Abstract
Barbara McClintock reported that the Ac/Ds transposable element system can generate major chromosomal rearrangements (MCRs), but the underlying mechanism has not been determined. Here, we identified a series of chromosome rearrangements derived from maize lines containing pairs of closely linked Ac transposable element termini. Molecular and cytogenetic analyses showed that the MCRs in these lines comprised 17 reciprocal translocations and two large inversions. The breakpoints of all 19 MCRs are delineated by Ac termini and characteristic 8-base-pair target site duplications, indicating that the MCRs were generated by precise transposition reactions involving the Ac termini of two closely linked elements. This alternative transposition mechanism may have contributed to chromosome evolution and may also occur during V(D)J recombination resulting in oncogenic translocations.
Keywords: V(D)J recombination, chromosome rearrangements, hAT elements, transposition
In the 1940s, Barbara McClintock reported that the maize Activator (Ac) element could induce transposition of the nonautonomous Dissociation (Ds) element, which she identified as a locus of chromosome breakage. She also showed, using cytogenetic methods, that transposition of Ds was sometimes accompanied by major chromosomal rearrangements (MCRs) including deletions, duplications, inversions, reciprocal translocations, and ring chromosomes (McClintock 1949, 1950a,b, 1951). Because these rearrangements only occurred in the presence of Ac and they retained Ds at their breakpoints, McClintock concluded that Ac/Ds transposition is somehow responsible for their origin. Although several hypotheses have been advanced to explain the origin of McClintock's MCRs, it is still unclear precisely how these large chromosome rearrangements were generated.
Standard Ac/Ds transposition only changes the position of the transposon in the genome, hence the MCRs isolated by McClintock must have originated by some other mechanism involving transposition. A type of aberrant Ds transposition that results in fusion of sister chromatids, chromosome breakage, and formation of deletions has been described (English et al. 1993, 1995; Weil and Wessler 1993). Previously, we showed that a pair of Ac termini in tandem orientation produced reciprocal deletion/duplication alleles via a mechanism in which Ac transposase interacts with Ac termini on sister chromatids; thus it was termed sister chromatid transposition (SCT) (Zhang and Peterson 1999). However, SCT cannot fully explain the origin of McClintock's MCRs because the only heritable products it generates are deletions and inverted duplications.
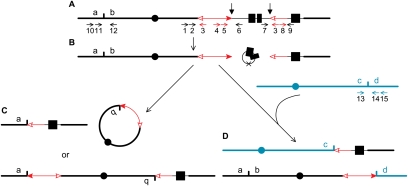
Recently, we and others showed that a pair of Ac termini in reversed orientation can also undergo transposition (reversed Ac ends transposition) (Zhang and Peterson 2004; Huang and Dooner 2008). In this reaction, Ac transposase acts on a pair of Ac termini on the same sister chromatid (Fig. 1A). Insertion of the excised Ac termini into nearby sites can produce relatively small rearrangements, including deletions and inversions (Zhang and Peterson 2004; Huang and Dooner 2008). However, the excised Ac ends could also insert into distant sites on the same or different chromosomes to generate a variety of MCRs. For example, insertion into the same chromatid would generate an acentric fragment and a ring chromosome (the top portion of Fig. 1C; Supplemental Movie 1), or an inversion (the bottom portion of Fig. 1C; Supplemental Movie 1), depending on the orientation of insertion of the transposon ends. In addition, insertion into another chromosome would generate a reciprocal translocation (Fig. 1D; Supplemental Movie 1), or a dicentric chromosome and an acentric fragment (Supplemental Movie 1), again depending on the orientation of insertion of the transposon ends.
Figure 1.
Model for generation of major chromosome rearrangements by reversed Ac ends transposition. The lines depict maize chromosomes, with centromeres indicated by black and green circles. The red arrows indicate Ac (double-headed arrow) and fAc, a ∼2.0-kb fragment containing the 3′ end of Ac (single arrowhead). The open and solid arrowheads indicate the 3′ and 5′ ends, respectively, of Ac/fAc. The fAc element is inserted into the second intron of the maize p1 gene, whose exons are indicated by solid boxes. The small vertical arrows indicate the Ac transposase cleavage sites. (A) Ac transposase cleaves at the 5′ end of Ac and the 3′ end of fAc. (B) Following transposase cleavage at the junctions of Ac/p1 and fAc/p1, the internal p1 genomic sequences are joined to form a circle. The “×” on the circle indicates the site where joining occurred, marked by a transposon footprint. The Ac 5′ and fAc 3′ ends are competent for insertion anywhere in the genome. C and D depict the outcomes of insertion into two possible target sites (short vertical lines). (C) The Ac/fAc termini insert into a site on the opposite arm of the same sister chromatid; in the top figure, the Ac 5′ end joins to the proximal side of the target site to form a ring chromosome, and the fAc 3′ end joins to the distal side of the target site to form an acentric fragment. (Bottom) Alternatively, ligation of the Ac 5′ end to the distal side of the target site and the fAc 3′ end to the proximal side of the target site would generate a pericentric inversion. (D) The transposon ends insert into a site in another chromosome; the Ac 5′ end joins to the distal side of the target site, and the fAc 3′ end joins to the proximal side of the target site to generate a reciprocal translocation. The short, horizontal arrows indicate the orientations and approximate positions of PCR primers. Primers are identified by numbers above or below the arrows. The structures of P1-rr11 and P1-rr910 are similar to that of the p1 allele in 1A; i.e., the Ac element is located upstream of the p1 gene; in P1-ovov454, however, the Ac element is inserted in intron 2 of p1. Primers 1, 2, and 6 differ for alleles P1-rr11, P1-rr910, and P1-ovov454 due to differences in the Ac insertion sites; see the Materials and Methods for details.
Some MCRs, including acentric fragments (Fig. 1C), acentric rings, and dicentric chromosomes, would likely be highly unstable or result in cell lethality. However, we predicted that certain rearrangements including duplications, inversions, and reciprocal translocations should be transmitted to the next generation. In previous studies, rearrangements generated by reversed-ends transposition were relatively small and could not be directly visualized. We have now confirmed, through cytogenetic methods, the formation of 19 MCRs (17 reciprocal translocations and two large inversions) generated by reversed Ac ends transposition.
Results
Detection of transposon-induced MCRs
The p1 gene encodes a Myb-homologous transcriptional activator required for the production of red phlobaphene pigments in maize floral tissues including kernel pericarp (the outermost layer of the maize seeds) and cob glumes (Grotewold et al. 1991, 1994). The standard P1-rr allele specifies red pericarp and red cob, and the kernels in a P1-rr ear are uniformly red. In contrast, three alleles (P1-rr11, P1-rr910, and P1-ovov454) exhibit unstable pigmentation in which kernels are predominantly red or orange but also exhibit frequent colorless stripe(s) and sectors. All three of these alleles have similar structures, with a fractured Ac element (fAc, 2039 base pairs [bp] of the 3′ portion of Ac) inserted in intron 2 of the p1 gene, and a full-length Ac element inserted nearby, upstream of the fAc element. In each allele, the full-length Ac element is oriented with its 5′ terminus closest to the 3′ terminus of fAc; the distances between Ac and fAc in P1-rr11, P1-rr910, and P1-ovov454 are 13,175 bp, 8919 bp and 823 bp, respectively (Fig. 1A). We reasoned that transposition reactions involving reversed Ac ends—i.e., the 5′ end of the full-length Ac element and the 3′ end of fAc—could be responsible for the colorless kernel pericarp sectors exhibited by these alleles. As shown in Figure 1 and the Supplemental Movie 1, most of the predicted products of reversed Ac ends transposition would have suffered loss of p1 gene exons 1 and 2, or separation of exons 1 and 2 from exon 3. Either outcome would prevent p1 gene function and result in colorless sectors. Therefore, we selected seeds with colorless kernel pericarp as candidate carriers of transposon-induced MCRs (see the Materials and Methods for details).
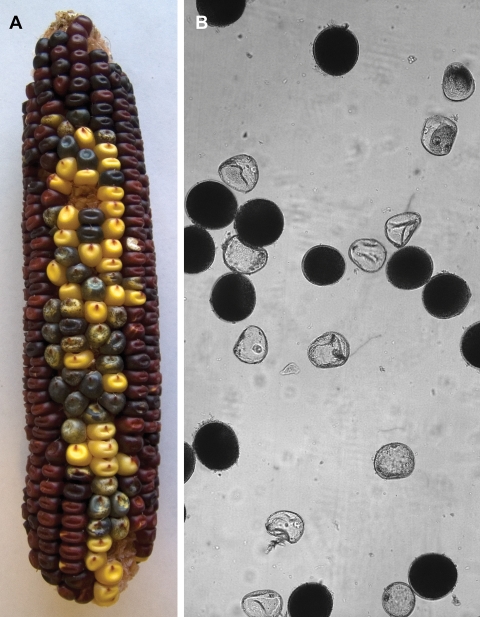
Plants heterozygous for MCRs including large deletions, inversions, or reciprocal translocations produce a significant proportion of inviable meiotic products. In maize, the resulting gametophytic lethality results in semisterile ears with irregular rows and defective pollen grains. In some cases, rearrangement events occurred sufficiently early in development to produce large, multikernel sectors in which the area of colorless pericarp coincides exactly with a region of female semisterility. For example, Figure 2A shows a mature ear from a plant heterozygous for progenitor allele P1-rr11 (red pericarp kernels); the ear has a large sector of kernels with colorless pericarp (source of translocation p1-wwB966). The red pericarp area has normal seed set and regular rows of kernels, while the colorless pericarp sector has semisterile seed set and irregular kernel placement. Most of the rearrangement alleles described here were obtained from smaller colorless-pericarp sectors within which semisterility could not be scored reliably. However, when these colorless pericarp kernels were grown into plants, their ears and pollen were screened for significant levels of sterility. Figure 2B shows pollen grains from a plant heterozygous for a large inversion; ∼50% of the pollen grains are small, misshapen, and devoid of starch. Plants that produced at least 20% defective pollen and irregular seed set were selected for further characterization. Their progeny were then examined to determine if the pollen abortion and irregular kernel set are inherited. PCR analysis was performed to test whether the candidate plants contain structures indicative of the presence of an MCR; four pairs of oligonucleotide primers were designed to detect the junctions between Ac/fAc and its flanking sequences (see Fig. 1 for the orientations and approximate positions of the primers). According to the model shown in Figure 1, inversion and reciprocal translocation alleles should produce specific PCR products with primer pairs 1 + 3 and 8 + 9, but not with primer pairs 5 + 6 and 7 + 3. Based on the combined genetic screening and PCR results, we identified 25 candidate alleles with characteristics consistent with the presence of large inversions or reciprocal translocations.
Figure 2.
Phenotypic effects of reciprocal translocations and large inversions. (A) Mature ear of progenitor allele P1-rr11 (red pericarp kernels), with origination sector of translocation p1-wwB966 (large sector of kernels with colorless pericarp). Note that kernels in the colorless pericarp sector are larger and irregularly spaced because ∼50% of the eggs have aborted (cf. smaller, evenly spaced kernels in red pericarp portion of ear). Kernels with purple-spotted aleurone indicate Ac-induced excision of Ds from r1-m3∷Ds allele; see the Materials and Methods for details. (B) Pollen from a large inversion heterozygote exhibiting ∼50% semisterility. The larger, starch-filled pollen grains (dark) carry a complete haploid genome, whereas the smaller, irregular, translucent pollen grains carry an imbalanced chromosome set following meiosis in the inversion heterozygote.
Sequence analysis of MCR breakpoints
The breakpoints flanking Ac in 24 of the 25 MCR candidates were cloned via Ac casting (see the Materials and Methods for details) (Singh et al. 2003), and the breakpoints flanking fAc in all the MCR candidates were cloned via PCR as described in the Materials and Methods. If the MCR candidates were indeed generated via reversed Ac ends transposition, we expect that the 8-bp sequence flanking the Ac 5′ end should match the 8 bp flanking the fAc 3′ end (target site duplication, TSD). Indeed, 8-bp TSDs were observed for all of the 19 confirmed MCR alleles (sequence data available in Supplemental Material file MCRsequences.doc). In the case of p1-wwB966, molecular and genetic analyses suggested that the Ac element had excised from the MCR breakpoint and inserted nearby in the genome; its MCR breakpoint sequences were isolated by ligation-mediated PCR (LM-PCR) and found to share only 7 bp instead of the expected 8-bp common sequence at the junctions of Ac/fAc with their flanking sequences. The base immediately adjacent to the Ac 5′ end is different from the corresponding base flanking the fAc 3′ end; this result is consistent with formation of a TSD by Ac insertion, followed by a 1-bp transversion (G to C) upon Ac excision.
Mapping chromosomal locations of MCR breakpoints
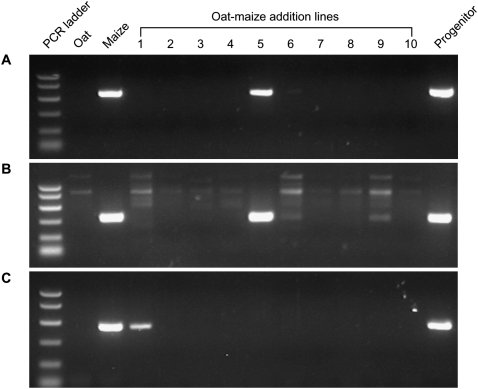
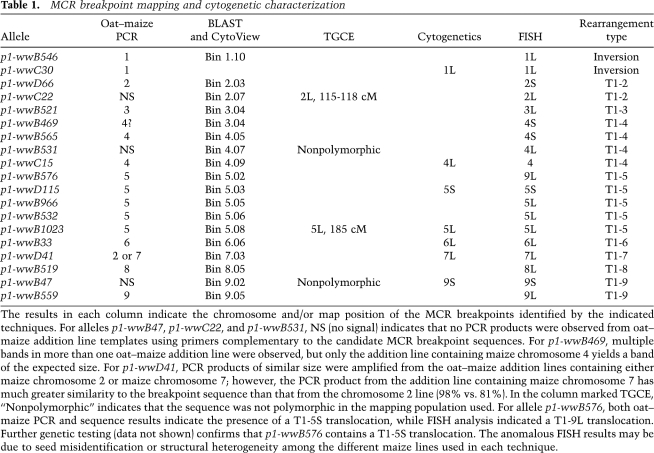
To identify the types of structural rearrangements in the MCR lines, we needed to determine the chromosomal locations of the breakpoints. Most breakpoint sequences were mapped to one of the maize chromosomes by PCR using the genomic DNA of a series of oat–maize chromosome addition lines. Each oat–maize line contains all of the oat chromosomes, and one of the 10 maize chromosomes (Ananiev et al. 1997; Kynast et al. 2002). Thus, primers for unique maize sequences will only amplify PCR products from the oat–maize line that contains the corresponding chromosome (Fig. 3). In this way, the breakpoints of 14 MCR candidates were unambiguously mapped to specific chromosomes. In five cases, PCR using oat–maize addition lines gave negative or ambiguous results. These data are summarized in Table 1.
Figure 3.
MCR breakpoint mapping by PCR using oat–maize addition lines as templates. PCR was performed using primers complementary to the sequences at the predicted MCR breakpoints. Sources of template genomic DNA are indicated at the top of each lane. The lanes marked 1–10 indicate DNA from oat–maize addition lines containing maize chromosomes 1–10, respectively. The lane marked Progenitor contains DNA from the maize allele that was the progenitor of the MCR (P1-rr11, P1-rr910, and P1-ovov454). (A) p1-wwB966. (B) p1-wwB1023. (C) p1-wwC30.
Table 1.
MCR breakpoint mapping and cytogenetic characterization
The results in each column indicate the chromosome and/or map position of the MCR breakpoints identified by the indicated techniques. For alleles p1-wwB47, p1-wwC22, and p1-wwB531, NS (no signal) indicates that no PCR products were observed from oat–maize addition line templates using primers complementary to the candidate MCR breakpoint sequences. For p1-wwB469, multiple bands in more than one oat–maize addition line were observed, but only the addition line containing maize chromosome 4 yields a band of the expected size. For p1-wwD41, PCR products of similar size were amplified from the oat–maize addition lines containing either maize chromosome 2 or maize chromosome 7; however, the PCR product from the addition line containing maize chromosome 7 has much greater similarity to the breakpoint sequence than that from the chromosome 2 line (98% vs. 81%). In the column marked TGCE, “Nonpolymorphic” indicates that the sequence was not polymorphic in the mapping population used. For allele p1-wwB576, both oat–maize PCR and sequence results indicate the presence of a T1-5S translocation, while FISH analysis indicated a T1-9L translocation. Further genetic testing (data not shown) confirms that p1-wwB576 contains a T1-5S translocation. The anomalous FISH results may be due to seed misidentification or structural heterogeneity among the different maize lines used in each technique.
Although the maize genome sequence is not yet complete, many maize BAC clones have been partially sequenced and their chromosome positions tentatively mapped. The sequences of candidate MCR breakpoints obtained here were used in BLAST alignments against the maize high-throughput genomic sequences (HTGS; http://www.ncbi.nlm.nih.gov/blast/). BACs with high homology with 18 of 19 MCR candidates were identified, and tentative chromosomal positions of homologous BAC clones were obtained from http://www.maizesequence.org. In most cases, the putative MCR map positions identified by oat–maize PCR agreed with the BAC map positions assigned by the Maize Genome Sequencing Project. Three cases gave ambiguous or conflicting results (see Supplemental Table 1). Finally, PCR and Temperature Gradient Capillary Electrophoresis (TGCE) (Fu et al. 2005) were also used to map the MCR breakpoint sequences of some candidate alleles. In all cases in which mapping results were obtained, the map positions were in good agreement with the results obtained by at least one independent method.
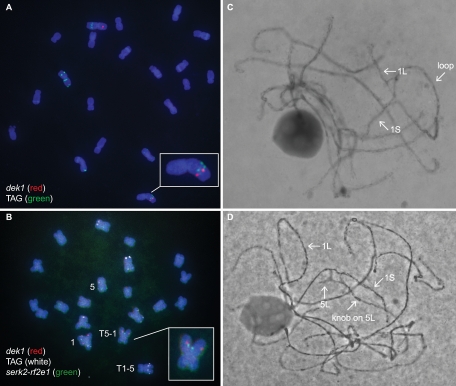
Visualization of MCRs by fluorescence in situ hybridization (FISH) in somatic cells and at pachytene in meiotic cells
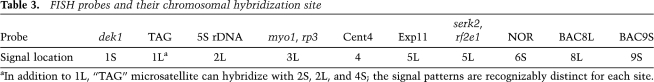
To confirm the molecular results indicating the presence of chromosomal inversions and translocations, we performed cytogenetic analysis of maize mitotic chromosomes using FISH). Maize seedlings heterozygous for putative MCR and normal chromosomes were pretreated with nitrous oxide as a spindle poison, and root tip cell chromosome preparations were hybridized with fluorescently labeled single gene or gene cluster probes for somatic chromosome identification (Kato et al. 2006). Two probes were used to detect the presence of pericentric inversions involving chromosome 1: One probe was complementary to the dek1 gene, which is located ∼700 kb distal to the p1 gene on the short arm of maize chromosome 1 (http://www.maizesequence.org). A second probe (“TAG”) hybridizes with the TAG-microsatellite locus on chromosomes 1L, 2S, 2L, and 4S of most genotypes. The results of FISH using these probes confirm that p1-wwC30 contains a pericentric inversion of chromosome 1 (Fig. 4A). Similar results were obtained for allele p1-wwB546 (data not shown).
Figure 4.
Cytogenetic analysis of MCR alleles. Cells of plants heterozygous for a rearrangement and corresponding normal chromosomes were characterized by FISH of mitotic metaphase chromosomes (A,B), and by propiocarmine staining of meiotic pachytene chromosomes (C,D). Arrows in C and D indicate sites of characteristic features. (A) Chromosome 1 pericentric inversion heterozygote (p1-wwC30) hybridized with dek1 (red, detects 1S) and TAG (green, detects 1L; also 2S, 2L, and 4S). Inversion chromosome is enlarged in the inset. (B) Translocation chromosome T1-5 heterozygote (p1-wwB1023) hybridized with dek1 (red, detects 1S), TAG (white, detects 1L; also 2S, 2L, and 4S), and serk2-rf2e1 (green; detects 5L). The T5-1 translocation chromosome is enlarged in the inset. (C) Chromosome 1 pericentric inversion heterozygote (p1-wwC30). (D) Translocation chromosome T1-5 heterozygote (p1-wwB1023).
Using the dek1 and “TAG” probes for chromosome 1 together with probes from other chromosomes, FISH was performed to determine whether the other putative MCRs contain reciprocal translocations. The result from p1-wwB1023 (a putative T1-5 reciprocal translocation) (Fig. 4B) is especially interesting. The probe serk2-rf2e1 hybridizes to a site on chromosome 5L. In addition to the normal chromosomes 1 and 5 (left side of panel 4B), these cells contain a translocation chromosome in which the dek1 and serk2-rf2e1 signals are juxtaposed (Fig. 4B, inset). These results confirm the presence of a translocation involving chromosomes 1S and 5L. Pachytene analysis (see below) also demonstrated the presence of a T1-5 translocation with a breakpoint very close to the end of chromosome 5 (Fig. 4D). This conclusion is consistent with the TGCE mapping data indicating that the translocation breakpoint is only 5 cM from the telomere of 5L; i.e., <3% of the total length of chromosome 5 from the telomere. In addition, the frequency of aborted pollen in p1-wwB1023 heterozygotes is ∼25%, much less than the 50% abortion frequency expected for translocations that involve chromosome segments containing genes essential for pollen viability. All of these data support the conclusion that p1-wwB1023 contains a T1-5 translocation whose breakpoint is located very close to the tip of chromosome 5L.
The results of oat-maize PCR indicated that p1-wwB521 contains a T1-3 translocation; sequencing and BLAST analysis (CytoView, http://www.maizesequence.org) placed the breakpoint on chromosome 3S, bin 3.04 (for a description of maize chromosome bin designations, see http://www.maizegdb.org/cgi-bin/bin_viewer.cgi). In contrast, FISH analysis using probes myo1 on chromosome 3L and TAG on chromosome 1L showed clearly that the 1S breakpoint is attached to a large segment of 3L (see Supplemental Fig. 1). Together, these results suggest that the breakpoint is located on chromosome 3L, very close to the centromere.
As described above, oat–maize PCR results for p1-wwB469 were ambiguous, with bands present in multiple addition lines. The best match obtained from BLAST of the breakpoint sequences is located on chromosome 3, bin 3.04 (Table 1). However, no rearrangements were detected by FISH using probes dek1 (1S), TAG (1L), and rp3 on chromosome 3, whereas evidence of a T1-4 reciprocal translocation was obtained using probes dek1, TAG, and cent4. The cent4 probe hybridizes to a site near centromere 4 (Jin et al. 2004). Cells from p1-wwB469 contain a presumptive translocation chromosome that hybridizes with both the dek1 (1S) and cent4 (4S) probes. In addition, a TAG-microsatellite locus is present on chromosome 4S, and the TAG probe hybridized in two positions on a presumptive translocation chromosome containing both the 1L and 4S loci. Overall, these results suggest that p1-wwB469 contains a T1-4 translocation, with the breakpoint on the short arm of chromosome 4 (Supplemental Fig. 2).
Cytogenetic analysis of maize pachytene chromosomes
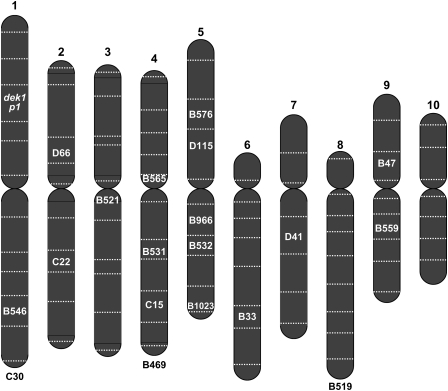
Although FISH of root tip cells is useful for confirming the identity of chromosomes involved in rearrangements, the resolution is quite low because the metaphase chromosomes are highly condensed. In contrast, maize chromosomes are much less condensed at the meiotic pachytene stage of microsporogenesis, and the homologous chromosomes are synapsed. In favorable preparations, each of the 10 maize chromosomes can be distinguished by their relative lengths, distinctive chromomere patterns, centromere positions, and the presence of deep-staining knobs at specific positions. In reciprocal translocation heterozygotes, pairing of the rearranged chromosome(s) and the corresponding normal chromosome(s) at pachytene forms characteristic cross-shaped configurations. Similarly, pairing of inversion and corresponding normal chromosomes in inversion heterozygote forms an inversion loop. The positions of the breakpoints of translocations and inversions can be determined in such configurations with a much higher degree of resolution than in FISH mitotic preparations. Examples of pachytene figures from a pericentric inversion heterozygote and a reciprocal translocation heterozygote are shown in Figure 4, C and D, respectively. One breakpoint in each rearrangement is at the p1 locus on chromosome 1S. In pericentric inversion p1-wwC30 (Fig. 4C), the other breakpoint is at ∼40% of the length of chromosome 1L. In translocation p1-wwB1023 (Fig. 4D), the other breakpoint is near the telomere on 5L. The results obtained from these and other samples are summarized in Table 1. Overall, the results of pachytene analysis confirm the other mapping data and FISH analysis. The distribution of Ac/fAc-induced MCR breakpoints is summarized in Figure 5.
Figure 5.
Distribution of chromosomal rearrangement breakpoints. Schematic diagram of maize chromosomes 1–10 (left to right) divided into genetic bins by horizontal lines. In each rearrangement, one breakpoint is in bin 1.03, at the site of Ac/fAc insertions in or near the p1 and dek1 genes (italics); chromosome bin positions of the other breakpoint in each rearrangement allele are shown. Notations below chromosomes indicate those rearrangements that map to particular chromosomes, but whose position within the chromosome is unknown. Evidence supporting the map positions shown here is summarized in Table 1.
Discussion
We showed here that certain types of transposition reactions involving the Ac/Ds transposable element system can generate MCRs, including inversions and reciprocal translocations. Sequencing of the rearrangement breakpoints shows that these occur precisely at the termini of either Ac or fAc, a terminally deleted derivative element. Moreover, the junctions of both reciprocal translocations and inversions contain complementary 8-bp sequences that most likely represent the 8-bp TSDs generated during Ac transposition. Taken together, these results strongly support the hypothesis that MCRs are produced by alternative transposition reactions; i.e., transposition involving the termini of different Ac/fAc elements (Fig. 1; see Supplemental Movie 1). According to this model, excision of the Ac/fAc termini followed by insertion at a chromosomal target site leads directly to a rearrangement of the sequences flanking the transposon termini. Previous studies have identified other products predicted by the alternative transposition model, including (1) local rearrangement, or permutation of the sequences located between the Ac/fAc termini that may or may not be accompanied by inversion (Zhang and Peterson 2004), and (2) deletions, some of which have fused the coding sequences of two linked paralogous genes to generate a new chimeric gene (Zhang et al. 2006). Similarly, rearrangements including inversion, deletion, and local rearrangement have been generated by transposition reactions of Ac and Ds at the maize bronze1 locus (Dooner and Weil 2007; Huang and Dooner 2008). Another study has shown that chromosomal rearrangements including deletion, inversion, and somatic translocation can also arise through transposition reactions involving Ds elements in Arabidopsis (Krishnaswamy et al. 2008). In all of these cases, the rearrangement junctions have been precise or nearly so. Thus, these results show that MCRs are produced directly by the Ac/Ds transposable element system and need not involve post-excision repair functions such as nonhomologous end joining (NHEJ). An independent study has shown the occurrence of large deletions in Arabidopsis associated with Ds transposition, but in these cases, the rearrangement junction sequences were not precise and likely resulted following host repair functions (Page et al. 2004).
Frequency of MCRs and standard versus alternative transposition
We isolated MCRs using a phenotypic screen for losses of maize kernel pericarp pigmentation; the progenitor alleles specify colored pericarp, while generation of an MCR is accompanied by disruption of the p1 gene, leading to a sector of colorless pericarp. Screening is done using one of the progenitor alleles (P1-rr11, P1-rr910, or P1-ovov454) heterozygous with an allele for colorless pericarp (p1-ww or p1-wr), so that a loss of p1 function is immediately apparent as a colorless pericarp sector. Disruption of p1 function by any means could cause a colorless sector; among ∼100 colorless pericarp mutants derived from P1-rr11, molecular analysis indicates that ∼90% of these are the result of the alternative Ac transposition mechanism, and the overall frequency of alternative transposition is ∼0.3% (data not shown). Previous studies have shown that excision of a single Ac element from the maize p1 locus occurs at a significantly higher frequency (∼5%, depending on genetic background) (Greenblatt and Brink 1962). Recently, Huang and Dooner (2008) reported that a pair of closely linked Ac/Ds elements at the maize bronze1 locus underwent standard transposition approximately 5.4 times more frequently than alternative transposition reactions. The observed differences in frequency of standard versus alternative transposition may be attributed to several factors, including the greater physical distance separating the Ac 5′ and 3′ termini involved in alternative transposition. Consistent with this idea, the number of colorless pericarp sectors we observed in our study was inversely correlated with the distances between the Ac and fAc elements present in each allele; i.e., the P1-ovov454 allele has the least distance between Ac and fAc (823 bp) and produces the highest frequency of colorless pericarp sectors (approximately three times as many as in P1-rr11). An additional consideration is that the recovery of balanced reciprocal translocations requires joining of the Ac 5′ end to the proximal side of the insertion target site (Fig. 1). Insertion in the opposite orientation (i.e., the Ac 5′ end inserted to the distal side of the target site) is expected to occur half of the time, but this would produce nontransmissable acentric and dicentric chromosomes. Even though alternative transposition may occur somewhat less frequently than standard transposition and not all products will be viable, it is clear that alternative transposition has the potential to significantly impact chromosome structure over evolutionary time (Huang and Dooner 2008).
Alternative transposition and the formation of oncogenic chromosome rearrangements
In vertebrates, V(D)J recombination produces a vast repertoire of B-cell receptor (BCR) and T-cell receptor (TCR) proteins. The V(D)J recombination reaction is catalyzed by RAG1 and RAG2, lymphocyte-specific enzymes encoded by two tightly linked genes, recombination activator gene 1 (rag1) and 2 (rag2). RAG1 can bind to recombination signal sequences (RSS) that flank the variable (V) segments, diversity (D) segments, and joining (J) segments (Chatterji et al. 2004; Rhodes et al. 2004; Jung et al. 2006). Kapitonov and Jurka (2005) proposed that the RAG1 core and RSSs evolved from the Transib transposon family. Interestingly, the excised DNA segments flanked by two RSSs can insert into other DNA sequences in vitro and in vivo, generating characteristic TSDs (Agrawal et al. 1998; Hiom et al. 1998; Reddy et al. 2006). These findings suggest that V(D)J recombination is mechanistically related to DNA transposition.
RSSs are composed of highly conserved heptamer and nonamer sequences, separated by a relatively nonconserved spacer of either 12 bp (12 RSS) or 23 bp (23 RSS). V(D)J recombination involving the 12 RSS flanking the 3′ end of the D segment and the 23 RSS flanking the 5′ end of the J segment results in joining of the D and J segments, with concomitant loss of the intervening sequence; this process, termed deletional rearrangement, resembles the excision step of conventional transposition. Recombination involving the 12 RSS flanking the 5′ end of the D segment and the 23 RSS flanking the 5′ end of the J segment can also occur, resulting in an inversion of the intervening sequence (inversional rearrangement). This latter reaction resembles the first step of Ac/Ds alternative transposition in which the 5′ and 3′ termini of different Ac/Ds elements are used as substrates. In V(D)J recombination, inversional rearrangement is estimated to occur at a frequency ∼1/20 that of deletional rearrangement (Shuh and Hixson 2005; Jung et al. 2006). The excised RSSs produced by either deletional or inversional rearrangements can undergo reinsertion into the genome approximately once per 50,000 V(D)J recombination events (Reddy et al. 2006); hence the frequency of reinsertion of RSS following inversional rearrangement would be approximately once per 106 V(D)J recombination events. Reinsertion of the RSS-containing segments produced by deletional rearrangement resembles a standard transposition reaction and hence would not lead to formation of an MCR. Whereas, RSS reinsertion following inversional rearrangement is analogous to Ac/Ds alternative transposition and should generate MCRs including inversions and reciprocal translocations. Considering that hundreds of millions of V(D)J recombination events occur daily during development of human lymphocytes, several hundred MCRs are likely to be generated each day in the cells of the immune system as a result of RSS insertion following inversional rearrangement. While most of these MCRs would likely be innocuous, insertions in the vicinity of proto-oncogenes could activate oncogene expression resulting in a potential lymphoid neoplasia (Marculescu et al. 2006). Our model predicts that the MCRs generated by this type of V(D)J recombination can be identified by characteristic sequence features, including a single RSS at each breakpoint, flanked by complementary 4- to 5-bp sequences representing the TSD formed upon RSS insertion.
Alternative transposition and chromosomal evolution
Alternative transposition reactions are not unique to the Ac/Ds system. In the fungus Fusarium, transposition involving termini of two adjacent impala elements, a member of the Tc1-mariner family, can generate deletions and inversions (Hua-Van et al. 2002). In Drosophila, transposition involving the termini of different P-elements can induce a variety of chromosomal rearrangements including deletions and inversions (Gray et al. 1996; Preston et al. 1996; Tanaka et al. 1997). In mice, concatemers of transgenes that contain Sleeping Beauty transposons exhibit high-frequency chromosome instability and generate chromosomal rearrangements including deletions and inversions (Geurts et al. 2006); these results are consistent with the generation of rearrangements via alternative transposition reactions, although the actual mechanism involved in each case is not yet known. In each of these systems, alternative transposition reactions may occur when transposase acts on transposon termini present in a “nonstandard” configuration; i.e., in either reversed or tandem orientation. It is well-known that Ac/Ds and some other transposable elements exhibit a preference for local transposition (Dooner et al. 1994); the resulting clusters of elements would have a high probability of undergoing subsequent alternative transposition reactions. Thus, the MCRs generated by alternative transposition events could represent an important evolutionary mechanism for genome evolution (Huang and Dooner 2008).
In general, closely related species exhibit karyotypic differences that can be accounted for by specific MCRs including inversions and translocations. Chromosomal evolution involves considerable rearrangements within chromosomes and between nonhomologs (Devos 2005; Tang et al. 2008). These rearrangements are particularly clear among the grasses, for which many comparative maps are available (Devos 2005). In addition, rearrangements following allopolyploidization events, such as with maize, scramble the gene order from the contributing genomes. These rearrangements contribute to chromosomal evolution and likely result in part from MCRs. Also, we recently demonstrated that alternative transposition reactions can produce deletions that generate new chimeric genes and alter the expression of genes near the breakpoint (Zhang et al. 2006); similar effects on gene coding and/or expression could be expected from the translocations and inversions reported here. These genetic changes are a direct and immediate outcome of the alternative transposition reaction and thus could promote the fixation of chromosomal rearrangements during evolution.
Materials and methods
Genetic stocks
Alleles of the p1 gene are identified by a two-letter suffix that indicates their expression pattern in pericarp and cob: e.g., P1-rr (red pericarp and red cob), p1-wr (white pericarp, red cob), and p1-ww (white pericarp and white cob). The standard p1-vv (variegated pericarp and variegated cob) allele described by Emerson (1917) contains an Ac insertion in the second intron of a P1-rr gene. From p1-vv, we obtained a spontaneous derivative termed P1-ovov1114 (orange-variegated pericarp and orange-variegated cob), in which the Ac element had undergone an intragenic transposition to a site 153 bp upstream in p1 gene intron 2 and inserted in the opposite orientation (Peterson 1990). From P1-ovov1114, we obtained a spontaneous derivative termed p1-vv9D9A (Zhang and Peterson 1999); this allele contains an Ac element, a 112-bp rearranged p1 gene fragment, and the terminally deleted Ac element fAc. From p1-vv9D9A, we obtained the alleles P1-rr11, P1-rr910, and P1-ovov454 described here; these alleles were generated from p1-vv9D9A by transposition of the full-length Ac element from its location in intron 2 of p1 to sites 13,175 bp, 8919 bp, and 823 bp, upstream of fAc, respectively (Fig. 1A).
Screening candidate transposition-induced MCRs
Most products of reversed Ac ends transposition are predicted to have a deletion of p1 gene exon(s) 1 and 2, or separation of exons 1 and 2 from exon 3, resulting in a loss of p1 gene function and the appearance of colorless sectors. The size of sector generated reflects the time of development at which the particular event occurred; early events give rise to large multikernel sectors, whereas later events give rise to single-kernel sectors and colorless stripes. To identify putative MCRs, we screened the mature ears produced by plants carrying one of the parental alleles (P1-rr11, P1-rr910, or P1-ovov454) for large colorless pericarp sectors (Fig. 2A) and single colorless kernel sectors. Reversed Ac ends transposition is predicted to generate a wide variety of possible products (see Supplemental Movie 1). In this study, we focused on the detection and characterization of large inversions (Fig. 1C) and reciprocal translocations (Fig. 1D), both of which are expected to carry an active Ac element. Ac can be detected by its ability to induce excision of Ds from the r1-m3∷Ds allele of the maize r1 gene, which is required for kernel aleurone pigmentation. Hence we selected kernels with colorless pericarp and spotted aleurone as containing candidate rearrangement events (Fig. 2A).
PCR amplifications
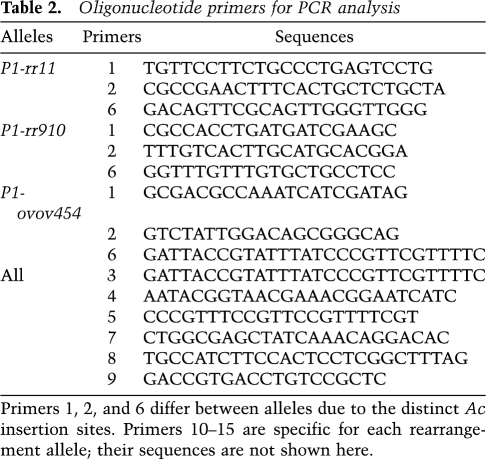
PCR amplifications were performed as described (Saiki 1989) using the oligonucleotide primers shown in Table 2. HotMaster Taq polymerase from Eppendorf was used in the PCR reaction. Reactions were heated for 3 min at 94° and then cycled 35 times for 20 sec at 94°, for 30 sec at 60°, and for 1 min at 65° per 1-kb length of expected PCR product and then for 8 min at 65°. In most of the PCR reactions, 1 M betaine and 4%–8% DMSO were added. The band amplified was purified from an agarose gel and sequenced directly. Sequencing was done by the DNA Synthesis and Sequencing Facility, Iowa State University.
Table 2.
Oligonucleotide primers for PCR analysis
Primers 1, 2, and 6 differ between alleles due to the distinct Ac insertion sites. Primers 10–15 are specific for each rearrangement allele; their sequences are not shown here.
LM-PCR
2.5 μg of Genomic DNA was digested with 20 units of HpyCH4IV (New England Biolabs) in a total 20-μL volume for 3 h at 37°C, then a mix containing the corresponding adaptor, HpyCH4IV buffer, ATP, and T4 DNA ligase (New England Biolabs) was added to the restriction digestion mix. The total volume was 40 μL; the final concentration of the restriction digestion buffer was 1×, the final concentration of the adapter was 0.5 pmol/μL, the final concentration of ATP was 0.5 mM, and the final concentration of T4 DNA ligase was 20 cohesive end units per microliter. Ligation was carried out overnight at 22°C; 2 μL of the ligation mix was used as template in the first round of the nested PCR reaction, and 1 μL of the mix of the first round PCR was used as template in the second round of PCR. PCR was performed as described above.
HpyCH4IV Adaptor, GTATCACCACCAGAGGAGCAAGCGAGTTCACAGAATCACACGAGTAGAGT and CTCATCTCAGC; primer in the first-round PCR, CCACCAGAGGAGCAAGCGAGTTCACAG; primer in the second-round PCR, GCAAGCGAGTTCACAGAATCACACGAG.
The above two primers are complementary to the adapter sequence. Other primers used in this study are summarized in Table 2.
Cloning the breakpoints of MCR candidates
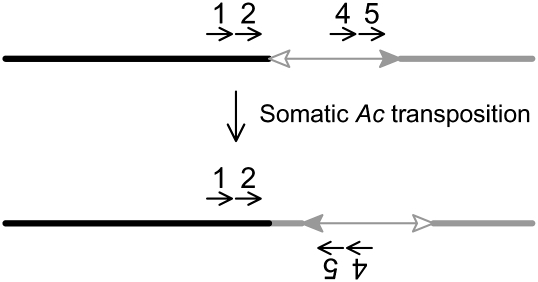
Ac casting was performed as described by Singh et al. (2003) to clone the breakpoint sequences flanking Ac. As shown in Figure 6, the sequences upstream of Ac (black line) are known, whereas the sequences downstream from Ac (shaded line) are unknown. Because Ac tends to transpose locally, Ac may excise and reinsert at a site close to the breakpoint in opposite orientation. Such short-range transposition events can be detected in DNA prepared from somatic tissues by amplification with nested PCR using primers 1/4 and 2/5 (Fig. 6). To avoid the negative Ac dosage effect (the higher Ac dose, the less frequent Ac transposition), DNA templates were made from rearrangement heterozygous plants carrying only a single copy of Ac in the genome. The first round of PCR was performed as described above, and 1 μL of the product of the first-round PCR was used as template for the second-round PCR. To obtain the precise MCR breakpoint junctions, we performed additional PCR using primer 5 paired with a primer (primer 11 or 14) (Figure 1) complementary to the flanking sequence (obtained via Ac casting). In some cases, the Ac element had undergone a germinal excision from the rearrangement breakpoint; these breakpoints were cloned by LM-PCR using primers 1 and 2 (Fig. 1) as described above.
Figure 6.
Local somatic Ac transposition as the basis for Ac casting (Singh et al. 2003). All the symbols have the same meaning as those in Figure 1.
The model for generation of MCRs predicts that the breakpoint sequences flanking the Ac/fAc termini were derived from the Ac/fAc insertion target site, and thus should be present as contiguous sequences in the progenitor genome (a/b or c/d in Fig. 1). Therefore, we used BLAST to compare the sequences adjacent to the 5′ Ac termini in MCRs (obtained from Ac casting or LM-PCR, above) with the maize sequence database (http://www.plantgdb.org) in order to identify the sequences predicted to flank the 3′ fAc element. These sequences were used to design a new primer (primer 12 or 13 in Fig. 1) that was used in PCR with primer 3 to amplify the breakpoint sequence flanking the fAc element (Fig. 1). If the BLAST analysis of the sequence flanking Ac did not produce a highly similar match, then two additional primers (primers 10 and 11 or 14 and 15 in Fig. 1) complementary to the breakpoint sequence flanking Ac were used in LM-PCR to amplify the target site sequence from the progenitor alleles (P1-rr11, P1-rr910, or P1-ovov454). Then, a primer (primer 12 or 13 in Fig. 1) was designed based on this new sequence and paired with primer 3 to amplify the breakpoint sequence flanking the fAc element.
Chromosome preparation and FISH
Somatic chromosome spreads were produced as described previously (Kato et al. 2004) except the concentration of cellulase was increased from 2% to 4% in the enzymatic mixture for root-tip digestion and slides were used within 4 h of preparation. Probe hybridization was carried out for 12–24 h at 55° and washed in 2× SSC for 20 min (Kato et al. 2004). Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI) containing Vectashield mounting media (Vector Laboratories). Signals were captured with an Olympus BX61 microscope using Applied Spectral Imaging (ASI) software and CCD camera Cool-1300QS (Table 3; Kato et al. 2004; Lamb et al. 2007).
Table 3.
FISH probes and their chromosomal hybridization site
Cytological analysis of male inflorescences
Cytological analysis of male inflorescences undergoing meiosis was carried out in plants heterozygous for the rearrangements to determine the breakpoints in the rearrangements. The immature tassels were fixed in a 3:1 mixture of 95% ethanol:propionic acid (v:v) at room temperature for 1 d and then maintained at −20°C. Cells at the pachytene stage of prophase I were stained with a propio-carmine solution (Sharma and Sharma 1965).
Acknowledgments
We thank Ronald L. Phillips (University of Minnesota) and members of his laboratory for providing genomic DNA from oat–maize addition lines; Pat Schnable (Iowa State University) and members of his laboratory for the TGCE genetic mapping of MCR breakpoints (http://magi.plantgenomics.iastate.edu/index.html); and Tim Bruihler, Lisa Coffey, Chris Cosgrove, Ryan Dietz, Peter Howe, Klint Kersten, Amanda Kopp, and Aaron Newell for field and laboratory assistance. This research was supported by NSF MCB 0450243 to T.P. and J.Z., NSF MCB 0450215 to D.W., and NSF DBI 0423898 to J.B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1776909.
Supplemental material is available at http://www.genesdev.org.
References
- Agrawal A., Eastman Q.M., Schatz D.G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Ananiev E.V., Riera-Lizarazu O., Rines H.W., Phillips R.L. Oat–maize chromosome addition lines: A new system for mapping the maize genome. Proc. Natl. Acad. Sci. 1997;94:3524–3529. doi: 10.1073/pnas.94.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji M., Tsai C.L., Schatz D.G. New concepts in the regulation of an ancient reaction: Transposition by RAG1/RAG2. Immunol. Rev. 2004;200:261–271. doi: 10.1111/j.0105-2896.2004.00167.x. [DOI] [PubMed] [Google Scholar]
- Devos K.M. Updating the ‘crop circle’. Curr. Opin. Plant Biol. 2005;8:155–162. doi: 10.1016/j.pbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Dooner H.K., Weil C.F. Give-and-take: Interactions between DNA transposons and their host plant genomes. Curr. Opin. Genet. Dev. 2007;17:486–492. doi: 10.1016/j.gde.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Dooner H.K., Belachew A., Burgess D., Harding S., Ralston M., Ralston E. Distribution of unlinked receptor sites for transposed Ac elements from the bz-m2(Ac) allele in maize. Genetics. 1994;136:261–279. doi: 10.1093/genetics/136.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson R.A. Genetical studies of variegated pericarp in maize. Genetics. 1917;2:1–35. doi: 10.1093/genetics/2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J., Harrison K., Jones J.D. A genetic analysis of DNA sequence requirements for Dissociation state I activity in tobacco. Plant Cell. 1993;5:501–514. doi: 10.1105/tpc.5.5.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J.J., Harrison K., Jones J. Aberrant transpositions of maize double Ds-like elements usually involve Ds ends on sister chromatids. Plant Cell. 1995;7:1235–1247. doi: 10.1105/tpc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Emrich S.J., Guo L., Wen T.J., Ashlock D.A., Aluru S., Schnable P.S. Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc. Natl. Acad. Sci. 2005;102:12282–12287. doi: 10.1073/pnas.0503394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts A.M., Collier L.S., Geurts J.L., Oseth L.L., Bell M.L., Mu D., Lucito R., Godbout S.A., Green L.E., Lowe S.W., et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray Y.H., Tanaka M.M., Sved J.A. P-element-induced recombination in Drosophila melanogaster: Hybrid element insertion. Genetics. 1996;144:1601–1610. doi: 10.1093/genetics/144.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt I.M., Brink R.A. Twin mutations in medium variegated pericarp maize. Genetics. 1962;47:489–501. doi: 10.1093/genetics/47.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Athma P., Peterson T. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc. Natl. Acad. Sci. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond B.J., Bowen B., Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Hiom K., Melek M., Gellert M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Huang J.T., Dooner H.K. Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell. 2008;20:2019–2032. doi: 10.1105/tpc.108.060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A., Langin T., Daboussi M.J. Aberrant transposition of a Tc1-mariner element, impala, in the fungus Fusarium oxysporum . Mol. Genet. Genomics. 2002;267:79–87. doi: 10.1007/s00438-002-0638-9. [DOI] [PubMed] [Google Scholar]
- Jin W., Melo J.R., Nagaki K., Talbert P.B., Henikoff S., Dawe R.K., Jiang J. Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–581. doi: 10.1105/tpc.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Lamb J.C., Birchler J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. 2004;101:13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Albert P.S., Vega J.M., Birchler J.A. Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech. Histochem. 2006;81:71–78. doi: 10.1080/10520290600643677. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy L., Zhang J., Peterson T. Reversed end Ds element: A novel tool for chromosome engineering in Arabidopsis . Plant Mol. Biol. 2008;68:399–411. doi: 10.1007/s11103-008-9377-6. [DOI] [PubMed] [Google Scholar]
- Kynast R.G., Okagaki R.J., Rines H.W., Phillips R.L. Maize individualized chromosome and derived radiation hybrid lines and their use in functional genomics. Funct. Integr. Genomics. 2002;2:60–69. doi: 10.1007/s10142-002-0052-8. [DOI] [PubMed] [Google Scholar]
- Lamb J.C., Danilova T., Bauer M.J., Meyer J.M., Holland J.J., Jensen M.D., Birchler J.A. Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics. 2007;175:1047–1058. doi: 10.1534/genetics.106.065573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marculescu R., Vanura K., Montpellier B., Roulland S., Le T., Navarro J.M., Jager U., McBlane F., Nadel B. Recombinase, chromosomal translocations and lymphoid neoplasia: Targeting mistakes and repair failures. DNA Repair (Amst.) 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- McClintock B. Carnegie Institution of Washington Year Book. Vol. 48. 1949. Mutable loci in maize; pp. 142–154. [PubMed] [Google Scholar]
- McClintock B. Carnegie Institution of Washington Year Book. Vol. 49. 1950a. Mutable loci in maize; pp. 157–167. [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. 1950b;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Page D.R., Kohler C., Da Costa-Nunes J.A., Baroux C., Moore J.M., Grossniklaus U. Intrachromosomal excision of a hybrid Ds element induces large genomic deletions in Arabidopsis . Proc. Natl. Acad. Sci. 2004;101:2969–2974. doi: 10.1073/pnas.0400089101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. Intragenic transposition of Ac generates a new allele of the maize P gene. Genetics. 1990;126:469–476. doi: 10.1093/genetics/126.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C.R., Sved J.A., Engels W.R. Flanking duplications and deletions associated with P-induced male recombination in Drosophila . Genetics. 1996;144:1623–1638. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy Y.V., Perkins E.J., Ramsden D.A. Genomic instability due to V(D)J recombination-associated transposition. Genes & Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes G., Parkhill J., Bird C., Ambrose K., Jones M.C., Huys G., Swings J., Pickup R.W. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 2004;70:7497–7510. doi: 10.1128/AEM.70.12.7497-7510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R.K. The design and optimization of the PCR. In: Erlich H.A., editor. PCR technology. Stockton Press; New York: 1989. pp. 7–16. [Google Scholar]
- Sharma A.K., Sharma A. Chromosome techniques: Theory and practice. Butterworths; London: 1965. [Google Scholar]
- Shuh M., Hixson D.C. V(D)J recombination of chromosomally integrated, wild-type deletional and inversional substrates occur at similar frequencies with no preference for orientation. Immunol. Lett. 2005;97:69–80. doi: 10.1016/j.imlet.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Singh M., Lewis P.E., Hardeman K., Bai L., Rose J.K., Mazourek M., Chomet P., Brutnell T.P. Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell. 2003;15:874–884. doi: 10.1105/tpc.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M.M., Liang X.M., Gray Y.H., Sved J.A. The accumulation of P-element-induced recombinants in the germline of male Drosophila melanogaster . Genetics. 1997;147:1769–1782. doi: 10.1093/genetics/147.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Weil C.F., Wessler S.R. Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell. 1993;5:515–522. doi: 10.1105/tpc.5.5.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T. Genome rearrangements by nonlinear transposons in maize. Genetics. 1999;153:1403–1410. doi: 10.1093/genetics/153.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peterson T. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167:1929–1937. doi: 10.1534/genetics.103.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang F., Peterson T. Transposition of reversed Ac element ends generates novel chimeric genes in maize. PLoS Genet. 2006;2:e164. doi: 10.1371/journal.pgen.0020164. [DOI] [PMC free article] [PubMed] [Google Scholar]