Scholar]
- Bell G. Croon Helm; London, UK: 1982. The masterpiece of nature. [Google Scholar]
- Bell G. Uniformity and diversity in the evolution of sex. In: Michod R.E., Levin B.R., editors. The evolution of sex. Sinauer Associates; Sunderland, MA: 1988. pp. 126–138. [Google Scholar]
- Bendall K.E., Macaulay V.A., Baker J.R., Sykes B.C. Heteroplasmic point mutations in the human mtDNA control region. Am. J. Hum. Genet. 1996;59:1276–1287. [PMC free article] [PubMed] [Google Scholar]
- Bergstrom C.T., Pritchard J. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics. 1998;149:2135–2216. doi: 10.1093/genetics/149.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C.W., Jr The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. doi:10.1146/annurev.genet.35.102401.090231 [DOI] [PubMed] [Google Scholar]
- Birky C.W., Jr, Walsh J.B. Effects of linkage on rates of molecular evolution. Proc. Natl Acad. Sci. USA. 1988;85:6414–6418. doi: 10.1073/pnas.85.17.6414. doi:10.1073/pnas.85.17.6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C.W., Jr, Maruyama T., Fuerst P.A. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts and some results. Genetics. 1983;103:513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J.L., Lynch M. Organellar genes: why do they end up in the nucleus? Trends Genet. 2000;16:315–320. doi: 10.1016/s0168-9525(00)02053-9. doi:10.1016/S0168-9525(00)02053-9 [DOI] [PubMed] [Google Scholar]
- Brown D.T., Samuels D.C., Michael E.M., Turnbull D.M., Chinnery P.F. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am. J. Hum Genet. 2000;68:533–536. doi: 10.1086/318190. doi:10.1086/318190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyński A., Zbawicka M., Skibinski D.O.F., Wenne R. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the baltic. Mol. Biol. Evol. 2003;20:388–392. doi: 10.1093/molbev/msg058. doi:10.1093/molbev/msg058 [DOI] [PubMed] [Google Scholar]
- Bustamante C., Nielsen R., Sawyer S.A., Olsen K.M., Purugganan M.D., Hartl D.L. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. doi:10.1038/416531a [DOI] [PubMed] [Google Scholar]
- Butlin R. The costs and benefits of sex: new insights from old asexual lineages. Nat. Rev. Genet. 2002;3:311–317. doi: 10.1038/nrg749. doi:10.1038/nrg749 [DOI] [PubMed] [Google Scholar]
- Cao L., Shitara H., Horii T., Nagao Y., Imai H., Abe K., Hara T., Hayashi J.-I., Yonekawa H. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 2007;39:386–390. doi: 10.1038/ng1970. doi:10.1038/ng1970 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Effects of inbreeding on the genetic diversity of populations. Phil. Trans. R. Soc. B. 2003;358:1051–1070. doi: 10.1098/rstb.2003.1296. doi:10.1098/rstb.2003.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Wright S.I. Mating systems and genome evolution. Curr. Opin. Genet. Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. doi:10.1016/S0959-437X(00)00254-9 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Morgan M.T., Charlesworth B. Mutation accumulation in finite outbreeding and inbreeding populations. Genet. Res. 1993;61:39–56. [Google Scholar]
- Chinnery P.F., Turnbull D.M. Mitochondrial DNA mutations in the pathogenesis of human disease. Mol. Med. Today. 2000;6:425–432. doi: 10.1016/s1357-4310(00)01805-0. doi:10.1016/S1357-4310(00)01805-0 [DOI] [PubMed] [Google Scholar]
- Chinnery P.F., Thorburn D.R., Samuels D.C., White S.L., Dahl H.H.M., Turnbull D.M., Lightowlers R.N., Howell N. The inheritance of mitochondrial heteroplasmy: random drift, selection or both? Trends Genet. 2000;16:500–505. doi: 10.1016/s0168-9525(00)02120-x. doi:10.1016/S0168-9525(00)02120-X [DOI] [PubMed] [Google Scholar]
- Colegrave N. Sex releases the speed limit on evolution. Nature. 2002;420:664–666. doi: 10.1038/nature01191. doi:10.1038/nature01191 [DOI] [PubMed] [Google Scholar]
- Comeron J.M., Kreitman M., Aguadé M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree L.M., Samuels D.C., de Sousa Lopes S.C., Rajasimha H.K., Wonnapinij P., Mann J.R., Dahl H.-H. M., Chinnery P.F. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. doi:10.1038/ng.2007.63 [DOI] [PubMed] [Google Scholar]
- Crow J.F., Kimura M. Harper and Row; New York, NY: 1970. An introduction to population genetics theory. [Google Scholar]
- Elson J.L., Turnbull D.M., Howell N. Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am. J. Hum. Genet. 2004;74:229–238. doi: 10.1086/381505. doi:10.1086/381505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. doi:10.1126/science.1147786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D., Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl Acad. Sci. USA. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. doi:10.1073/pnas.95.4.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W., Lynch M., Bürger R. Muller's ratchet and mutational meltdowns. Evolution. 1993;47:1744–1757. doi: 10.1111/j.1558-5646.1993.tb01266.x. doi:10.2307/2410218 [DOI] [PubMed] [Google Scholar]
- Gantenbein B., Fet V., Gantenbein-Ritter I.A., Balloux F. Evidence for recombination in scorpion mitochondrial DNA (Scorpiones: Buthidae) Proc. R. Soc. B. 2005;272:697–704. doi: 10.1098/rspb.2004.3017. doi:10.1098/rspb.2004.3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell N.J., Metcalf V.J., Allendorf F.W. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol. 2004;19:238–244. doi: 10.1016/j.tree.2004.02.002. doi:10.1016/j.tree.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Gillespie J.H. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics. 2000;155:909–919. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J.H. Is the population size of a species relevant to its evolution? Evolution. 2001;55:2161–2169. doi: 10.1111/j.0014-3820.2001.tb00732.x. doi:10.1554/0014-3820(2001)055[2161:ITPSOA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Glémin S. Mating systems and the efficacy of selection at the molecular level. Genetics. 2007;177:905–916. doi: 10.1534/genetics.107.073601. doi:10.1534/genetics.107.073601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S., Bazin E., Charlesworth D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc. R. Soc. B. 2006;273:3011–3019. doi: 10.1098/rspb.2006.3657. doi:10.1098/rspb.2006.3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke C.D., Benko F.A., Rogan P.K. Transmission of mitochondrial DNA heteroplasmy in normal pedigrees. Hum. Genet. 1998;102:182–186. doi: 10.1007/s004390050674. doi:10.1007/s004390050674 [DOI] [PubMed] [Google Scholar]
- Graustein A., Gasper J.M., Walters J.R., Palopoli M.F. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics. 2002;161:99–107. doi: 10.1093/genetics/161.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.F., Noakes D.L.G. Is a little of bit of sex as good as a lot? J. Theor. Biol. 1995;174:87–96. doi:10.1006/jtbi.1995.0081 [Google Scholar]
- Guo X., Liu S., Liu Y. Evidence for recombination of mitochondrial DNA in triploid crucian carp. Genetics. 2006;172:1745–1749. doi: 10.1534/genetics.105.049841. doi:10.1534/genetics.105.049841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P.R., Halligan D.L., Tomaras D., Charlesworth B. Reduced efficiency of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. doi:10.1186/gb-2007-8-2-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Cao Y., Yang Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: nonsyonymous/synonymous rate ratio is much higher within species than between species. Mol. Biol. Evol. 1998;15:1499–1505. doi: 10.1093/oxfordjournals.molbev.a025877. [DOI] [PubMed] [Google Scholar]
- Hauswirth W.W., Laipis P.J. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc. Natl Acad. Sci. USA. 1982;79:4686–4690. doi: 10.1073/pnas.79.15.4686. doi:10.1073/pnas.79.15.4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R., Maynard Smith J. Does Muller's ratchet work with selfing? Genet. Res. 1979;32:289–293. [Google Scholar]
- Hill W.G. The effect of linkage on limits to artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Howell N. Mutational analysis of the human mitochondrial genome branches into the realm of bacterial genetics. Am. J. Hum. Genet. 1996;59:749–755. [PMC free article] [PubMed] [Google Scholar]
- Howell N. mtDNA recombination: what do in vitro data mean? Am. J. Hum. Genet. 1997;61:18–22. doi: 10.1086/513910. doi:10.1086/513915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R. Gene genealogies and the coalescent process. Oxford Surv. Evol. Biol. 1990;7:1–44. [Google Scholar]
- Innan H., Nordborg M. Recombination or mutational hot spots in human mtDNA? Mol. Biol. Evol. 2002;19:1122–1127. doi: 10.1093/oxfordjournals.molbev.a004170. [DOI] [PubMed] [Google Scholar]
- Jansen R.P.S. Germline passage of mitochondria: quantitative considerations and possible embryological sequelae. Hum. Reprod. (Suppl. 2) 2000;15:112–128. doi: 10.1093/humrep/15.suppl_2.112. [DOI] [PubMed] [Google Scholar]
- Jansen R.P.S., de Boer K. The bottleneck: mitochondrial imperatives in oogenesis and ovarian follicular fate. Mol. Cell. Endocrinol. 1998;145:81–88. doi: 10.1016/s0303-7207(98)00173-7. doi:10.1016/S0303-7207(98)00173-7 [DOI] [PubMed] [Google Scholar]
- Jarne P., Städler T. Population genetic-structure and mating system evolution in fresh-water pulmonates. Experientia. 1995;51:482–497. doi:10.1007/BF02143200 [Google Scholar]
- Jenuth J.P., Peterson A.C., Fu K., Shoubridge E.A. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. doi:10.1038/ng1096-146 [DOI] [PubMed] [Google Scholar]
- Johnson K.P., Seger J. Elevated rates of nonsynonymous substitutions in island birds. Mol. Biol. Evol. 2001;18:874–881. doi: 10.1093/oxfordjournals.molbev.a003869. [DOI] [PubMed] [Google Scholar]
- Johnson L.J., Koufopanou V., Goddard M.R., Hetherington R., Schafer S.M., Burt A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. doi:10.1534/genetics.166.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander O.A., Karhunen P.J., Holt I.J., Jacobs H.T. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2:1007–1012. doi: 10.1093/embo-reports/kve233. doi:10.1093/embo-reports/kve233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A.S. Sex and U. Trends Genet. 2001;17:75–78. doi: 10.1016/s0168-9525(00)02188-0. doi:10.1016/S0168-9525(00)02188-0 [DOI] [PubMed] [Google Scholar]
- Krakauer D.C., Mira A. Mitochondria and germ-cell death. Nature. 1999;400:125–126. doi: 10.1038/22026. doi:10.1038/22026 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Schwartz M., Brown T.A., Ebralidse K., Kunz W.S., Clayton D.A., Vissing J., Khrapko K. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. doi:10.1126/science.1096342 [DOI] [PubMed] [Google Scholar]
- Kujoth G.C., Bradshaw P.C., Haroon S., Prolla T.A. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:0161–0173. doi: 10.1371/journal.pgen.0030024. doi:10.1371/journal.pgen.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis E., Zouros E. Recombination in animal mitochondrial DNA: evidence from published sequences. Mol. Biol. Evol. 2001;18:2127–2131. doi: 10.1093/oxfordjournals.molbev.a003755. [DOI] [PubMed] [Google Scholar]
- Li W.-H., Wu C.-I., Luo C.-C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood and nucleotide and codon changes. Mol. Biol. Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Linnane A.W., Ozawa T., Marzuki S., Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. The Lancet. 1989;333:642–645. doi: 10.1016/s0140-6736(89)92145-4. doi:10.1016/S0140-6736(89)92145-4 [DOI] [PubMed] [Google Scholar]
- Loewe L. Quantifying the genomic decay paradox due to Muller's ratchet in human mitochondrial DNA. Genet. Res. 2006;87:133–159. doi: 10.1017/S0016672306008123. doi:10.1017/S0016672306008123 [DOI] [PubMed] [Google Scholar]
- Lunt D.H., Hyman B.C. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. doi:10.1038/387247a0 [DOI] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in transfer RNAs: molecular evidence for Muller's ratchet in mitochondrial genomes. Mol. Biol. Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol. Biol. Evol. 1997;14:914–925. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- Lynch M. Sinauer Associates; Sunderland, MA: 2007. The origins of genome architecture. [Google Scholar]
- Lynch M., Blanchard J.L. Deleterious mutation accumulation in organelle genomes. Genetica. 1998;103:29–39. doi:10.1023/A:1017022522486 [PubMed] [Google Scholar]
- Lynch M., Bürger R., Butcher D., Gabriel W. The mutational meltdown in asexual populations. J. Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- Marais G., Charlesworth B. Genome evolution: recombination speeds up adaptive evolution. Curr. Biol. 2003;13:R68–R70. doi: 10.1016/s0960-9822(02)01432-x. doi:10.1016/S0960-9822(02)01432-X [DOI] [PubMed] [Google Scholar]
- Marchington D.R., Macaulay V., Hartshorne G.M., Barlow D., Poulton J. Evidence from human oocytes for a genetic bottleneck in an mtDNA disease. Am. J. Hum. Genet. 1998;63:769–775. doi: 10.1086/302009. doi:10.1086/302009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G.A.T., Charlesworth B. The effects of Hill–Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C.D., Montooth K.L., Rand D.M. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007;23:259–263. doi: 10.1016/j.tig.2007.03.008. doi:10.1016/j.tig.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Moran N.A. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl Acad. Sci. USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. doi:10.1073/pnas.93.7.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H.J. The relation of recombination to mutational advance. Mut. Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. doi:10.1016/0027-5107(64)90047-8 [DOI] [PubMed] [Google Scholar]
- Munte A., Aguadé M., Segarra C. Changes in the recombinational environment affect divergence in the yellow gene of Drosophila. Mol. Biol. Evol. 2001;18:1045–1056. doi: 10.1093/oxfordjournals.molbev.a003876. [DOI] [PubMed] [Google Scholar]
- Nachman M.W. Deleterious mutations in animal mitochondrial DNA. Genetica. 1998;102/103:61–69. doi:10.1023/A:1017030708374 [PubMed] [Google Scholar]
- Navarro-Sabaté A., Aguadé M., Segarra C. Excess of nonsynonymous polymorphism at Acph-1 in different gene arrangements of Drosophila subobscura. Mol. Biol. Evol. 2003;20:1833–1843. doi: 10.1093/molbev/msg196. doi:10.1093/molbev/msg196 [DOI] [PubMed] [Google Scholar]
- Nordborg M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark B.B., Moran N.A. Testing for the accumulation of deleterious mutations in asexual eukaryotic genomes using molecular sequences. J. Nat. Hist. 2000;34:1719–1729. doi:10.1080/00222930050122147 [Google Scholar]
- Ohta T., Kimura M. On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1971;1:18–25. doi: 10.1007/BF01659391. doi:10.1007/BF01659391 [DOI] [PubMed] [Google Scholar]
- Ort B.S., Pogson G.H. Molecular population genetics of the male and female mitochondrial DNA molecules of the California sea mussel Mytilus californianus. Genetics. 2007;177:1087–1099. doi: 10.1534/genetics.107.072934. doi:10.1534/genetics.107.072934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T. Mitochondrial genome mutation in cell death and aging. J. Bioenerget. Biomemb. 1999;31:377–390. doi: 10.1023/a:1005479920097. doi:10.1023/A:1005479920097 [DOI] [PubMed] [Google Scholar]
- Paland S., Lynch M. Transitions to asexuality result in excess amino acid substitutions. Science. 2006;311:990–992. doi: 10.1126/science.1118152. doi:10.1126/science.1118152 [DOI] [PubMed] [Google Scholar]
- Pamilo F., Nei M., Li W.H. Accumulation of mutations in sexual and asexual populations. Genet. Res. 1987;49:135–146. doi: 10.1017/s0016672300026938. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome. 2001;12:318–322. doi: 10.1007/s003350020005. doi:10.1007/s003350020005 [DOI] [PubMed] [Google Scholar]
- Piganeau G., Gardner M., Eyre-Walker A. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 2004;21:2319–2325. doi: 10.1093/molbev/msh244. doi:10.1093/molbev/msh244 [DOI] [PubMed] [Google Scholar]
- Pollak E. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics. 1987;117:353–360. doi: 10.1093/genetics/117.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl Acad. Sci. USA. 2001;98:13 757–13 762. doi: 10.1073/pnas.241370698. doi:10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D.M. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 2001;32:415–448. doi:10.1146/annurev.ecolsys.32.081501.114109 [Google Scholar]
- Rand D.M. Mitigating mutational meltdown in mammalian mitochondria. PLoS Biol. 2008;6:0229–0232. doi: 10.1371/journal.pbio.0060035. doi:10.1371/journal.pbio.0060035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D.M., Harrison R.G. Mitochondrial DNA transmission genetics in crickets. Genetics. 1986;114:955–970. doi: 10.1093/genetics/114.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D.M., Haney R.A., Fry A.J. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. doi:10.1016/j.tree.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Rispe C., Moran N.A. Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Am. Nat. 2000;156:425–441. doi: 10.1086/303396. doi:10.1086/303396 [DOI] [PubMed] [Google Scholar]
- Rokas A., Ladoukakis E.D., Zouros E. Animal mitochondrial DNA recombination revisited. Trends Ecol. Evol. 2003;18:411–417. doi:10.1016/S0169-5347(03)00125-3 [Google Scholar]
- Roze D., Rousset F., Michalakis J. Germline bottlenecks, biparental inheritance and selection on mitochondrial variants: a two-level selection model. Genetics. 2005;170:1385–1399. doi: 10.1534/genetics.104.039495. doi:10.1534/genetics.104.039495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge E.A., Wai T. Sidestepping mutational meltdown. Science. 2008;319:914–915. doi: 10.1126/science.1154515. doi:10.1126/science.1154515 [DOI] [PubMed] [Google Scholar]
- Stewart D.T., Kenchington E.R., Singh R.K., Zouros E. Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics. 1996;143:1349–1357. doi: 10.1093/genetics/143.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.B., Freyer C., Elson J.L., Larsson N.-G. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 2008;9:657–662. doi: 10.1038/nrg2396. doi:10.1038/nrg2396 [DOI] [PubMed] [Google Scholar]
- Takahata N., Slatkin M. Evolutionary dynamics of extranuclear genes. Genet. Res. 1983;42:257–265. [Google Scholar]
- Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. doi:10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.R., Zeyl C., Cooke E. Conflicting levels of selection in the accumulation of mitochondrial defects in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2002;99:3690–3694. doi: 10.1073/pnas.072660299. doi:10.1073/pnas.072660299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaousis A.D., Martin D.P., Ladoukakis E.D., Posada D., Zouros E. Widespread recombination in published animal mtDNA sequences. Mol. Biol. Evol. 2005;22:925–933. doi: 10.1093/molbev/msi084. doi:10.1093/molbev/msi084 [DOI] [PubMed] [Google Scholar]
- Ujvari B., Dowton M., Madsen T. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 2007;3:189–192. doi: 10.1098/rsbl.2006.0587. doi:10.1098/rsbl.2006.0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.P., Gabriel W. Quantitative variation in finite parthenogenetic populations: what stops Muller's ratchet in the absence of recombination? Evolution. 1990;44:715–731. doi: 10.1111/j.1558-5646.1990.tb05950.x. doi:10.2307/2409447 [DOI] [PubMed] [Google Scholar]
- Wai T., Teoli D., Shoubridge E.A. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. doi:10.1038/ng.258 [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Rand D.M. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics. 2000;156:385–399. doi: 10.1093/genetics/156.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.A., Lively C.M., Read A.F. A pluralistic approach to sex and recombination. J. Evol. Biol. 1999;12:1003–1012. doi:10.1046/j.1420-9101.1999.00119.x [Google Scholar]
- White D.J., Wolff J.N., Pierson M., Gemmell N.J. Revealing the hidden complexities of mitochondrial inheritance. Mol. Ecol. 2008;17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. doi:10.1111/j.1365-294X.2008.03982.x [DOI] [PubMed] [Google Scholar]
- Woolfit M., Bromham L. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol. Biol. Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. doi:10.1093/molbev/msg167 [DOI] [PubMed] [Google Scholar]
- Woolfit M., Bromham L. Population size and molecular evolution on islands. Proc. R. Soc. B. 2005;272:2277–2282. doi: 10.1098/rspb.2005.3217. doi:10.1098/rspb.2005.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]