Abstract
Xenopus oocytes provide an excellent model system for understanding the cis-elements and protein factors that carry out mRNA localization in vertebrate cells. More than 20 mRNAs have been identified that localize to the vegetal cortex during stages II–IV of oogenesis. The earliest localizing RNAs are presorted to a subcellular structure, the Balbiani body (also called the mitochondrial cloud in Xenopus), of stage I oocytes prior to entering the vegetal cortex. While some evidence has suggested that diffusion drives RNA localization to the Balbiani body, a role for temperature and metabolic energy in this process has not been explored. To address this issue, we developed a quantitative assay to monitor RNA localization in stage I oocytes. Here we show that the rate of RNA accumulation to the Balbiani body is highly dependent on temperature and the intracellular concentration of ATP. In fact, while ATP depletion severely impairs RNA localization, increasing the intracellular concentration of ATP by a factor of two doubles the localization rate, indicating that ATP is limiting under normal conditions. We also show that RNA localization in stage I oocytes is reduced by inhibition of kinesin II, and that the Xcat-2 RNA localization element recruits kinesin II to the Balbiani body. We conclude from these studies that the energy state of the cell regulates the rate of RNA localization to the Balbiani body and that this process, at least to some extent, involves kinesin II.
Keywords: RNA localization, ATP, temperature, diffusion, kinesin
INTRODUCTION
The subcellular localization of specific mRNAs to distinct regions of a cell provides an important mechanism for polarizing gene expression (St Johnston 2005). For example, the localization of mRNAs in neurons serves to distribute mRNAs to processes where local protein synthesis modulates synaptic activity (Kiebler and Bassell 2006). Early studies in Drosophila showed that the localization of several key mRNAs such as gurken, bicoid, oskar, and nanos generates protein gradients that set up cascades of gene expression required for establishing the basic structure of the embryo (St Johnston 2005). While it generally had been thought that mRNA localization only occurs for a small subset of mRNAs in the egg that drive embryonic polarity, a recent study in Drosophila has shown that 71% of more than 2000 mRNAs expressed in embryos are localized to subcellular regions where their encoded proteins are translated (Lecuyer et al. 2007). Likewise, microarray and computational analyses suggest that thousands of mRNAs are localized in mammalian neurons (Andken et al. 2007; Matsumoto et al. 2007; Suzuki et al. 2007). Thus, mRNA localization appears to be a common mechanism for regulating gene expression on a global scale in animals.
Xenopus oocytes provide an excellent model system for RNA localization studies in vertebrates. The value of this system has been illustrated most clearly by the computational identification and experimental verification of RNA localization sequences in human genes that possess a sequence composition similar to those that direct localization to the vegetal cortex of Xenopus oocytes (Andken et al. 2007). Vg1 was one of the first mRNAs shown to localize in Xenopus oocytes (Rebagliati et al. 1985), and since its discovery, more than 20 additional mRNAs have been identified (King et al. 2005) that can be generally categorized into three groups based on the time at which they fully accumulate at the vegetal cortex. The earliest group of vegetal RNAs localizes to the cortex during stage II of oogenesis, whereas intermediate RNAs, such as Xlerk, show only partial accumulation at the cortex during stage II, but become fully localized during stage III (Betley et al. 2002). The latest pathway is characterized by Vg1 mRNA, which becomes enriched toward the vegetal region during stage II of oogenesis followed by its accumulation at the vegetal cortex during stages III and IV. Interestingly, several lines of evidence suggest that some basal RNA localization factors function throughout all pathways (Claussen et al. 2004; Choo et al. 2005).
The primary feature that distinguishes early localizing RNAs from later ones in Xenopus is that early RNAs show a robust localization pattern in stage I oocytes, whereas late RNAs appear uniformly distributed at this stage (Forristall et al. 1995; Kloc and Etkin 1995). Early RNAs, such as Xcat-2, Xlsirt, and Xwnt, become localized to a large subcellular structure called the Balbiani body or mitochondrial cloud. The Balbiani body is highly enriched in membranes of the endoplasmic reticulum (ER), Golgi, and mitochondria and has been identified in the developing oocytes of invertebrates as well as vertebrates, including humans (Kloc and Etkin 2005) and mice (Pepling et al. 2007). Thus, the Balbiani body may provide evolutionarily conserved functions for oocyte specification and development of the egg. Double labeling experiments show that different localized RNAs occupy distinct subregions of this structure. For example, Xcat-2, Xlsirts, and Xwnt all localize to the Balbiani body; however, these RNAs have a discreet order in their distribution such that Xcat-2 is positioned closest to the future vegetal cortex of late stage I oocytes, followed by Xlsirt and then Xwnt (Kloc and Etkin 1995). This indicates that there is a continuum along the developing animal–vegetal axis upon which individual mRNAs adopt distinct positions during oogenesis. The consequence of such a hierarchical system may be that each mRNA accumulates in a specific layer within the vegetal cortex as oocytes grow. This highly organized pattern of gene expression within the oocytes of adult females is established relatively slowly, since oogenesis requires several months to complete (Keem et al. 1979; Green 2002). Thus, even though molecular motors are thought to mediate this process in stage III and IV oocytes (Betley et al. 2004; Yoon and Mowry 2004), motor-dependent movements are likely to be quite short and function in a probabilistic and bidirectional fashion, as has been proposed from imaging the transport of RNA granules in other types of living cells (Fusco et al. 2003; Bullock et al. 2006).
While functional studies have implicated kinesin II (Betley et al. 2004), staufen I (Yoon and Mowry 2004), and Vg1RBP/Vera (Kwon et al. 2002) in the localization of late pathway RNAs during stage III/IV of oogenesis in Xenopus, no protein inhibition studies have yet been performed in stage I oocytes. Consequently, the molecular mechanism responsible for localizing early RNAs to the Balbiani body is poorly understood. However, it has been suggested that transcripts are localized to the Balbiani body by a diffusion and entrapment mechanism, since microtubule-disrupting drugs have no detectable effect on the localization of endogenous or microinjected early RNAs (Kloc et al. 1996; Zhou and King 1996b; Chang et al. 2004). Moreover, fluorescence recovery after photobleaching (FRAP) experiments showed that the 228-nucleotide Xcat-2 mitochondrial cloud RNA localization element (MCLE) exhibits dispersion kinetics in the cytoplasm that are similar to a nonlocalizing RNA (Chang et al. 2004).
Even though a diffusion-mediated mechanism has been proposed to mediate RNA localization to the Balbiani body of stage I oocytes (Chang et al. 2004), and molecular motors drive localization directly to the vegetal cortex of later-staged oocytes (Betley et al. 2004), a number of parallels have been observed between early and late RNA localization mechanisms which suggest that a common cellular machinery mediates RNA localization throughout stages I–IV of oogenesis. First, many early and late RNA localization elements (LEs) are enriched in short CAC-containing motifs that are essential for localization, indicating that the sequence composition of early and late RNA LEs is similar (Betley et al. 2002; Bubunenko et al. 2002; Kwon et al. 2002). In addition, early LEs can utilize later pathways if injected into stage III/IV oocytes (Zhou and King 1996b; Allen et al. 2003). Early and late RNA LEs also compete for common localization factors in vivo (Choo et al. 2005) and are bound specifically by some of the same RNA binding proteins, such as Vg1RBP/Vera (Claussen et al. 2004; Choo et al. 2005). Interestingly, both the late Vg1 and early Xcat-2 RNA localization elements recruit the molecular motor kinesin II when microinjected into stage II oocytes (Betley et al. 2004). Finally, the ER, which originally was implicated in localization in stage III/IV oocytes (Deshler et al. 1997), subsequently has been shown also to be involved in the process that drives RNA localization to the Balbiani body (Kloc and Etkin 1998; Chang et al. 2004).
Taken together, these data indicate that early and late localizing RNAs associate with common localization machinery that operates throughout early and mid-oogenesis. Differences in the localization patterns of early and late RNAs thus may be due to the kinetics and efficiencies with which each RNA interacts with this machinery (Choo et al. 2005). To gain a better understanding of the underlying mechanism(s) that determine whether an RNA localizes to the Balbiani body during stage I of oogenesis or to the vegetal cortex during stages II–IV of oogenesis, it is essential to characterize the mechanism that drives RNA localization in stage I oocytes.
Prior to the studies presented here, it was not known whether the process of RNA accumulation to the Balbiani body requires metabolic energy or depends on temperature, two major characteristics that substantially can distinguish a mechanism that is driven solely by diffusion from one that potentially involves enzymatic processes. In this study, we developed a quantitative assay to monitor the localization of RNAs microinjected into stage I oocytes. We find that the rate of RNA localization in stage I oocytes is highly dependent on the intracellular ATP concentration and increases by approximately twofold with a temperature increase of 12°C. We also show that expression of a dominant negative kinesin II polypeptide reduces localization to the Balbiani body. Therefore, one factor that could account for the observed ATP requirement during this localization process could be the ATP-dependent molecular motor kinesin II. Consistent with this, the Xcat-2 MCLE is able to recruit kinesin II to the Balbiani body. These data support a model in which localization of RNAs to the Balbiani body in Xenopus is most likely an enzymatically driven process, with one factor contributing to this process being kinesin II.
RESULTS
A quantitative assay to monitor RNA localization to the Balbiani body
Previous assays characterizing the early pathway have relied on qualitative assessments of localization and often employed a (+, +/−, −) scoring scheme to characterize the enrichment of RNAs in the mitochondrial cloud. Such scoring systems have been used successfully to map sequences, which confer the localization pattern of early pathway RNAs (Zhou and King 1996a; Kloc et al. 2000). While this type of assay can be reliable for detecting “all or none” localization phenomenon, it is less practical for detecting subtle changes in the localization rate. To determine if specific treatments of oocyte groups result in a slight reduction/enhancement in the localization process, it is essential to establish a more sensitive assay with which the effects on RNA localization can be compared in a quantitative fashion.
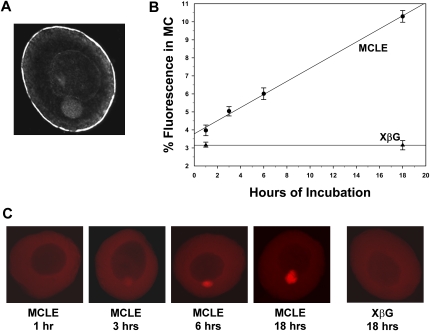
To establish such a method, we developed a protocol that gave reproducible linear plots of RNA localization to the Balbiani body (mitochondrial cloud) of stage I oocytes over a 3- to 18-h period. This procedure involved microinjecting stage I oocytes with Alexa Fluor-546 labeled Xcat-2 mitochondrial cloud localization element (MCLE) or a standard control RNA (XβG) that does not localize in oocytes. Oocytes were cultured at 18°C following injection of the fluorescent RNAs. After 2, 4, 6, or 18 h, groups of oocytes were placed in fixative to terminate the localization process. Confocal images were acquired from an optical section through the center of the mitochondrial cloud, and localization was quantified by measuring the integrated optical density (IOD) within the mitochondrial cloud (IODMC) and dividing that value by the IOD of the entire optical section (IODtotal) of the oocyte. To identify reliably the mitochondrial cloud, a dark-field image was taken from each oocyte in addition to the fluorescent image (Fig. 1A). The percentage of fluorescent signal observed in the mitochondrial cloud from each group of oocytes then was plotted as an average value of ∼30 oocytes per group. When localization is quantified in this manner, a linear increase in localization is observed from 3 to 18 h in oocytes injected with MCLE RNA (Fig. 1B). Oocytes injected with XβG RNA showed consistent values and no change in localization over an 18-h time period. Oocytes injected with MCLE RNA showed an increase in fluorescent signal that was easily detectable by eye over an 18-h time period (Fig. 1C). After 18 h, the signal in the mitochondrial cloud generally begins to become saturated, and the data are no longer linear (data not shown). This is similar to observations made from single-oocyte recordings of localization in living stage I oocytes, where localization appeared to be linear for at least the first 10 h of observation (Chang et al. 2004). Therefore, all subsequent experiments involved incubation times from 3 to 8 h to ensure that we were examining localization in the linear range.
FIGURE 1.
A quantitative assay for RNA localization in stage I oocytes. Stage I oocytes were injected with either fluorescently labeled MCLE or XβG RNA. At the indicated times after injection, groups of oocytes were placed in fixative. Confocal images were acquired and localization was quantified as described in the Materials and Methods. (A) In order to reliably locate the mitochondrial cloud for quantification, a dark-field image was collected for each stage I oocyte. In these images the mitochondrial cloud (arrow) and germinal vesicle (GV) are readily visualized. (B) The average percentage of fluorescent signal that accumulated in the mitochondrial cloud (MC) from each group of oocytes is shown on the y-axis. The time that each group of oocytes was cultured following injection is given along the x-axis. This plot represents a single experiment in which all oocytes were prepared from a single frog. The numbers of oocytes used to generate the average localization values for each data point in this experiment were n = 15, 28, 33, and 33 for the MCLE RNA at the 1, 3, 6, and 18 h time points, respectively. Error bars represent standard error of the mean for each group. For the XβG RNA, n = 16 and 12 for the 3 and 18 h time points, respectively. (C) Confocal images of representative oocytes from each group. The mitochondrial cloud is oriented toward the bottom in each image.
RNA localization to the Balbiani body is temperature dependent
It has been suggested previously that RNA localization to the Balbiani body of stage I oocytes is mediated by a diffusion and entrapment mechanism (Zhou and King 1996a; Chang et al. 2004). This conclusion is based primarily on two observations. First, drugs that depolymerize microtubules or actin filaments have little effect on RNA localization to the Balbiani body. Secondly, FRAP experiments show that the nonlocalizing XβG RNA and the Xcat-2 MCLE RNA recover with similar rates into photobleached regions of the cytoplasm in stage I oocytes. To explore further the role of diffusion in the localization of RNAs in stage I oocytes, we used our quantitative assay to determine if RNA localization to the Balbiani body is affected by temperature. Such an approach is often used to distinguish between diffusion and enzymatically driven mechanisms because average displacement rates mediated solely by diffusion depend on absolute temperature and are, therefore, relatively unaffected by 10°C–15°C temperature shifts. In contrast, enzymatic processes generally display a temperature coefficient (Q10) of 1.5–2.5, such that reaction rates increase approximately twofold when the temperature is increased by 10°C. Thus, if no detectable difference is observed in the rate of a particular localization process with a 10°C temperature shift, it generally is concluded that diffusion drives the process. For example, a 10°C temperature shift has no detectable effect on the redistribution of several proteins within the nucleus (Phair and Misteli 2000), consistent with diffusion mediating their localization from one compartment to another. Similar results have been reported for mRNA movement in the nucleus (Politz et al. 2006), though this depends to some extent on the technique used to measure diffusion of mRNAs (Vargas et al. 2005).
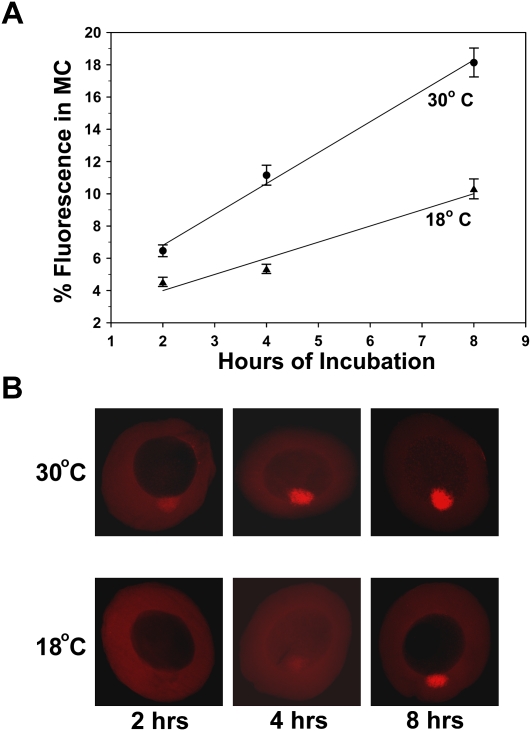
To determine if the rate-limiting step(s) of RNA localization to the Balbiani body is affected by temperature, we injected Alexa Fluor-546 labeled MCLE RNA into stage I oocytes and incubated the oocytes at either 18°C or 30°C. Groups of oocytes from each temperature were fixed after 3, 6, or 9 h of incubation, and localization to the mitochondrial cloud was quantified. Figure 2 shows that at each time point, localization was more extensive in oocytes incubated at 30°C than in those incubated at 18°C. Importantly, the slope of the 30°C line (1.92) is approximately two times that of the 18°C line (1.00), suggesting that the rate of localization at 30°C is twice as fast as at 18°C (Fig. 2A). In other words, the Q10 for RNA localization to the Balbiani body is in the range of most known enzymatic processes. Corresponding fluorescent images taken at each time point were consistent with the quantitative assessment of localization and demonstrated increased fluorescence in the mitochondrial cloud that was surprisingly easy to detect by visual inspection of oocytes incubated at higher temperature (Fig. 2B). These data imply for the first time that RNA localization to the Balbiani body in stage I oocytes is an enzymatically driven process.
FIGURE 2.
The rate of RNA localization to the mitochondrial cloud is affected by temperature. Stage I oocytes were injected with fluorescently labeled MCLE RNA and were then incubated at either 18°C or 30°C. At 2, 4, and 8 h after injection, groups of oocytes from each temperature were fixed and localization was quantified. (A) The average percentage of fluorescent signal that accumulated in the mitochondrial cloud from each group of oocytes is shown on the y-axis. The time that each group of oocytes was cultured following injection is given along the x-axis. This plot represents a single experiment in which all oocytes were prepared from a single frog. The numbers of oocytes used to generate each data point are as follows: n = 37, 32, and 34 (18°C) and 40, 29, and 31 (30°C) for the 2, 4, and 8 h time points, respectively. The slope of the 18°C line is 1.00%/h, whereas that of the 30°C line is 1.92%/h. Thus, these slopes vary by a factor of about two, indicating that the rate of RNA localization at 30°C is much faster than at 18°C. Error bars represent standard error of the mean for each group. (B) Confocal images of representative oocytes from each group.
Depletion of endogenous ATP in stage I oocytes impairs localization
Another method that typically is used to distinguish between enzymatically and diffusion-mediated processes is to test whether metabolic energy is required. For example, the movement of proteins to nucleoli, interchromatin granules, and within nucleoplasm, which is driven by diffusion, is unaffected by depletion of ATP (Phair and Misteli 2000). While ATP depletion can create structural artifacts that reduce the movement of RNA molecules in the nucleus (Shav-Tal et al. 2004), similar effects have not been observed in the cytoplasm (Vargas et al. 2005). Therefore, we performed ATP depletion experiments in stage I oocytes to complement the temperature experiments above and to establish whether RNA localization to the Balbiani body requires metabolic energy.
In order to test whether the rate of RNA localization in stage I oocytes depends on the intracellular concentration of ATP, we employed a previously reported procedure involving the injection of apyrase into oocytes to deplete endogenous ATP (Newmeyer et al. 1986). One group of oocytes was injected with fluorescent MCLE RNA alone and another group was co-injected with fluorescent MCLE RNA plus the ATP hydrolyzing enzyme apyrase (∼1 unit/mL final intracellular concentration). After 6 h of culturing, endogenous ATP levels were quantified in oocytes isolated from each group using a luciferase-based assay. In oocytes injected with MCLE RNA alone, the endogenous ATP concentration was determined to be ∼2.4 mM. This is similar to the intracellular concentration reported for larger stage VI oocytes (Newmeyer et al. 1986; Gribble et al. 2000). In oocytes co-injected with the MCLE RNA and apyrase, the ATP concentration was reduced to ∼0.3 mM. In stage VI oocytes, the same procedure reduced the ATP concentration to undetectable levels (Newmeyer et al. 1986). Our inability to reduce intracellular ATP below 0.3 mM in stage I oocytes is likely due to their smaller size, which technically limits our capacity to inject apyrase to a final intracellular concentration of ∼20 units/mL, as was obtained in larger oocytes (Newmeyer et al. 1986).
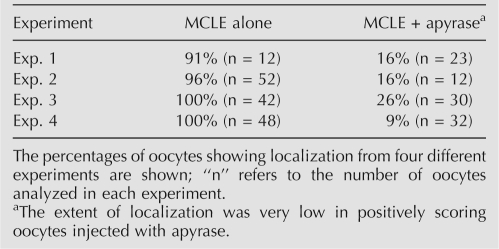
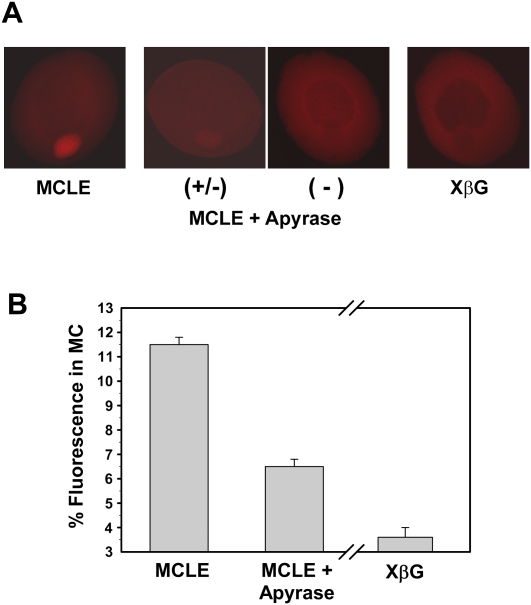
Even though we could not decrease endogenous ATP to background or undetectable levels, we sought to determine if a ∼90% reduction affects RNA localization to the Balbiani body. Qualitative assessment revealed that the effects of ATP reduction on localization are extremely robust. While >90% of stage I oocytes injected with the MCLE alone routinely showed strong enrichment in the mitochondrial cloud after 6 h of incubation, localization was severely impaired in oocytes co-injected with the MCLE and apyrase, with only ∼20% of oocytes showing detectable localization (Table 1). In most oocytes injected with apyrase and the MCLE, no enrichment could be visualized in the mitochondrial cloud (Fig. 3A). In the ∼20% of oocytes where localization was apparent, the amount of MCLE RNA that reached the Balbiani body was low compared with oocytes injected with the MCLE alone (Fig. 3A). When the signal from these oocytes was quantified, the data were consistent with the qualitative assessment. Figure 3B shows the quantification of RNA localization from experiment 4 of Table 1. In oocytes injected with the MCLE alone, 11.5% of the fluorescent signal was detected within the mitochondrial cloud. However, only 6.5% of signal was present in the mitochondrial cloud of oocytes co-injected with the MCLE and apyrase (Fig. 3B). To control for background localization, we consistently found that ∼3.6% of the fluorescent signal from the XβG RNA was present in the mitochondrial cloud under a variety of conditions (data not shown). Subtracting this value from the localization values obtained for the MCLE and MCLE/apyrase injected oocytes indicates that depleting intracellular ATP levels to ∼0.3 mM reduces RNA localization by ∼63% (Fig. 3B). A similar calculation for each of the four ATP depletion experiments shown in Table 1 indicates that localization is consistently reduced by ∼60% in oocytes co-injected with apyrase and MCLE RNA. Together, these data demonstrate that decreasing intracellular ATP by ∼10-fold substantially reduces the rate of localization in stage I Xenopus oocytes. We also attempted to utilize nonhydrolyzable analogs of ATP to inhibit the localization process, but at concentrations similar to endogenous ATP they were toxic to oocytes (data not shown).
TABLE 1.
Qualitative assessment of localization in ATP-depleted oocytes
FIGURE 3.
RNA localization to the mitochondrial cloud is severely impaired in ATP-depleted stage I oocytes. (A) Qualitative assessment of oocytes injected with fluorescently labeled MCLE alone or co-injected with MCLE and apyrase (intracellular concentration of 1 U/mL) and incubated for 6 h at 25°C. An oocyte injected with a nonlocalizing RNA (XβG) is shown as a negative control. Oocytes that were depleted of ATP showed either no apparent localization to the mitochondrial cloud (−) or weak signal in the cloud (+/−). (B) Quantification of MCLE localization. Since the value for XβG localization stays relatively constant from experiment to experiment, it was averaged from 11 groups of oocytes in five different experiments (range 2.7%–5%, mean 3.6% signal in the mitochondrial cloud) and subtracted as background. The break in the x-axis is shown to indicate that the MCLE values come from a single experiment, whereas the XβG measurement is an average value from multiple experiments. Localization is reduced by ∼60% in ATP-depleted oocytes (P < 10−17).
The rate of RNA localization to the Balbiani body is enhanced by increasing intracellular ATP
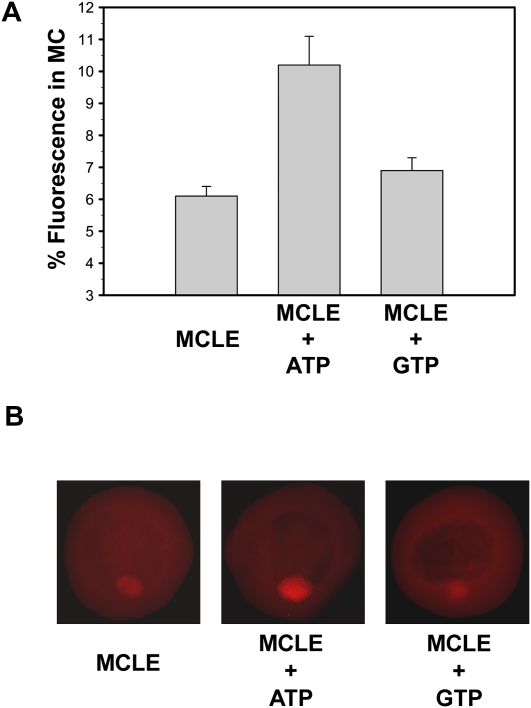
It has been observed that ATP depletion can decrease diffusion-based movements of RNA in the nucleus by forming intranuclear barriers that apparently constrain Brownian motion (Shav-Tal et al. 2004; Vargas et al. 2005). Even though this has not been observed in the cytoplasm of somatic cells (Vargas et al. 2005), we were somewhat concerned that the impairment of localization we observed might have been due to similar artifacts created by ATP depletion. Thus, another way to determine if RNA localization to the Babliani body is affected by intracellular ATP levels is to raise the intracellular ATP concentration. In fact, since only a 90% reduction of intracellular ATP in stage I oocytes was sufficient to extensively impair localization of the MCLE RNA (Fig. 3; Table 1), we hypothesized that the rate of RNA localization may be extremely sensitive to intracellular levels of ATP, such that ATP itself may be limiting for localization. In such a case, we speculated that it should be possible to enhance the rate of localization by increasing intracellular levels of ATP. Therefore, we injected fluorescent MCLE RNA by itself or co-injected it with ATP such that the final intracellular ATP concentration was approximately doubled (∼5 mM). As a negative control, we co-injected an identical amount of GTP. Whereas only 6% of the fluorescent RNA was seen in the Balbiani body of oocytes injected with the MCLE RNA alone, >10% of the RNA was localized in oocytes co-injected with the MCLE and ATP (Fig. 4A). In fact, localization was increased by ∼250% in oocytes co-injected with ATP compared with control oocytes when corrected for background. Interestingly, this dramatic increase was specific for ATP since GTP did not promote localization to such an extent (Fig. 4A). Fluorescent images of the injected oocytes correlated with the quantitative assessment and showed distinctive higher amounts of fluorescence in the mitochondrial cloud of oocytes injected with MCLE plus ATP compared with control oocytes injected with MCLE alone or oocytes injected with MCLE and GTP (Fig. 4B). These data show clearly that the rate of RNA localization in stage I oocytes is limited by the intracellular concentration of ATP in vivo. Moreover, this indicates that the inhibition of localization observed in our ATP depletion experiments (Fig. 3) probably does not arise from structural artifacts that form under ATP-limited conditions, as appears to happen in the nucleus (Shav-Tal et al. 2004; Vargas et al. 2005).
FIGURE 4.
The rate of RNA localization is increased substantially by co-injection of ATP, but not GTP. (A) Stage I oocytes were injected with fluorescently labeled MCLE RNA or co-injected with MCLE and ATP or GTP such that the final intracellular ATP concentration was ∼5 mM, and the endogenous GTP concentration was increased by 2.5 mM. Oocytes were incubated for 6 h at 25°C, fixed, and localization was quantified. RNA localization was increased to higher levels with co-injection of ATP (P < 0.001) than GTP. (B) Confocal images of representative oocytes from each group are shown.
The Xcat-2 RNA localization element recruits kinesin II to the Balbiani body
Since RNA localization to the Balbiani body is temperature and ATP dependent, it is possible that an ATPase directly contributes to this process. Molecular motors are one type of ATPase that mediate a multitude of intracellular transport and localization processes, and we have shown previously that the Xcat-2 MCLE RNA, like the Vg1 mRNA localization element, recruits kinesin II to the vegetal region of stage II oocytes. It is interesting to note that kinesin II is more efficiently recruited by the Xcat-2 MCLE than it is by the Vg1 RNA localization element (Betley et al. 2004). Moreover, localization of the Xcat-2 MCLE to the vegetal region of stage III oocytes is dependent on kinesin II (Betley et al. 2004). Since a diffusion and entrapment mechanism is thought to mediate localization of the Xcat-2 MCLE to the Balbiani body, it is somewhat surprising that these previous studies demonstrated such motor-dependent activities in vivo. We wondered whether the recruitment observed in stage II oocytes (Betley et al. 2004) is a stage-specific phenomenon or whether the MCLE RNA is also able to recruit kinesin II in stage I oocytes. Since protein blot analysis has shown that kinesin II is highly expressed in stage I oocytes (Betley et al. 2004), we first assessed the distribution of kinesin II in uninjected stage I oocytes by immunocytochemistry (Fig. 5). As can be seen in Figure 5B, kinesin II is distributed uniformly throughout stage I oocytes both within the mitochondrial cloud as well as in the surrounding cytoplasm. However, no enrichment is apparent in the Balbiani body region. In fact, the kinesin II signal appears weaker within the Balbiani body than it does in the surrounding cytoplasm of stage I oocytes. This relative distribution of kinesin II is ideal for testing recruitment because an ectopic enrichment of kinesin II within the Balbiani body can be visualized easily.
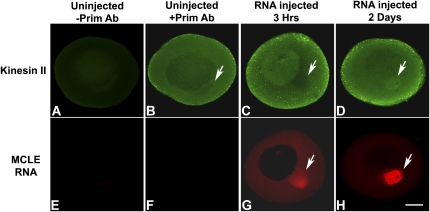
FIGURE 5.
MCLE RNA can recruit kinesin II to the Balbiani body. Low-resolution images of stage I oocytes injected with fluorescently labeled MCLE RNA (red) and immunolabeled with a kinesin II specific antibody (green). (Arrows) Balbiani bodies. (A,E) Uninjected stage I oocyte minus primary antibody control; (B,F) Uninjected stage I oocyte immunolabeled with primary and secondary antibodies; (C,G) Stage I oocyte 3 h after injection of fluorescent MCLE RNA; (D,H) Stage I oocyte 2 d after injection of MCLE RNA. After 2 d, high amounts of MCLE RNA have accumulated in the Balbiani body and little RNA remains in the cytoplasm; enrichment of kinesin II is clearly detected in the mitochondrial cloud (cf. D and C). Scale bar is 30 μm.
To determine if the MCLE RNA is able to recruit kinesin II to the Balbiani body, we injected fluorescently labeled RNA into stage I oocytes, incubated them for various amounts of time, and evaluated the distribution of endogenous kinesin II in the Balbiani body. Low levels of kinesin II were observed in the Balbiani body of uninjected oocytes (Fig. 5B) as well as oocytes injected with the MCLE RNA and incubated for only 3 h (Fig. 5C). However, after 2 d of incubation, during which time localization of the injected MCLE RNA continued to increase, a clear enrichment of kinesin II was observed in the Balbiani body (Fig. 5D,H). This enrichment was most apparent under high magnification (Fig. 6), where kinesin II clearly was enriched in the Balbiani body (Fig. 6B) and co-localized extensively with the injected RNA (Fig. 6C). After only 3 h of incubation, little RNA had accumulated in the mitochondrial cloud (Figs. 5G; 6D), and even under high magnification, little if any enrichment (Fig. 6E) or co-localization (Fig. 6F) of kinesin II was detectable. These data demonstrate that kinesin II can be recruited by the Xcat-2 MCLE RNA to the Balbiani body during the localization process in stage I oocytes (Figs. 5, 6). Thus, the ability to recruit kinesin II is an inherent property of this RNA localization element and occurs throughout all stages of oogenesis in which RNA localization takes place (Betley et al. 2004). Since a number of in vivo competition studies indicate that RNA localization factors in Xenopus oocytes are in a large excess relative to microinjected RNA (Kwon et al. 2002; Choo et al. 2005; Czaplinski and Mattaj 2006), it is expected that detecting the enrichment of such factors in the Balbiani body would require more time than detecting enrichment of the RNA that recruits it (see Materials and Methods).
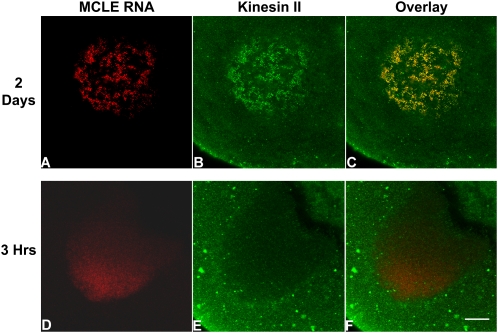
FIGURE 6.
MCLE RNA co-localizes with kinesin II in the Balbiani body. High-resolution images of stage I oocytes injected with the MCLE RNA and incubated for 2 d (A–C) show extensive co-localization of the MCLE RNA (red) and kinesin II protein (green) in the mitochondrial cloud. In contrast, after 3 h of localization (D–F), when only low amounts of RNA have accumulated in the mitochondrial cloud, no enrichment of kinesin II is detected. Scale bar, 10 μm.
Inhibition of kinesin II reduces RNA localization to the Balbiani body
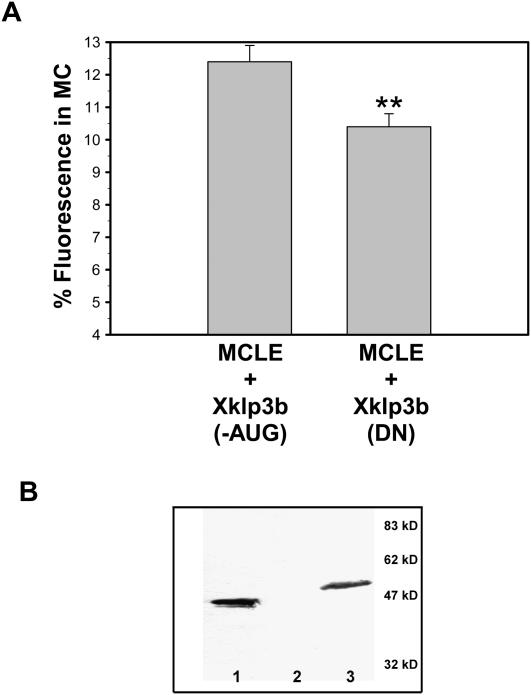
The recruitment of kinesin II to the Balbiani body suggested that Xcat-2 MCLE is indeed able to interact with this molecular motor in stage I oocytes. Since inhibition of kinesin II blocks localization of the MCLE RNA in stage III oocytes (Betley et al. 2004) and localization to the Balbiani body is dependent on ATP, we wondered whether kinesin II may contribute also to the localization process in stage I oocytes. To test this, we expressed a Flag-tagged dominant negative kinesin II polypeptide in stage I oocytes that previously has been shown to impair kinesin II function in Xenopus (Le Bot et al. 1998; Gelfand et al. 2001; Betley et al. 2004). Kinesin II is a heterotrimeric complex, consisting of two kinesin-like motor protein subunits (Xklp3a and Xklp3b) that interact through their coiled-coil tail regions, and a third nonmotor subunit called KAP (kinesin-associated polypeptide) (Cole et al. 1993; Wedaman et al. 1996). The dominant negative construct (Xklp3bDN) used to inhibit kinesin II encodes a truncated Xklp3b subunit, which lacks the motor domain but maintains its ability to incorporate into the heterotrimeric complex, thereby inactivating it (Le Bot et al. 1998).
The Flag-tagged Xklp3bDN polypeptide was expressed in stage I oocytes by co-injecting in vitro transcribed, capped, and polyadenylated Xklp3bDN mRNA along with fluorescently labeled MCLE RNA. An identical Xklp3b construct containing a defective AUG start codon (Xklp3b-AUG) was analyzed in parallel. Stage I oocytes co-injected with fluorescently labeled MCLE RNA and either the Xklp3bDN or Xklp3b-AUG mutant mRNA were incubated for 7 h, and localization was quantified. We found that localization to the Balbiani body was impaired significantly in oocytes injected with the Xklp3bDN mRNA, but not with the Xklp3b-AUG mRNA (Fig. 7A). Expression of the Xklp3bDN polypeptide fragment was confirmed by Western blot analysis using an anti-Flag antibody (Fig. 7B, lane 3). In the multiple experiments conducted, localization was consistently reduced by ∼20%–30%, when subtracting for background localization of the XβG RNA. We also used a transcript consisting of Escherichia coli-derived plasmid sequences as a negative mRNA control and obtained similar results (data not shown).
FIGURE 7.
Expression of a dominant negative kinesin II construct impairs localization in stage I oocytes. Stage I oocytes were injected with an mRNA encoding either a dominant negative kinesin II polypeptide (Xklp3bDN) or with an identical mRNA that contained a mutant start codon (Xklp3b-AUG). (A) Quantification of localization shows inhibition of kinesin II impairs localization compared with control oocytes (P < 0.01) (n = 54 or 56 oocytes for the AUG mutant and DN construct, respectively). (B) Western blot confirming expression of dominant negative Flag-tagged kinesin II protein (lane 3). (Lane 1) Detection of 50 ng of a Flag-BAP fusion protein as a control, (lane 2) extract of oocytes injected with the control Xklp3b-AUG mRNA.
These findings demonstrate that inhibition of endogenous kinesin II reduces RNA localization to the Balbiani body and are consistent with the observation that kinesin II becomes recruited by early pathway RNAs in stage I oocytes (Fig. 5, 6). Our attempts to reduce RNA localization by >30% with the Xklp3bDN construct have not yet been successful in stage I oocytes, even though the same approach reduces localization by ∼75% in stage III oocytes (Betley et al. 2004). This could be explained by the fact that endogenous kinesin II is more highly expressed in stage I oocytes than in later-staged oocytes (Betley et al. 2004). In addition, the small size of stage I oocytes limits the amount of Xklp3bDN mRNA that we can inject. Therefore, we may not be producing sufficient amounts of the dominant negative polypeptide to inhibit endogenous kinesin II completely (see Materials and Methods). Alternatively, kinesin II may play only a partial role in the localization of RNAs to the Balbiani body, working together with other molecular mechanisms. In either case, these data suggest that kinesin II plays a role in the overall localization process in stage I oocytes (see Discussion).
DISCUSSION
The selective localization of specific mRNAs to the vegetal cortex of Xenopus oocytes is important for organizing gene expression in the fertilized egg such that proper development of the embryo can occur. The vegetal RNAs that localize earliest during oogenesis, such as Xcat2, Xdazl, and Xpat, first associate with the Balbiani body of stage I oocytes before this entire subcellular structure localizes to the vegetal cortex during stage II of oogenesis. The Balbiani body (also called the mitochondrial cloud in Xenopus) is an evolutionarily conserved structure present in early oocytes and is found throughout the animal kingdom (Kloc and Etkin 2005; Pepling et al. 2007). Previous work suggested that RNAs are sorted to the Balbiani body in Xenopus by a diffusion and entrapment mechanism (Zhou and King 1996a) that functions independently of microtubules (Kloc et al. 1996; Chang et al. 2004). Here, we show that localization of the Xcat-2 mitochondrial cloud localization element (MCLE) to the Balbiani body is coupled strongly to the energy state of the cell and that this process has a Q10 of ∼2 in vivo. Since these data suggest that ATPases are required for RNA localization in stage I oocytes, we tested whether kinesin II, a microtubule-dependent ATPase, is involved in this process. Indeed, kinesin II is recruited to the Balbiani body by the MCLE RNA, and inhibition of kinesin II activity reduces RNA localization in stage I oocytes.
The quantitative assay developed in this work allowed us to ask fundamental questions about the mechanism that regulates the localization of CAC-rich RNA localization elements to the Balbiani body. Interestingly, the cis-elements that target specific mRNAs to the Balbiani body appear to be conserved in vertebrates. For example, the UGCAC motifs essential for localization of the Xcat-2 MCLE in Xenopus (Betley et al. 2002; Chang et al. 2004) also are required for RNA localization to the Balbiani body in zebrafish (Kosaka et al. 2007). With our quantitative assay we addressed whether the RNA localization process in stage I oocytes is sensitive to intracellular ATP concentrations and temperature. Indeed, our ATP depletion and temperature shift experiments suggest that most RNA localization in stage I oocytes is driven by enzymatic processes. While it is possible that the ATP effects observed on RNA localization rates are indirect, it is more likely that ATPases are essential for the overall localization process. For example, the requirement for ATP possibly could reflect the involvement of a specific RNA helicase(s) that facilitates RNP remodeling essential for interactions with RNA localization factors, as has been suggested recently (Lewis and Mowry 2007; Lewis et al. 2008). In fact, some RNA helicases have been shown to have a Km in the mM range for ATP (Lam et al. 2003), consistent with the levels we found in stage I oocytes. The identification of such an enzyme would represent an important breakthrough in our understanding of the mechanism that sorts RNAs to the Balbiani body.
Kinesin II is one ATPase shown here to be involved in localizing RNAs to the Babliani body. This finding is somewhat surprising since numerous studies have shown that localization of early RNAs to the Balbiani body is not blocked by drugs that depolymerize microtubules. In fact, sensitivity to microtubule depolymerizing drugs has been used in the past to distinguish between putative mechanisms that drive RNA localization during distinct stages of oogenesis in Xenopus. Treatment of stage I oocytes with nocodazole does not inhibit the localization of RNAs to the Balbiani body (Kloc and Etkin 1995; Kloc et al. 1996; Chang et al. 2004), but partially disrupts localization to the vegetal cortex of larger oocytes (Yisraeli et al. 1990; Kloc and Etkin 1995; Zhou and King 1996b; Kloc et al. 1998). We have repeated these experiments many times and observed no quantitative reduction in RNA localization in stage I oocytes when long microtubules, detectable by confocal microscopy, were eliminated by a combination of cold treatment and incubation with nocodazole (data not shown). However, we also have observed that RNA localization in stage IV oocytes is reduced to a greater extent by inhibiting kinesin II (Betley et al. 2004) than it is by nocodazole (data not shown). This suggests that the interpretation of data involving microtubule depolymerizing drugs may be complicated in vivo and that complete disassembly of microtubules in oocytes may not occur following drug treatment. A similar series of seemingly contradictory results exists in the field of membrane trafficking, where microtubule depolymerizing drugs do not necessarily block protein secretion even though secretory vesicles are large and should not be able to diffuse significant distances within the cell (Bloom and Goldstein 1998; Thyberg and Moskalewski 1999).
To explain why kinesin II inhibition reduces RNA localization in stage I oocytes but microtubule drugs do not, we envision two possible scenarios. First, diffusion-mediated localization of mRNA particles and their directed transport on microtubules may be operating simultaneously, with diffusion being the prevalent mechanism of localization. It has been suggested previously that diffusion accounts for the movement and localization of the MCLE RNA in the stage I oocytes because photobleaching experiments show that localizing and nonlocalizing RNAs redistribute at similar rates in the cytoplasm (Chang et al. 2004). If diffusion and directed transport both contribute to the localization process, kinesin II-dependent transport may only account for ∼25% of overall localization, since blocking kinesin II decreases RNA localization to the Balbiani body by about this amount (Fig. 7). The remaining 75% of RNA localization could be mediated primarily by diffusion. In nocodazole-treated oocytes, the overall rate of localization could be relatively unaffected because disruption of the microtubule network may release a number of nonmotile RNA complexes from microtubules, enabling them to diffuse to the mitochondrial cloud, thereby compensating for any loss of localization due to motor-directed transport.
Alternatively, most RNA localization in stage I oocytes may be similar to that in later-staged oocytes (Messitt et al. 2008) and mediated by molecular motors acting either sequentially or redundantly, with diffusion playing less of an explicit role in the localization process. This also may explain why expression of the dominant negative kinesin II polypeptide does not inhibit localization completely. In this scenario, the extreme dependence of the localization process on metabolic energy could indicate that microtubule-directed transport mediates most RNA localization to the Balbiani body. While it has been suggested that molecular motors may facilitate RNA localization in invertebrate systems indirectly by promoting cytoplasmic streaming (Forrest and Gavis 2003), cytoplasmic streaming does not occur in Xenopus oocytes (Sehy et al. 2002). Thus, if most RNA localization is directed on microtubules in stage I oocytes, a subpopulation of stable microtubules must exist in nocodazole-treated oocytes. Using electron microscopy, we have observed an abundance of short microtubule profiles both in nocodazole-treated (∼63 microtubule profiles/μm2) and untreated stage I oocytes (∼50 microtubule profiles/μm2) (data not shown); this is about nine times higher than the number of microtubules reported for stage VI oocytes using the same method (Pfeiffer and Gard 1999). Moreover, a large number of MCLE RNA-containing particles co-localize with tubulin foci in nocodazole-treated (data not shown) and untreated oocytes (Choo et al. 2005) analyzed by confocal microscopy. Therefore, a large number of short microtubule tracks may be sufficient to mediate motor-dependent localization in nocodazole-treated stage I oocytes.
While the specific kinetics of RNA movements along microtubules are likely to be complicated and have not yet been characterized in the Xenopus oocyte system, it has become evident in other systems that bidirectional, probabilistic transport, consisting of numerous short movements along microtubules, may be the prevalent mechanism by which many cellular cargos, including mRNAs, become localized by molecular motors in vivo (Welte 2004; Bullock et al. 2006; Ross et al. 2006; Dynes and Steward 2007; Zimyanin et al. 2008). If the short microtubules present in nocodazole-treated oocytes are capable of supporting RNA localization, it may be that the frequency and direction with which particular RNAs move are more important than the distance an RNA actually travels along a single microtubule. This scenario could also explain why nocodazole has no effect on the localization of the Vg1 RNA localization element in stage II oocytes (Kloc and Etkin 1998), even though the RNA interacts with kinesin II at this stage (Betley et al. 2004). Interestingly, a probabilistic mechanism has been proposed previously to explain why RNA localization in oocytes requires days to occur (Fusco et al. 2003). If RNA localization was driven by a processive mechanism, localization of the RNA should require less than an hour given that RNA localization factors are present in large excess relative to the RNA cargos they carry (Kwon et al. 2002; Choo et al. 2005). Finally, a recent microarray study of maternal mRNAs in Xenopus suggests that localized mRNAs have a higher affinity for microtubules than nonlocalized mRNAs. Endogenous mRNAs, such as Xpat and Xcat-2, which localize to the Balbiani body of stage I oocytes, are enriched on microtubules to a greater extent than the late localizing Vg1 mRNA (Blower et al. 2007), consistent with the finding that early localizing mRNAs contain stronger RNA localization elements than late localizing ones (Choo et al. 2005). Real-time imaging studies of oocytes will be required to resolve exactly how these localizing RNA molecules move to their final destinations in vivo.
MATERIALS AND METHODS
Quantitative RNA localization assay in stage I oocytes
Stage I oocytes ranging in size from 160 to 220 μm were prepared from a single 6.25- to 7.5-cm female Xenopus laevis as previously described (Choo et al. 2005). Oocytes in this size range virtually always have a single mitochondrial cloud that is more closely associated with the germinal vesicle than the vegetal cortex. Approximately 0.04 nL of a 4-5 fmol/nL solution containing the Xcat-2 mitochondrial cloud RNA localization element (MCLE) (Zhou and King 1996a; Kloc et al. 2000) fluorescently labeled by the incorporation of Alexa Fluor-546-14-UTP (Invitrogen) during in vitro transcription was injected into the cytoplasm of these stage I oocytes as previously described (Choo et al. 2005). A PV83 pneumatic picopump (World Precision Instruments) was used to microinject the RNA. Oocytes were then cultured in 0.5× Leibovitz L-15 medium (Sigma) containing 15 mM HEPES at pH 7.6, 0.1 mg/mL tetracycline (Sigma), and 0.1 mg/mL gentamycin (Roche). The temperature and duration of the incubations were varied as indicated for distinct experiments. Occasionally the RNA was inadvertently injected into the nucleus, but such oocytes were easily recognized during image acquisition since much of the RNA remained in the nucleus throughout the culturing period. Since the kinetics of nuclear export could affect the rate at which injected RNAs localized to the Balbiani body, we discarded the few oocytes that had been injected into the nucleus. Because a large percentage (∼75%) of oocytes die upon injection and large numbers of oocytes are required to generate statistically significant localization data, we typically inject at least 200 oocytes for each experimental condition. Consequently, it is difficult to assess more than three experimental conditions in a single experiment.
Oocytes were mounted in Vectashield (Vector Laboratories, Inc.), and confocal images were acquired on a Zeiss LSM 510 confocal laser scanning microscope with a Helium-Neon laser (543 nm) for detection of Alexa Fluor-546-14-UTP labeled RNA. Both dark-field and a fluorescent images were collected for each oocyte from an optical section through the center of the Balbiani body. Since we could not specify the orientation of oocytes during mounting, this optical section included the nucleus in some oocytes, but not in others. For quantification, the intensity of the laser was adjusted slightly for each oocyte such that the nonlocalized RNA signal in the region of cytoplasm outside the Balbiani body was set to 20% of maximum detection levels. Both the dark-field and fluorescent images were exported for each oocyte, and a stack of the two images was created using ImageJ software. A circle was drawn around the mitochondrial cloud of the dark-field image, which was then used to measure signal in the fluorescent image. The IOD within the mitochondrial cloud (IODmc) and the entire oocyte section (IODTotal) from the fluorescence layer were determined using ImageJ software. Percent localization was defined as:
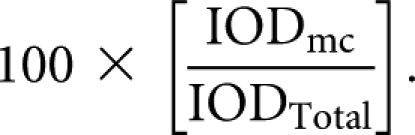 |
The standard deviation for each value typically was ∼25% of the actual value for localization. Therefore, ∼30–40 oocytes were used for each data point, and error bars represent the standard error of the mean in all histograms. Each experiment was repeated a minimum of three times, and similar results were obtained in all experiments. However, the amount of localization observed with the MCLE at a given time point could vary by as much as twofold between experiments that were run on different days with oocytes isolated from different frogs.
Nucleotide depletions
To reduce the endogenous concentration of ATP, apyrase (Grade VI, Sigma-Aldrich) was co-injected with fluorescently labeled MCLE RNA (final intracellular concentration 0.8 units/mL). The integrity of the fluorescent RNA was verified by electrophoresis of the apyrase–RNA injection mixes in denaturing polyacrylamide gels run just after the injections were performed. In addition, the viability of oocytes with apyrase appeared viable was assessed by overall morphology and tensegrity of the plasma membrane.
The ATP concentration in stage I oocytes was measured using an ATP determination kit (Invitrogen-Molecular Probes), which is based on ATP-dependent luciferase activity. Individual stage I oocytes were homogenized by pipetting and boiling in 50 μL, 20 mM HEPES at pH 7.5 (buffer), and the ATP concentration was determined following the protocol provided by the manufacturer. The average ATP concentration was found to be ∼2.4 mM in stage I oocytes, which is similar to that reported for stage VI oocytes (∼2.3 mM) in two previous studies (Newmeyer et al. 1986; Gribble et al. 2000). Similar levels of intracellular levels of ATP were found when measurements were obtained from pooled oocytes.
Kinesin II recruitment and immunocytochemistry
Stage I oocytes injected with Alexa Fluor-546-UTP-labeled MCLE RNA were fixed after the indicated culturing times in MEMFA for 1 h, washed twice in 1× PBS (pH 7.4) and stored in 1× PBS until oocytes of different time points were collected. Oocytes were then washed twice for 20 min each in 1× PBS containing 0.1% Tween followed by one wash in 1× PBS containing 0.05% Tween. The oocytes were incubated in blocking solution (1 mg/mL BSA in 1× PBS, 0.05% Tween) for 30 min. Oocytes were then incubated overnight at 4°C with the monoclonal anti-kinesin II K2.4 antibody (Covance) in blocking solution and washed twice the next day for 30 min in 1× PBS containing 0.05% Tween. A second overnight incubation at 4°C with Cy2-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch) in blocking solution was followed by two washes in 1× PBS containing 0.05% Tween. Whole oocytes were mounted in Vectashield (Vector Laboratories, Inc.) and subjected to confocal imaging. This analysis showed that kinesin II levels only became higher in the Balbiani bodies than in the surrounding cytoplasm after a day (data not shown) or two (Figs. 4, 5) of culturing following injection of the MCLE RNA. This is a longer time period than is required for detecting enrichment of the MCLE RNA itself in the Balbiani body; ∼3 h are required to detect a ∼1.2-fold enrichment of RNA in the Balbiani body relative to that of surrounding cytoplasm. At the 3-h time point, this ∼1.2-fold enrichment is the result of only ∼0.44% of the RNA becoming localized to the Balbiani body. Since RNA localization factors appear to be in excess of injected RNAs (Kwon et al. 2002; Choo et al. 2005; Czaplinski and Mattaj 2006), and kinesin II is most highly concentrated in stage I oocytes (Betley et al. 2004), it would be expected that a higher percentage of MCLE RNA would need to be localized before enrichment of kinesin II could be detected in the Balbiani body. For example, if the concentration of kinesin II was five times higher than that of the injected MCLE RNA in the cytoplasm, five times as much of the RNA/motor complex would need to become localized in order to detect a higher concentration of kinesin II in the Balbiani body. Interestingly, approximately five times as much of the MCLE RNA, or 2%, localized to the Balbiani body after 2 d when compared with 3 h (0.44 percent). This estimation is based on diameters of 200, 100, and 30 μm for the oocyte, nucleus, and Balbiani body, respectively. In addition, the average enrichment of the MCLE RNA in the Balbiani body is ∼1.1- and ∼4.5-fold for oocytes cultured for 3 h or 2 d, respectively.
Dominant negative kinesin II construct and mRNA transcription
For expression of the dominant negative Xklp3b polypeptide in Xenopus oocytes, a PCR amplification product derived from the plasmid HA-Xklp3-ST* (Le Bot et al. 1998) was cloned into the T7 transcription vector p18.polyA.BX. The transcription vector p18.polyA.BX contains a stretch of 30 adenine base pairs, near the polylinker such that a poly-A tail is incorporated during in vitro transcription. The upstream primers used to generate the amplified products were:
5′-ATAGGCGTTAGAGCTC GCCACCATG GATTACAAGGAC GACGACGATAAGGCAGGCCATCATCATCATC-3′; or
5′-ATAGGCGTTAGAGCTC GCCACCACG GATTACAAGGAC GACGACGATAAGGCAGGCCATCATCATCATC-3′.
These primers contain a SacI restriction site (underlined), a Flag-tag (italics), and start codon or mutated start codon inside a Kozak sequence (bold). The downstream primer contained an XmaI restriction site (underlined) and a stop codon (square):
5′-CTCGCTATATCCCGGGCCGGTGGATCCTTATTTGG-3′.
During in vitro transcription, the mRNA was capped using 7mG(5′)ppp(5′)G according to the manufacturer's recommendations (New England Biolabs). This mRNA was co-injected with fluorescently labeled MCLE RNA, and in six of six independent experiments performed, a reduction was always observed.
Western blot analysis
Expression of the Flag-tagged Xklp3DN kinesin II polypeptide was verified by Western blot analysis using monoclonal anti-Flag antibody (Sigma, F1804). Extracts for detecting the Flag-tagged protein were prepared via homogenization of RNA-injected stage II oocytes in SDS sample buffer, and each lane contained 15 oocytes. A Flag-BAP fusion protein (50 ng) was used as a positive control (Sigma, P7457) for detection. For technical reasons it is difficult to determine the relative amounts of endogenous kinesin II and the ectopically expressed dominant negative Flag-tagged polypeptide in microinjected stage I oocytes. However, we estimate that the ratio of the dominant negative polypeptide to endogenous kinesin II is five times less in stage I oocytes than previously has been achieved in stage III oocytes, where 75% inhibition of RNA localization was achieved (Betley et al. 2004). This estimate is based on an injection volume equal to ∼3% of the oocyte volume, and the fact that the concentration of endogenous kinesin II is approximately seven times higher in stage I oocytes when compared with stage III oocytes (Betley et al. 2004).
ACKNOWLEDGMENTS
We thank Sid Tamm and Gary Borisy for critical comments during this work and Todd Blute for his assistance with confocal imaging and electron microscopy. This work was supported by the National Institutes of Health and the National Science Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.975309.
REFERENCES
- Allen L., Kloc M., Etkin L.D. Identification and characterization of the Xlsirt cis-acting RNA localization element. Differentiation. 2003;71:311–321. doi: 10.1046/j.1432-0436.2003.7106003.x. [DOI] [PubMed] [Google Scholar]
- Andken B.B., Lim I., Benson G., Vincent J.J., Ferenc M.T., Heinrich B., Jarzylo L.A., Man H.Y., Deshler J.O. 3′-UTR SIRF: A database for identifying clusters of short interspersed repeats in 3′-untranslated regions. BMC Bioinformatics. 2007;8:274. doi: 10.1186/1471-2105-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley J.N., Frith M.C., Graber J.H., Choo S., Deshler J.O. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- Betley J.N., Heinrich B., Vernos I., Sardet C., Prodon F., Deshler J.O. Kinesin II meditates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 2004;14:219–224. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Bloom G.S., Goldstein L.S. Cruising along microtubule highways: How membranes move through the secretory pathway. J. Cell Biol. 1998;140:1277–1280. doi: 10.1083/jcb.140.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Feric E., Weis K., Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubunenko M., Kress T.L., Vempati U.D., Mowry K.L., King M.L. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 2002;248:82–92. doi: 10.1006/dbio.2002.0719. [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Nicol A., Gross S.P., Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr. Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- Chang P., Torres J., Lewis R.A., Mowry K.L., Houliston E., King M.L. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol. Biol. Cell. 2004;15:4669–4681. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo S., Heinrich B., Betley J.N., Chen Z., Deshler J.O. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev. Biol. 2005;278:103–117. doi: 10.1016/j.ydbio.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Claussen M., Horvay K., Pieler T. Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development. 2004;131:4263–4273. doi: 10.1242/dev.01283. [DOI] [PubMed] [Google Scholar]
- Cole D.G., Chinn S.W., Wedaman K.P., Hall K., Vuong T., Scholey J.M. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Mattaj I.W. 40LoVe interacts with Vg1RBP/Vera and hnRNP I in binding the Vg1-localization element. RNA. 2006;12:213–222. doi: 10.1261/rna.2820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler J.O., Highett M.I., Schnapp B.J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Dynes J.L., Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J. Comp. Neurol. 2007;500:433–447. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- Forrest K.M., Gavis E.R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Forristall C., Pondel M., Chen L., King M.L. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development. 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- Fusco D., Accornero N., Lavoie B., Shenoy S.M., Blanchard J.M., Singer R.H., Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand V.I., Le Bot N., Tuma M.C., Vernos I. A dominant negative approach for functional studies of the kinesin II complex. Methods Mol. Biol. 2001;164:191–204. doi: 10.1385/1-59259-069-1:191. [DOI] [PubMed] [Google Scholar]
- Green S.L. Factors affecting oogenesis in the South African clawed frog (Xenopus laevis) Comp. Med. 2002;52:307–312. [PubMed] [Google Scholar]
- Gribble F.M., Loussouarn G., Tucker S.J., Zhao C., Nichols C.G., Ashcroft F.M. A novel method for measurement of submembrane ATP concentration. J. Biol. Chem. 2000;275:30046–30049. doi: 10.1074/jbc.M001010200. [DOI] [PubMed] [Google Scholar]
- Keem K., Smith D., Wallace R.A., Wolf D. Growth rate of oocytes in laboratory-maintained Xenopus laevis. Mol. Reprod. Dev. 1979;2:125–135. [Google Scholar]
- Kiebler M.A., Bassell G.J. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- King M.L., Messitt T.J., Mowry K.L. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- Kloc M., Etkin L.D. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- Kloc M., Etkin L.D. Apparent continuity between the messenger transport organizer and late RNA localization pathways during oogenesis in Xenopus. Mech. Dev. 1998;73:95–106. doi: 10.1016/s0925-4773(98)00041-0. [DOI] [PubMed] [Google Scholar]
- Kloc M., Etkin L.D. RNA localization mechanisms in oocytes. J. Cell Sci. 2005;118:269–282. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- Kloc M., Larabell C., Etkin L.D. Elaboration of the messenger transport organizer pathway for localization of RNA to the vegetal cortex of Xenopus oocytes. Dev. Biol. 1996;180:119–130. doi: 10.1006/dbio.1996.0289. [DOI] [PubMed] [Google Scholar]
- Kloc M., Larabell C., Chan A.P., Etkin L.D. Contribution of METRO pathway localized molecules to the organization of the germ cell lineage. Mech. Dev. 1998;75:81–93. doi: 10.1016/s0925-4773(98)00086-0. [DOI] [PubMed] [Google Scholar]
- Kloc M., Bilinski S., Pui-Yee Chan A., Etkin L.D. The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3′UTR. Dev. Biol. 2000;217:221–229. doi: 10.1006/dbio.1999.9554. [DOI] [PubMed] [Google Scholar]
- Kosaka K., Kawakami K., Sakamoto H., Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech. Dev. 2007;124:279–289. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Kwon S., Abramson T., Munro T.P., John C.M., Kohrmann M., Schnapp B.J. UUCAC and Vera dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 2002;12:558–564. doi: 10.1016/s0960-9822(02)00740-6. [DOI] [PubMed] [Google Scholar]
- Lam A.M., Keeney D., Eckert P.Q., Frick D.N. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 2003;77:3950–3961. doi: 10.1128/JVI.77.7.3950-3961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot N., Antony C., White J., Karsenti E., Vernos I. Role of xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the Golgi apparatus. J. Cell Biol. 1998;143:1559–1573. doi: 10.1083/jcb.143.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lewis R.A., Mowry K.L. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007;75:507–518. doi: 10.1111/j.1432-0436.2007.00159.x. [DOI] [PubMed] [Google Scholar]
- Lewis R.A., Gagnon J.A., Mowry K.L. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol. Cell Biol. 2008;28:678–686. doi: 10.1128/MCB.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Setou M., Inokuchi K. Transcriptome analysis reveals the population of dendritic RNAs and their redistribution by neural activity. Neurosci. Res. 2007;57:411–423. doi: 10.1016/j.neures.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Messitt T.J., Gagnon J.A., Kreiling J.A., Pratt C.A., Yoon Y.J., Mowry K.L. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev. Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer D.D., Lucocq J.M., Burglin T.R., De Robertis E.M. Assembly in vitro of nuclei active in nuclear protein transport: ATP is required for nucleoplasmin accumulation. EMBO J. 1986;5:501–510. doi: 10.1002/j.1460-2075.1986.tb04239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling M.E., Wilhelm J.E., O'Hara A.L., Gephardt G.W., Spradling A.C. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc. Natl. Acad. Sci. 2007;104:187–192. doi: 10.1073/pnas.0609923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer D.C., Gard D.L. Microtubules in Xenopus oocytes are oriented with their minus-ends towards the cortex. Cell Motil. Cytoskeleton. 1999;44:34–43. doi: 10.1002/(SICI)1097-0169(199909)44:1<34::AID-CM3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Politz J.C., Tuft R.A., Prasanth K.V., Baudendistel N., Fogarty K.E., Lifshitz L.M., Langowski J., Spector D.L., Pederson T. Rapid, diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Mol. Biol. Cell. 2006;17:1239–1249. doi: 10.1091/mbc.E05-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati M.R., Weeks D.L., Harvey R.P., Melton D.A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- Ross J.L., Wallace K., Shuman H., Goldman Y.E., Holzbaur E.L. Processive bidirectional motion of dynein–dynactin complexes in vitro. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Sehy J.V., Ackerman J.J., Neil J.J. Apparent diffusion of water, ions, and small molecules in the Xenopus oocyte is consistent with Brownian displacement. Magn. Reson. Med. 2002;48:42–51. doi: 10.1002/mrm.10181. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., Singer R.H. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Tian Q.B., Kuromitsu J., Kawai T., Endo S. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci. Res. 2007;57:61–85. doi: 10.1016/j.neures.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Thyberg J., Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Vargas D.Y., Raj A., Marras S.A., Kramer F.R., Tyagi S. Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman K.P., Meyer D.W., Rashid D.J., Cole D.G., Scholey J.M. Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 1996;132:371–380. doi: 10.1083/jcb.132.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M.A. Bidirectional transport along microtubules. Curr. Biol. 2004;14:R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Yisraeli J.K., Sokol S., Melton D.A. A two-step model for the localization of maternal mRNA in Xenopus oocytes: Involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- Yoon Y.J., Mowry K.L. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and Kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- Zhou Y., King M.L. Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development. 1996a;122:2947–2953. doi: 10.1242/dev.122.9.2947. [DOI] [PubMed] [Google Scholar]
- Zhou Y., King M.L. RNA transport to the vegetal cortex of Xenopus oocytes. Dev. Biol. 1996b;179:173–183. doi: 10.1006/dbio.1996.0249. [DOI] [PubMed] [Google Scholar]
- Zimyanin V.L., Belaya K., Pecreaux J., Gilchrist M.J., Clark A., Davis I., St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]