Abstract
Cytosine methylation in DNA is a major epigenetic signal, and plays a central role in propagating chromatin status during cell division. However the mechanistic links between DNA methylation and histone methylation are poorly understood. A multi-domain protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is required for DNA CpG maintenance methylation at replication forks, and mouse UHRF1-null cells show enhanced susceptibility to DNA replication arrest and DNA damaging agents. Recent data demonstrated that the SET and RING associated (SRA) domain of UHRF1 binds hemimethylated CpG and flips 5-methylcytosine out of the DNA helix, whereas its tandom tudor domain and PHD domain bind the tail of histone H3 in a highly methylation sensitive manner. We hypothesize that UHRF1 brings the two components (histones and DNA) carrying appropriate markers (on the tails of H3 and hemimethylated CpG sites) ready to be assembled into a nucleosome after replication.
Keywords: DNA methylation, histone modifications, base flipping, SRA domain, inter-domain interactions
Introduction
In mammals and other vertebrates, DNA methylation occurs at the C5 position of cytosine (5mC), mostly within CpG dinucleotides. The mammalian DNA nucleotide methyltransferases (Dnmts) include four members, in two families that are structurally and functionally distinct.1 The Dnmt3 family establishes the initial CpG methylation pattern de novo, whereas Dnmt1 maintains this pattern during chromosome replication2 and repair.3 These enzymes utilize a conserved base flipping mechanism4 that has been studied best in the bacterial 5mC methyltransferase (MTase) M.HhaI.5 Briefly, this mechanism involves MTase binding to the DNA, eversion of the target nucleotide so that it projects out of the double helix (“base flipping”), covalent attack of a conserved Cys nucleophile on cytosine C6, transfer of the methyl group from S-adenosyl-l-methionine to the activated cytosine C5, and the various release steps. This methylation, together with histone modifications, plays an important role in modulating chromatin structure, thus controlling gene expression and many other chromatin-dependent processes.6
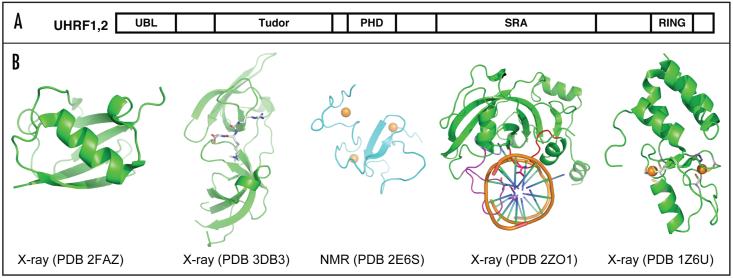
As befits a maintenance MTase, Dnmt1 has a 30-40-fold preference for hemimethylated sites (reviewed in refs. 7 and 8). However, Dnmt1 is necessary but not sufficient for proper maintenance methylation. An accessory protein UHRF1 targets Dnmt1 to hemimethylated replication forks (and presumably repair sites).9-11 The murine ortholog of this protein is also known as NP95 (nuclear protein of 95 kDa);12-15 the human ortholog is called ICBP90 (inverted CCAAT binding protein of 90 kDa).16 UHRF1 harbors at least five recognizable functional domains: an ubiquitin-like domain (UBL) at the N terminus, followed by a tandem tudor domain, a plant homeodomain (PHD), a SET and RING associated (SRA) domain, and really interesting new gene (RING) domain at the C terminus (Fig. 1A). At the time of this writing (October, 2008), structures are available for all five individual domains, either from human or mouse UHRF1 or its homolog UHRF2 (Fig. 1B). However, we do not know whether these domains act independently or in a cooperative fashion.
Figure 1.
UHRF1—a multi-domain protein. (A) Schematic representation of UHRF1 and its homolog UHRF2. (B) Five domain structures are currently available.
SRA Domain Structure
The crystal structure of the SRA domain of UHRF1 in complex with DNA containing a hemimethylated CpG site was recently determined.17-19 It reveals that the SRA domain contains two twisted β sheets packed together forming a crescent moon-like structure, whose basic inner surface binds DNA (Fig. 2A). The inner surface of the crescent contains two loops, responsible for CpG recognition (R496-associated loop) and promotion of base flipping (V451-associated loop). The tips of these loops approach the DNA from opposite directions contacting major (R496 loop) and minor grooves (V451 loop), and are stabilized at the center of the DNA helix by van der Waals contacts between the R496 and V451 side chains (Fig. 2B). The 5-methylcytosine (5mC) flips completely out of the DNA helix and positions in a binding pocket with planar stacking contacts, and both Watson-Crick polar hydrogen bonds and van der Waals interactions specific for 5mC (Fig. 2C). The methyl group at the C5 position in 5mC is in van der Waals contact with Cα and Cβ atoms of S486 (see insert in Fig. 2C), make tight-fit shape recognition that distinguishes 5mC from cytosine. The structure also suggests an explanation for the preference for hemimethylated sites. In the major groove side, the backbone carbonyl oxygen of N494 is so close to the C5 ring carbon of the unmethylated cytosine, that addition of a methyl group to the C5 of the cytosine would cause a steric clash.
Figure 2.
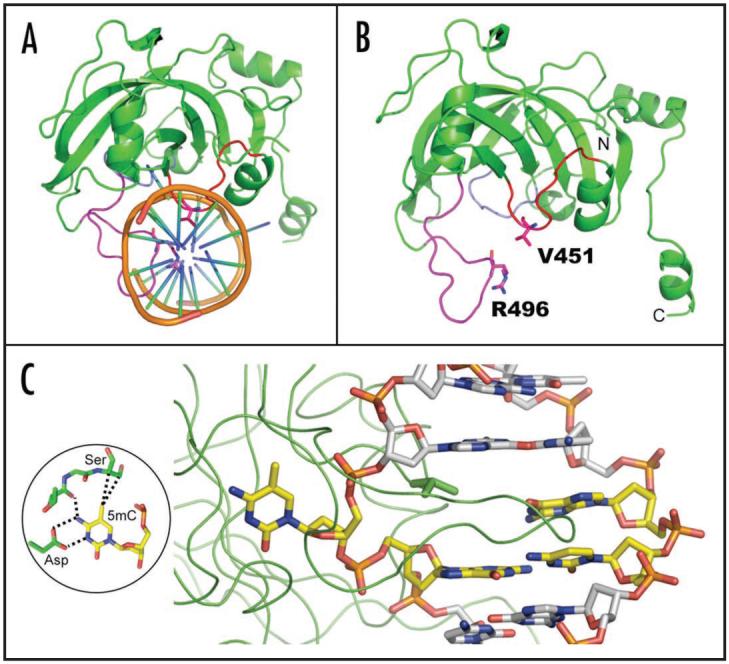
Structure of SRA-DNA complex. (A) The two loops—CpG recognition and base flipping—penetrate into the DNA helix from opposite directions. Adapted from ref.19 (B) The side chains of V451 of base flipping loop and R496 of CpG recognition loop are in direct van der Waals contact. Adapted from ref.19 (C) The 5mC flips out and is bound in a cage-like pocket.
Base Flipping Mechanism
Base flipping is a conserved mechanism that is widely used by nucleotide modifying enzymes, including DNA MTases,4,5 DNA repair enzymes,20-22 and RNA modification enzymes.23 This mechanism, first discovered in the bacterial 5mC MTase M.HhaI,5 involves enzyme binding to the DNA and eversion of the target nucleotide so that it projects out of the double helix and into the active-site pocket. The SRA domain is the first-discovered non-enzymatic sequence-specific DNA binding protein domain that uses the base flipping mechanism in its interaction with DNA.
One unexpected finding from our original structure19 is that the adenine (Ade) two bases 3′ to the 5mC, also adopts an extrahelical conformation and stacks against a hydrophobic surface patch. Concurrently, the end of DNA near the flipped Ade is destabilized, due to the lack of interactions with SRA domain. To address (1) whether this second flipping outside of CpG dinucleotide is Ade specific and (2) how the crystallographic restraining forces influence the stability of DNA ends, we determined three additional crystal structures using an oligonucleotide containing a thymine (Thy) at the position corresponding to flipped Ade, i.e., two bases 3′ to the 5mC (Table 1). The complexes crystallized either in the space group C2221 (with two different cell dimensions, crystal forms 1 and 2) or P21 (crystal form 3), with resolution ranging from 1.41 Å to 2.29 Å. Survey of the three structures revealed three modes of protein-DNA crystallographic packing forces (Fig. 3A-C). The high packing mode involves two DNA molecules stacking head-to-head surrounded by eight total SRA molecules, two interact specifically and six non-specifically (Fig. 3A) (crystal forms 1 and 2). The intermediate packing mode involves two head-to-head DNA molecules bound specifically by two SRA molecules plus two SRA capping on two ends of the DNA molecules, respectively (Fig. 3B) (crystal form 3). The low packing mode (only observed in crystal form 1) does not involve DNA-DNA stacking interactions. Instead, each DNA has one SRA molecule specifically bound at CpG site and another one non-specifically bound only at one end of DNA molecule, leaving the other end unattended (Fig. 3C)—a situation very similar to the crystal structure in P41212 space group where Ade flipping occurred.19 In contrast to the previously described, the Thy is intrahelical with normal base paring under all three modes of protein-DNA crystallographic packing forces.
Table 1.
New crystal structures of SRA-DNA complexes#
| Data collection | Crystal 1 | Crystal 2 | Crystal 3 |
| Space group | C2221 | P21 | |
| Cell dimensions | (α = β = γ = 90°) | α = γ = 90°, β = 98.9° | |
| a (Å) | 84.678 | 61.67 | 50.08 |
| b (Å) | 103.691 | 91.59 | 62.42 |
| c (Å) | 149.838 | 118.51 | 113.45 |
| Asymmetric unit | 2 complexes | 1 complex | 2 complexes |
| DNA | 5′-CCATGMGCTGAC-3′ 3′-GGTACGCGACTG-5′ |
||
| Beamline (SERCAT) | APS 22-ID | ||
| Wavelength (Å) | 1.00000 | ||
| Resolution (Å)* | 32.79-1.41 (1.45-1.41) |
29.91-1.99 (2.06-1.99) |
34.53-2.29 (2.37-2.29) |
| Rsym or Rmerge* | 0.095/0.771 | 0.102/0.568 | 0.102/0.352 |
| I/σI* | 10.9/1.0 | 9.0/3.6 | 10.5/2.3 |
| Completeness (%)* | 97.0/69.9 | 99.8/99.1 | 97.5/90.5 |
| Redundancy* | 5.1/1.7 | 9.6/6.5 | 3.4/2.9 |
| Observed reflections | 662,990 | 224,631 | 102,763 |
| Unique reflections* | 121,098 | 23,488 | 30,041 |
| Refinement | |||
| Resolution (Å) | 1.41 | 1.99 | 2.29 |
| No. reflections | 111,836 | 22,237 | 27,777 |
| Rwork/Rfree | 0.149/0.186 | 0.198/0.232 | 0.222/0.265 |
| Number of atoms | |||
| Protein | 3220 | 1609 | 3208 |
| DNA | 974 | 487 | 974 |
| Heterogen | 64 (ethylene glycol) |
54 (glycerol) |
- |
| Water | 596 | 162 | 150 |
| B-factors (Å2) | 17.8 | 28.8 | 35.1 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | 0.005 | 0.006 |
| Bond angles (°) | 1.1 | 1.1 | 1.1 |
| Dihedral angles (°) | 22.0 | 22.2 | 22.7 |
| Improper angles (°) | 0.99 | 0.96 | 1.08 |
| Estimated coordinate error | |||
| From Luzzati plot (Å) | 0.20 | 0.22 | 0.31 |
Highest resolution shell is shown in parenthesis.
Protein production, crystallization, X-ray data collection, structure determination and refinement were made as described {in ref. 19}.
Figure 3.
Three new X-ray structures in which the thymine two bases 3′ to the 5mC are intrahelical (see Table 1). (A and B) Crystal packing force involving two DNA molecules stacking head-to-head. The SRA domain molecules involved in specific interactions are shown in blue, and the SRA domain in non-specific interactions in green. (C) Crystal packing force involving a single DNA molecule. (D) Structural comparison of DNA containing an extrahelical Ade (magenta) and intrahelical Thy (green). (E) Structure of E. coli β clamp-DNA shows a flipped-out nucleotide at the junction of double-strand (ds) and single-strand (ss) DNA.
Superimposition of the intrahelical Thy and the extrahelical Ade indicates that the flipped Ade is associated with increased interstrand phosphate-phosphate distance (Fig. 3D). Trapping the flipped Ade is possible due to the existence of a hydrophobic surface patch nearby. Base flipping of an Ade nucleotide outside of recognition sequence (or non-target base) has been observed in HinP1I endonuclease,24 EcoP15I DNA MTase,25 EcoRV DNA MTase26 and E. coli MutY adenine glycosylase.27 Interestingly, the interaction between a hydrophobic surface patch and a flipped Ade is reminiscent of a structure of the E. coli β clamp on a primed DNA template,28 where the nucleotide (an Ade) of the terminal base pair flips out (Fig. 3E).
Comparison with HhaI MTase and MBD Domains
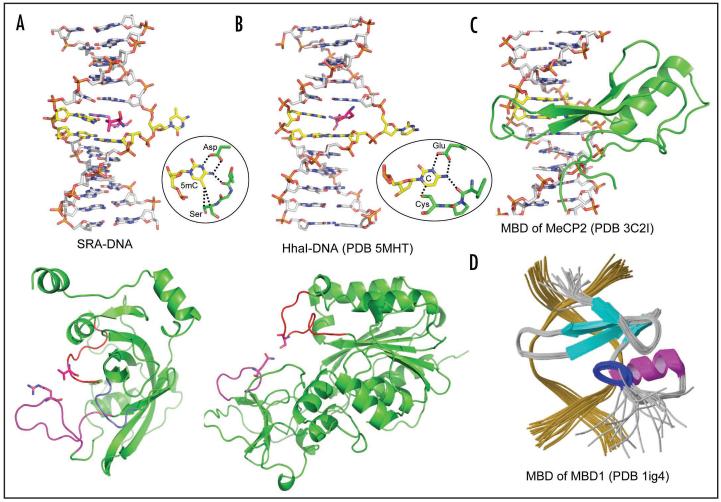
There is no apparent sequence or structural similarity between SRA and enzymes that flip base such as DNA MTase domains or DNA repair enzymes. However, several features are shared among these base-flipping proteins: the phosphodiester backbone pinching29 due to extensive protein-phosphate contacts surrounding the flipped nucleotide, the use of two loops to approach DNA from the major and minor grooves simultaneously, and the binding of the flipped base in a concave pocket (Fig. 4A and B).30 Whereas enzymes use base flipping to gain access to a DNA base to perform chemistry on it, the SRA domain probably uses base flipping to increase its protein-DNA interface 19 and to prevent the SRA domain from linear diffusion away from the target site on DNA. This may be particularly important for the SRA domain, as its recognition sequence is only two base pairs. In comparison, the surface area buried at the SRA-DNA interface is approximately 70% more than the buried surface at the methyl-binding domain (MBD) of MeCP2-DNA (Fig. 4C) and MBD1-DNA interfaces (Fig. 4D) that do not involve base flipping31,32 but also recognize only CpG sequence. We therefore suggest that the SRA-DNA interaction (through recognition and flipping of the 5mC) serves as an anchor to keep UHRF1 at a hemimethylated CpG site where it recruits Dnmt1 for maintenance methylation.
Figure 4.
Comparison of base flipping by the SRA domain and HhaI MTase. (Top) DNA structures bound by SRA (A) and HhaI (B) show a flipped nucleotide. The intercalating amino acids are shown in each case. (Bottom) Structures of SRA (A) and HhaI (B) show the two opposite-side DNA-approaching loops. Inserted are the major determinants of 5mC (A) and cytosine (B). (C) X-ray structure of MeCP2-DNA and (D) the NMR structure of MBD1-DNA shows MBD domain inserts a beta-hairpin through the DNA major groove. The methyl-binding domains of MBD1,31 and MeCP2,32 instead of using a base-flipping mechanism, recognize changes in hydration of the major groove of a fully methylated CpG rather than detecting methyl groups directly.
UHRF1-DNMT1 Interactions
There are two studies on the direct interaction between UHRF1 and Dnmt1, but they differ in the regions involved in the interaction.10,11 One study involves the SRA domain,11 whereas the other involves the PHD domain.10 From the side of Dnmt1, three regions have been implicated, residues 1-446, or 401-615, or 1081-1408.10,11 Additional data will be required to settle this point. It remains to be shown whether the binding of UHRF1 affects the affinity and/or specificity of DNMT1 to its substrate, hemi-methylated DNA.
UHRF1-histone Interactions
Besides the SRA domain, the Structural Genomics Consortium at Toronto has solved three additional domain structures by X-ray crystallography (Fig. 1B): the N-terminal ubiquitin-like domain (PDB 2FAZ33) and the tandem tudor domain with and without bound histone H3K9me3 (PDB 3DB3 and 3DB4,34) for human UHRF1, and the C-terminal RING domain (PDB 1Z6U35) of human UHRF2, whereas the RIKEN Structural Genomics/Proteomics Initiative at Japan solved an NMR structure for the PHD domain of human UHRF2 (PDB 2E6S36). Individual domain functions have been suggested for the PHD that may be involved in histone H3 tail binding,12,16 and for the RING domain that may confer E3 ubiquitin ligase activity on histones.12
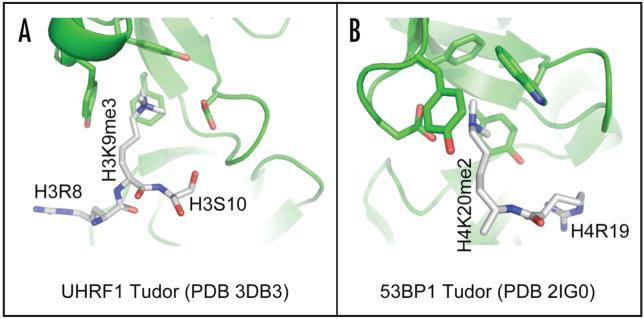
The new structure of the tandem tudor domain bound with histone H3 tail contains three structured H3 residues, H3R8-H3K9me3-H3S10 (PDB 3DB3) (Fig. 5A). Similarly only two structured histone residues, H4R19 and H4K20me2, were observed in the structure of another tandem tudor domain of 53BP1,37 (Fig. 5B). Structural comparison of two tandem tudor domains reveal that (1) the histone peptide runs along the shortest dimension of the domain, a possible reason that only 2-3 substrate peptide residues are structured; (2) the histone peptide is bound between the two individual tudor domains with the methyl-lysine binding aromatic cage formed by residues from a single tudor domain; and (3) a glutamate or aspartate residue in (or near) the binding cage seems to be a common mode of interaction involving methyl-lysine.38
Figure 5.
Methyllysine binding cage in tandom todor domain of (A) UHRF1 and (B) 53BP1.
Conclusion and Perspectives
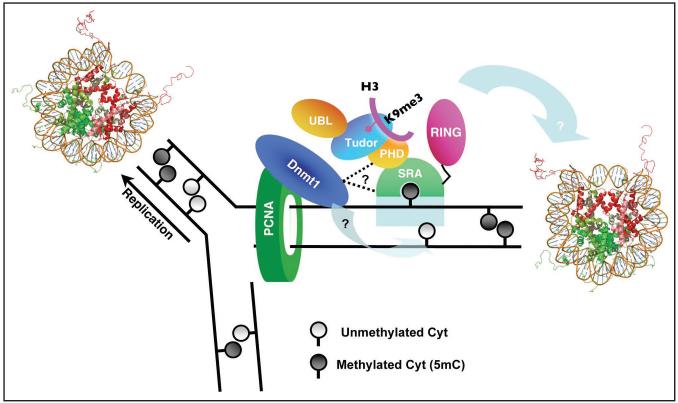
The methylation of mammalian DNA, primarily at CpG dinucleotides, has long been recognized to play a major role in controlling gene expression among other functions. Given their importance, many basic questions remain to be answered about the proteins responsible for this methylation, particularly how DNA methylation coordinates with the parallel chromatin-marking system that modifies histones. The fact that UHRF1 contains modules, within the same polypeptide, recognizing both DNA silencing mark (via the SRA) and histone silencing marks (via the Tudor and/or PHD) suggests it maybe a key component to couple the preservation of histone-modification through the cell cycle with maintenance DNA methylation (Fig. 6). In this context the missing key experiment is whether these domains act independently or in a cooperative fashion. For example, does binding of UHRF1 SRA domain to hemi-methylated DNA improve the binding of UHRF1 Tudor and/or PHD domains to histone tail? Furthermore, the E3 ubiquitin ligase activity,12 residing in the C-terminal RING domain, may add an additional level of modification, such as H2A ubiquitylation that is enriched with inactive genes.39 Without understanding the interactions and spatial relationships between such modular domains, or whether inter-domain interactions contribute to target specificity, it is not possible to construct a temporal sequence of events or causal relationships in gene silencing.
Figure 6.
Hypothetical model of UHRF1-mediated replication-coupled crosstalk between DNA methylation and histone modifications. Existence of both silencing mark readers recognizing DNA (via the SRA) and histone (via the Tudor and/or PHD) facilitates the idea of maintenance and conversion of epigenetic silencing marks on both DNA and histone modifications.
The experimental characterizations of Dnmts and their associated protein factors are providing a rapidly and convergent picture of the kinetic mechanisms (activities of oligomers40), binding partners (UHRF1-Dnmt1,9,10 and Dnmt3L-Dnmt3a41), chromatin recognition42 (histone binding such as H3K4me0,43 and H3K9me3,44), RNA-directed DNA methylation.45,46 However, understanding the basis for establishing, maintaining and disturbing DNA methylation patterns will require a much better understanding of the union between structure and function in the Dnmts and their associated protein factors than we currently possess.
Acknowledgements
We thank Dr. Ashok Bhagwat for critical comments. The Emory University School of Medicine supported the use of SER-CAT beamlines. This work was supported by grant GM049245 to X.C. from the National Institutes of Health (NIH). X.C. is a Georgia Research Alliance Eminent Scholar. The X-ray structures (coordinates and structure factor files) of mouse UHRF1 SRA with bound DNA have been submitted to PDB as 3FDE (crystal form 1), 3F8J (crystal form 2) and 3F8I (crystal from 3) respectively.
References
- 1.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–50. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci USA. 2005;102:8905–9. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–95. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–69. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–6. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- 8.Ooi SK, Bestor TH. Cytosine methylation: remaining faithful. Curr Biol. 2008;18:174–6. doi: 10.1016/j.cub.2007.12.045. [DOI] [PubMed] [Google Scholar]
- 9.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 10.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 11.Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, Schini-Kerth VB, Bronner C. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–97. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 12.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–35. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem. 2002;277:34549–55. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- 14.Papait R, Pistore C, Grazini U, Babbio F, Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada F, Leonhardt H, McBlane F, Oudet P, Bonapace IM. The PHD Domain of Np95 (mUHRF1) Is Involved in Large-Scale Reorganization of Pericentromeric Heterochromatin. Mol Biol Cell. 2008;19:3554–63. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol Biol Cell. 2007;18:1098–106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–17. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–21. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 18.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–5. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–9. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–5. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–5. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 22.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–7. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–48. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy YV, Rao DN. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J Mol Biol. 2000;298:597–610. doi: 10.1006/jmbi.2000.3673. [DOI] [PubMed] [Google Scholar]
- 26.Gowher H, Jeltsch A. Molecular enzymology of the EcoRV DNA-(Adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J Mol Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 27.Bernards AS, Miller JK, Bao KK, Wong I. Flipping duplex DNA inside out: a double base-flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J Biol Chem. 2002;277:20960–4. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- 28.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werner RM, Jiang YL, Gordley RG, Jagadeesh GJ, Ladner JE, Xiao G, Tordova M, Gilliland GL, Stivers JT. Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase. Biochemistry. 2000;39:12585–94. doi: 10.1021/bi001532v. [DOI] [PubMed] [Google Scholar]
- 30.Cheng X, Blumenthal RM. Finding a basis for flipping bases. Structure. 1996;4:639–45. doi: 10.1016/s0969-2126(96)00068-8. [DOI] [PubMed] [Google Scholar]
- 31.Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–97. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 32.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29:525–31. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 33.Walker JR, Wybenga-Groot L, Doherty RS, Finerty PJ, Jr, Newman E, Mackenzie FM, Weigelt J, Sundstrom M, Arrowsmith C, Edwards A, Bochkarev A, Dhe-Paganon S. Ubiquitin-Like Domain of Human Nuclear Zinc Finger Protein NP95. 2005 [Google Scholar]
- 34.Walker JR, Avvakumov GV, Xue S, Dong A, Li Y, Bountra C, Weigelt J, Arrowsmith CH, Edwards AM, Bochkarev A, Dhe-Paganon S. Cryptic tandem tudor domains in UHRF1 interact with H3K9ME and are important for pericentric heterochromatin replication. 2008 [Google Scholar]
- 35.Walker JR, Avvakumov GV, Xue S, Newman EM, Mackenzie F, Sundstrom M, Arrowsmith C, Edwards A, Bochkarev A, Dhe-Pagnon S. 2.1 Angstrom Crystal Structure of the Human Ubiquitin Liagse NIRF. 2005 [Google Scholar]
- 36.Kadirvel S, He F, Muto Y, Inoue M, Kigawa T, Shirouzu M, Terada T, Yokoyama S. Solution structure of the PHD domain in RING finger protein 107. 2006 [Google Scholar]
- 37.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brent MM, Marmorstein R. Ankyrin for methylated lysines. Nat Struct Mol Biol. 2008;15:221–2. doi: 10.1038/nsmb0308-221. [DOI] [PubMed] [Google Scholar]
- 39.Lee N, Zhang Y. Chemical answers to epigenetic crosstalk. Nat Chem Biol. 2008;4:335–7. doi: 10.1038/nchembio0608-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurkowska RZ, Anspach N, Urbanke C, Jia D, Reinhardt R, Nellen W, Cheng X, Jeltsch A. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 45.Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–5. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 46.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]