Abstract
Spinal Muscular atrophy is a prevalent genetic disease caused by mutation of the SMN1 gene, which encodes the SMN protein involved in assembly of small nuclear ribonucleoprotein (snRNP) complexes. A paralog of the gene, SMN2, cannot provide adequate levels of functional SMN because exon 7 is skipped in a significant fraction of the mature transcripts. A C to T transition located at position 6 of exon 7 is critical for the difference in exon skipping between SMN1 and SMN2. Here we report that this nucleotide difference results in increased ultraviolet light-mediated crosslinking of the splicing factor U2AF65 with the 3′ splice site of SMN1 intron 6 in HeLa nuclear extract. U2 snRNP association, analyzed by native gel electrophoresis, is also more efficient on SMN1 than on SMN2, particularly under conditions of competition, suggesting more effective use of limiting factors. Two trans-acting factors implicated in SMN regulation, SF2/ASF and hnRNP A1, promote and repress, respectively, U2 snRNP recruitment to both RNAs. Interestingly, depending on the transcript and the regulatory factor, the effects on U2 binding not always correlate with changes in U2AF65 crosslinking. Furthermore, blocking recognition of a Tra2-β1-dependent splicing enhancer located in exon 7 inhibits U2 snRNP recruitment without affecting U2AF65 crosslinking. Collectively, the results suggest that both U2AF binding and other steps of U2 snRNP recruitment can be control points in SMN splicing regulation.
Keywords: SMN, U2 snRNP, U2AF, splicing
INTRODUCTION
Spinal muscular atrophy (SMA) is the most common genetic cause of infant mortality, with an incidence of approximately one in 6000 live births. This neuromuscular disorder progressively affects motor neurons in the anterior horns of the spinal cord, leading to muscular atrophy and paralysis (for review, see Russman 2007). Homozygous loss-of-function mutations in the SMN1 gene are the cause of SMA (Lefebvre et al. 1995). SMN1 encodes the SMN protein, which forms, together with seven Gemin proteins, the SMN complex (for review, see Gubitz et al. 2004). The SMN complex is involved in the assembly of Sm proteins on most U-rich small nuclear RNAs (U snRNAs), an essential step in U-rich small nuclear ribonucleoprotein particle (U snRNP) biogenesis (for review, see Will and Lührmann 2005). SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing that are believed to underpin the molecular pathology of SMA (Zhang et al. 2008).
The SMN1 gene is located in the complex chromosomal region 5q13 (Brzustowicz et al. 1990; Melki et al. 1990) which in humans contains a large inverted duplication, with a second, more centromeric copy of SMN1, the SMN2 gene. Of relevance for SMA, the SMN2 gene cannot fully substitute for SMN1 because exon 7 is frequently skipped in SMN2, but not in SMN1 transcripts (Monani et al. 1999). Exon 7 skipping generates a truncated, SMN2Δ7 protein isoform, which can only provide limited SMN function (Le et al. 2005). Several nucleotide differences exist between the genomic sequence of SMN1 and SMN2 genes, and two of them have been implicated in increased skipping of exon 7 in SMN2 transcripts, a critical translationally silent C to T transition at position 6 of exon 7 (Lorson et al. 1999) and an A to G transition at position 100 of intron 7 (Kashima et al. 2007a). Because the extent of SMN2 exon 7 skipping, which is variable in SMA patients, has been correlated with disease severity (Mailman et al. 2002), modulation of this splicing event has been proposed as a potential therapy for the disease. Indeed, various strategies have been utilized to harness SMN2 exon 7 inclusion that led to restoration of SMN function in cells in culture and correction of SMN2 splicing in transgenic mice. These include methods that enhance exon 7 recognition using synthetic exon-specific peptide–nucleic acid activators or bifunctional antisense oligonucleotides (Cartegni and Krainer 2003; Skordis et al. 2003), or antisense oligonucleotides that antagonize a silencer sequence (Hua et al. 2008). Additional strategies have included reducing the competitiveness of the exon 8 3′ splice site (Madocsai et al. 2005) or employing trans-splicing to redirect SMN2 splicing (Coady et al. 2007).
Given this potential, understanding the molecular mechanisms underlying differential splicing of SMN1 and SMN2 is of considerable interest. The sequence elements required for exon 7 inclusion in the SMN1 and SMN2 genes have been the subject of intensive studies, including systematic mutagenesis of exonic sequences and antisense oligonucleotide tiling methods (for review, see Singh 2007; Hua et al. 2008). One concept emerging from these studies is that the sequences in and around SMN exon 7 are dense in splicing regulatory elements. Thus, several splicing enhancers have been defined in exon and intron 7, including a Tra2-β1 enhancer in the central region of the exon which binds Tra2-β1, SRp30c, and hnRNP G (Hofmann et al. 2000; Young et al. 2002; Hofmann and Wirth 2002). Several splicing silencers have also been identified in exon 7 and flanking introns (Singh et al. 2004, 2006; Hua et al. 2007, 2008). One key question is how the interplay between these regulatory elements is affected by the nucleotide differences between SMN1 and SMN2 that cause more extensive skipping of exon 7 in SMN2 transcripts (Lorson et al. 1999; Kashima et al. 2007a).
In this study we focused on how the C to T nucleotide substitution at position +6 of SMN exon 7 influences 3′ splice site recognition by the basic splicing machinery. It has been proposed that the T +6 substitution weakens a splicing enhancer recognized by the splicing factor SF2/ASF (Cartegni and Krainer 2002; Cartegni et al. 2006). SF2/ASF belongs to the SR family of splicing regulatory factors characterized by the presence of one or two RNA-recognition motifs and a C-terminal domain with multiple alternating arginine and serine-dipeptides (for review, see Lin and Fu 2007). One classical function of SR proteins bound to exonic sequences is to stimulate recognition of the flanking splice sites (for review, see Singh and Valcárcel 2005). 3′ splice sites are initially recognized by the branchpoint binding protein (BBP/SF1) and the 65 and 35 kDa subunits of the U2 Auxiliary Factor U2AF (for review, see Will and Lührmann 2005). Enhancer-bound SR proteins can promote U2AF65 binding to the polypyrimidine tract that precedes the conserved AG dinucleotide present at the 3′ end of the intron (Zuo and Maniatis 1996; Zhu and Krainer 2000). The results from other experiments, including tethering of RS domains through heterologous RNA–protein interactions, suggest that other steps in U2 snRNP assembly and even late steps of the splicing process can be stimulated by exonic enhancers and RS domains (Chew et al. 1999; Guth et al. 1999; Kan and Green 1999; Shen and Green 2004; Shen et al. 2004). Splicing enhancers can also stimulate splicing by modulating the activity of inhibitory complexes (Zhu et al. 2001).
Another model proposes that the T +6 substitution creates a splicing silencer sequence in SMN2 exon 7, which is recognized by hnRNP A1 (Kashima and Manley 2003; Cartegni et al. 2006; Kashima et al. 2007b). Binding of hnRNP proteins to intronic and exonic sequences has been correlated with inhibition of neighboring (or intervening) splice sites (for review, see Singh and Valcárcel 2005). hnRNP A1 has been shown to reduce the early association of splicing factors (Eperon et al. 2000), but can also target later events in spliceosome assembly, depending on the pre-mRNA substrate (Tange et al. 2001). The inhibitory function of hnRNP A1 and possibly other hnRNP proteins often requires homotypic cooperative interactions between RNA-bound molecules, and may be based upon looping out the intervening region of the RNA and/or multimerization of the hnRNP proteins from a high-affinity site to create a silenced region in the pre-mRNA (Blanchette and Chabot 1999; Zhu et al. 2001; Wagner and Garcia-Blanco 2001). Decreased occupancy of a regulated 3′ splice site by U2AF has been reported upon phosphorylation of the splicing regulator Sam68 by the ERK–MAP kinase pathway (Tisserant and König 2008).
The combinatorial effects of enhancers and silencers, as well as their mutual influences, determine the extent of association of splicing factors with the pre-mRNA. In this manuscript we have tested whether differential recognition of SMN1 and SMN2 exon 7 can be correlated with differential recruitment of U2AF and U2 snRNP to the 3′ splice site of this exon, as well as the effects of SF2/ASF, hnRNP A1, and a Tra2-β1-dependent exonic enhancer on these events. Our results suggest that both U2AF and U2 snRNP binding can be control points in SMN splicing regulation.
RESULTS
U2AF binding to SMN1 and SMN2 RNAs
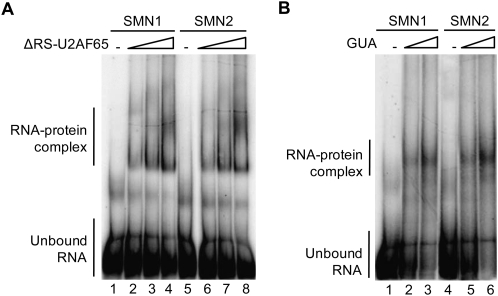
It has been proposed that the higher level of exon 7 inclusion in SMN1 transcripts compared with SMN2 transcripts is due to more efficient recruitment of U2AF to the SMN1 intron 6 3′ splice site (Kashima and Manley 2003; Cartegni et al. 2006). To test this hypothesis, we first confirmed that recognition of SMN1 and SMN2 intron 6 by U2 snRNP requires U2AF (Supplemental Fig. 1), as is the case for most, but not all 3′ splice sites (MacMillan et al. 1997; Lutzelberger et al. 2005; Sridharan and Singh 2007). Second, we tested whether the U2AF intrinsic binding affinity is higher for SMN1 than for SMN2 transcripts. Radioactively labeled RNAs comprising the 3′ 68 nucleotides (nt) of SMN intron 6, SMN1, or SMN2 exon 7 (54 nt) and 25 nt of intron 7 were incubated with purified recombinant U2AF65, or partially purified U2AF heterodimer from HeLa cells, in the presence of an excess of unrelated RNA (tRNA). RNA–protein complexes were then resolved by electrophoresis in native gels. To avoid nonspecific RNA–protein interactions mediated by the positively charged RS domain expressed in Escherichia coli (Lee et al. 1993; Singh et al. 1995), a U2AF65 derivative lacking the RS domain region was used in these experiments. As expected from the reported binding specificity of U2AF for polypyrimidine tracts (Singh et al. 1995), U2AF65 and the partially purifed U2AF heterodimer displayed comparable apparent binding affinities for the SMN1 and SMN2 RNAs utilized, which differ only at position 6 of exon 7 (Fig. 1A,B).
FIGURE 1.
Intrinsic U2AF affinity is similar for SMN1 and SMN2. 32P-radioactively labeled SMN1 or SMN2 exon 7 pre-mRNAs containing the 3′ 68 nt of intron 6, exon 7 sequences, and 25 nt of intron 7, were incubated with increasing concentrations (30, 90, and 270 nM) of recombinant U2AF65 lacking the RS domain (ΔRS–U2AF65) (A) or endogenous U2AF partially purified from HeLa nuclear extract (GUA-guanidine eluate from oligo-dT cellulose columns used to prepare U2AF-depleted extracts) (B). Complexes were assembled on ice and RNA–protein complexes fractionated by electrophoresis on native polyacrylamide gels at 4°C. The positions of unbound–RNA and RNA–protein complexes are indicated.
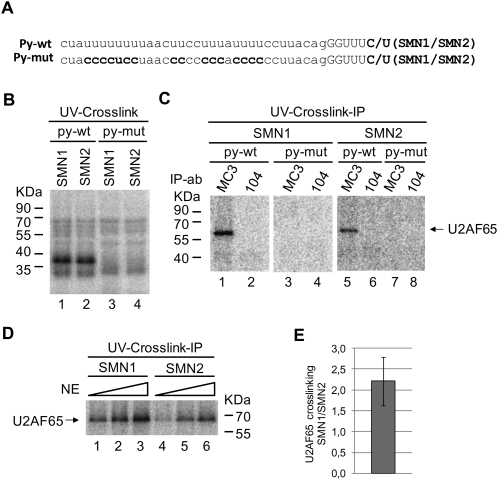
Next, the possibility that additional factors, differentially recruited to SMN1 and SMN2 RNAs, influence U2AF binding was addressed using ultraviolet light (UV)-induced crosslinking/immunoprecipitation. The 32P-uridine labeled RNAs described above were incubated with HeLa nuclear extract, irradiated with short wavelength UV-light, and after RNase A digestion, U2AF65 was immunoprecipitated using the specific MC3 monoclonal antibody (Gama-Carvalho et al. 1997). Radioactively labeled U2AF65 was identified by electrophoresis on SDS gels and PhosphorImager analysis. First, the specificity of U2AF65 crosslinking for the polypyrimidine tract of intron 6 was assessed using mutant versions of the RNAs in which uridine residues in the region upstream of exon 7 were replaced by cytidines (Fig. 2A, Py-mut). The results of Figure 2C indicate that U2AF65 crosslinking to both SMN1 and SMN2 wild-type transcripts was readily detectable, while crosslinking to the py-mut RNAs was not, indicating that the crosslinking signal reflects specific interaction of U2AF65 with the Py-tract region of SMN1 and SMN2 intron 6. To verify that the crosslinking/immunoprecipitation assay was quantitative, we performed the assay using increasing concentrations of nuclear extract. The results of Figure 2D show that increasing concentrations of nuclear extract resulted in increased levels of U2AF65 crosslinking to SMN1 and SMN2 RNAs, indicative of the quantitative nature of the assay. Importantly, the extent of U2AF65 crosslinking to SMN1 was consistenly higher than to SMN2 RNA (Fig. 2D, cf. lanes 1 and 4, cf. lanes 2 and 5, cf. lanes 3 and 6). Because the overall pattern of protein crosslinking in nuclear extract is comparable for both RNAs (Fig. 2B), the difference observed in U2AF65 crosslinking is specific for the interaction between this protein and SMN1 and SMN2 transcripts. Figure 2E quantifies the results of several independent experiments confirming the approximately twofold higher levels of crosslinking of U2AF65 to the SMN1 than to the SMN2 intron 6 3′ splice site. Time course experiments indicated that the differences in U2AF65 crosslinking are not due to different kinetics of association of U2AF65 with SMN1 and SMN2 transcripts (data not shown).
FIGURE 2.
Higher levels of U2AF65 crosslinking to SMN1 intron 6 polypyrimidine-tract. (A) Sequences of the SMN intron 6 3′ splice site region and a mutant derivative (Py-mut) in which uridine residues at the polypyrimidine tract (Py-tract) were replaced by cytidines. Exon 7 position 6, different between SMN1 (C) and SMN2 (T) transcripts is also indicated. (B) Pattern of proteins from HeLa nuclear extracts crosslinked to SMN transcripts upon irradiation with UV light. 32P-uridine-labeled RNAs (SMN1 or SMN2, wild type or Py-mut, as indicated) containing the 3′ 68 nt of intron 6, exon 7 sequences, and 25 nt of intron 7 were incubated with nuclear extracts, and after UV irradiation and RNAse treatment, 10% of the crosslinked material was fractionated on a SDS-polyacrylamide gel and analyzed by PhosphorImager. (C) U2AF65 immunoprecipitation. 90% of the products of crosslinking obtained as in B were immunoprecipitated using the anti-U2AF65 antibody MC3, or a control monoclonal antibody (104). Immunoprecipitated materials were fractionated on SDS denaturing gels and analyzed by PhosporImager. The positions of molecular weight markers and U2AF65 are indicated. (D) UV-crosslinking and U2AF65 immunoprecipitation with increasing amounts of nuclear extracts (NE). Assays were carried out as in (C) using 1.3%, 4%, and 12% of nuclear extracts. (E) Ratio of U2AF65 crosslinking to SMN1 versus SMN2 transcripts, quantified by PhosphorImager analyses, corresponding to three independent experiments. Error bar represents standard deviation.
We note that differences in U2AF65 crosslinking to SMN1 and SMN2 transcripts were detected in extracts that also showed differences in the relative inclusion of exon 7 between SMN1 and SMN2 in in vitro splicing assays (using minigenes spanning SMN genomic sequences between exon 6 and exon 8) (Supplemental Fig. 2; Cartegni and Krainer 2002). Nuclear extracts that, for unknown reasons (but unrelated to the relative levels of U2AF, SF2/ASF, or hnRNP A1), failed to recognize SMN1 or SMN2 exon 7, displayed equivalent levels of U2AF65 crosslinking to SMN1 and SMN2 RNAs (Supplemental Figs. 2, 3). This correlation suggests that differential U2AF recruitment to intron 6 is relevant for differential exon 7 recognition in SMN transcripts.
U2 snRNP assembly on SMN1 and SMN2 RNAs
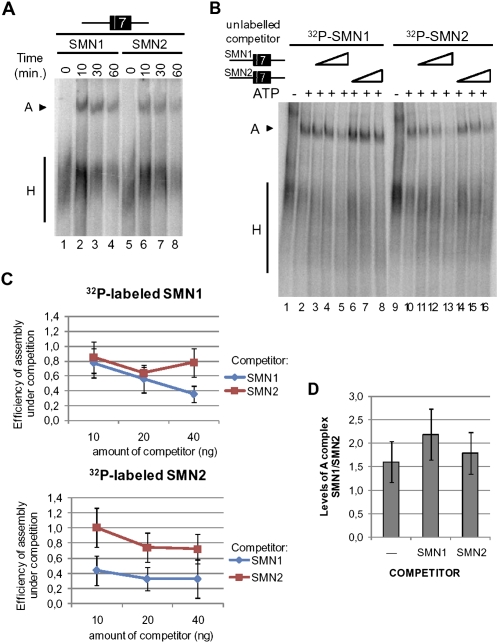
To investigate whether the differential binding of U2AF65 to SMN1 and SMN2 transcripts has subsequent effects on the assembly of other spliceosomal components around SMN exon 7, the same radioactively labeled RNAs were incubated with nuclear extract under splicing conditions and after heparin treatment, splicing complexes were resolved by electrophoresis on native polyacrylamide/agarose gels. The extent and kinetics of ATP-dependent complex A formation (which reflects stable binding of U2 snRNP on to the 3′ splice site region) showed an average difference of 1.6-fold between SMN1 and SMN2 (Fig. 3A, D, lane 1, P value=0.03). This difference is somewhat lower than the difference observed for U2AF65 crosslinking to these transcripts (Fig. 2).
FIGURE 3.
U2 snRNP recruitment to SMN1 and SMN2 RNAs. (A) Time-course of spliceosome assembly on SMN1 and SMN2 exon 7 pre-mRNAs. RNAs containing the 3′ 68 nt of intron 6, exon 7, and 25 nt of intron 7 were incubated with HeLa nuclear extracts under splicing conditions and after 30 min complexes were analyzed by electrophoresis on native polyacrylamide/agarose gels. The positions of A complex (U2 snRNP binding to intron 6 3′ splice site region) and H complex (hnRNP proteins bound to the same RNA) are indicated. (B) Spliceosome assembly under conditions of competition. Assays were set up as in A in the absence or presence of 10, 20, or 40 ng of the indicated unlabeled RNAs. (C) Quantification of the results of independent competition experiments carried out as in B. The efficiency of complex A formation (A/H ratios) on 32P-labeled SMN1 or SMN2 in the presence of competitor RNAs relative to assembly in the absence of competitor is represented for four independent experiments. (D) Relative levels of A complex formation on SMN1 versus SMN2. Ratios of A/H complex formation on SMN transcripts in the absence or presence of competitor RNAs are represented. Bars indicate standard deviations. P values for the null hypothesis that complex A assembly is identical on SMN1 and SMN2 are 0.03 (in the absence of competitor) and lower than 0.001 (in the presence of either competitor).
We next considered the possibility that differences in spliceosome assembly could become more prominent under conditions of competition for factors involved in U2 snRNP binding, which may mimic more accurately the conditions of splice-site competition operating during splice-site selection. Spliceosome assembly assays were therefore carried out in the presence of increasing amounts of unlabeled competitor. The results of Figure 3B and the quantifications of Figure 3C indicate that SMN1 RNA is a better competitor than SMN2 RNA in this assay: unlabeled SMN1 competed more efficiently than SMN2 the assembly of complexes on either radiolabeled-SMN1 or -SMN2 RNAs (Fig. 3B, levels of A complex, cf. lanes 5 and 8, cf. lanes 11 and 14, cf. lanes 12 and 15, cf. lanes 13 and 16; Fig. 3C, stronger reduction in assembly caused by competition, cf. SMN1 [blue line] and SMN2 [red line]). Under conditions of competition with SMN1 RNA, the intrinsic difference in U2 snRNP recruitment between SMN1 and SMN2 RNAs was enhanced and became more significant (Fig. 3D, P < 0.001). We conclude that U2 snRNP assembly is more efficient on SMN1 than on SMN2 transcripts, particularly under conditions of limited availability of assembly factors.
Regulation of U2 snRNP recruitment after U2AF binding
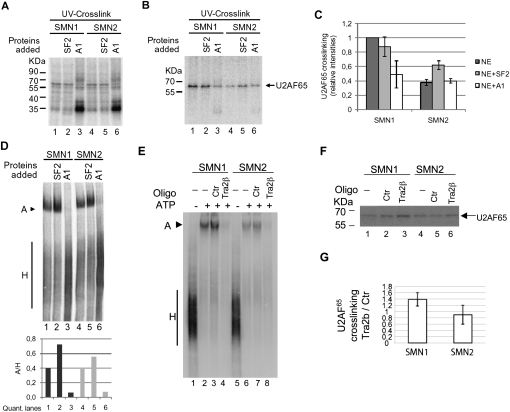
Previous work implicated the splicing regulatory factors SF2/ASF and hnRNP A1 in the differential use of exon 7 in SMN1 and SMN2: the C to T nucleotide difference at exonic position 6 has been proposed to cause the loss of a SF2/ASF-bound enhancer or/and the gain of a hnRNP A1-bound silencer in SMN2 transcripts, consequently leading to enhanced exon skipping (Cartegni and Krainer 2002; Cartegni et al. 2006; Kashima and Manley 2003; Kashima et al. 2007b). We therefore tested the effect of increasing the levels of each of these splicing regulators on U2AF65 crosslinking to SMN1 and SMN2 RNAs. Figure 4A shows the overall pattern of protein crosslinking. Addition of recombinant purified SF2/ASF did not increase U2AF65 crosslinking to the SMN1 intron 6 3′ splice site region (Fig. 4B, lanes 1,2), while it slightly increased crosslinking of U2AF65 to SMN2 transcripts (Fig. 4B, lanes lanes 4,5). In contrast, hnRNP A1 reduced U2AF65 crosslinking to SMN1 (Fig. 4B, cf. lanes 1 and 3), but not to SMN2 RNAs (Fig. 4B, cf. lanes 4 and 6). These effects were reproduced in independent experiments and the quantification of U2AF65 crosslinking, normalized to the levels of SMN1 crosslinking in the absence of added factors, is presented in Figure 4C. We conclude that SF2/ASF and hnRNP A1 display differential effects on U2AF65 crosslinking, and that these effects depend on the nature of the 3′ splice site of SMN1 and SMN2 intron 6 (see Discussion).
FIGURE 4.
Differential effects of SF2/ASF and hnRNPA1 on U2AF65 crosslinking and spliceosome assembly to SMN transcripts. (A) Pattern of proteins crosslinked to SMN transcripts upon addition of recombinant purified SF2/ASF or hnRNP A1 (0.4 pmol/μL). (B) Crosslinking/immunoprecipitation of U2AF65 in assays as in A, carried out as in Figure 2C. (C) PhosphorImager quantification of signals corresponding to U2AF65 precipitates obtained in independent experiments as in B. (D) Spliceosomal complexes assembled on SMN transcripts in the absence or presence of recombinant SF2/ASF or hnRNP A1, as in B. Complexes assembled on SMN1 and SMN2 RNAs after incubation with nuclear extract and the indicated proteins for 30 min were resolved by electrophoresis on native polyacrylamide/agarose gels. Signals from A and H complexes were quantified by PhosphorImager analysis and the ratios represented at the bottom of the figure. (E) Spliceosomal complexes assembled on SMN transcripts in the presence of 125 nM of a 2′-O-methyl oligonucleotide complementary to the Tra2-β1 enhancer located in exon 7 positions 21–35 (5′-GAGCACCTTCCTTCT-3′) or a control (Ctr) oligonucleotide (5′-ATTAGTGGAATTGGC-3′), analyzed as in D. (F) Crosslinking/immunoprecipitation of U2AF65 in assays as in E, carried out as in Figure 2C. (G) Ratio between U2AF65 crosslinking in the presence of Tra2-β1 enhancer antisense and control oligos from PhosphorImager quantification of signals corresponding to U2AF65 precipitates obtained in three independent experiments as in F.
Next, we tested the effects of increasing the levels of SF2/ASF and hnRNP A1 on the complexes assembled on SMN1 and SMN2 transcripts. Addition of recombinant SF2/ASF resulted in increased levels of A complex on both SMN1 and SMN2 transcripts, while addition of recombinant hnRNP A1 resulted in decreased levels of A complex on both transcripts (Fig. 4D).
Taken together, the results above indicate that regulatory factors can target both U2AF65 binding as well as subsequent steps in U2 snRNP assembly. To further explore this concept, we used an antisense 2′-O-methyl oligonucleotide complementary to SMN exon 7 positions 21–35; antisense oligonucleotides bound to this region have been shown to block the function of a splicing enhancer (Hua et al. 2007) recognized by Tra2-β1, SRp30C, hnRNP G, and possibly other factors (Hofmann et al. 2000; Young et al. 2002; Hofmann and Wirth 2002). Addition of this oligonucleotide, but not of a control, caused efficient inhibition of U2 snRNP binding to both SMN1 and SMN2 RNAs (Fig. 4E). This was not accompanied by reduced U2AF65 crosslinking (Fig. 4F, G), once again suggesting that certain regulatory factors and sequences modulate U2 snRNP recruitment to SMN transcripts at a step subsequent to U2AF binding to the Py-tract.
DISCUSSION
Despite extensive analysis of cis-acting elements and trans-acting factors relevant for SMN splicing, little was known about how the critical nucleotide difference at position 6 of exon 7 influences recognition of the upstream 3′ splice site of SMN1 and SMN2 transcripts by the basic splicing machinery. This is, however, a relevant question to understand the more efficient inclusion of exon 7 in SMN1 transcripts and eventually to design strategies that can stimulate exon inclusion in SMN2 transcripts. Such strategies have significant therapeutic promise based upon results obtained in vitro and in animal models of SMA (Cartegni and Krainer 2003; Skordis et al. 2003; Madocsai et al. 2005; Coady et al. 2007; Hua et al. 2008).
Our results indicate that the C to U transition at exon 7 position 6 decreases U2AF association to the intron 6 3′ splice site in SMN2 transcripts by about twofold (Fig. 2). This difference in U2AF recruitment in vitro may represent a significant alteration in the balance of splice-site choice under in vivo conditions of 3′ splice site cotranscriptional competition, and therefore be responsible for the substantial difference in exon 7 inclusion between SMN1 and SMN2 transcripts observed in vivo. Differential U2AF recruitment correlates with a 1.6-fold lower efficiency of U2 snRNP assembly on SMN2 transcripts (Fig. 3). The difference in the fold reduction of U2AF65 crosslinking and A complex formation may reflect the different set up of the two assays: U2 snRNP assembly is monitored by the retarded electrophoretic mobility of heparin-treated complexes, while U2AF association is monitored by UV-light-induced protein–RNA crosslinking, which is likely to provide a snapshot of the occupancy of the RNA by U2AF65.
Alternatively, U2AF65 binding may not be rate-limiting for U2 snRNP recruitment to the SMN intron 6 3′ splice site (Hastings et al. 2007), and therefore the observed differences in U2AF65 binding may not fully determine the extent of U2 snRNP recruitment. U2 snRNP binding involves base-pairing interactions between U2 snRNA and the branch point sequence that require U2AF (Valcárcel et al. 1996). Stable U2 binding, however, requires a number of additional factors and ATP-driven conformational changes that may not directly depend on the levels of U2AF binding. In this model, the higher efficiency of SMN1 exon 7 inclusion could be related to differences in later events in spliceosome assembly or even catalysis, consistent with observations in other systems (Lallena et al. 2002; Lim and Hertel 2004; House and Lynch 2006; Bonnal et al. 2008; Kotlajich et al. 2008; Sharma et al. 2008).
The differences in U2 snRNP association to the SMN1 and SMN2 intron 6 3′ splice sites became more obvious under conditions of limited availability of splicing factors (using an excess of competing RNA substrates) (Fig. 3B,C). Although it is unclear how these conditions relate to the physiological situation in the cell nucleus, it is possible that distinct features of U2 snRNP assembly on SMN1 and SMN2 revealed by the competition assays of Figure 3C play a role in the efficiency of exon 7 utilization under conditions of cotranscriptional assembly in vivo. Taken together, the results of UV crosslinking and spliceosome assembly assays suggest that both U2AF binding and U2 snRNP assembly on SMN1 and SMN2 transcripts are distinguishable, at least under particular experimental conditions.
Previous work implicated the SR protein SF2/ASF and the hnRNP A1 protein in the differential splicing of SMN transcripts. One model proposed that the C to T +6 transition leads to loss of a SF2/ASF-dependent splicing enhancer in SMN2 exon 7 RNAs (Cartegni and Krainer 2002; Cartegni et al. 2006). The second, not necessarily mutually exclusive, model proposed the gain of a hnRNP A1-dependent splicing silencer (Kashima and Manley 2003; Cartegni et al. 2006; Kashima et al. 2007b). One simple mechanism for the function of these factors would be that their association with SMN1 or SMN2 exon 7 promotes or represses recognition of the 3′ splice site by U2AF. This model predicts that SF2/ASF would stimulate U2AF binding to SMN1, but not to SMN2 transcripts, while hnRNP A1 would decrease U2AF binding to SMN2, but not to SMN1 RNAs. The results of Figure 4, however, show the opposite trend: increasing the levels of SF2/ASF increases U2AF65 crosslinking to SMN2, but not to SMN1 transcripts; and increased levels of hnRNP A1 reduce crosslinking of U2AF65 to SMN1, but not to SMN2 RNAs. One possible explanation for these results is that SF2/ASF binding to SMN1, and that of hnRNP A1 to SMN2, are saturated (Cartegni et al. 2006), and therefore, that increasing their levels in the assays does not further enhance the difference in U2AF65 association to SMN1 and SMN2 detected in our crosslinking assays under normal conditions. Increasing the levels of SF2/ASF could, however, enhance its association with SMN2 transcripts (for which the presence of uridine at position +6 represents a suboptimal SF2/ASF binding site), perhaps decreasing hnRNP A1 binding, and thus increasing U2AF65 recruitment. Conversely, increasing the levels of hnRNP A1 could enhance its association with SMN1 transcripts (for which the cytidine at position 6 represents a suboptimal binding site), perhaps decreasing SF2/ASF binding and decreasing U2AF65 recruitment to these RNAs.
The results of Figure 4D indicate that SF2/ASF stimulates, while hnRNP A1 decreases, U2 snRNP binding to both SMN1 and SMN2. SF2/ASF-dependent stimulation of U2 recruitment to SMN2 transcripts (Fig. 4D) correlates with enhanced U2AF binding (Fig. 4B,C), whereas, stronger U2 recruitment to SMN1 transcripts does not. These results suggest that an excess of SF2/ASF stimulates other steps in SMN1 3′ splice site recognition by U2 snRNP. Similarly, decreased U2 recruitment to SMN1 transcripts by hnRNP A1 (Fig. 3C) correlates with reduced U2AF binding (Fig. 3B,C), while the decrease in U2 binding to SMN2 does not. The latter result suggests that other steps in U2 snRNP binding would be decreased by hnRNP A1 in SMN2 RNAs, as observed for other transcripts (Tange et al. 2001). A possible common explanation for these observations would be that the effects of SF2/ASF and hnRNP A1, when mediated through sequences involving position +6, affect steps in U2 snRNP recruitment other than U2AF binding. In contrast, effects mediated through other SF2/ASF and hnRNP A1 binding sites within exon 7 or flanking sequences influence U2 snRNP recruitment through modulation of U2AF binding.
The observation that an antisense oligonucletide that blocks the function of a Tra2-β1-dependent exonic enhancer inhibits U2 snRNP recruitment without affecting U2AF65 binding (Fig. 4E–G) is consistent with similar observations made for SR protein-dependent exonic enhancers (Shen et al. 2004).
Collectively, the results presented in this manuscript suggest that both U2AF binding and other steps required for stable assembly of U2 snRNP at the branch site can be affected by the single nucleotide difference in exon 7 position 6 between SMN1 and SMN2 transcripts. They also suggest that trans-acting factors like SF2/ASF and hnRNP A1 can target these different steps in a transcript-specific manner.
MATERIALS AND METHODS
RNAs
pCISMNxΔ-wt or pCISMNxΔ-c6t (Cartegni and Krainer 2002) were used to transcribe SMN1 and SMN2 RNAs. Py-mut constructs were obtained by replacing the thymidine residues contained in the polypyrimidine tract by cytidines via PCR-based site-directed mutagenesis of pCI plasmids as described by Hemsley et al. (1989) using 5′-phosphorylated oligonucleotides. Templates used to generate transcripts containing the 3′ 68 nt of intron 6, the complete sequence of exon 7 and 26 nt of intron 7 were generated by PCR using primers SMNCXLFor 5′-CGTAATACGACTCACTATAGGGCTATCTATATATAGCTATC-3′ and SMNCXLBack 5′-CACTTTCATAATGCTGGCAG-3′. RNAs were synthesized from 400 ng of PCR templates in 25 μL transcription reactions containing 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2, 2 mM spermidine, 0.8 mM DTT, 0.4mM ATP, CTP, and GTP, 0.08 mM UTP, 36 U of T7 RNA polymerase and 50 μCi [α-32P]UTP (Amersham). Full-length substrates were transcribed in the presence of 1.9 mM CAP-analog (m7G[5])ppp[5′]G) (New England Biolabs) and GTP was at 0.04 mM final concentration. Reactions were incubated for 2 h at 37°C and transcription products were gel-purified.
Protein expression
GST-U2AF65, SF2/ASF, and hnRNP A1 were expressed in and purified from E. coli using previously published procedures (Mayeda and Krainer 1992; Valcárcel et al. 1996) and gel-quantified by comparing dilutions of the preparations to a BSA standard.
In vitro splicing and spliceosome assembly assays
Pre-mRNA substrates (10 fmol) were incubated in 10 μL reaction mixtures containing HeLa nuclear extract, 32 mM HEPES (pH 7.9), 1.56 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate, and 2.6% polyvinyl alcohol. For in vitro splicing assays, reactions were incubated at 30°C for 2 h; RNA was extracted and analyzed on 9% denaturing polyacrylamide gels. Gels were exposed to a PhosphorImager screen. For spliceosome assembly analysis, reactions were incubated at 30°C for 30 min, unless otherwise stated, and then treated with heparin (5 μg/μL). Complexes were separated on 4% acrylamide:bisacrylamide (80:1)–0.5% agarose gels in 50 mM Tris base-50 mM glycine buffer. Dried gels were exposed to a PhosphorImager screen.
UV crosslinking and immunoprecipitation of U2AF65
Pre-mRNAs were incubated under the same conditions as described for in vitro splicing and spliceosome assembly assays, except that the reaction volume was increased to 25 μL. After 30 min of incubation at 30°C, samples were UV-irradiated (254 nm, 0.4J, 1.7 cm from light source) and treated with RNase A (1 μg/μL, final concentration) at 37°C for 30 min. A 10% aliquot of the crosslinked samples was loaded onto a 10% SDS-PAGE. To immunoprecipitate U2AF65, 50 μL of anti-U2AF65 monoclonal antibody, MC3 (Gama-Carvalho et al. 1997), was added to 90% of the crosslinked material, and 0.1 M KCl Buffer D (20 mM HEPES [pH 8.0], 0.5 mM EDTA, 20% glycerol, 1 mM DTT, 0.05% NP-40) was added to a final volume of 120 μL. Monoclonal antibody 104 (Roth et al. 1990) was used for mock-immunoprecipitation. Antibody and antigen were allowed to react on ice for 1 h with periodic swirling. After addition of 50 μL of a 1:1 slurry of Protein A + Protein G Sepharose 4 Fast Flow (at 1:1 proportion; Amersham) in low salt buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40), samples were incubated at 4°C with rotation for 1 h Beads were washed three times with high salt buffer (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1% NP-40) and three times with low salt buffer and resuspended in 10 μL of SDS-loading dye and boiled for 5 min. After centrifugation, the supernatant was loaded on a 10% SDS-polycrylamide gel. Dried gels were exposed to a PhosphorImager screen and signals quantified with ImageQuant TL software (Amersham Biosciences).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
M.M.d.A. was supported by a fellowship from the Portuguese Foundation of Science and Tecnhology (FCT), from the Graduate Programme in Areas of Basic and Applied Biology (GABBA). Work in J.V.’s laboratory is supported by grants from AICR, Ministerio de Educación y Ciencia, EURASNET and Fundación Marcelino Botín. Work in A.R.K.’s laboratory was supported by NIH Grant GM42699.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1273209.
REFERENCES
- Blanchette M., Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S., Martínez C., Förch P., Bachi A., Wilm M., Valcárcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol. Cell. 2008;32:81–95. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Brzustowicz L.M., Lehner T., Castilla L.H., Penchaszadeh G.K., Wilhelmsen K.C., Daniels R., Davies K.E., Leppert M., Ziter F., Wood D., et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2–13.3. Nature. 1990;344:540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Hastings M.L., Calarco J.A., De Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew S.L., Liu H.X., Mayeda A., Krainer A.R. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc. Natl. Acad. Sci. 1999;96:10655–10660. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady T.H., Shababi M., Tullis G.E., Lorson C.L. Restoration of SMN function: Delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol. Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- Eperon I.C., Makarova O.V., Mayeda A., Munroe S.H., Caceres J.F., Hayward D.G., Krainer A.R. Selection of alternative 5′ splice sites: Role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M., Krauss R.D., Chiang L., Valcárcel J., Green M.R., Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz A.K., Feng W., Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Guth S., Martinez C., Gaur R.K., Valcárcel J. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol. Cell. Biol. 1999;19:8263–8271. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M.L., Allemand E., Duelli D.M., Myers M.P., Krainer A.R. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS One. 2007;2:e538. doi: 10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M.D., Cortopassi G., Galas D.J. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y., Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-β1. Hum. Mol. Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl. Acad. Sci. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House A.E., Lynch K.W. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat. Struct. Mol. Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan J.L., Green M.R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes & Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- Kashima T., Rao N., Manley J.L. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. 2007a;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T., Rao N., David C.J., Manley J.L. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 2007b;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- Kotlajich M.V., Crabb T.L., Hertel K.J. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in A complex. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.01071-08. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena M.J., Chalmers K., Llamazares S., Lamond A.I., Valcárcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNδ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Lee C.G., Zamore P.D., Green M.R., Hurwitz J. RNA annealing activity is intrinsically associated with U2AF. J. Biol. Chem. 1993;268:13472–13478. [PubMed] [Google Scholar]
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lim J.R., Hertel K.J. Commitment to splice site pairing coincides with A complex formation. Mol. Cell. 2004;15:477–483. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Lin S., Fu X.D. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzelberger M., Backstrom E., Akusjarvi G. Substrate-dependent differences in U2AF requirement for splicing in adenovirus-infected cell extracts. J. Biol. Chem. 2005;280:25478–25484. doi: 10.1074/jbc.M413737200. [DOI] [PubMed] [Google Scholar]
- MacMillan A.M., McCaw P.S., Crispino J.D., Sharp P.A. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. 1997;94:133–136. doi: 10.1073/pnas.94.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madocsai C., Lim S.R., Geib T., Lam B.J., Hertel K.J. Correction of SMN2 pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol. Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Mailman M.D., Heinz J.W., Papp A.C., Snyder P.J., Sedra M.S., Wirth B., Burghes A.H., Prior T.W. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Melki J., Sheth P., Abdelhak S., Burlet P., Bachelot M.F., Lathrop M.G., Frezal J., Munnich A. Mapping of acute (type I) spinal muscular atrophy to chromosome 5q12–q14. The French Spinal Muscular Atrophy Investigators. Lancet. 1990;336:271–273. doi: 10.1016/0140-6736(90)91803-i. [DOI] [PubMed] [Google Scholar]
- Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Roth M.B., Murphy C., Gall J.G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J. Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russman B.S. Spinal muscular atrophy: Clinical classification and disease heterogeneity. J. Child Neurol. 2007;22:946–951. doi: 10.1177/0883073807305673. [DOI] [PubMed] [Google Scholar]
- Sharma S., Kohlstaedt L.A., Damianov A., Rio D.C., Black D.L. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Green M.R. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Shen H., Kan J.L., Green M.R. Arginine-serine-rich domains bound at splicing enhancers contact the branch-point to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.N., Androphy E.J., Singh R.N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcárcel J., Green M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Singh R.N. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- Skordis L.A., Dunckley M.G., Yue B., Eperon I.C., Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan V., Singh R. A conditional role of U2AF in splicing of introns with unconventional polypyrimidine tracts. Mol. Cell. Biol. 2007;27:7334–7344. doi: 10.1128/MCB.00627-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T.O., Damgaard C.K., Guth S., Valcárcel J., Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 2001;20:5748–5758. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserant A., König H. Signal regulated pre-mRNA occupancy by the general splicing factor U2AF. PLoS One. 2008;3:e1418. doi: 10.1371/journal.pone.00001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J., Gaur R.K., Singh R., Green M.R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosome structure and function. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. RNA world III. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. pp. 369–400. [Google Scholar]
- Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 β1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Krainer A.R. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes & Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mayeda A., Krainer A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- Zuo P., Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]