Abstract
Understanding the structural adaptation and signaling of adhesion sites in response to mechanical stimuli requires in situ characterization of the dynamic activation of a large number of adhesion components. Here, we review high resolution live cell imaging approaches to measure forces, assembly and interaction of adhesion components, and the activation of adhesion-mediated signals. We conclude by outlining computational multiplexing as a framework for the integration of these data into comprehensive models of adhesion signaling pathways.
Introduction
Adhesion of cells to extracellular matrix (ECM) and neighboring cells plays an essential role in the development and function of multicellular organisms. In addition to providing mechanical support, adhesions operate as dynamic signaling hubs which transmit information in and out of cells. On one hand environmental signals, including mechanical stresses, are transmitted from ECM to the intracellular domain via adhesion receptors, where they contribute to the regulation of cellular responses such as proliferation, differentiation, migration, or death. On the other hand activation and affinity of adhesion receptors for binding extracellular ligands, and thus for adhering to ECM, is regulated by intracellular signaling mechanisms. This transduction of outside-in and inside-out signals is modulated via dynamic feedback interactions among adhesion receptors, cytoskeletal elements, and a wide variety of adaptor proteins. A recent study has identified as many as 156 component molecules that cooperate through manifold interactions to mediate the link between cell and ECM [1].
Attaining a systems-level understanding of adhesion signaling pathways requires accurate in situ characterization of adhesion components in the context of ever-changing cell functions. Importantly, to resolve feedback interactions among these components, time-resolved measurements of their activation and deactivation are required. Such data allows the inference of input – output relationships between components and the identification of loops therein. Classical biochemical detection methods average cell behavior across large populations and are limited to the acquisition of snapshots of the signaling state at fixed time points. Thus, they largely fail to report on the transient dynamics of molecular interactions and activities, which varies between cells and between adhesions within a single cell. At best these methods allow the detection of adhesion signals in response to acute and global stimulations [2] that are strong enough to homogenize the signaling state within cells and across a cell populations. However, it remains difficult to identify the dynamics of feedback interactions based on such data and there is the concern that such stimulation activates pathways outside their normal range of operation.
As an alternative, high-resolution live cell imaging allows the study of spatially and temporally heterogeneous adhesion signals at the sub-cellular scale. Furthermore, live cell microscopy is often sensitive enough to capture the dynamics of constitutive adhesion signaling and structural adaptation at a steady state. In situ measurements of the timing among molecular activities can be further processed mathematically to determine the cause-and-effect cascades and the kinetics of signal transduction. Here we review emerging visualization and image analysis techniques that will advance a quantitative understanding adhesion dynamics.
Visualizing force response in adhesions
Focal adhesions are the primary locations at which cells sense the mechanical properties of the environment. Therefore, measuring cellular traction forces on substrates is an essential step in deciphering the mechanism of signal transduction in adhesions. Traction force microscopy is a standard procedure for the reconstruction of cellular traction forces. Cellular forces exerted on the ECM are measured via the displacement of marker micro-beads embedded in an elastic substrate. Displacement maps are then computationally converted into force maps with the assumption that the substrate deforms elastically under the force impact [3, 4]. Using traction force microscopy of migrating fibroblasts expressing green fluorescence protein (GFP)-zyxin Beningo et al. showed that nascent focal adhesions near the leading edge transmit strong propulsive forces, whereas the large mature focal adhesions behind the leading edge mainly serve as passive anchor points for maintaining a spread cell morphology [5]. One limitation of these early applications of traction force microscopy is the spatial resolution of a few µm. Thus, it has been difficult to differentiate in more detail the force responses of different adhesion types in different cellular locations, and to monitor how the spatially heterogeneous responses alter as the cell migrates. Some of these problems have been alleviated by culturing cells on arrays of micro-fabricated elastic pillars that deform under traction forces [6]. It was shown that each pillar acts as a localized force sensor, allowing the read out of an independent force response for each adhesion. Also, Sabass et. al. recently optimized substrate-based traction force measurements by seeding fluorescent nano-beads in the substrate at the density of the light-optical diffraction limit [7]. In combination with improved computational algorithms traction force maps were obtained with a resolution of ~1 µm, sufficient to analyze force transduction at individual adhesion sites. Another limitation inherent to the measurement of substrate-coupled forces is that only extracellular force components are detected. Complementary approaches are emerging deriving the intracellular force levels from the transient deformation of cytoskeletal structures [8]. A combination of these methods with high-resolution traction force microscopy will open the possibility to define the hierarchy of force transduction at individual adhesion sites in different states.
Visualizing and measuring structural interactions within adhesions
Transmission of forces between the cytoskeleton and ECM is mediated by a chain of dynamically interacting adhesion molecules linking integrin receptors to the actin filament (F-actin) system. Identification of the molecular basis of the ECM-integrin-actin linkage requires direct measurement of the dynamics of these molecules in living cells. This has been achieved recently by two very similar live cell imaging approaches:
Correlational fluorescence speckle microscopy
Fluorescence speckle microscopy (FSM) relies on the stochastic variation in the spatial density of fluorophores that are incorporated at very low abundance into a macromolecular assembly, such as an array of actin filaments [9]. In a high resolution image of the assembly, regions with a higher fluorophore density than the immediate surroundings generate a local intensity maximum above a dim background, referred to as a speckle. Speckle movements indicate the translocation and deformation of the labeled assembly, while fluctuations in the speckle intensity indicate the exchange of molecular subunits within the assembly. The correlated movements of speckles in two molecular assemblies labeled with spectrally distinct fluorophores can indicate the interaction between the assemblies, as demonstrated for components of focal adhesions and F-actin [10].. Multi-spectral correlational FSM was combined with total internal reflection microscopy (TIRFM) to visualize and quantify the dynamic interaction of focal adhesion proteins with the actin cytoskeleton specifically at the ventral cell-ECM contacts. Speckles were generated by expressing in epithelial cells very low amounts of GFP-conjugates to three classes of adhesion proteins: fibronectin-binding integrins (GFP-αv integrin coexpressed with untagged β3 integrin); proteins binding F-actin directly (α-actinin, vinculin, and talin); and adhesion core proteins (paxilin, zyxin, and focal adhesion kinase (FAK)) that bind neither F-actin nor ECM directly. The movement of adhesion speckles was correlated with the movement of F-actin speckles generated by injecting low amounts of X-rhodamine-labeled actin monomers into the same cells. The data revealed a decreasing motion correlation from actin-binding proteins to core proteins and further to integrins, indicating the hierarchical transmission of F-actin flow through focal adhesions. The study suggested that interactions between vinculin, talin, and F-actin constitute a slippage interface between the cytoskeleton and integrins, referred to as a molecular friction clutch. It was shown that variations in the level of friction are directly coupled to local variations in the protrusion efficiency of epithelial cells. Thus, it was concluded that the variable state of structural interactions among adhesion proteins defines the morphological transitions required for directed cell migration [10].
Spatio-temporal image correlation spectroscopy
Similar to correlational FSM, spatio-temporal image correlation spectroscopy (STICS) [11], a derivative of fluorescence correlation microscopy (FCS) [12] detects directed movements of fluorescently tagged proteins by calculating the spatial correlation of the image signal as a function of time lag between image pairs. Persistent directed motion of molecules will result in a correlation peak that begins at zero time lag and moves in the direction opposite to the molecular flow. Thus, high-resolution velocity maps of adhesion components have been obtained and compared to flow maps of actin [13]. These measurements were used to examine the efficiency of the linkage between integrin and F-actin among different cell types and the dependence on ligand density. The study suggested that the efficiency increases as F-actin and adhesions become more organized showing the importance of signaling activities that control the structural dynamics within individual adhesion sites.
Visualizing signaling activities using biosensors
Activation of integrins leads to the activation of intracellular signals including the Rho family of small GTPases, protein tyrosine and serine/threonine kinases, lipid kinases and membrane transporters [1,14]. In turn, integrin–ligand binding is regulated by intracellular signals [15]. To study the dynamics of these processes in living cells, fluorescent reporters called biosensors have been designed to monitor the activation state of signaling molecules in real-time. The design of current biosensors can be coarsely classified in three groups [16,17]: 1) Fluorescence-resonance-energy-transfer (FRET)-based biosensors measure inter- and intra molecular distance changes that occur with conformational changes during activation/deactivation, or with the binding of a fluorescent reporter to a specific signaling molecule in either the activated or deactivated state. 2) Environment-sensitive fluorophores change their fluorescence properties when the signaling molecule or substrate undergoes a conformational change, or during the binding of the signaling molecule to an upstream activator or downstream effector. 3) Biosensors that report changes in the localization or expression level of a signaling molecule via redistribution of the fluorescent probe. Table 1 lists a representative selection of biosensors for detecting adhesion-related protein activities.
Table 1.
Live cell biosensors of adhesive signaling: protein conformational change, posttranslational modification, and second messengers.
| Vinculin [49] | Intramolecular FRET of FPs | Insertion of a FRET pair in vinculin | |
| α4 [50] | Intermolecular FRET of FDs | LDV-Fluorescein; octadecyl rhodamine B | |
| αIIbβ3 [38] | Intermolecular FRET of FDs | Dye-Fab fragments of two different antibodies | |
| αLβ2 [51,52] | Intermolecular FRET of FPs | αL-mCFP; αL-mYFP | |
| Myosin II [26,53] | Intermolecular FRET of FDs | Dye-RLC and -HC of Myosin II | |
| N-WASP [54,55] | Intramolecular FRET of FPs | YFP- N-WASP-CFP (Stinger) or CFP- N-WASP-YFP | |
| Profilin [56] | Probe redistribution | Dye-actin with increased affinity to profilin | |
| RhoA [57] | Intramolecular FRET of FPs | YFP-RBD (PKN)-RhoA-CFP-Farnesyl moiety (Raichu) | |
| RhoA [29] | Intramolecular FRET of FPs | RBD (rhotekin)-CFP-YFP-RhoA | |
| Arf6 [41] | Intermolecular FRET of FPs | Arf6-CFP; YFP-GGA3 effector domain | |
| Cdc42 [27] | Environment-sensing FD | Dye-CBD from WASP (MeroCBD) | |
| Cdc42 [58] | Intermolecular FRET of FP and FD | GFP-Cdc42; Alexa-PBD | |
| Cdc42/Rac [59] | Intramolecular FRET of FPs | EBFP-PBD [PAK]-EGFP | |
| heterotrimeric G-proteins [60] | Intermolecular FRET of FPs | α-CFP; β-YFP | |
| Rac1/Cdc42 [61] | Intramolecular FRET of FPs | YFP-PBD (PAK)-Rac1/Cdc42-CFP-Farnesyl moiety (Raichu) | |
| Rac1 [24] | Intermolecular FRET of FP and FD | GFP-Rac1; Alexa546-PBD from PAK (FLAIR) | |
| Ran [62] | Intramolecular FRET of FPs | YFP-RBD (Yrb1)-CFP; YFP-IBB (importin alpha)-CFP | |
| Ras/Rap1 [28] | Intramolecular FRET of FPs | YFP-Ras/Rap1-RBD (Raf)-CFP-Farnesyl moiety (Raichu) | |
| Akt/PKB [63] | Intramolecular FRET of FPs | CFP-14-3-3-substrate-YFP | |
| Akt/PKB [64] | Intramolecular FRET of FPs | CFP-FHA2-Akt substrate-YFP | |
| CaMKII [65] | Intramolecular FRET of FPs | YFP-CaMKII-CFP | |
| EGFR [66] | Intermolecular FRET using FLIM | GFP-EGFR; dye-pTyr antibody | |
| Erk [45] | Intramolecular FRET of FPs | CFP-Erk-YFP | |
| FAK [37] | Intermolecular FRET of FPs | CFP-FAK; dSH2 (Src)-YFP | |
| FAK [37] | Intramolecular FRET of FPs | Insertion of a FRET pair in FAK | |
| MK2 [67] | Intramolecular FRET of FPs | CFP-MK2-YFP | |
| MLCK [68] | Intramolecular FRET of FPs | MLCK-GFP-CaMBD-BFP (MLCK-FIP) | |
| phospho-CrkII [69] | Intramolecular FRET of FPs | CFP-CrkII-YFP | |
| PKA [70] | Intramolecular FRET of FPs | CFP-KID of CREB-YFP | |
| PKA [71] | Intramolecular FRET of FPs | CFP-14-3-3tau-PKA substrate-YFP | |
| PKC [25] | Intermolecular FRET using FLIM | FP-PKC; dye-phospho antibody | |
| PKC [30] | Intramolecular FRET of FPs | CFP-FHA2-PKC substrate-YFP | |
| PKC [39] | Environment-sensing FD | Caged dye-substrate | |
| PKD [43] | Intramolecular FRET of FPs | CFP-FHA2-PKD substrate-YFP | |
| PTK [72] | Environment-sensing FD | caged dye-substrate | |
| pTyr [73] | Probe redistribution | YFP-dSH2 (Src) | |
| pTyr [31] | Intramolecular FRET of FPs | CFP-SH2-Tyr substrate-YFP | |
| Src [31,34] | Intramolecular FRET of FPs | CFP-SH2-Tyr substrate-YFP | |
| Lipid rafts [74] | Intermolecular FRET of FPs | MyrPalm/GerGer/PalmPalm/Caveolin-mCFP/mYFP | |
| PI(3,4,5)P3 [75] | Probe redistribution | PH (ARNO or GRP1)-GFP | |
| PI(3,4,5)P3 [18] | Probe redistribution | PH (BtK)-GFP | |
| PI(3,4,5)P3 [76] | Intramolecular FRET of FPs | YFP-binding domain of IP3R1-CFP | |
| PI(4)P [19] | Probe redistribution | GFP-PH of OSBP | |
| PI(4,5)P2 [20,21] | Probe redistribution | GFP-PH of PLCdelta1 | |
| PI3P [22] | Probe redistribution | GFP-RING FYVE domains | |
| PIP3/PIP2 [23] | Probe redistribution | PH (PKB or GRP1)-GFP |
FRET, fluorescence resonance energy transfer
FLIM, fluorescence Lifetime Imaging
FP, fluorescent protein
FD, fluorescent dye
Intram., intramolecular
Interm., intermolecular
LDV, Leu-Asp-Val (integrin binding peptide)
CFP, cyan fluorescent protein
YFP, yellow fluorescent protein
RLC, (myosin) regulatory light chain
HC, (myosin) heavy chain
N-WASP, Neural Wiskott-Aldrich syndrome protein
PKN, protein kinase N
RBD, Rho, Ras or Rap1-binding domain
CBD, Cdc42-binding domain
PBD, p21-binding domain
Yrb1, yeast Ran-binding protein 1
IBB, importin-β-binding domain
FHA2, forkhead-associated 2 domain
CaMKII, calcium/calmodulin-dependent protein kinase II
SH2, Src homology 2 domain
dSH2, dual Src homology 2 domains
MyrPalm, myristoylated and palmitoylated
GerGer, geranylgeranylated
PalmPalm, tandemly palmitoylated
MLCK, myosin light chain kinase
MK2, MAPK-activated protein kinase 2
PH, pleckstrin homology domain
ARNO, ARF nucleotide-binding site opener (Arf GEF)
GRP1, general receptor for phosphoinositides-1
BtK, Bruton’s tyrosine kinase
IP3R1, inositol 1,4,5-triphosphate receptor 1
CaMBD, calmodulin-binding domain
EBFP or BFP, enhanced blue fluorescent protein
OSBP, oxysterol binding protein
RING, zinc finger related domain named after Really Interesting New Gene
FYVE, zinc finger domain named after Fab1, YOTB, Vac1 and EEA1
PKA, Akt/PKB, PKC, PKD, PKN, PTK, EGFR and FAK are not listed.
The many valuable biosensors now available for adhesion research are based largely on a few design themes. To design and interpret experiments, it is important be aware of the caveats, appropriate applications and controls for these designs. Most commonly a biosensor is based on an interaction between the molecule being studied and some ‘affinity reagent’, a protein fragment that binds only to the active form of the targeted molecule. In some simple and therefore robust designs, an affinity reagent fused to GFP simply translocates to regions where activated protein has built up [18–23]. This straightforward visualization of activity minimizes artifacts due to image analysis, but it can be difficult to produce quantitative readouts using simple translocation. Furthermore, translocation biosensors often do not reveal subtle gradients of activity. Because accumulation of the biosensor must be visualized over a background of the same fluorescence “color”, such biosensors work best when the activated target concentrates in a clearly demarcated subcellular region, producing stark contrast between the localized and unlocalized states.
There are very real practical concerns that limit the utility of any biosensor, including the expression level needed to produce useful signal/noise. Higher biosensor concentrations lead to brighter signal, but overexpression can mask activation signals if the biosensor saturates binding sites producing localization, or saturates upstream pathways that produce activation. Biosensors can act as dominant negative inhibitors, binding their targets and preventing interactions with downstream effectors; this is alright provided the biosensor is used at a suitable concentration (i.e. measured as brightness / unit area) where physiological effects are not seen and where the biosensor shows normal localization. Biosensors that compete with upstream, activating targets are more problematic as they mask the very activation they are meant to report.
Biosensors where activation produces changes in fluorescence excitation or emission (i.e. using FRET, solvent sensitive dyes, or lifetime imaging) can reveal subtle gradients in activation and can be more readily quantified [24,25]. Clearly, among these, genetically encoded designs have a clear advantage in convenience of use, and access to cells, tissues and animals. Designs requiring microinjection (i.e. dye-labeled proteins [26,27]) are used when warranted for enhanced sensitivity, multiplex imaging etc. The great majority of genetically encoded biosensors are based on FRET, though other techniques such as BiFC are finding a niche. When comparing advantages and caveats, genetically encoded FRET biosensors can be grouped into three broad categories: intramolecular FRET between a linked target and affinity reagent, intermolecular FRET when the target and affinity reagent are not linked, and biosensors that report modifications of a substrate peptide. In intramolecular FRET biosensors, a single chain incorporates the target protein, the affinity reagent, and two fluorescent proteins that undergo FRET. When activation occurs, the affinity reagent interacts with the target protein, changing the orientation and/or distance between the fluorescent proteins to affect FRET. With all components linked in one chain, image analysis is greatly simplified because it is not possible for the fluorescent proteins to have different subcellular distributions [28]. However, design of such biosensors can be more difficult, as linking multiple components can affect biological activity [29]. For intermolecular biosensors, the affinity reagent, bearing one of the fluorescent proteins, is expressed separately from the target protein, which is fused to the other fluorescent protein. In recent years this design has been used less frequently because it requires careful bleedthrough corrections to avoid artefacts, and because the response of the biosensor may be influenced by differences in the local concentrations of the two species. However, it has also recently become apparent that such intermolecular designs can provide greatly superior sensitivity, as there is greater fluorescence change between the active and inactive state. Furthermore, this design is less prone to artefacts in which normal activities of the target protein are reduced. Importantly, either design works well when one is simply monitoring the kinetics of activation on a single cell basis, or in suspended cell populations, without attempting to map spatial variations. Other designs are based on substrate peptides modified so that phosphorylation affects intramolecular FRET [30,31]. Here, the advantages of intramolecular FRET imaging apply, but specificity can be an issue, as the substrate can be affected by more than one protein. When comparing biosensors, it is valuable to focus on in vitro studies showing fluorescence spectra. Carefully note methods of background subtraction or normalization. The many different methods used to quantify extent of change in vivo can readily lead to false conclusions. Using different methods in the published literature, the same biosensor can be shown producing a cell signal that varies 40%, or >100 fold.
Mechanical stimulation of signaling activities in adhesions
Adhesion sites respond to mechanical stimuli both by structural adaptation and by activation of signaling cascades. A large body of literature, reviewed comprehensively elsewhere [32,33], reports responses of adhesions to global changes in tension induced by e.g. altering substratum stiffness, exposing cells to shear flow, or inhibition and activation of intracellular contraction. More recently, by combining the power of high-resolution live cell microscopy and micro-manipulation techniques it has become possible to measure the dynamics of structural adaptation and signal activation in response to very locally applied loads. Src is known to regulate the integrin-cytoskeleton interaction. Measurement of FRET signals reporting on activation of Src in response to stimulation by laser-tweezer traction on fibronectin-coated beads adhering to human endothelial cells shows a rapid distal Src activation and a slower Src activation wave that propagates directionally along the cell membrane [34]. The long range waves were destroyed by disruption of actin filaments with cytochalasin D or perturbation of microtubules with nocodazole indicating the critical role of the cytoskeleton in transmission of mechanically induced signals to specific destinations. Another study used optical tweezers to apply local forces to fibroblasts, inducing focal complex formation, as observed by monitoring GFP-vinculin localization. This work was seminal in demonstrating the reinforcement of cell-ECM adhesions in response to force increase [35]. The results obtained from a FRET based assay of Rac1 activation in fibroblast cells incubated with fibronectin-coated beads for local mechanical stimulation showed that interaction of activated Rac1 with cytoplasmic effectors is enhanced in specific regions near cell edges and is induced locally by integrin stimulation [36].
Multiplexed imaging of signaling and structural component activities
For a complete understanding of the functions of adhesion signaling, the dynamic activities of all relevant adhesion components should be related to structural responses at adhesion sites and to morphological responses at the cellular level. To date quantification of the activation dynamics of multiple signaling molecules and/or the assembly dynamics of multiple structural components in the same cell has been hindered by the spectral overlap of fluorescent proteins, allowing simultaneous observation of at most 4 – 6 colors, but often only 2 – 3 colors. Whereas this has provided the possibility to monitor structural interactions among several adhesion proteins [10, 13, 37–46], the imaging of multiple biosensors is not yet as well established. Many of the existing biosensors use fluorophores with similar wavelengths and thus cannot be combined. More fundamentally, most biosensors rely on interactions between two fluorophores, or spectral shifts of a single fluorophore. This uses a relatively broad portion of the wavelength spectrum and limits detection to one or two biosensors at a time. Even if these limits are pushed in the future, it seems unlikely that more than a few of the ~150 relevant adhesion components can be imaged simultaneously. Therefore, we conclude this review by outlining a new approach that will enable the integrated analysis of many adhesion components using “computational multiplexing” to correlate the activities of individual components measured independently in separate experiments.
Computational multiplexing relies on the repeated sequential observation of pairs of activities. Each experiment defines whether the two observed activities are related, i.e. the associated components are directly or indirectly connected in a pathway. The hierarchy of multiple components within a pathway is then inferred by compiling the pairwise relationships that account for the strengths of the measured connections between components. Using high-resolution live cell imaging, the strength of a connection and the upstream-downstream relationship between two components can be estimated from constitutive pathway fluctuations at steady state ([47] and unpublished data). Constitutive pathway fluctuations are stimulated, for example, by random binding of a ligand to a surface receptor, or a local increase in the concentration of a signaling molecule. Fluctuations propagate through the pathway, resulting in correlated fluctuations of connected components, while fluctuations in unconnected components are uncorrelated.
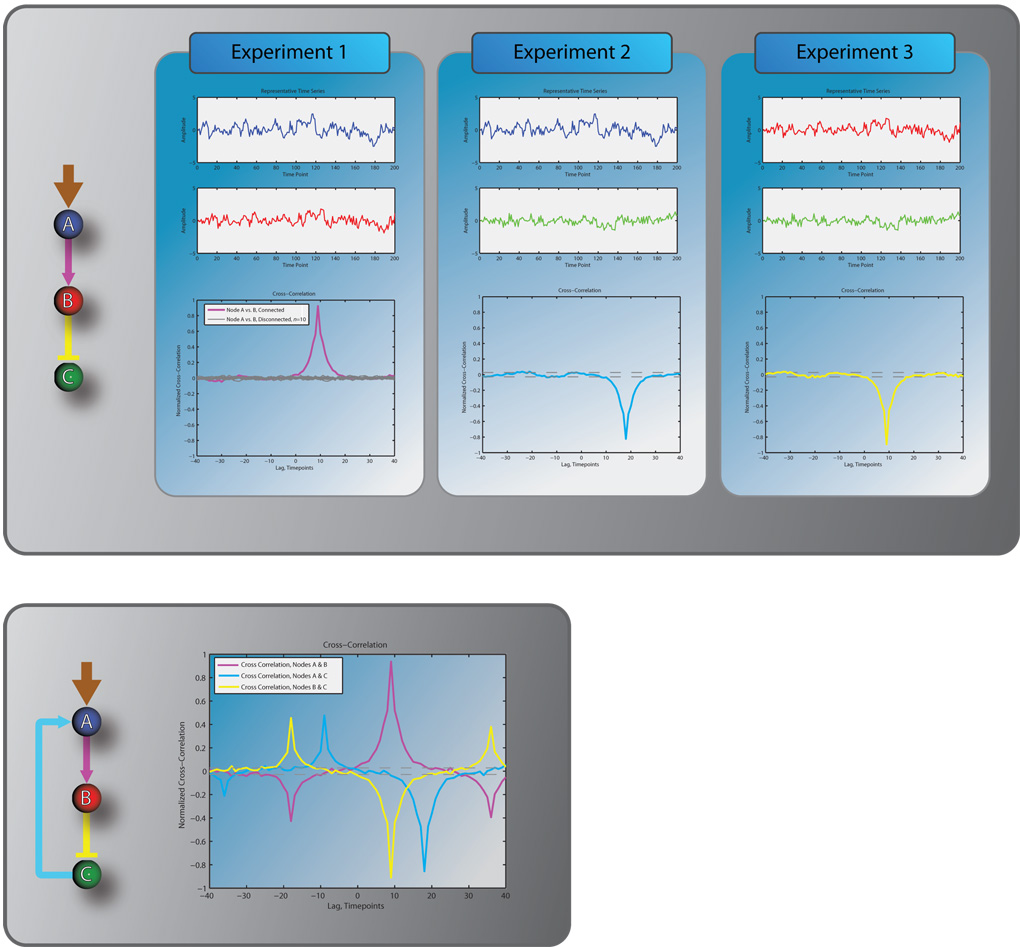
Figure 1 illustrates, for a hypothetical pathway with three nodes (A, B, C), how the pairwise correlation between constitutive fluctuations in different component activities can be exploited to resolve the hierarchy of nodes within the pathway. In experiment 1, time courses of the component activity in node A and in node B are measured. The cross-correlation of the two time courses displays a significant positive peak with a positive time lag, suggesting that node A is an upstream activator of node B. If the two nodes were not connected, their component activities would exhibit insignificant correlation at any time lag (see cross-correlation functions in gray). In experiment 2, time courses of the component activity in node A and in node C are measured. The cross-correlation of the two time courses displays a weaker, yet still significant negative peak with a positive time lag, suggesting that node A is also upstream of node C (because of the positive time lag), but that A and C are in an inhibitory relationship (because of the negative peak). Furthermore, since the magnitude of the correlation between A and C is lower than that between A and B, and because the time lag between A and C is larger than that between A and B, the two experiments together provide evidence for a cascade in which A activates B, which inhibits C. Further confirmation for this prediction could be gained from a third experiment in which the component activities in nodes B and C are measured. However, simple pairwise correlation analysis can not distinguish this model from the alternative model that A activates B, and directly inhibits C independent of B. Experimentally this could be resolved by molecular intervention where the function of B is blocked. In addition, complementary mathematical approaches are emerging which can distinguish between these competing models by detailed analysis of the signal fluctuations [48]. Evidently, the quantitative integration of cross-correlation data from many experiments into an unambiguous pathway graph is technically more involved and goes beyond the scope of this review. However, the example indicates that sequential accumulation of evidence for the coupling of component activities contains critical information for the reconstruction of pathways.
Figure 1.
Resolving the hierarchy of pathway components by pairwise cross correlations of fluctuations in time courses.
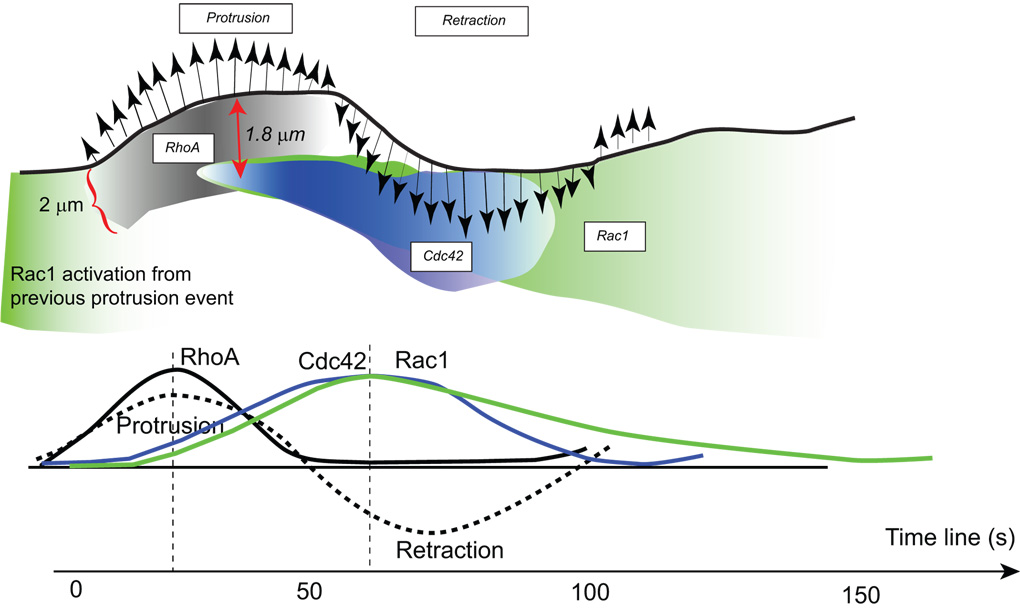
By computational multiplexing it was possible, for example, to establish the spatiotemporal coordination of the Rho GTPases Rac1, Cdc42, and RhoA at the leading edge of migrating mouse embryonic fibroblasts [47]. Experiments were combined in which RhoGTPases were imaged individually. Their activities were related to the shape fluctuations of the cell edge, serving as a common “timer” between all experiments. The data showed that RhoA is activated concurrently and specifically during edge protrusion, while Rac1 and Cdc42 are activated with a delay of ~40 s (Figure 2). Importantly, in a control experiment where the level of biosensor expression or microinjection was varied these timing relations remained unaffected [47]. Both expression level as well as the combination of different biosensor designs needs to be carefully calibrated for the extraction of kinetics. By analyzing the correlation between activities at various distances from the cell edge, it was also established that Rac1 and Cdc42 activities are initiated ~1.8 µm from the cell edge. Rac1 and Cdc42 maintain significant levels of activity during retraction phases following the protrusion, such that low level Rac1 activation overlapped with the onset of a next protrusion phase. Based on these results it was proposed that RhoA acts as an initiator of actin polymerization leading to cell edge protrusion while the roles of Rac1 and Cdc42 may be involved in reinforcing protrusion at a later time point through the control of adhesion dynamics.
Figure 2.
Model of spatiotemporal activation of Rac1, Cdc42, and RhoA at the leading edge of migrating mouse embryonic fibroblasts as determined by computational multiplexing. Adapted from [47].
Correlation-based computational multiplexing relies on the assumption that pathway nodes are connected in linear cascades. This model breaks down in the presence of significant feedback interactions between nodes. For the simple linear feedforward pathway in Figure 1, addition of a positive feedback from node C to node A results in additional features in pairwise cross-correlations indicative of nonlinear oscillatory behavior. Thus, the relationships between component activities are no longer unambiguous. Novel mathematical approaches for computational multiplexing are being developed to identify such feedback interactions from time courses of constitutive fluctuations in many molecular activities.
Acknowledgement
We thank Hunter Elliott for simulations of pathway fluctuations in Figure 1. This research is funded by the Cell Migration Consortium (NIH grant U54 GM64346 to G.D. and K.M.H) and by the P50 GM68762 (to G.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1. Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858.. A new approach in study of molecular basis of integrin-mediated signaling which is based on detailed data mining of published experimental studies. It provides a visual perspective of the entire integrin signaling network known so far. Further insights are provided through simulations detecting common network motifs.
- 2.Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao GY, DeLisser H, Schwartz MA. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 3.Oliver T, Jacobson K, Dembo M. Design and use of substrata to measure traction forces exerted by cultured cells. Methods Enzymol. 1998;298:497–521. doi: 10.1016/s0076-6879(98)98042-9. [DOI] [PubMed] [Google Scholar]
- 4.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881.. An outstanding work on visualization and analysis of the role of focal adhesions in force transduction during cell migration. This study showed small nascent adhesions near the leading edge are more important for transmission of strong propulsive tractions than large, mature focal adhesions. They showed mature focal adhesions act as passive anchorage points responsible for maintaining a spread cell morphology.
- 6.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. From the Cover: Cells lying on a bed of microneedles: An approach to isolate mechanical force. PNAS. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophysical Journal. 2008;94:207–220. doi: 10.1529/biophysj.107.113670.. A thorough discussion of the latest improvements in experimental and computational techniques used in high resolution traction force microscopy. Strength and limitations of established and newly developed procedures for reconstruction of cellular traction force on flat elastic substrate is thoroughly discussed.
- 8.Ji L, Loerke D, Gardel M, Danuser G. Methods in Cell Biology. vol 83. 2007. Probing Intracellular Force Distributions by High-Resolution Live Cell Imaging and Inverse Dynamics; pp. 200–237. Edited by: Academic Press; [DOI] [PubMed] [Google Scholar]
- 9.Danuser G, Waterman-Storer CM. Quantitative Fluorescent Speckle Microscopy of Cytoskeleton Dynamics. Ann. Rev. of Biophys. Biomol. Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- 10.Hu K, Ji L, Applegate K, Danuser G, Waterman-Storer CM. Differential Transmission of Actin Motion within Focal Adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 11.Hebert B, Costantino S, Wiseman PW. Spatiotemporal image correlation Spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophysical Journal. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiti S, Haupts U, Webb WW. Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc Natl Acad Sci U S A. 1997;94:11753–11757. doi: 10.1073/pnas.94.22.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CM, Hebert B, Kolin DL, Zareno J, Whitmore L, Horwitz AR, Wiseman PW. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 14.Shattil SJ. Integrins and Src: dynamic duo of adhesion signaling. Trends in Cell Biology. 2005;15:399–403. doi: 10.1016/j.tcb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- 16.Giuliano KA, Post PL, Hahn KM, Taylor DL. Fluorescent protein biosensors: measurement of molecular dynamics in living cells. Annu Rev Biophys Biomol Struct. 1995;24:405–434. doi: 10.1146/annurev.bb.24.060195.002201. [DOI] [PubMed] [Google Scholar]
- 17. Gaits F, Hahn K. Shedding light on cell signaling: interpretation of FRET biosensors. Sci STKE. 2003;2003:PE3. doi: 10.1126/stke.2003.165.pe3.. This is a concise yet authoritative review of recent advances in detection and interpretation of spatiotemporal activities of intact proteins in live cells using biosensors.
- 18.Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- 19.Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 20. Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6.. In this paper, translocation of a domain that binds to the activated form of the target protein is used as a robust and simple readout of protein activity.
- 21.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 23.Gray A, Van Der Kaay J, Downes CP. The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem J. 1999;344(Pt 3):929–936. [PMC free article] [PubMed] [Google Scholar]
- 24. Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333.. Because of the importance of the Rho family GTPases, biosensors for this protein family have undergone continually evolving designs (ref 24, 28, 29). This paper describes the prototype FRET biosensor for GTPase activity, showing that FRET between a GTPase protein and a fragment of a downstream effector could be used to study GTPase nucleotide state in living cells. The paper revealed gradients of Rac activity controlling cell polarization.
- 25.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, Hanby A, Harris W, Barnes D, Schmidt S, Mellor H, et al. Imaging protein kinase Calpha activation in cells. Science. 1999;283:2085–2089. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 26. Post PL, Trybus KM, Taylor DL. A genetically engineered, protein-based optical biosensor of myosin II regulatory light chain phosphorylation. J Biol Chem. 1994;269:12880–12887.. One of the first biosensors demonstrating that cytoskeletal protein conformation, rather than simple localization, could be visualized in living cells. This paper supported models of polarized cell movement and the role of myosin activity.
- 27. Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367.. This biosensor demonstrated that activation of endogenous Cdc42 could be studied in living cells using bright dyes. A fragment of the Cdc42 effector WASP was derivatized with a dye whose fluorescence changed upon binding to the Cdc42 target. The use of a fluorescent dye provides enhanced signal and the ability to image multiple activities at different wavelengths, but to date requires injection of the biosensor.
- 28. Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594.. Here the advent of fluorescent proteins undergoing FRET permitted the construction of a GTPase biosensor that was completely genetically encoded. Linking all components in a single chain produced a biosensor that was free of many of the image processing corrections required for intermolecular FRET.
- 29. Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665.. This paper describes a RhoA biosensor in which the genetically encoded FRET pair was moved to the inside of the biosensor chain, leaving the C terminus free for normal interaction with GDI. GDI is an important regulator of RhoA membrane localization. This paper revealed substantial RhoA activation at the leading edge of motile cells.
- 30.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598.. This and reference 71 demonstrated a broadly applicable biosensor design, reporting modification of enzyme substrates.
- 32. Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annual Review of Cell and Developmental Biology. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011.. A comprehensive review of cellular mehcanosensitivity.
- 33. Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nature Reviews Molecular Cell Biology. 2006;7:265–275. doi: 10.1038/nrm1890.. An insightful review of cell responses to local force and rigidity sensing.
- 34. Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469.. Src regulates integrin-cytoskeleton interaction. By applying external mechanical stimulation localized at adhesion sites and sing a FRET biosensor this study showed a rapid Src activation at stimulation sites followed by a wave of Src activation that propagates along the plasma membrane. The force induced directional and long-range Src activation wave is destroyed by disruption of actin network or microtubules indicating the role of cytoskeleton on force induced Src activation.
- 35.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.del Pozo MA, Kiosses WB, Alderson NB, Meller N, Hahn KM, Schwartz MA. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat Cell Biol. 2002;4:232–239. doi: 10.1038/ncb759. [DOI] [PubMed] [Google Scholar]
- 37. Cai X, Lietha D, Ceccarelli DF, Karginov AV, Rajfur Z, Jacobson K, Hahn KM, Eck MJ, Schaller MD. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28:201–214. doi: 10.1128/MCB.01324-07.. In this work the relationship between the maturation of an initial adhesion into a focal complex and its ability to exert migration force was probed by the application of mechanical force to fibronectin receptors from inside and outside the cell. This study showed that cells use mechanical force as a signal to strengthen initial adhesion complexes into focal complexes and regulate the amount of traction force applied to adhesion sites localized in different regions of lamella.
- 38.Coutinho A, Garcia C, Gonzalez-Rodriguez J, Lillo MP. Conformational changes in human integrin alphaIIbbeta3 after platelet activation, monitored by FRET. Biophys Chem. 2007;130:76–87. doi: 10.1016/j.bpc.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Dai Z, Dulyaninova NG, Kumar S, Bresnick AR, Lawrence DS. Visual snapshots of intracellular kinase activity at the onset of mitosis. Chem Biol. 2007;14:1254–1260. doi: 10.1016/j.chembiol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrett SC, Hodgson L, Rybin A, Toutchkine A, Hahn KM, Lawrence DS, Bresnick AR. A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall B, McLean MA, Davis K, Casanova JE, Sligar SG, Schwartz MA. A fluorescence resonance energy transfer activation sensor for Arf6. Anal Biochem. 2008;374:243–249. doi: 10.1016/j.ab.2007.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaper T, Looger LL, Takanaga H, Platten M, Steinman L, Frommer WB. Nanosensor detection of an immunoregulatory tryptophan influx/kynurenine efflux cycle. PLoS Biol. 2007;5:e257. doi: 10.1371/journal.pbio.0050257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunkel MT, Toker A, Tsien RY, Newton AC. Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J Biol Chem. 2007;282:6733–6742. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 45.Sato M, Kawai Y, Umezawa Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Anal Chem. 2007;79:2570–2575. doi: 10.1021/ac062171d. [DOI] [PubMed] [Google Scholar]
- 46.Zou J, Hofer AM, Lurtz MM, Gadda G, Ellis AL, Chen N, Huang Y, Holder A, Ye Y, Louis CF, et al. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry. 2007;46:12275–12288. doi: 10.1021/bi7007307. [DOI] [PubMed] [Google Scholar]
- 47.Machacek M, Hodgson L, Nalbant P, Pertz O, Shen F, Abell A, Johsnson GL, Hahn KM, Danuser G. Coordination of multiple Rho GTPase activities during cell protrusion. doi: 10.1038/nature08242. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nykamp DQ. Revealing pairwise coupling in linear-nonlinear networks. Siam Journal on Applied Mathematics. 2005;65:2005–2032. [Google Scholar]
- 49. Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100.. By using a reporter of vinculin conformational changes this report provided direct evidence that vinculin is activated at focal adhesions and led to a spatio-temporal map of vinculin conformational changes.
- 50.Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular alpha4-integrin conformational activation. Biophys J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 52.Kim M, Carman CV, Yang W, Salas A, Springer TA. The primacy of affinity over clustering in regulation of adhesiveness of the integrin {alpha}L{beta}2. J Cell Biol. 2004;167:1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Post PL, DeBiasio RL, Taylor DL. A fluorescent protein biosensor of myosin II regulatory light chain phosphorylation reports a gradient of phosphorylated myosin II in migrating cells. Mol Biol Cell. 1995;6:1755–1768. doi: 10.1091/mbc.6.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 55.Ward ME, Wu JY, Rao Y. Visualization of spatially and temporally regulated N-WASP activity during cytoskeletal reorganization in living cells. Proc Natl Acad Sci U S A. 2004;101:970–974. doi: 10.1073/pnas.0306258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giuliano KA, Taylor DL. Fluorescent actin analogs with a high affinity for profilin in vitro exhibit an enhanced gradient of assembly in living cells. J Cell Biol. 1994;124:971–983. doi: 10.1083/jcb.124.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshizaki H, Ohba Y, Kurokawa K, Itoh RE, Nakamura T, Mochizuki N, Nagashima K, Matsuda M. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol. 2003;162:223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tzima E, Kiosses WB, del Pozo MA, Schwartz MA. Localized cdc42 activation, detected using a novel assay, mediates microtubule organizing center positioning in endothelial cells in response to fluid shear stress. J Biol Chem. 2003;278:31020–31023. doi: 10.1074/jbc.M301179200. [DOI] [PubMed] [Google Scholar]
- 59.Graham DL, Lowe PN, Chalk PA. A method to measure the interaction of Rac/Cdc42 with their binding partners using fluorescence resonance energy transfer between mutants of green fluorescent protein. Anal Biochem. 2001;296:208–217. doi: 10.1006/abio.2001.5306. [DOI] [PubMed] [Google Scholar]
- 60.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 61.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 63.Sasaki K, Sato M, Umezawa Y. Fluorescent indicators for Akt/protein kinase B and dynamics of Akt activity visualized in living cells. J Biol Chem. 2003;278:30945–30951. doi: 10.1074/jbc.M212167200. [DOI] [PubMed] [Google Scholar]
- 64.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J Biol Chem. 2005;280:5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takao K, Okamoto K, Nakagawa T, Neve RL, Nagai T, Miyawaki A, Hashikawa T, Kobayashi S, Hayashi Y. Visualization of synaptic Ca2+ /calmodulin-dependent protein kinase II activity in living neurons. J Neurosci. 2005;25:3107–3112. doi: 10.1523/JNEUROSCI.0085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wouters FS, Bastiaens PI. Fluorescence lifetime imaging of receptor tyrosine kinase activity in cells. Curr Biol. 1999;9:1127–1130. doi: 10.1016/s0960-9822(99)80484-9. [DOI] [PubMed] [Google Scholar]
- 67.Neininger A, Thielemann H, Gaestel M. FRET-based detection of different conformations of MK2. EMBO Rep. 2001;2:703–708. doi: 10.1093/embo-reports/kve157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chew TL, Wolf WA, Gallagher PJ, Matsumura F, Chisholm RL. A fluorescent resonant energy transfer-based biosensor reveals transient and regional myosin light chain kinase activation in lamella and cleavage furrows. J Cell Biol. 2002;156:543–553. doi: 10.1083/jcb.200110161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurokawa K, Mochizuki N, Ohba Y, Mizuno H, Miyawaki A, Matsuda M. A pair of fluorescent resonance energy transfer-based probes for tyrosine phosphorylation of the CrkII adaptor protein in vivo. J Biol Chem. 2001;276:31305–31310. doi: 10.1074/jbc.M104341200. [DOI] [PubMed] [Google Scholar]
- 70.Nagai Y, Miyazaki M, Aoki R, Zama T, Inouye S, Hirose K, Iino M, Hagiwara M. A fluorescent indicator for visualizing cAMP-induced phosphorylation in vivo. Nat Biotechnol. 2000;18:313–316. doi: 10.1038/73767. [DOI] [PubMed] [Google Scholar]
- 71. Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798.. This and reference 31 demonstrated a broadly applicable biosensor design, reporting modification of enzyme substrates.
- 72.Wang Q, Dai Z, Cahill SM, Blumenstein M, Lawrence DS. Light-regulated sampling of protein tyrosine kinase activity. J Am Chem Soc. 2006;128:14016–14017. doi: 10.1021/ja065852z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirchner J, Kam Z, Tzur G, Bershadsky AD, Geiger B. Live-cell monitoring of tyrosine phosphorylation in focal adhesions following microtubule disruption. J Cell Sci. 2003;116:975–986. doi: 10.1242/jcs.00284. [DOI] [PubMed] [Google Scholar]
- 74.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 75.Oatey PB, Venkateswarlu K, Williams AG, Fletcher LM, Foulstone EJ, Cullen PJ, Tavare JM. Confocal imaging of the subcellular distribution of phosphatidylinositol 3,4,5-trisphosphate in insulin- and PDGF-stimulated 3T3-L1 adipocytes. Biochem J. 1999;344(Pt 2):511–518. [PMC free article] [PubMed] [Google Scholar]
- 76.Matsu-ura T, Michikawa T, Inoue T, Miyawaki A, Yoshida M, Mikoshiba K. Cytosolic inositol 1,4,5-trisphosphate dynamics during intracellular calcium oscillations in living cells. J Cell Biol. 2006;173:755–765. doi: 10.1083/jcb.200512141. [DOI] [PMC free article] [PubMed] [Google Scholar]