Abstract
The hepatitis C virus (HCV) is a flavivirus replicating in the cytoplasm of infected cells. The HCV genome is a single-stranded RNA encoding a polyprotein that is cleaved by cellular and viral proteases into 10 different products. While the structural proteins core protein, envelope protein 1 (E1) and E2 build up the virus particle, most nonstructural (NS) proteins are required for RNA replication. One of the least studied proteins is NS2, which is composed of a C-terminal cytosolic protease domain and a highly hydrophobic N-terminal domain. It is assumed that the latter is composed of three trans-membrane segments (TMS) that tightly attach NS2 to intracellular membranes. Taking advantage of a system to study HCV assembly in a hepatoma cell line, in this study we performed a detailed characterization of NS2 with respect to its role for virus particle assembly. In agreement with an earlier report (Jones, C. T., Murray, C. L., Eastman, D. K., Tassello, J., and Rice, C. M. (2007) J. Virol. 81 ,8374 -8383), we demonstrate that the protease domain, but not its enzymatic activity, is required for infectious virus production. We also show that serine residue 168 in NS2, implicated in the phosphorylation and stability of this protein, is dispensable for virion formation. In addition, we determined the NMR structure of the first TMS of NS2 and show that the N-terminal segment (amino acids 3-11) forms a putative flexible helical element connected to a stable α-helix (amino acids 12-21) that includes an absolutely conserved helix side in genotype 1b. By using this structure as well as the amino acid conservation as a guide for a functional study, we determined the contribution of individual amino acid residues in TMS1 for HCV assembly. We identified several residues that are critical for virion formation, most notably a central glycine residue at position 10 of TMS1. Finally, we demonstrate that mutations in NS2 blocking HCV assembly can be rescued by trans-complementation.
The hepatitis C virus (HCV)4 is a major causative agent of acute and chronic liver diseases, including liver cirrhosis and hepatocellular carcinoma. About 170 million individuals worldwide are infected with HCV (1), but despite the obvious high medical need, current therapy is limited because of side effects and insufficient efficacy. HCV has been classified as the Hepacivirinae genus within the family Flaviviridae and includes a group of enveloped RNA viruses with a single-stranded genome of positive polarity (2). The genome has a length of about 9,600 nucleotides and encodes a polyprotein of about 3,000 amino acids in a single open reading frame. It is flanked at the 5′ and 3′ ends by nontranslated regions (NTRs) that are required for RNA translation and replication (reviewed in Refs. 3-5). An internal ribosome entry site (IRES), present in the 5′-untranslated region, directs translation of the HCV genome in a cap-independent manner. The polyprotein precursor is processed co- and post-translationally by cellular and viral proteases presumably at the membrane of the endoplasmic reticulum giving rise to 10 mature proteins (6). The structural proteins include Core (C), which forms the viral nucleocapsid, and glycoproteins E1 and E2 that are embedded into the lipid envelope. The structural region is separated from the nonstructural (NS) region by a small membrane-bound peptide (p7) that at least in vitro can form an ion channel (7). p7 is required for virus assembly and release both in cell culture and presumably also in vivo (8-10). The NS region is composed of six proteins as follows: NS2, NS3, NS4A, NS4B, NS5A, and NS5B (from the N to the C terminus). NS3 to NS5B in synergy with host cell factors and the NTRs are sufficient for RNA replication and thus constitute the minimal HCV replication unit (11).
NS2 is a 217-aa-long cysteine protease that cleaves the NS2-3 junction in cooperation with the N-terminal ∼180 aa of NS3, forming the serine-type protease, in a rapid presumably intramolecular reaction (12-14). Liberation of NS2 from NS3 is essential for RNA replication in cell culture, presumably because the authentic N terminus of NS3 is required to generate an active replicase complex. This assumption is based on analogy to pestiviruses (15) and on the fact that NS2 itself is not required for RNA replication (11).
NS2 is an integral membrane protein that is tightly bound to intracellular membranes via a highly hydrophobic N-terminal domain (16), whereas the cytosolic protease domain appears to contribute to membrane association only to a minor extent (17). The x-ray crystal structure of the protease domain revealed a dimer with two composite active sites (17). Whereas His-143 and Glu-163 of the catalytic triad are provided by the N-terminal part of one monomer, Cys-184 is provided by the C-terminal part of the other monomer (17). This surprising structural feature is supported by the observation that cotransfection of cysteine and histidine active site mutants, which on their own do not have protease activity, rescues cleavage activity because of restoration of one functional active site in the NS2 dimer (17).
The exact membrane topology of NS2 so far is not known. A signal of reinitiation of translocation (signal-like sequence) in the C-terminal half of p7, presumably in conjunction with two internal signal-like sequences that are able to target NS2 cotranslationally to membranes, appear to orchestrate NS2 topology during/after translation (18, 19). Studies by Yamaga and Ou (19) suggest that NS2 has four trans-membrane segments (TMS) with both the N and the C terminus of the protein residing in the endoplasmic reticulum lumen. However, the x-ray crystal structure of the NS2 protease domain and structure modeling revealed a globular cytosolic subdomain (17). It is therefore assumed that the N-terminal domain of NS2 is composed of one or three TMS followed by the cytosolic C-terminal protease domain.
Numerous interactions between NS2 and viral or cellular proteins have been reported. These include, for example, interactions between NS2 and NS3 (20), NS2 and NS4A (21), or NS2 and a p7-E1-E2 complex (10, 22). In addition, NS2 may have direct or indirect effects on cyclic AMP-dependent pathways (23), induce cell cycle arrest (24), or interfere with apoptosis (25). Finally, it was suggested that NS2 degradation via the proteasome is regulated by phosphorylation of serine residue 168 by casein kinase II (26). However, the in vivo relevance of these observations is unknown especially because in most cases individually (over) expressed NS2 has been studied.
Apart from acting as a protease, NS2 appears to play a very important role for the production of infectious HCV particles. First, we identified a site in the N-terminal NS2 domain as most relevant for the construction of highly assembly-competent intra- and intergenotypic HCV chimeras (27). This site resides right C-terminal of TMS1 of NS2 and allows the construction of replication- and assembly-competent chimeras by exchanging the core to the TMS1 region of NS2 of the highly replication-competent HCV isolate JFH1 against the analogous region of any other HCV isolate (Fig. 1A). Similar sites within NS2 have also been found with natural HCV recombinants in patients (28-30) Second, cell culture adaptation experiments aimed to increase the titer of either inefficient HCV chimeras or the JFH1 isolate itself identified mutations within NS2 as major contributors to assembly (31, 32). Finally, by analyzing a set of NS2 mutants Jones et al. (9) obtained evidence that NS2 appears to play an essential role for HCV particle production. Thus, NS2 has a dual function in the HCV replication cycle.
FIGURE 1.
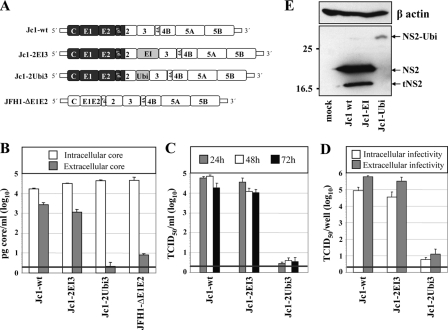
Insertion of a heterologous IRES between NS2 and NS3 does not affect virus production. A, schematic diagram of basic HCV constructs used in this study. The region encoding core to the first putative TMS of NS2 originates from the HCV isolate J6 (dark gray); the remainder of the chimeric Jc1 genome originates from the JFH1 isolate (white region). JFH1-ΔE1E2 carries an in-frame deletion of 350 codons within the E1-E2 coding region. In case of the bicistronic Jc1-2EI3, the EMCV IRES (EI) was inserted between NS2 and NS3. In Jc1-2Ubi3, the ubiquitin coding region is inserted in-frame between NS2 and NS3. B, Huh7.5 cells were transfected with constructs specified at bottom, and the amounts of intracellular and extracellular core were determined 48 h post-transfection by using a core-specific ELISA. Note that JFH1-ΔE1E2 still releases core protein but no infectivity (59). A representative result of two independent experiments with error ranges is shown. C, kinetics of release of infectious particles into culture supernatants were quantified at given time points by TCID50 assay. D, intracellular and extracellular infectivities were determined 48 h post-transfection by using TCID50 assay. C and D, representative results of three independent experiments with error ranges are shown. Background of the assays was determined by using JFH-ΔE1E2 (black line). E, NS2 expression patterns in Huh7.5 cells transfected with constructs specified at bottom. Cells were harvested 48 h post-transfection, and NS2 proteins were detected by Western blot. β-Actin detected on the same blot was used as an internal loading control. The positions of molecular mass marker proteins (in kDa) are indicated at left, and NS2 proteins are specified at right. In addition to full-length NS2, a truncated NS2-specific protein was consistently detected (tNS2).
In this study we confirm and largely extend the observation that NS2 is a critical HCV assembly determinant. In addition, we provide a structure-activity study of TMS1 of NS2 and demonstrate that assembly defects caused by mutations in NS2 can be rescued by trans-complementation.
EXPERIMENTAL PROCEDURES
Sequence Analyses and Structure Predictions—Sequence analyses were performed using the website tools of the European HCV data base (euHCVdb (33)) and Network Protein Sequence Analysis (NPSA, (34)) available at the Institut de Biologie et Chimie des Protéines. Multiple sequence alignments and aa conservations were carried out with the ClustalW program using default parameters (35). The repertoire of residues at each aa position and their frequencies observed in natural sequence variants were computed by using the tool “Extract” in the euHCVdb. Various methods were combined for the prediction of trans-membrane sequences as follows: PHDhtm (36), TMpred (37), TMHMM (38), SOSUI (39), TopPred (40), and HMMTOP (41).
Cell Culture—Monolayers of the highly permissive cell line Huh7.5 (42) were grown in Dulbecco's modified minimal essential medium (DMEM; Invitrogen) supplemented with 2 mm l-glutamine, nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum.
Plasmids and DNA Cloning—All nucleotide and aa numbers refer to the JFH1 genome (GenBank™ accession number AB047639). The chimeras Con1/C3 and Jc1 as well as the JFH1/ΔE1-E2 construct have been described previously (27, 43). Single aa substitutions were introduced into plasmids pFK-JFH1/wt and the chimera pFK-Con1/C3 by PCR-based site-directed mutagenesis using standard procedures. Construct pFK-Jc1-2Ubi3 in which the ubiquitin coding sequence was inserted in-frame between NS2 and NS3 was generated by using an overlap PCR method. To generate the bicistronic RNA genome pFK-Jc1-2EI3, the internal ribosome entry site (IRES) of the encephalomyocarditis virus (EMCV) was inserted into pFK-Jc1 right after nt 3431. An additional stop codon was inserted at the 3′ end of the NS2 coding region as well as an additional start codon at the 5′ end of the NS3 coding region. PFK-Jc1-2EI3-ΔProt contains a deletion of the NS2 protease domain (nt 3066-33431, corresponding to codon 97-217 of NS2). Plasmid pFK-Jc1-2EI3-Δ12 contains a deletion of the two putative N-terminal TMS of NS2 (nt 2805-2966, corresponding to codon 10-62 of NS2) (19). PFK-Jc1-2EI3-Δ23 contains a deletion of the putative TMS2 and TMS3 of NS2 (nt 2868-3065, corresponding to codon 32-96 of NS2). All three deletion mutations were introduced into pFK-Jc1-2EI3. Single aa substitutions were generated by site-directed mutagenesis using standard procedures. For trans-complementation analyses, the basic construct pFKI341PI-EI/NS3-3′_Δ2328-2435_dg_JFH was generated. This plasmid contains the T7 promoter sequence fused to nucleotides 1-341 of the JFH1 consensus sequence, the poliovirus IRES, a multiple cloning site containing restriction sites for BglII and SwaI, the EMCV IRES, the NS3 to NS5B coding sequence, and the 3′ NTR of JFH1, the genomic ribozyme of the hepatitis delta virus (dg), and the T7 polymerase terminator sequence. To discriminate in TCID50 assays between the virus genome and the helper RNA, a 324-nt-long deletion (nt 7322-7645 of JFH1) was introduced into domain III of NS5A. This deletion removes the epitope that is recognized by the NS5A-specific monoclonal antibody 9E10 (44). The following HCV coding sequences were inserted into the multiple cloning site of pFKI341PI-EI/NS3-3′_Δ2328-2435_dg_JFH via the BglII and SwaI restriction sites as follows: SpE2-p7-NS2, Spp7-NS2, and SpNS2 with “Sp” referring to the authentic signal-like peptide of each protein.5 Amplified DNA fragments were analyzed by automated nucleotide sequencing by using an ABI 310 sequencer (Applied Biosystems). Further details of the cloning strategies are available upon request.
In Vitro Transcription—To obtain in vitro transcripts of the individual constructs, 10 μg of plasmid DNA was treated for 1 h with either MluI (in case of JFH1 or Jc1-derived constructs) or AseI (in case of the Con1/C3 chimera). Linearized plasmid DNA was extracted with phenol and chloroform and after precipitation with ethanol was dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mm HEPES (pH 7.5), 12 mm MgCl2, 2 mm spermidine, 40 mm dithiothreitol, 3.125 mm of each nucleotide triphosphate, 1 unit of RNasin (Promega) per μl, 0.1 μg of plasmid DNA/μl, and 0.6 units of T7 RNA polymerase (Promega) per μl of reaction mixture. After incubation for 2 h at 37 °C,0.3 units of T7 RNA polymerase/μl reaction mixture were added, followed by additiona l2 h of incubation at 37 °C. Transcription was terminated by addition of 1.2 units of RNase-free DNase (Promega) per μg of plasmid DNA and 30 min of incubation at 37 °C. The RNA was extracted with acidic phenol and chloroform, precipitated with isopropyl alcohol, and dissolved in RNase-free water. Denaturing agarose gel electrophoresis was used to check RNA integrity, and the concentration was determined by measurement of the absorbance at 260 nm.
Electroporation of HCV RNAs—Single-cell suspensions were prepared by trypsinization, and phosphate-buffered saline (PBS)-washed Huh7.5 cells were resuspended at a density of 1.5 × 107 cells per ml in Cytomix (45) containing 2 mm ATP and 5 mm glutathione. Unless otherwise stated, 5 μg of in vitro transcribed RNA were mixed with 400 μl cell suspension and electroporated with a Gene Pulser system (Bio-Rad, Munich, Germany) in a cuvette with a gap width of 0.4 cm (Bio-Rad) at 960 μF and 270 V. For trans-complementation assays a mixture of 5 μg of the NS2 mutant and 5 μg of helper RNA was used. After electroporation, cells were immediately transferred to 20-25 ml complete DMEM and seeded as required for the assay.
Quantification of HCV Core Protein—The Ortho® trak-C™ ELISA kit (Ortho Clinical Diagnostics, Germany) was used for quantification of intra- and extracellular HCV core protein amounts 48 h after transfection. Cell culture supernatants were filtered through 0.45-μm-pore-size filters and used directly for core ELISA, whereas for determination of intracellular core amounts, cells were harvested by addition of 0.5 ml of PBS containing 1% Triton X-100, 1:10,000 volume of aprotinin (1 unit/ml), 1:1000 volume of leupeptin (4 mg/ml), and 1:100 volume of phenylmethylsulfonyl fluoride (100 mm) and cleared at 18,000 × g for 5 min. Depending on the construct and the time point of harvest, samples were diluted 1:10 or higher and processed for ELISA according to the manufacturer's protocol. Colorimetric measurements were performed using a Sunrise colorimeter (Tecan Trading AG, Switzerland).
Determination of Virus Titers in Cell Culture Supernatants and Cell Lysates—Virus titers were determined as described elsewhere with slight modifications (46). In brief, Huh7.5 target cells were seeded at a concentration of 1.1 × 104 cells per well of a 96-well plate in a total volume of 200 μl of complete DMEM. Twenty four hours later, serial dilutions of virus containing supernatant were added with 8 wells per dilution. Two to 3 days later, cells were washed with PBS, fixed for 20 min with ice-cold methanol at -20 °C, washed three times with PBS, permeabilized, and blocked for 1 h with PBS containing 0.5% saponin, 1% bovine serum albumin, 0.2% dried skim milk, and 0.02% sodium azide. Endogenous peroxidases were blocked by 5 min of incubation at room temperature with PBS containing 0.3% H2O2 (v/v). After three washes with PBS and one with PBS containing 0.5% saponin (PBS/saponin), NS5A was detected with a 1:2000 dilution of hybridoma supernatant 9E10 (kindly provided by C. M. Rice and T. Tellinghuisen, New York) in PBS/saponin for 1 h or overnight incubation at room temperature at 4 °C. Cells were washed again three times with PBS and once with PBS/saponin, and bound antibody was detected by incubation with peroxidase-conjugated anti-mouse antibody (Sigma) diluted 1:200 in PBS/saponin. After 1 h of incubation at room temperature, cells were washed three times with PBS and once with PBS/saponin, and peroxidase activity was detected by using the Vector NovaRED substrate kit (Linaris Biologische Produkte GmbH, Germany). Virus titers (50% tissue culture infective dose (TCID50/ml)) were calculated as described recently (46). Infectivity in all cell lysates was measured in the same way by using lysates obtained by three cycles of freezing and thawing (47). Cells were washed once with PBS, scraped into PBS, and collected by centrifugation at 400 × g for 5 min. The cell pellet was resuspended in 500 μl of complete DMEM, and cells were lysed by three rapid freeze-thaw cycles. Cell debris was removed by centrifugation for 10 min at 10,000 × g, and the supernatant was used for titration as described above.
Antisera—The polyclonal NS2 antiserum was raised by immunization of rabbits with two peptides spanning amino acid residues 148-163 (TPMSDWAASGLRDLAV) and 202-217 (LLGPADGYTSKGWKLL) of JFH1 NS2. Monoclonal antibody 18/7 (Ma18/7; kindly provided by W. Gerlich, Giessen, Germany) is a mouse monoclonal antibody recognizing the epitope DPAF in the large surface protein of the hepatitis B virus. HCV core protein was detected by using mouse monoclonal antibody C7/50 (kindly provided by D. Moradpour, Lausanne, Switzerland) at a dilution of 1:1000. The E2 protein of the HCV isolate J6CF was detected by using a 1:500 dilution of a rabbit polyclonal antiserum that was raised by immunization with a recombinant E2 protein derived from the same HCV strain. NS5A was detected by immunoblotting using a 1:1000 dilution of a JFH1 NS5A-specific rabbit polyclonal antiserum.
SDS-PAGE and Western Blotting—Huh7.5 cells were mock-treated or transfected with HCV RNA, and samples were harvested after given time points (24, 48, or 72 h) by scraping into SDS loading buffer (400 mm Tris (pH 8.8), 10 mm EDTA, 3% (w/v) SDS, 20% (v/v) Sucrose, 2% β-mercaptoethanol, 0.2% (w/v) bromphenol blue), and SDS-PAGE was performed. Prestained molecular weight standards (broad range, 6-175 kDa) were used for calibration (New England Biolabs). Proteins were blotted onto polyvinylidene difluoride or nitrocellulose membrane (PerkinElmer Life Sciences) for 1 h with an electric current of 0.8 mA/cm2. Membranes were blocked for 1 h in 5% blocking buffer (5% dried milk, 0.5% Tween 20 in PBS) prior to incubation for 1 h with primary antibody diluted in 2% blocking buffer (2% dried milk, 0.5% Tween 20 in PBS). Membranes were washed for 30 min with three changes of washing buffer (0.5% Tween 20 in PBS) and incubated with horseradish peroxidase-conjugated secondary antibody (anti rabbit, 1:25,000; anti mouse, 1:10,000 dilution; both from Sigma) for 1 h. An additional 30-min wash with three changes of washing buffer was performed, and samples were incubated in ECL solution (50 ml of 100 mm Tris (pH 9.35), 2.25 mm 3-aminophtalhydrazide (Luminol, Sigma A8511), 0.43 mm p-coumaric acid (Sigma C9008), mixed with 3 μl of 30% H2O2). Luminescence signals were detected on Kodak BioMax MS films.
ATP Viability Assay—The ATP levels of Huh7.5 cells were measured 48 h after transfection by using the glow-type CellTiter-Glo Luminescent Assay (Promega) according to the instructions of the manufacturer. Transfected cells were cultured in opaque-walled 96-well plates and equilibrated to room temperature for 30 min. CellTiter-Glo reagent was added to each well, and the plates were incubated at room temperature for 10 min to stabilize the luminescent signal, which was then measured with a Mithras LB 940 (Berthold Technologies, Germany) microplate reader. Values obtained with cells after transfection with the subgenomic replicon were set to 100% and used for normalization.
Peptide Synthesis and Purification—Peptide NS2[1-27] (MDREMAASAGGAVFVGLVLLTLSPHYK) was synthesized by Clonestar Biotech s.r.o. (Brno, Czech Republic) and purified by reverse-phase RP-HPLC on a Vydac C8 column (300 Å, 10 μm, 10 × 250 mm) using a water/acetonitrile gradient containing 0.1% trifluoroacetic acid. The purity of the peptide (>98%) was checked by RP-HPLC, electrospray mass spectroscopy (calculated mass, 2834.36 Da; observed mass, 2833.84 ± 0.48 Da), and NMR spectroscopy. To avoid any unexpected disulfide bond formation during the CD and NMR experiments, the cysteine residue at position 9 in NS2 of genotype 1b (see Fig. 4) was replaced by alanine. This residue is often found at this position in NS2 of other genotypes, and thus should not disturb the folding, at least in the isolated peptide.
FIGURE 4.
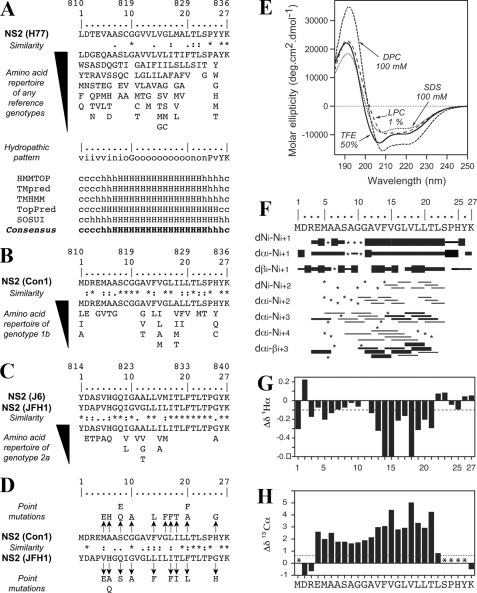
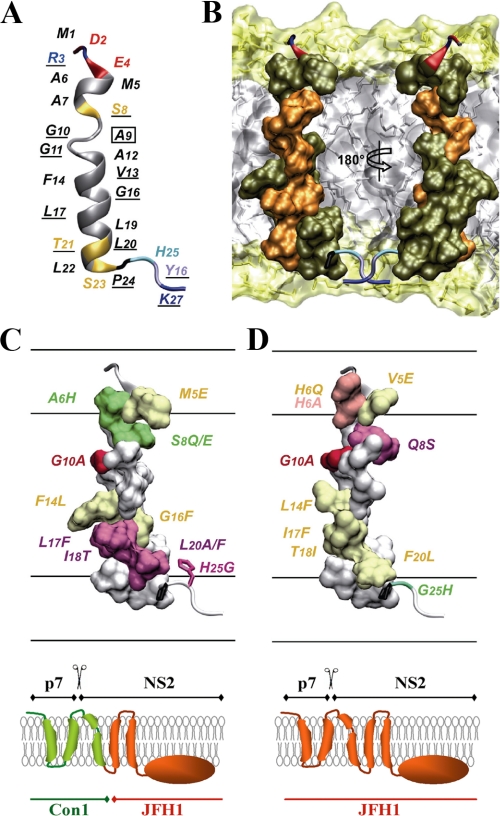
Sequence and structure analyses of TMS 1 of NS2. A, amino acid repertoire and prediction of the trans-membrane segment for the N-terminal NS2 segment residing between aa 1 and 27. The NS2 sequence of the HCV H77 consensus clone (GenBank™ accession number AF009606) is shown at the top. Amino acids are numbered with respect to NS2 and the HCV polyprotein (top row). The aa repertoire of 27 representative NS2 sequences from confirmed HCV genotypes and subtypes, including the recently described genotype 7a (accession number EF108306; see the European HCV Data base (33) for details), is given below. The degree of aa and physicochemical conservation at each sequence position can be inferred from the extent of variability (with the observed aa listed in decreasing order of frequency from top to bottom) together with the similarity index according to ClustalW convention (asterisk, invariant; colon, highly similar; dot, similar (35)) and the consensus hydropathic pattern deduced from the aa repertoire. o, hydrophobic position (F, I, W, Y, L, V, M, P, and C); n, neutral position (G, A, T, and S); i, hydrophilic position (K, Q, N, H, E, D, and R); v, variable position (i.e. when both hydrophobic and hydrophilic residues are observed at a given position). Fully conserved residues 10, 24, 26, and 27 in any genotype are indicated by their one-letter code. Predictions of a putative trans-membrane helix in the NS2 segment 1-27 (bottom) were made by using the available web-based algorithms HMMTOP, TMpred, TMHMM, TopPred, and SOSUI. Trans-membrane helix prediction for each residue position is indicated as strong (H), probable (h), or undetermined (coil, c). For each method, the reported prediction corresponds to the consensus deduced from the analysis of the 27 representative NS2 sequences from confirmed HCV genotypes and subtypes (see above). The consensus prediction of the trans-membrane segment deduced from all these methods is reported in boldface at bottom. B, amino acid repertoire deduced from the ClustalW multiple alignments of 334 NS2 sequences of genotype 1b. Residues observed at a given position with a frequency <1% were not included. Amino acids are numbered with respect to NS2 and the HCV polyprotein of the Con1 isolate (accession number AJ238799). C, amino acid repertoire deduced from the ClustalW multiple alignments of 21 NS2 sequences of genotype 2a. Amino acids are numbered with respect to NS2 and the HCV polyprotein from the HCV J6 and JFH1 clones (accession numbers AF177036 and AB047639, respectively). D, sequence comparison of the NS2 segment 1-27 from Con1 and JFH1 used to design the NS2 mutants. Arrows point to the substituting residues. E, far UV circular dichroism analyses of the NS2[1-27] synthetic peptide (Con1) in various membrane mimetic environments. CD spectra were recorded in either 50% TFE (solid line), or in 100 mm SDS (dotted line), or 100 mm DPC (small dashed line), or 1% α-lysophosphatidylcholine (LPC)(large dashed line). F and G, NMR structure of the NS2[1-27] synthetic peptide in 50% TFE-d2. F, summary of sequential (i, i + 1) and medium (i, i + 2 to i + 4) range NOEs deduced from the analysis of NOESY spectra. Intensities of NOEs are reflected by bar thickness; asterisks indicate that the presence of a NOE is not confirmed because of resonance overlap. For Pro residue, NOE intensities to its Hδ protons were substituted for NOE intensities to amide protons. G and H, chemical shift differences for 1Hα and 13Cα at each position, as calculated by subtraction of the experimental values to the random coil conformation values (52). The dashed horizontal line indicates the standard threshold values for an α-helix (Δδ1Hα ≤ -0.1 ppm; Δδ13Cα ≥ 0.7 ppm). Stars in H indicate that the corresponding 13Cα were not identified.
Circular Dichroism—CD spectra were recorded on an Applied Photophysics Chirascan spectrometer calibrated with 1S-(+)-10-camphorsulfonic acid. Measurements were carried out at room temperature in a 0.1-cm path length quartz cuvette (Hellma), with peptide concentrations ranging from 54 to 60 μm. Spectra were recorded in the 180-260-nm wavelength range with 0.2-nm increments, 0.5-nm bandwidth, and 1-s integration time. The raw spectra were smoothed after base-line correction, and the spectral units were expressed in molar ellipticity per residue using peptide concentrations determined by measurements of UV light absorbance of tyrosine at 280 nm (molar extinction coefficient of 1490 m-1·cm-1). The secondary structure content was estimated with the CDSSTR program (48) by using the DICHROWEB server (49). CD experiments were performed on the platform “Production et Analyses de Protéines” from the IFR 128 BioSciences Gerland Lyon-Sud.
NMR Spectroscopy—Purified NS2[1-27] was dissolved at 1.22 mm in a mixture of 50% 2,2,2-trifluoroethanol (TFE)-d2 (>99%) in H2O (v/v), and 2,2-dimethyl-2-silapentane-5-sulfonate was added to the NMR samples as an internal 1H chemical shift reference. All NMR spectra were acquired at 25 and 35 °C. Multidimensional experiments were performed on a Varian Unity-plus 500-MHz using standard homonuclear pulse sequences such as NOESY (mixing times between 100 and 250 ms) and clean total TOCSY (isotropic mixing time of 80 ms), as detailed previously (Refs. 50, 51 and references therein). Water suppression was achieved by presaturation. Varian VNMR software was used to process all data, and Sparky was used for spectra analyses.6 Intraresidue backbone resonances and aliphatic side chains were identified from homonuclear 1H TOCSY experiments and confirmed with 1H-13C heteronuclear single quantum correlation spectroscopy in 13C natural abundance. Sequential assignments were determined by correlating intraresidue assignments with inter-residues cross peaks observed in two-dimensional 1H NOESY. NMR derived 1Hα and 13Cα chemical shifts are reported relative to the random coil chemical shifts in TFE (52).
NMR-derived Constraints and Structure Calculation—NOE intensities used as input for structure calculations were obtained from the NOESY spectrum recorded with a 100-ms mixing time and checked for spin diffusion on spectra recorded at lower mixing times (50 ms). NOEs were partitioned into three categories of intensities that were converted into distances ranging from a common lower limit of 1.8 Å to upper limits of 2.8, 3.9, and 5.0 Å, respectively. Protons without stereospecific assignments were treated as pseudoatoms, and the correction factors were added to the upper distance constraints (53). Neither additional dihedral angle nor hydrogen bond restraints were introduced. Three-dimensional structures were generated from NOE distances by the dynamic simulated annealing protocol with the XPLOR-NIH 2.9.7 program (54) by using the standard force fields and default parameter sets. A set of 50 structures was initially calculated to widely sample the conformational space, and the structures of low energy with no distance restraint violations (>0.5 Å) were retained. The selected structures were compared by pairwise root mean square deviation (r.m.s.d.) over the backbone atom coordinates (N, C-α, and C′). Local analogies were analyzed by calculating the local r.m.s.d. of a tripeptide window sliding along the sequence. Statistical analyses, superimposition of structures, and structural analyses were performed with MOLMOL version 2.6 (55), AQUA version 3.2, and PROCHECK-NMR version 3.5.4 (56).
RESULTS
Construction and Characterization of HCV Genomes Replicating Independently from NS2-3 Cleavage—It has been shown that NS2 protease activity is required for HCV genome replication in cell culture and for viability of a functional HCV genome in vivo (57, 58). Therefore, studies of the assembly function of NS2 required a system in which alterations of NS2 could be introduced without affecting viral RNA replication. As a starting point we used the highly assembly competent Jc1 chimeric genome in which the region from core to the first putative TMS of NS2 of the HCV isolate J6 is fused with the remainder of the JFH1 polyprotein (Fig. 1A) (27). Uncoupling of NS2-3 processing from RNA replication was attempted in two different ways. First, we inserted the EMCV IRES between NS2 and NS3 (construct Jc1-2EI3) resulting in a bicistronic RNA with two translation units (core to NS2 and NS3 to NS5B). Second, we inserted the coding region for ubiquitin in-frame between NS2 and NS3 (Jc1-2Ubi3). In this case, cleavage at the NS2-3 site is mediated by the host cell proteasome liberating the authentic N terminus of NS3 (Fig. 1A).
To characterize the engineered virus genomes, Huh7.5 cells were transfected with these viral RNAs in parallel with a mutant in which most of the envelope glycoproteins had been deleted and which was used as a negative control in infectivity assays throughout this study (JFH1-ΔE1E2; Fig. 1A). Cells and culture media were harvested 48 h after transfection, and core protein amounts were determined by ELISA. As shown in Fig. 1B, intracellular core protein amounts in transfected cells were comparable arguing for similar replication efficiency of the constructs. Core release (extracellular core), which is a marker for virus production, was only slightly reduced in case of Jc1-2EI3, as compared with the wild type, whereas an ∼500-fold reduction of core release was found with the mutant carrying the ubiquitin insertion (Jc1-2Ubi3). This result indicates that the latter construct has a defect in virus production. Note that the envelope deletion mutant releases low amounts of core protein and therefore cannot be used as a negative control in core ELISA (59).
Kinetics of release of infectious virus was determined by using TCID50 assays with culture supernatants harvested 24, 48, and 72 h after transfection. In agreement with the core-ELISA data, the insertion of the EMCV IRES between NS2 and NS3 only slightly reduced infectivity titers and had no effect on the overall release kinetics in comparison with the wild type (Fig. 1C). Very low infectivity, close to the background, was detected with Jc1-2Ubi3 corroborating the core-ELISA data. Finally, we wanted to know whether the insertions between NS2 and NS3 had an effect on the accumulation of intracellular infectivity. To this end we determined intracellular infectivity in cell lysates that had been prepared by three repetitive cycles of freezing and thawing. The results displayed in Fig. 1D show that the ratio of extracellular over intracellular infectivity was not changed between Jc1-2EI3 as compared with the wild type, whereas extremely low amounts of cell-associated infectivity were detected with Jc1-2Ubi3 arguing for a general assembly defect of this construct.
Western blot analysis of NS2 amounts in cells 48 h after transfection revealed no gross difference between Jc1wt and Jc1-2EI3 (Fig. 1E). In addition to full-length NS2, we consistently observed low and somewhat variable amounts of an NS2-reactive protein that migrated with an apparent molecular mass of about 17 kDa. The nature of this protein that we designate truncated NS2 (tNS2) throughout this study is not clear, but because of its immunoreactivity with our NS2-specific antiserum that targets two linear epitopes in the protease domain, it may correspond to an N-terminally truncated NS2 protein. In the case of Jc1-2Ubi3, no authentic NS2 was detected but instead were low amounts of an NS2-ubiquitin fusion protein with an apparent molecular mass of about 32 kDa (Fig. 1E). This observation suggests that even though NS2-Ubi does not interfere with RNA replication, it is either non-functional for HCV assembly or that the amounts of NS2 (or NS2-Ubi) obtained with this construct are too low to support assembly. In summary these data show that Jc1-2EI3 is fully assembly competent thus providing the ideal tool to study the assembly function of NS2 independent from its role in RNA replication.
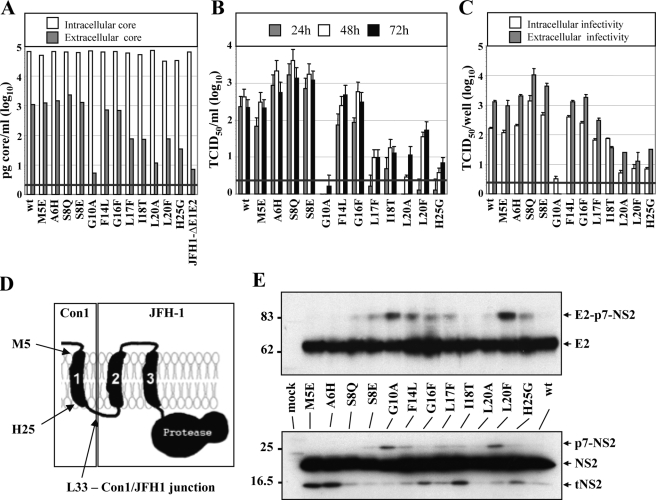
Full Length NS2 Protein Is Required for HCV Assembly—To investigate which part of NS2 is required for particle assembly, we constructed several deletion mutants. As a guideline for mutagenesis, we used a membrane topology model of NS2, which assumes three TMS in the N-terminal half, similar to a recent report (19), and a cytosolic NS2 protease domain (17) (Fig. 2A). Two constructs were generated in which we deleted either the putative TMS1 and TMS2 (construct Jc1-2EI3-Δ1,2) or TMS2 and TMS3 (construct Jc1-2EI3-Δ2,3) to preserve the overall topology (Fig. 2A). In addition we constructed Jc1-2EI3-ΔProt, in which the NS2 protease domain had been deleted and TMS3 was fused to either a FLAG tag or the MA18/7 epitope. All mutations were inserted into Jc1-2EI3 to avoid defects in RNA replication because of these NS2 alterations (Fig. 2A).
FIGURE 2.
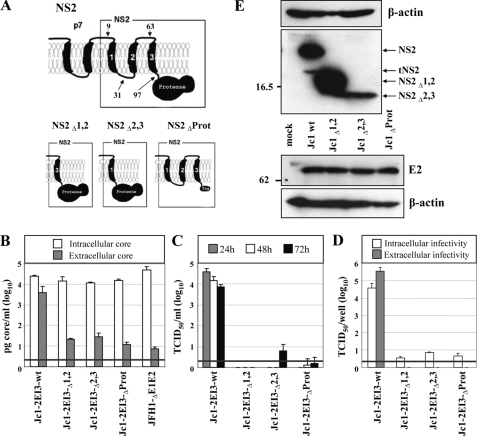
Full-length NS2 protein is essential for production of infectious HCV particles. A, schematic diagram of the putative membrane topology of NS2 according to Refs. 17, 19 in conjunction with p7. The putative membrane topology of the NS2 deletion mutants is shown below. NS2Δ1,2 and NS2Δ2,3 lack the corresponding putative TMS (aa 10-62 and 32-96 of NS2, respectively), and in NS2ΔProt the protease domain is deleted (aa 97-217). All NS2 mutations were introduced into Jc1-2EI3. B, Huh7.5 cells were transfected with constructs specified at bottom, and the amounts of intracellular and extracellular core were determined by core-specific ELISA 48 h post-transfection. A representative result of two independent experiments with error ranges is shown. C, kinetics of release of infectious particles was measured by using TCID50 assay. D, intracellular and extracellular infectivity 48 h post-transfection as determined by TCID50 assay. Representative results of three independent experiments with error ranges are shown in C and D. E, Western blot analysis of NS2 (upper panel) and E2 proteins (lower panel) expressed in Huh7.5 cells that had been transfected with constructs specified between the panels. Cells were harvested 48 h post-transfection. Note that the tagged NS2 protein lacking the protease domain could not be detected with a tag-specific antibody (data not shown), whereas E2 amount was comparable with the other constructs. β-Actin was used as an internal loading control. The positions of molecular weight marker proteins (in kDa) are indicated at left, and HCV proteins are specified at right. tNS2, truncated NS2 protein.
All NS2 mutants replicated with comparable efficiency as indicated by the intracellular core protein levels (Fig. 2B). However, with all deletion mutants core release was reduced about 100-fold. This correlated well with the absence of infectivity in the culture supernatant at every time point after transfection (Fig. 2C). Moreover, intracellular infectivity was also not detectable arguing that the deletions introduced into NS2 did not affect release of virus particles but rather assembly (Fig. 2D).
To investigate whether these deletions affected polyprotein processing, lysates of cells that had been transfected with the NS2 mutants were analyzed by Western blot (Fig. 2E). In all cases we detected proteins with the expected molecular weights: NS2 (21 kDa) and tNS2 (17 kDa) in case of the wild type, NS2Δ1,2 (17 kDa), and NS2Δ2,3 (15 kDa). No bands corresponding to p7-NS2 or E2-p7-NS2 precursors were detected (not shown). Although we cannot rule out that the different signal intensities obtained for the variant NS2 proteins are because of different reactivities with the antibody used for Western blot, this is very unlikely because the antibody targets two linear epitopes in the C-terminal region of NS2. We therefore assume that the signal intensities rather reflect protein stabilities. Despite intensive efforts, the NS2 protein lacking the protease domain (NS2ΔProt) could not be detected, neither when using the C-terminal FLAG tag nor the MA18/7 epitope tag arguing that the N-terminal trans-membrane domain is unstable in the absence of the protease domain. However, there was no alteration in the processing of E2, which was detected at levels comparable with the wild type (Fig. 2E, lower panels).
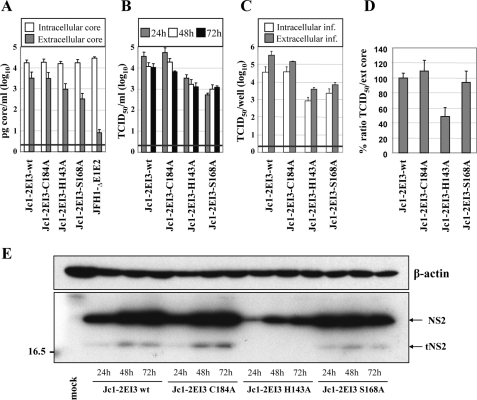
Important Role of NS2 Protease Active Site His Residue and Ser-168 for HCV Assembly—According to the x-ray crystal structure of the NS2 protease domain, NS2 forms a homodimer with each monomer having a composite active site (17). Although His-143 and Glu-163 are provided by the N-terminal subdomain of one monomer, Cys-184 is provided by the C-terminal subdomain of the other monomer. Recently it was shown that an alanine substitution of the active site cysteine residue does not affect infectious HCV production (9). Moreover, Franck et al. (26) provided evidence that NS2, at least when expressed on its own, is rapidly degraded via the proteasome and that NS2 stability is regulated by phosphorylation at serine residue 168. To corroborate and expand these analyses and to determine whether Ser-168 plays a role in HCV assembly, we generated three alanine substitutions in the context of Jc1-2EI3 as follows: two affecting the active site residues Cys-184 and His-143 and one affecting the putative phosphorylation site Ser-168 (Fig. 3). These mutants were transiently transfected into Huh7.5 cells, and intraas well as extracellular core protein accumulation was determined (Fig. 3A). Although intracellular core protein levels 48 h after transfection were similar, indicating comparable RNA replication, core release was reduced about 3-fold in case of the H143A mutant and up to 5-fold with the S168A mutant. An analogous reduction was found when we analyzed the kinetics of release of extracellular infectivity by using TCID50 assays (Fig. 3B). As already expected from the impaired core release, this reduction was not caused by a block in release of infectious virus, because intracellular infectivity was reduced to the same extent (Fig. 3C). The specific infectivity (ratio between extracellular infectivity and released core protein) was comparable with wild type in case of the C184A and S168A mutants, but about 2-fold reduced with the alanine substitution affecting the active site histidine residue (H143A; Fig. 3D).
FIGURE 3.
Role of active site residues of the NS2 protease domain and serine 168 for HCV particle production. A, Huh7.5 cells were transfected with the constructs specified at bottom, and the amounts of intracellular and extracellular core were determined by ELISA 48 h post-transfection. A representative result of two independent experiments with error ranges is shown. B, kinetics of release of infectious particles was measured 24, 48, and 72 h post-transfection by TCID50 assay. C, intracellular and extracellular infectivity as determined by TCID50 assay 48 h post-transfection. Representative results of three independent experiments with standard deviations are shown in B and C. D, specific infectivity was calculated as the ratio of TCID50 units (infectivity) divided by core amounts (release) and normalized to the ratio obtained with the wild type, which was set to 100%. E, Western blot analysis of NS2 proteins expressed in Huh7.5 cells after transfection with constructs specified at bottom. For further details see legend to Fig. 1.
Because the mutations may affect NS2 stability, we analyzed the accumulation of the variant NS2 proteins in cells at different time points after transfection (Fig. 3E). Increasing amounts of NS2 and tNS2 were observed in Huh7.5 cells transfected with Jc1-2EI3 wild type. The same pattern was found with Jc1-2EI3-C184A that was fully assembly-competent. In contrast, a much lower NS2-specific signal was detected with the H143A mutant. This could be because of lower reactivity of the NS2 protein with the primary antibody used for detection or, more likely, to reduced stability of this protein. Alternatively, the mutation itself may have disrupted an assembly function of NS2. To our great surprise, in cells transfected with the S168A mutant NS2 accumulated to levels very comparable with wild type arguing that at least in the context of Jc1, and thus a chimeric NS2 protein, this highly conserved serine residue does not affect NS2 stability. With the exception of H143A, where the NS2-specific signals were much lower, tNS2 was consistently detected.
Structure Analyses of TMS1 of NS2 by CD and NMR—We have recently described the construction of intra- and intergenotypic HCV chimeras that support assembly of infectious virus particles (27). In most cases, highest infectivity titers were achieved with chimeric genomes in which the junction was positioned into the loop region connecting the putative TMS1 of NS2 with TMS2 (Fig. 2A). This result suggested that NS2, especially TMS1, may play a very important role in infectious particle assembly. We therefore used CD and NMR to determine the structure of this TMS in membrane mimetic environments and to use the structure as a guideline for subsequent mutational analyses and phenotypic assays.
The aa repertoire derived from the ClustalW alignment of 27 reference sequences, representative of all major HCV genotypes and subtypes (33, 60), revealed a large inter-genotypic variability of the N-terminal NS2 sequence (Fig. 4A, top). This inter-genotypic aa variability contrasts with the relatively high intra-genotypic aa conservation as illustrated in Fig. 4, B and C, for genotypes 1b and 2a, respectively. Nevertheless, the conservation of the physicochemical character of residues at most positions (exemplified by the hydropathic pattern in the middle of Fig. 4A) indicates that the overall structure of the NS2 N-terminal sequence is conserved among the different HCV genotypes. This is also supported by the TMS predictions that almost invariably predicted a consensus TMS for aa ∼7-23, irrespective of the analyzed HCV genotypes and subtypes (Fig. 4A, bottom).
For the structural analysis of NS2 TMS1, a peptide corresponding to the first 27 aa of NS2 from the genotype 1b isolate Con1, designated NS2[1-27], was chemically synthesized and purified. As expected from its hydrophobic nature, the NS2[1-27] peptide aggregated in water, and therefore its secondary structure was examined by CD spectroscopy in various membrane mimetic media. The CD spectra of the peptide in a TFE/water mixture containing 30-80% TFE displayed the same intensity as well as the typical shape of an α-helix with two minima at 208 and 222 nm and one maximum around 193 nm (the spectrum recorded in 50% TFE is shown in Fig. 4E). An α-helical content of about 35% was estimated in this TFE/water mixture range (30-80%). Below 30% TFE, the peptide underwent a transition to a β-sheet-like structure that ultimately led to aggregation. In detergent such as dodecylphosphocholine (DPC) or SDS, the peptide also folded into an α-helix. The estimated α-helix content appears to be more important in the zwitterionic detergent DPC (49%) than in the anionic detergent SDS (25%). Although it is less typical of helical folding, the deconvolution of the CD spectrum of NS2[1-27] in the zwitterionic α-lysophosphatidylcholine indicates a higher α-helix content (31%) than in SDS (25%). In a neutral detergent such as dodecyl maltopyranoside, the peptide was unstable and precipitated. In summary, CD spectral analyses indicated the high propensity of the N-terminal NS2 region (aa 1-27) to adopt an α-helix structure upon lipid binding. In addition, the level and/or stabilization of the helical fold appears to depend on the nature of the polar head of the phospholipids.
Deuterated micellar SDS and DPC are popular membrane mimetics for structure analyses of membrane peptides by liquid NMR (61). Unfortunately, samples of NS2[1-27] prepared in SDS and DPC displayed broad, poorly resolved NMR spectra. As the conformation of the peptide, as determined by CD, in 50% TFE is comparable with that observed in SDS and DPC, we probed the structure of NS2[1-27] dissolved at a concentration of 1.22 mm in 50% TFE-d2 and obtained well resolved NMR spectra. Sequential attribution of all spin systems was complete, and an overview of the sequential and medium range NOE connectivities is shown in Fig. 4F. The NOE connectivity patterns between nonadjacent aa clearly indicate that the segment Ala-12 to Thr-21 displays typical characteristics of an α-helix conformation, including strong dNN(i,i + 1) and medium dαN(i,i + 1) sequential connectivities, weak dαN(i,i + 2), medium dαN(i,i + 3), medium or strong dαβ(i,i + 3), and weak dαN(i,i + 4) medium range connectivities. Concerning the N-terminal segment, several expected NOE cross-peaks could not be identified because of the overlapping of NMR resonances (indicated by asterisks in Fig. 4F). However, the presence of some connectivities (dαN(i,i + 3) and dαβ(i,i + 3)) in segment Arg-3 to Gly-11 indicates some nascent helical folding. In contrast, the C-terminal sequence (Ser-23 to Lys-27) is devoid of medium range NOEs and remains unstructured.
The NOE-based indications of α-helical conformation were supported by the deviation of the 1Hα and 13Cα chemical shifts from random coil values (Chemical Shift Index (62)). The long series of negative variation of 1Hα chemical shifts (Δδ1Hα ≤-0.1 ppm) as well as the positive variation of 13Cα chemical shifts (Δδ13Cα ≥0.7 ppm) observed for residues Ala-12 to Thr-21 (Fig. 4G) are indeed typical for an α-helical conformation. The weaker negative Δδ1Hα values observed for Arg-3 to Ser-8 indicate the presence of some helical folding, but the absence of a clear continuous negative pattern is in keeping with the flexibility of this region. In contrast, the continuous positive pattern of Δδ13Cα values for Glu-4 to Thr-21 is indicating that the overall segment could adopt a helical folding, including the two glycine residues 10 and 11.
Taken together these results show that the N-terminal NS2 sequence (residues 1-27) exhibits a clear propensity to adopt an overall helical fold approximately between residues 3 and 23, including a stable α-helix (residues 12-21) connected to a potential flexible helix in the N-terminal part (residues 3-11) by a flexible junction because of the two consecutive glycine residues at position 10 and 11. Based on the NOE-derived interproton distance constraints, a set of 50 structures was calculated with X-PLOR, and a final set of 40 low energy structures that fully satisfied the experimental NMR data were retained. The number and types of NOE constraints used for the structure calculations as well as the statistics for this final set of structures are given in supplemental Table 1. Superimposition of the 40 structures (supplemental Fig. 1) shows that the α-helix segment 13-21 is well defined (supplemental Fig. 1B), with an r.m.s.d. of 0.3 Å (supplemental Table 1). The structure of the N-terminal part of the peptide (supplemental Fig. 1A) is undetermined because of the lack of distance restraints because of the overlap of NMR resonances, which is indicative of some structural flexibility. In addition, the fraying of the N-terminal α-helix 3-9 observed in the isolated NS2[1-27] peptide could be explained by the absence of stabilizing interactions with the remaining NS2 membrane domain(s), and/or with other viral or cellular membrane proteins, and/or membrane lipids. The representative structure model of the putative TMS1 of NS2 shown in supplemental Fig. 1C and Fig. 5A was chosen because of its elongated shape, which is required to cross a membrane bilayer. Its tentative location in the membrane bilayer is shown in Fig. 5B. This putative trans-membrane α-helix displays several remarkable features, including an unusually high number of small glycine and alanine residues in the N-terminal part, and an absolutely conserved helix side in genotype 1b (Fig. 5B, residues highlighted in orange). These numerous well conserved small aa residues as well as the conservation of residues on one side of the helix point to intra-membrane protein-protein interactions.
FIGURE 5.
Structure of NS2 TMS1. A, ribbon representation of a representative experimental structure of the NS2[1-27] segment (Con1 isolate) selected from the final set of 40 calculated NMR structures (PDB entry 2JY0) for its elongated shape required to cross a membrane bilayer. Residue backbones are colored based on the chemical properties of their side chains: hydrophobic (gray) and polar (yellow). Pro is dark gray, and Gly is light gray. Acidic (Asp, Glu) and basic (Arg, Lys) residues are red and blue, respectively. His is cyan, and Tyr is ice-blue. Fully conserved residues in genotype 1b are underlined. The C9A substitution introduced into the peptide that was used for the structural studies is indicated by a box (see “Experimental Procedures”). B, amino acid van der Waals representation of NS2[5-22] helical structure and tentative position within a phospholipid bilayer. Fully and less conserved residues are colored orange and olive green, respectively. The orientation of the left structure is the same as in A, whereas the right structure is rotated by 180°. This panel shows that the majority of fully conserved residues in genotype 1b are located on one side of the trans-membrane α-helix. The membrane is represented as a simulated model of a 1-palmitoyl-2-oleoyl-3-sn-glycero-3-phospholcholine (POPC) bilayer (obtained from P. Tieleman, University of Calgary, Canada). Polar heads and hydrophobic tails of phospholipids (surface and stick structures) are light yellow and gray, respectively. C and D, surface representation of the NS2 helix 5-22 of Con1 and JFH1, respectively, is shown in the upper panel. The membrane interfaces and hydrophobic core are schematically represented. The overall orientation of both structures is the same as in A. The mutated residues in this study are indicated and color-coded according to their effect on infectious particle assembly: mutations without significant effect (group 1) are colored yellow; mutations exhibiting a moderate adaptive phenotype (group 2) are in green, and mutations inducing a reduction of infectious virus particles production (group 3) are in red and magenta for strong and medium effect, respectively. Residue 6 in D, for which the two tested mutations exhibited phenotypes of either group 1 or 3, is colored pink. As residue 25 does not belong to the helix part, its side chain is represented as sticks. The molecular model of NS2[1-27] of JFH1 shown in D was constructed by using the NMR structure of Con1 (this study) as template and the Swiss-PdbViewer program. Figures were generated from structure coordinates using VMD (69) and rendered with POV-Ray. For better comparison of the structure-function relationship, the p7-NS2 proteins expressed by either the Con1/C3 chimera or JFH1 that were used to study the effects of the mutations on HCV assembly are drawn below the corresponding panel (left and right panel, respectively).
Mutation Analysis of TMS1 of NS2, Rationale for the Selected Substitutions—Having established the NMR structure of TMS1, we next investigated the contribution of individual aa residues to HCV assembly by an extensive mutation analysis. To preserve the folding of NS2 TMS1, the mutations were designed by using both the NMR structure model and the natural inter- and intra-genotypic aa conservation and variability (Fig. 4, A-C) as a guideline. Substituting aa residues reported in Fig. 4D were chosen on the basis of conservation or difference between the Con1 and JFH1 isolate as well as size, charge, polarity, and hydrophobicity of the respective aa residue. The drastic change at aa position 5 of NS2 (M5E and V5E; Fig. 4D) was chosen because a Glu or other hydrophilic residues can be found at this position in other genotypes (Fig. 4A). The mutations A6H/H6A, I18T/T18I, and H25G/G25H, which change the physicochemical character of these residues, were chosen because of the presence of the corresponding residue in Con1 and JFH1, respectively, or in other genotypes in case of the A6Q mutation that was introduced into JFH1. The same rule was applied to the mutations S8Q/Q8S, F14L/L14F, and L20F/F20L, for which the physicochemical character of the substituting residues was kept constant. The additional mutation S8E in Con1 (small polar to charged residue) was chosen, because a Glu has been observed at this position in other genotypes (see Fig. 4A). The additional mutation L20A in Con1 was generated to study the role of the strong hydrophobicity at this position for HCV assembly. The invariance of Gly-10 among all genotypes (Fig. 4A) suggests that it could play a role in intra-membrane protein-protein interactions (63). It was therefore replaced by Ala in both the Con1 and the JFH1 context to fill up the “hole” resulting from the absence of a side chain with glycine and to hide the backbone polar groups potentially involved in intra-membranous interactions. Similarly, Gly-16 was replaced by the bulky hydrophobic Phe residue in Con1.
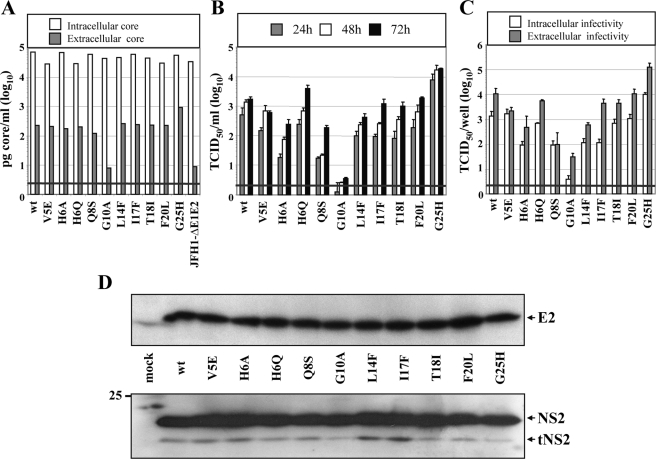
Mutation Analysis of TMS1 of NS2 in the Context of the Chimera Con1/C3—Because the structure determination was performed with TMS1 of the Con1 isolate, we initiated the mutation analysis by using the Con1/C3 chimera in which the region from core to TMS1 is derived from Con1 and the remainder from JFH1 (Fig. 6D). Single point mutations were introduced into TMS1, and the chimeric genomes were tested for core protein expression, core release, kinetic of infectivity release, as well as intra- and extracellular infectivity. As deduced from intracellular core protein accumulation, all mutants replicated to a level comparable with wild type in transfected Huh7.5 cells (Fig. 6A). However, when considering core release or the release of infectious virus particles, the mutants could be classified into three groups. The first group contains the M5E, F14L, and G16F mutants that have an assembly competence comparable with the wild type (Fig. 6, A and B). The second group includes mutants A6H, S8E, and S8Q that have a moderate adaptive phenotype and release slightly higher amounts of infectivity. These mutants replicate comparably with wild type but have a moderately enhanced production of infectious particles, most notably in case of the S8Q mutant (Fig. 6, B and C). The third group includes mutants G10A, L17F, I18T, L20A, L20F, and H25G. Despite intracellular core levels comparable with wild type, these mutations very much reduce the production of infectious virus particles. Interestingly, the most profound impairment is found with the G10A substitution affecting the central glycine residue that may act like a flexible kink in TMS1 and could be involved in intramembranous TMS interactions. It is also interesting to note that the mutations impairing virus production exclusively reside C-terminal of this glycine residue within TMS1 and proximal to the fusion site between the Con1 and the JFH1 sequences (Fig. 6D).
FIGURE 6.
Mutation analysis of TMS1 of NS2 in the context of the Con1/C3 chimera. A, Huh7.5 cells were transfected with Con1/C3 wt or mutants specified according to aa positions within NS2. Amounts of intracellular and extracellular core accumulated 48 h post-transfection were determined by using core-specific ELISA. B, kinetics of release of infectious particles were determined by analyzing culture supernatants at given time points by TCID50 assay. C, intracellular and extracellular infectivities were determined by TCID50 assay at 48 h post-transfection. A representative result of three independent experiments with error ranges is shown in B and C. D, composition of NS2 expressed in the Con1/C3 chimera. The first TMS originates from the Con1 isolate, and the remainder is derived from JFH1. E, Western blot analysis of NS2 and E2 proteins expressed from Con1/C3 constructs specified between both panels. Huh7.5 cells were harvested 48 h post-transfection, and cell lysates were analyzed by Western blot using NS2- or E2-specific antibodies (lower and upper panel, respectively). For further details see legend to Fig. 2.
Western blot analysis of cell lysates was performed to determine whether the alterations of virus titers were because of alterations of polyprotein processing, affecting in particular the E2-p7-NS2 precursor. As shown in Fig. 6E, comparable amounts of full-length NS2 and E2 proteins were obtained with all constructs. However, for some mutants we detected elevated levels of uncleaved p7-NS2 and E2-p7-NS2 precursors (G10A, L20F, and to a lesser extent F14L). The latter precursor was also detected with the NS2-specific antibody (data not shown). Although the presence of these precursors indicates some effect on polyprotein processing, the amounts of uncleaved p7-NS2 and E2-p7-NS2 did not correlate with virus titers. Moreover, some of the NS2 mutants expressed more tNS2 (M5E, A6H, G16F, I18T, and H25G) than wild type (not visible in the exposure shown), but also in this case no correlation with enhanced or reduced virus titers was found. It should be noted that for none of the mutants uncleaved NS2-3 was detected, supporting the notion that RNA replication was not affected.
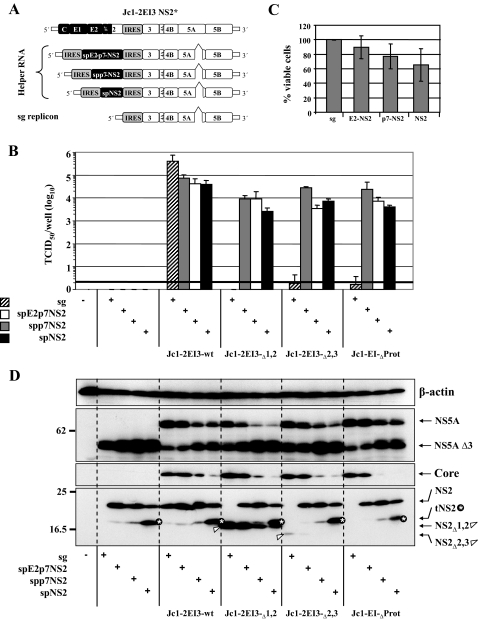
Mutation Analysis of TMS1 of NS2 in the Context of the JFH1 Wild Type—A possible limitation of the mutation analysis described above is its restriction to the Con1/C3 chimera. To exclude the possibility that the phenotypes we observed were because of the chimeric nature of the selected HCV genome rather than reflecting properties of NS2, we generated an analogous panel of mutations in TMS1 in the context of the JFH1 wild type genome. All mutants replicate to wild type level as deduced from the similar amounts of intracellular core protein detected in cells 48 h after transfection (Fig. 7A). Comparable with the results with the Con1/C3 chimera, also the JFH1 mutants could be grouped into three distinct phenotypes. The first group (V5E, H6Q, L14F, I17F, T18I, F20L) is comparable with wild type with respect to core release, assembly, and release of infectious virus particles (Fig. 7, A-C). Group two includes only the mutant in which the C-terminal glycine residue was replaced by a histidine residue (G25H) leading to an enhancement of assembly and release of infectious particles. Mutants of group three are either slightly (H6A, Q8S) or severely (G10A) impaired in the assembly of infectious virus particles.
FIGURE 7.
Mutation analysis of TMS1 of NS2 in the context of the JFH1 wild type. A, Huh7.5 cells were transfected with JFH1-wt or constructs containing single point mutations in the first TMS of NS2. Amounts of intracellular and extracellular core accumulated 48 h post-transfection were determined by using core-specific ELISA. B, kinetics of the release of infectious particles into the supernatants of transfected cells was quantified 24, 48, and 72 h post-transfection by TCID50 assay. C, intracellular and extracellular infectivities were determined by TCID50 assay. Representative results of three independent experiments with error ranges are shown in B and C. D, Western blot analysis of NS2 and E2 proteins in Huh7.5 cells transfected with constructs specified between the panels. Forty eight hours post-transfection cells were harvested, and lysates were analyzed as described in the legend to Fig. 2.
Western blot analysis of NS2 and E2 expressed in cells that had been transfected with the mutants revealed comparable amounts of full-length proteins (Fig. 7D). No additional bands corresponding to p7-NS2, E2-p7-NS2, or uncleaved NS2-3 were detected (not shown).
In summary the results obtained from this mutation analysis clearly identify the central glycine residue in TMS1 as a key element for virus assembly. The observation that mutations affecting residues within TMS1 C-terminal of this glycine were better tolerated in the context of JFH1 than the Con1/C3 chimera suggests that residues C-terminal of glycine 10 in TMS1 may be engaged in interaction with the other TMS of NS2 and/or p7. Because in the Con1/C3 chimera the TMS1 sequence is derived from Con1 and the remainder of NS2 from JFH1 (Fig. 6D), the freedom for mutational alterations in the C-terminal half of TMS1 may be more limited for this chimera.
Rescue of the NS2 Assembly Defect by Trans-complementation—We have recently developed a system that allows the trans-complementation of subgenomic replicons by structural proteins expressed in trans (64). Moreover, we have found that assembly defects caused by mutations in NS5A can be rescued by trans-complementation (59). To analyze whether assembly defects imposed by NS2 can be trans-complemented as well, bicistronic helper RNAs were created (Fig. 8A). These are composed of the JFH1 replicase module (NTRs and the NS3 to NS5B coding region) anda5′-terminal expression module that was used for the production of three different polyprotein fragments as follows: spE2-p7-NS2, spp7-NS2, or spNS2. In all cases the authentic N-terminal signal-like sequence (sp) was retained. The expression module is under control of the poliovirus IRES to allow high level RNA translation (Fig. 8A). All helper RNAs carried a deletion of domain III of NS5A that completely abrogates HCV assembly (59). This deletion removes the epitope that is recognized by the NS5A-specific monoclonal antibody 9E10 (44) that was used for TCID50 assays. In this way we avoided the detection of the helper RNA that can be trans-packaged into virus-like particles (59) and selectively detected the NS2 mutant that has an intact NS5A (detectable by TCID50 assay). A subgenomic (sg) replicon lacking an expression cassette was used as a negative control (Fig. 8A).
FIGURE 8.
Rescue of NS2 mutants defective in virus production by trans-complementation. A, schematic diagram of the NS2 mutants, the helper RNAs, and the subgenomic (sg) replicon. The 5′-terminal IRES is derived from the poliovirus, and the internal IRES is derived from EMCV (each indicated with gray boxes). The deletion of domain III in NS5A is indicated with a triangle. For further details see text. B, Huh7.5 cells were transfected with constructs specified at bottom. Kinetics of release of infectious particles were measured 48 h post-transfection by TCID50 assay. Representative results of three independent experiments with error ranges are shown. C, cytotoxicity assay of Huh7.5 cells transfected with sg replicon or helper RNAs specified at bottom. ATP content in samples was measured 48 h after transfection by using a luminescence-based cell viability assay. Values were normalized to the sg replicon, which was set to 100%. Representative results of three independent experiments with error ranges are shown. D, Western blot analysis of NS5A, core and NS2 proteins in lysates of cells that had been transfected with constructs specified at bottom. Lysates were prepared 48 h post-transfection. The truncated NS2 protein is labeled with a white asterisk and NS2 expressed from the mutants with a white arrowhead. Note that NS2Δ2,3 and NS2Δprot were barely or not at all detectable. β-Actin was used as an internal loading control.
For trans-complementation we used the three NS2 deletion mutants in NS2 (NS2Δ1,2; NS2Δ2,3; and NS2ΔProt) in parallel with the Jc1-2EI3 wild type (wt). In pilot experiments we found that the optimal molar ratio between helper RNA and mutant is 1:1.5. Huh7.5 cells were transfected with this RNA mixture, and supernatants were harvested 48 h later. TCID50 and Western blot assays were performed to determine trans-complementation efficiency and the expression levels of HCV proteins.
Helper and sg replicon RNA each transfected alone as well as cotransfection of sg replicon RNA with any of the NS2 mutants did not give rise to infectious HCV particles (Fig. 8B). Infectivity titers in culture supernatants of cells that had been cotransfected with Jc1-2EI3-wt and sg RNA were comparable with titers obtained with only the bicistronic genome excluding a competition between both replicating RNAs (not shown). Upon cotransfection of any of the helper RNAs with Jc1-2EI3-wt, infectivity titers dropped to 10-fold, most drastically in case of the spNS2 helper RNA. This is at least in part because of some cytotoxicity that we observed with Huh7.5 cells after transfection of only the helper RNA (Fig. 8C). Nevertheless, all three NS2 mutants (NS2Δ1,2; NS2Δ2,3; NS2ΔProt) could be rescued by trans-complementation. Best rescue was obtained with the helper expressing the E2-p7-NS2 protein, which also had the lowest cytotoxic effect. Infectivity titers in rescue experiments were about 10-fold lower as compared with the Jc1-2EI3 wild type (Fig. 8B).
To discriminate between recombination of NS2 mutant and helper RNA leading to a wild type genome that had lost the NS2 mutation, supernatants from primary transfected cells were passaged on naive Huh7.5 cells. Only virus harvested after cotransfection of Jc1-2EI3 wild type and helper RNA could be passaged, excluding the possibility that the infectivity obtained with the NS2 mutants after primary transfection was derived from recombinants (supplemental Fig. 2).
Western blot analysis with the NS2-specific antibody detected four types of NS2 proteins as follows: full-length NS2 (21 kDa), tNS2 (17 kDa), NS2Δ1,2 (17 kDa), and NS2Δ2,3 (15 kDa). In cell lysates containing only helper RNAs, NS2 and tNS2 were exclusively detected (Fig. 8D). In all cases, NS2 expressed from the helper RNAs was processed correctly. Nevertheless, an increase in the amounts of tNS2 (labeled with a star in Fig. 8D) was observed with helper RNAs expressing shorter complementing polyprotein fragments. Highest amounts of tNS2 were detected in cells that had been transfected with the spNS2 helper RNA. This increase was independent from the cotransfected NS2 mutant. The reason for the enhanced production of tNS2 remains to be determined.
The core-specific Western blot revealed a decrease of intracellular core protein (expressed from the NS2 mutant) that correlated with the cytotoxicity of the transfected helper RNA (Fig. 8D). In addition, replication of the genomic NS2 mutants appeared to be reduced by the coreplicating helper RNA. This is best visible in the NS5A-specific Western blot (Fig. 8D). Although the amounts of truncated NS5A expressed from the various helper RNAs increased from spE2-p7-NS2 to spNS2, the amounts of full-length NS5A (that is expressed from the NS2 mutant) decreased. Nevertheless, despite this competition, the results clearly show that assembly defects caused by mutations in NS2 can be rescued by trans-complementation.
DISCUSSION
In this study we characterized the role of NS2 for HCV particle production. We confirm and substantially extend the earlier observation that full-length NS2, but not its protease activity, is required for the production of infectious HCV (9). The structure of TMS1 was probed by CD, and NMR in membrane mimetics and TMS1 residues that are critical for assembly have been identified. Finally, we demonstrate that assembly defects caused by inactivating mutations in NS2 can be rescued by trans-complementation. These results provide the first profound insights into a novel function of NS2, and they will be discussed in more detail below.
Genetic Uncoupling of the Replication Function from the Assembly Function of NS2—With the aim to characterize the role of NS2 for HCV particle production, we generated two Jc1-derived constructs carrying an insertion of either the EMCV IRES or the ubiquitin coding region between NS2 and NS3. The failure of the latter construct to produce infectious HCV may be due to the fact that ubiquitin remained fused to the C terminus of NS2 and thus may interfere with proper folding of the NS2 dimer. In the x-ray crystal structure, the C terminus of NS2 is relocated to the active site of the protease (17), which may be blocked by the fusion with ubiquitin. Moreover, conformational disturbances induced by the C-terminal fusion may interfere with interactions between NS2 and host cell factors involved in HCV assembly. Finally, the NS2-ubiquitin fusion protein appears to be unstable as inferred from the low amounts detectable in transfected cells (Fig. 1E), and these low amounts may be insufficient to support HCV particle formation.
In agreement with an earlier report (9), we found that the insertion of the EMCV IRES between NS2 and NS3 did not interfere with RNA replication and reduced infectivity titers only very moderately (2-5-fold). In addition, we show here that processing of NS2 is also not affected. Thus, as discussed earlier (9), HCV assembly appears to be independent from uncleaved NS2-3 precursors, which is at variance to the replication and assembly properties described for pestiviruses (65).
Role of Individual NS2 Domains for HCV Particle Production—To identify the minimal domain within NS2 that is sufficient for HCV particle production, we used a deletion mapping study. Deletions were based on a modified topology model that we deduced from an earlier report (19) and the x-ray crystal structure of the protease domain (17). According to this model NS2 consists of a TMS1 at the very N terminus and two putative subsequent regions that may form TMS2 and TMS3 as proposed in Fig. 2A. We also assume that the protease domain forms a cytosolic dimeric domain (17). The deletions were designed such that the putative membrane topology should not be disturbed. The fact that processing of E2 was not altered is in keeping with this assumption (data not shown). However, the low amounts of the NS2Δ2,3 protein and the inability to detect even a tagged NS2 protein lacking the protease domain suggests that these proteins were unstable. Such instability could arise from defects in folding that appears to require hydrophobic interaction(s) with membrane lipids and/or hydrophobic regions within the same or another NS2 molecule (17). Although we cannot exclude these possibilities, the data provided in Fig. 2 and the observation that deletions in one domain of NS2 could not be trans-complemented by mutants carrying a deletion in another nonoverlapping region (data not shown) suggest that both the membrane domain and the protease domain of NS2 are required for HCV particle production.
The NS2 protease has a composite active site constituted by His-143 and Glu-163 from one monomer and Cys-184 by the other monomer (13, 17). In support of an earlier report, we found that proteolytic activity of NS2 is dispensable for HCV particle formation (9). However, although the NS2 expression level of the C184A mutant was comparable with wild type, it was much lower in case of the H143A mutant. According to the x-ray crystal structure of the protease domain, His-143 of one monomer is in close contact with Leu-216, Leu-217, and Ser-194 of the other monomer, and forms a salt bridge with Glu-163 of the same monomer (17). Based on structure modeling, we assume that the alanine substitution for His-143 would disturb these interactions and thus also destabilize binding of one monomer to the other (data not shown). As a consequence, dimer formation of NS2 is likely disturbed, which would explain the reduced stability of this NS2 protein. In case of Cys-184 that also resides at the dimer interface, structure modeling suggests that the alanine substitution at this site most likely affects the interactions within the same monomer and between the monomers only to a very minor extent (not shown). Moreover, the physicochemical properties of Cys and Ala are similar providing an explanation for the limited impact of the C184A mutation on HCV assembly and infectivity.
Processing and Stability of NS2—In addition to full-length NS2, we consistently detected a distinct NS2-specific protein with an apparent molecular mass of 17 kDa that we designate here truncated (t)NS2. This protein was observed both with the Con1/C3 chimera and with full-length JFH1 arguing that tNS2 is an authentic product. The origin of this protein is not known, but it likely arises from additional processing either by cytosolic proteases or, more likely, by membrane-associated enzymes. NS2 membrane topology appears to result from cotranslational membrane insertion guided by the C-terminal region of p7 as well as an internal signal-like sequence residing in the N-terminal region of NS2 (aa 810-883) (16, 18, 19). The latter may serve as an internal cleavage site utilized by cellular membrane-bound protease(s) such as signal peptide peptidase, signal peptidase, the rhomboid serine protease or the aspartic acid protease presenilin. It is interesting to note that higher amounts of tNS2 were observed upon expression of isolated NS2 in comparison with expression of the authentic E2-p7-NS2 precursor. This result suggests that proper folding of NS2 is influenced by interaction with E2 and p7. We note that tNS2 has not been observed in earlier studies even when NS2 was expressed on its own, which may be due to either the chosen HCV isolate (the H77 strain) or the lower expression level achieved with the utilized DNA constructs (26).
A recent study suggested that NS2 is unstable and rapidly degraded in a proteasome-dependent manner. This fast degradation appears to depend on phosphorylation of Ser-168 of NS2 by CKII (26). We found that a mutation affecting this serine residue did not affect stability of NS2 of Jc1 (Fig. 3E). The reason for this discrepancy to the earlier report may be due to the different HCV isolate that we chose for our study (JFH1 versus the genotype 1a H77 isolate). Moreover, interactions between NS2 with other structural proteins such as E2 (10) or core protein (66) may stabilize NS2, which could not be detected in the earlier study because the authors only expressed NS2 on its own (25). Nevertheless, even though we did not observe a decreased expression level of NS2, the Ser-168 mutant was impaired in infectious HCV production (about 10-fold reduced TCID50/ml) arguing that phosphorylation at this site directly or indirectly contributes to assembly. Ser-168 resides on the surface of the NS2 protease and presumably is oriented toward the membrane (17). Although the exact interaction of the protease domain with intracellular membranes is not known, we speculate that phosphorylation at this site may destabilize membrane association by introduction of a repulsive electrostatic interaction. By using structure modeling, we assume that the alanine substitution for Ser-168 most likely affects neither NS2 structure (dimerization) nor membrane interaction (not shown). Further studies will be required to determine the phosphorylation status of NS2 in a fully permissive culture system and the possible regulatory role of this NS2 modification.
Structure of TMS1 of NS2 and Its Role for HCV Particle Production—CD and NMR analyses of peptide NS2[1-27] corresponding to the N-terminal 27 aa of NS2 from genotype 1b (Con1 isolate) provide the first description of the structure of NS2 TMS1. Our data show that the core of TMS1 (aa 3-23) exhibits a clear propensity to adopt an overall helical fold in membrane mimetic media. TMS1 displays several remarkable features, including a putative flexible helix in the N-terminal segment (aa 3-11) connected to a stable α-helical segment (aa 12-21) via flexible glycine residues at positions 10 and 11. The numerous well conserved small aa as well as the conservation of residues on one side of the helix point to intra-membrane protein-protein interactions, which may stabilize the helical folding of the flexible N-terminal helix in combination with membrane lipids. The large conservation of aa within each genotype, as illustrated here for genotypes 1a and 2a (Fig. 4, B and C), contrasts with the large variability of aa observed between genotypes (Fig. 4A). This genotype-specific conservation indicates a coadaptation of residues to ensure the specific intra- or inter-molecular interactions between viral proteins required for NS2 processing and function. The full conservation of Gly-10 in all HCV genotypes, as well as that of Pro-24 that marks the C-terminal end of the TMS1 helix, points to key roles of these residues for TMS1 structure and function.
Guided by the NMR structure of TMS1 as well as aa conservation, we performed an extensive mutational analysis to determine the role of individual aa residues in this NS2 domain for HCV particle production. Because the NMR structure was generated with an NS2 peptide7 of genotype 1b, we utilized both the Con1/C3 chimera as well as the authentic JFH1 isolate for this approach. The first major observation we made is the key role of Gly-10 for virus assembly, which we found in the context of both genomes and which could be explained by the involvement of this residue in TM-TM helix interactions. Indeed, glycine and, to a lesser extent, alanine and serine residues allow very close contacts between TM helices. This proximity permits extensive interhelical van der Waals interactions (63, 67). The lack of tolerance of Gly-10 for the Ala replacement reinforces this hypothesis.
The second major observation we made is the high impact of mutations in the C-terminal segment of TMS1 in the context of the Con1/C3 chimera but not the JFH1 genome. It is possible that the N-terminal half of TMS1 (aa 3-10) interacts with protein sequences N-terminal of itself (core to p7), whereas the C-terminal half of TMS1 (aa 12-21) interacts with protein sequences C-terminal of itself. Thus, in case of the Con1/C3 chimera incompatibilities between these C-terminal sequences may exist that could render these interaction sites even more sensitive to mutations as compared with the wild type. In agreement with this assumption, Yi et al. (32) described cell culture adaptive mutations residing in p7 and NS2. These mutations that were identified in the context of a genotype 1a/2a chimera, in which the two genome segments were fused via a naturally occurring recombination site within the NS2 protease domain, enhanced primarily infectivity of the virus particles. Interestingly, these mutations were chimera-specific and appear to compensate for incompatibilities between the fused genome segments. Thus, the observed positions of the mutations may point to interaction(s) between p7 and NS2 (32). The slightly higher titer of infectious particles observed for the A6H and S8Q mutants in the context of the Con1/C3 chimera indicate an adaptive phenotype and suggest that this region likely interacts with the rest of NS2 and/or other NS proteins. The fact that the counterpart mutations H6A and Q8S in the JFH1 context reduced particle production is in keeping with this assumption. In addition, it was reported that the efficiency of p7-NS2 cleavage is limited by structural constraints existing at the N-terminal region of NS2, and that point mutations or aa linker insertions in this region could facilitate p7-NS2 cleavage (68). One could thus postulate that the slightly higher infectivity titer could be due to a release of these structural constraints and facilitation of p7-NS2 processing.
The reduced production of infectious particles observed for L17F, I18T, L20F, as well as the H25G mutant in the Con1/C3 chimera suggests that the corresponding positions are likely not involved in interactions of TMS1 with the remaining TM domain of NS2. Indeed, if it were the case, one could expect a positive or at least a neutral effect on particle production exerted by these aa substitutions, because they revert the aa sequence from Con1 into JFH1. In contrast, the I17F, T18L, and F20L substitutions introduced into the JFH1 genome have almost no detectable effect on infectivity titers. Moreover, the G25H mutation enhances particle production. The apparent tolerance to these mutations suggests a more complex interaction network between TMS1 and the remaining TM of NS2 and/or the structural proteins. Clearly, structural data on full-length NS2, especially its membrane domain, will be required to further elucidate the relationship between NS2 structure and HCV assembly. Finally, the absence of detectable effects on particle production for the mutations M5E, F14L, and G16F in the Con1/C3 chimera and V5E and L14F in the JFH1 genome indicates that these residues are not critical for NS2 interactions and function.
Restoration of HCV Particle Production by Trans-complementation of NS2—Recently it was shown that two assembly deficient genomes lacking either the envelope glycoproteins or p7 can complement each other leading to the production of infectious HCV particles (9). Moreover, we have shown that subgenomic HCV replicons can be packaged in trans (59, 64). In this study we extended this observation by demonstrating that defects in NS2 can also be rescued by trans-complementation. Most efficient rescue was achieved with a subgenomic helper replicon directing the expression of an E2-p7-NS2 precursor, even though replication efficiency was much lower as compared with the helper replicon expressing only NS2. At least two reasons may account for this observation. First, the NS2 helper replicon was more cytotoxic as compared with the replicon expressing the E2-p7-NS2 precursor; second, because of its high replication level the NS2 helper replicon competed much more with the Jc1 genome as compared with the other helper RNAs. Therefore, lower amounts of the genomic NS2 mutant were available resulting in a lower virus titer. For these reasons firm conclusions about the most efficient trans-complementing NS2 protein (E2-p7-NS2 or p7-NS2 or NS2) cannot be drawn. Nevertheless, we found that infectivity titers achieved with the Jc1 wild type genome were reduced about 5-10-fold less by the subgenomic replicon that did not express an NS2 protein (sg; Fig. 8A) as compared with the replicons expressing NS2 or NS2 containing precursors. This result argues for inhibitory interactions between the NS2 protein expressed from the helper RNA with proteins expressed from the Jc1 wild type genome.
In summary, we show that NS2 has a dual function in the HCV replication cycle by acting both as a protease and an essential assembly cofactor. Interestingly, the observation that mutations in NS2 can be rescued by trans-complementation opens new avenues to study the assembly function of NS2 in more detail, thus defining a possible new target for antiviral therapy.
Supplementary Material
Acknowledgments
We are grateful to Ulrike Herian and Stephanie Kallis for excellent technical assistance. We thank Charles M. Rice for providing Huh7.5 cells, Timothy L. Tellinghuisen and Charles M. Rice for the NS5A monoclonal antibody 9E10, Wolfram Gerlich for the Ma18/7 antibody, and Darius Moradpour for the C7/50 core-specific monoclonal antibody.
The atomic coordinates and structure factors (code 2JY0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The proton chemical shifts of all residues have been deposited in the BioMagResBank (BMRB) under the accession number 15579.
This work was supported by the Bioquant Ph.D. program, Ministry of Science, Research, and the Arts of Baden-Württemberg Grant Az 23-7532.24-22-21-12/1 (to R. B. and T. P.), German Research Council Grants BA 1505/2-1 (to N. A. and R. B.), SFB 638, Teilprojekt A5 (to R. B.), and grants from the CNRS (to F. P.) and the Agence Nationale pour la Recherche sur le SIDA et les Hépatites Virales (to F. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
Footnotes
The abbreviations used are: HCV, hepatitis C virus; aa, amino acid; DPC, n-dodecyl phosphocholine; NOE, nuclear Overhauser enhancement; NOESY, NOE spectroscopy; NS2, nonstructural protein 2; NS2[1-27], synthetic peptide representing aa 1-27 of NS2; r.m.s.d., root mean square deviation; RP-HPLC, reversed-phase high performance liquid chromatography; TFE, 2,2,2-trifluoroethanol; TOCSY, total correlation spectroscopy; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; DMEM, Dulbecco's modified Eagle's medium; TMS, trans-membrane segment; NTR, nontranslated region; EMCV, encephalomyocarditis virus; IRES, internal ribosome entry site; nt, nucleotide; sg, subgenomic.
C. Brohm, E. Steinmann, and T. Pietschmann, unpublished data.
T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco.
For consistency with standard protein chemistry nomenclature, the NS2 peptide will be referred to as NS2[1-27], and residues within this peptide will be numbered from the start of the NS2 protein (HCV polyprotein residues 810 to 836, strain Con1, genotype 1b; GenBank™ accession number AJ238799).
References
- 1.Poynard, T., Yuen, M. F., Ratziu, V., and Lai, C. L. (2003) Lancet 3622095 -2100 [DOI] [PubMed] [Google Scholar]
- 2.van Regenmortel, M. H. V., Fauquet, C. M., Bishop, D. H. L., Carstens, E. B., Estes, M. K., Lemon, S. M., Maniloff, J., Mayo, M. A., McGeoch, D. J., Pringle, C. R., and Wickner, R. B. (2000) Virus Taxonomy: The 7th Report of the International Committee on Taxonomy of Viruses, pp. 872-878, Academic Press, San Diego
- 3.Bartenschlager, R., Frese, M., and Pietschmann, T. (2004) Adv. Virus Res. 63 71-180 [DOI] [PubMed] [Google Scholar]
- 4.Appel, N., Schaller, T., Penin, F., and Bartenschlager, R. (2006) J. Biol. Chem. 2819833 -9836 [DOI] [PubMed] [Google Scholar]
- 5.Moradpour, D., Penin, F., and Rice, C. M. (2007) Nat. Rev. Microbiol. 5453 -463 [DOI] [PubMed] [Google Scholar]
- 6.Grakoui, A., Wychowski, C., Lin, C., Feinstone, S. M., and Rice, C. M. (1993) J. Virol. 671385 -1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin, S. D., Beales, L. P., Clarke, D. S., Worsfold, O., Evans, S. D., Jaeger, J., Harris, M. P., and Rowlands, D. J. (2003) FEBS Lett. 53534 -38 [DOI] [PubMed] [Google Scholar]
- 8.Sakai, A., Claire, M. S., Faulk, K., Govindarajan, S., Emerson, S. U., Purcell, R. H., and Bukh, J. (2003) Proc. Natl. Acad. Sci. U. S. A. 10011646 -11651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, C. T., Murray, C. L., Eastman, D. K., Tassello, J., and Rice, C. M. (2007) J. Virol. 818374 -8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinmann, E., Penin, F., Kallis, S., Patel, A. H., Bartenschlager, R., and Pietschmann, T. (2007) PLoS Pathog. 3e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann, V., Körner, F., Koch, J. O., Herian, U., Theilmann, L., and Bartenschlager, R. (1999) Science 285110 -113 [DOI] [PubMed] [Google Scholar]
- 12.Hijikata, M., Mizushima, H., Akagi, T., Mori, S., Kakiuchi, N., Kato, N., Tanaka, T., Kimura, K., and Shimotohno, K. (1993) J. Virol. 674665 -4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grakoui, A., McCourt, D. W., Wychowski, C., Feinstone, S. M., and Rice, C. M. (1993) Proc. Natl. Acad. Sci. U. S. A. 9010583 -10587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomei, L., Failla, C., Santolini, E., De Francesco, R., and La Monica, N. (1993) J. Virol. 674017 -4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrens, S. E., Grassmann, C. W., Thiel, H. J., Meyers, G., and Tautz, N. (1998) J. Virol. 722364 -2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santolini, E., Pacini, L., Fipaldini, C., Migliaccio, G., and Monica, N. (1995) J. Virol. 697461 -7471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz, I. C., Marcotrigiano, J., Dentzer, T. G., and Rice, C. M. (2006) Nature 442831 -835 [DOI] [PubMed] [Google Scholar]
- 18.Mizushima, H., Hijikata, M., Tanji, Y., Kimura, K., and Shimotohno, K. (1994) J. Virol. 682731 -2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaga, A. K., and Ou, J. H. (2002) J. Biol. Chem. 27733228 -33234 [DOI] [PubMed] [Google Scholar]
- 20.Kiiver, K., Merits, A., Ustav, M., and Zusinaite, E. (2006) Virus Res. 117264 -272 [DOI] [PubMed] [Google Scholar]
- 21.Flajolet, M., Rotondo, G., Daviet, L., Bergametti, F., Inchauspe, G., Tiollais, P., Transy, C., and Legrain, P. (2000) Gene (Amst.) 242369 -379 [DOI] [PubMed] [Google Scholar]
- 22.Selby, M. J., Glazer, E., Masiarz, F., and Houghton, M. (1994) Virology 204114 -122 [DOI] [PubMed] [Google Scholar]
- 23.Kim, K. M., Kwon, S. N., Kang, J. I., Lee, S. H., Jang, S. K., Ahn, B. Y., and Kim, Y. K. (2007) Biochem. Biophys. Res. Commun. 356948 -954 [DOI] [PubMed] [Google Scholar]
- 24.Yang, X. J., Liu, J., Ye, L., Liao, Q. J., Wu, J. G., Gao, J. R., She, Y. L., Wu, Z. H., and Ye, L. B. (2006) Virus Res. 121134 -143 [DOI] [PubMed] [Google Scholar]
- 25.Erdtmann, L., Franck, N., Lerat, H., Le Seyec, J., Gilot, D., Cannie, I., Gripon, P., Hibner, U., and Guguen-Guillouzo, C. (2003) J. Biol. Chem. 27818256 -18264 [DOI] [PubMed] [Google Scholar]
- 26.Franck, N., Le Seyec, J., Guguen-Guillouzo, C., and Erdtmann, L. (2005) J. Virol. 792700 -2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietschmann, T., Kaul, A., Koutsoudakis, G., Shavinskaya, A., Kallis, S., Steinmann, E., Abid, K., Negro, F., Dreux, M., Cosset, F. L., and Bartenschlager, R. (2006) Proc. Natl. Acad. Sci. U. S. A. 1037408 -7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreau, I., Hegarty, S., Levis, J., Sheehy, P., Crosbie, O., Kenny-Walsh, E., and Fanning, L. J. (2006) Virol. J. 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noppornpanth, S., Lien, T. X., Poovorawan, Y., Smits, S. L., Osterhaus, A. D., and Haagmans, B. L. (2006) J. Virol. 807569 -7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalinina, O., Norder, H., Mukomolov, S., and Magnius, L. O. (2002) J. Virol. 764034 -4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang, D. R., Lai, H. Y., Chang, M. L., Hsu, J. T., and Yeh, C. T. (2005) J. Virol. Methods 129170 -177 [DOI] [PubMed] [Google Scholar]
- 32.Yi, M., Ma, Y., Yates, J., and Lemon, S. M. (2007) J. Virol. 81629 -638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Combet, C., Garnier, N., Charavay, C., Grando, D., Crisan, D., Lopez, J., Dehne-Garcia, A., Geourjon, C., Bettler, E., Hulo, C., Le Mercier, P., Bartenschlager, R., Diepolder, H., Moradpour, D., Pawlotsky, J. M., Rice, C. M., Trepo, C., Penin, F., and Deleage, G. (2007) Nucleic Acids Res. 35D363 -D366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Combet, C., Blanchet, C., Geourjon, C., and Deleage, G. (2000) Trends Biochem. Sci. 25 147-150 [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 224673 -4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rost, B., Casadio, R., Fariselli, P., and Sander, C. (1995) Protein Sci. 4 521-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmann, K., and Stoffel, W. (1993) Biol. Chem. Hoppe-Seyler 374166. [DOI] [PubMed] [Google Scholar]
- 38.Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001) J. Mol. Biol. 305567 -580 [DOI] [PubMed] [Google Scholar]
- 39.Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998) Bioinformatics (Oxf.) 14 378-379 [DOI] [PubMed] [Google Scholar]
- 40.Claros, M. G., and Von Heijne, G. (1994) Comput. Appl. Biosci. 10 685-686 [DOI] [PubMed] [Google Scholar]
- 41.Tusnady, G. E., and Simon, I. (1998) J. Mol. Biol. 283489 -506 [DOI] [PubMed] [Google Scholar]
- 42.Blight, K. J., McKeating, J. A., and Rice, C. M. (2002) J. Virol. 7613001 -13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakita, T., Pietschmann, T., Kato, T., Date, T., Miyamoto, M., Zhao, Z., Murthy, K., Habermann, A., Krausslich, H. G., Mizokami, M., Bartenschlager, R., and Liang, T. J. (2005) Nat. Med. 11791 -796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindenbach, B. D., Evans, M. J., Syder, A. J., Wolk, B., Tellinghuisen, T. L., Liu, C. C., Maruyama, T., Hynes, R. O., Burton, D. R., McKeating, J. A., and Rice, C. M. (2005) Science 309623 -626 [DOI] [PubMed] [Google Scholar]
- 45.van-den Hoff, M. J., Labruyere, W. T., Moorman, A. F., and Lamers, W. H. (1990) Nucleic Acids Res. 18 6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koutsoudakis, G., Herrmann, E., Kallis, S., Bartenschlager, R., and Pietschmann, T. (2007) J. Virol. 81 588-598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gastaminza, P., Kapadia, S. B., and Chisari, F. V. (2006) J. Virol. 8011074 -11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manavalan, P., and Johnson, W. C., Jr. (1987) Anal. Biochem. 16776 -85 [DOI] [PubMed] [Google Scholar]
- 49.Whitmore, L., and Wallace, B. A. (2004) Nucleic Acids Res. 32W668 -W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penin, F., Geourjon, C., Montserret, R., Bockmann, A., Lesage, A., Yang, Y. S., Bonod-Bidaud, C., Cortay, J. C., Negre, D., Cozzone, A. J., and Deleage, G. (1997) J. Mol. Biol. 270496 -510 [DOI] [PubMed] [Google Scholar]
- 51.Favier, A., Brutscher, B., Blackledge, M., Galinier, A., Deutscher, J., Penin, F., and Marion, D. (2002) J. Mol. Biol. 317131 -144 [DOI] [PubMed] [Google Scholar]
- 52.Merutka, G., Dyson, H. J., and Wright, P. E. (1995) J. Biomol. NMR 514 -24 [DOI] [PubMed] [Google Scholar]
- 53.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York
- 54.Schwieters, C. D., Kuszewski, J. J., Tjandra, N., and Clore, G. M. (2003) J. Magn. Reson. 160 65-73 [DOI] [PubMed] [Google Scholar]
- 55.Koradi, R., Billeter, M., and Wüthrich, K. (1996) J. Mol. Graphics 1429 -32 [DOI] [PubMed] [Google Scholar]
- 56.Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 57.Welbourn, S., Green, R., Gamache, I., Dandache, S., Lohmann, V., Bartenschlager, R., Meerovitch, K., and Pause, A. (2005) J. Biol. Chem. 28029604 -29611 [DOI] [PubMed] [Google Scholar]
- 58.Kolykhalov, A. A., Mihalik, K., Feinstone, S. M., and Rice, C. M. (2000) J. Virol. 742046 -2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Appel, N., Zayas, M., Miller, S., Krijnse-Locker, J., Schaller, T., Friebe, P., Kallis, S., Engel, U., and Bartenschlager, R. (2008) PLoS Pathog. 4e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmonds, P., Bukh, J., Combet, C., Deleage, G., Enomoto, N., Feinstone, S., Halfon, P., Inchauspe, G., Kuiken, C., Maertens, G., Mizokami, M., Murphy, D. G., Okamoto, H., Pawlotsky, J. M., Penin, F., Sablon, E., Shin, I., Stuyver, L. J., Thiel, H. J., Viazov, S., Weiner, A. J., and Widell, A. (2005) Hepatology 42 962-973 [DOI] [PubMed] [Google Scholar]
- 61.Opella, S. J. (1997) Nat. Struct. Biol. 4S845 -S848 [PubMed] [Google Scholar]
- 62.Wishart, D. S., Sykes, B. D., and Richards, F. M. (1992) Biochemistry 311647 -1651 [DOI] [PubMed] [Google Scholar]
- 63.Senes, A., Ubarretxena-Belandia, I., and Engelman, D. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 989056 -9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steinmann, E., Brohm, C., Kallis, S., Bartenschlager, R., and Pietschmann, T. (2008) J. Virol. 827034 -7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agapov, E. V., Murray, C. L., Frolov, I., Qu, L., Myers, T. M., and Rice, C. M. (2004) J. Virol. 782414 -2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murray, C. L., Jones, C. T., Tassello, J., and Rice, C. M. (2007) J. Virol. 8110220 -10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacKenzie, K. R., and Engelman, D. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 953583 -3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carrere-Kremer, S., Montpellier, C., Lorenzo, L., Brulin, B., Cocquerel, L., Belouzard, S., Penin, F., and Dubuisson, J. (2004) J. Biol. Chem. 27941384 -41392 [DOI] [PubMed] [Google Scholar]
- 69.Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graphics 1433 -38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.