Abstract
Under stress, cells need to optimize the activity of a wide range of gene products during the response phases: shock, adaptation, and recovery. This requires coordination of several levels of regulation, including turnover and translation efficiencies of mRNAs. Mitogen-activated protein (MAP) kinase pathways are implicated in many aspects of the environmental stress response, including initiation of transcription, translation efficiency, and mRNA turnover. In this study, we analyze mRNA turnover rates and mRNA steady-state levels at different time points following mild hyperosmotic shock in Saccharomyces cerevisiae cells. The regulation of mRNA stability is transient and affects most genes for which there is a change in transcript level. These changes precede and prepare for the changes in steady-state levels, both regarding the initial increase and the later decline of stress-induced mRNAs. The inverse is true for stress-repressed genes, which become stabilized during hyperosmotic stress in preparation of an increase as the cells recover. The MAP kinase Hog1 affects both steady-state levels and stability of stress-responsive transcripts, whereas the Hog1-activated kinase Rck2 influences steady-state levels without a major effect on stability. Regulation of mRNA stability is a wide-spread, but not universal, effect on stress-responsive transcripts during transient hyperosmotic stress. By destabilizing stress-induced mRNAs when their steady-state levels have reached a maximum, the cell prepares for the subsequent recovery phase when these transcripts are to return to normal levels. Conversely, stabilization of stress-repressed mRNAs permits their rapid accumulation in the recovery phase. Our results show that mRNA turnover is coordinated with transcriptional induction.
Keywords: mRNA turnover, Saccharomyces cerevisiae, stress-activated MAP kinase, HOG pathway
INTRODUCTION
In order to maintain viability and proliferation during increasing turgor and concomitant molecular crowding, cells need to recognize and rapidly adapt to changes in extracellular osmolarity. In the yeast Saccharomyces cerevisiae, the required adaptation to hyperosmosis is mainly initiated by the high osmolarity glycerol (HOG) pathway. The HOG pathway consists of a stress-activated mitogen-activated protein (MAP) kinase (SAPK) core module upon which two independent upstream branches, the Sln1 and the Sho1 branches, converge. Activation of any of these branches by hyperosmotic stress leads to rapid phosphorylation of the MAP kinase kinase Pbs2, which in turn phosphorylates and activates the MAP kinase Hog1. In the initial shock phase following mild salt stress, a major fraction of phosphorylated Hog1 quickly (within 1 min) translocates to the nucleus (Maeda et al. 1994), where it resides for about 10 min. In the adaptation phase, nuclear phosphorylated Hog1 in turn induces, as well as represses, transcription of osmoresponsive genes (Gasch et al. 2000; Posas et al. 2000; Rep et al. 2000; Causton et al. 2001; Krantz et al. 2004) through interactions with different transcription factors, including Hot1, Sko1, and Smp1 (Rep et al. 2000; Proft et al. 2001; de Nadal et al. 2003), as well as through recruitment of the Rpd3 histone deacetylase (de Nadal et al. 2004). The genes induced by Hog1 are involved in stress defense processes, such as production of the osmolyte glycerol (Albertyn et al. 1994), ion homeostasis (Marquez and Serrano 1996), and redox metabolism (Schüller et al. 1994; Krantz et al. 2004). By contrast, genes repressed by Hog1 are mainly involved in translation, ribosome biogenesis, and amino acid synthesis (Mager and Varela 1993; Gasch et al. 2000). These responses essentially reflect a redirection of cellular resources from growth to stress adaptation. The levels of the stress-responsive mRNAs change transiently with a timing that depends on the severity of the stress (Rep et al. 1999). After treatment with 0.7 M NaCl, the levels of induced mRNAs peak after 45 min (Rep et al. 2000), while after 0.4 M of NaCl stress the peak appears as early as after 10 min (Posas et al. 2000). After adaptation, cells eventually resume growth in the recovery phase, which is characterized by lower levels of stress proteins and increased translational activity (Warner 1999).
Under hyperosmotic stress, Hog1 also phosphorylates substrates not known to have a role in transcription. Within 5 min of 0.4 M NaCl treatment, activated Hog1 targets the plasma membrane ion channels Nha1 and Tok1 (Proft and Struhl 2004). Cell cycle arrest is mediated by activated Hog1 after 10 min of treatment with 0.4 M NaCl in G1 through phosphorylation of the CDK inhibitor Sic1, or in G2 (Escote et al. 2004) through phosphorylation of the protein kinase Hsl1 (Clotet et al. 2006), respectively. Furthermore, activated Hog1 phosphorylates the MAP kinase activated kinase (MAPKAP kinase) Rck2 (Bilsland-Marchesan et al. 2000), which has been implicated in regulation of translation (Teige et al. 2001; Swaminathan et al. 2006). Both Hog1 and Rck2 contribute to cellular hyperosmotic and oxidative stress tolerance (Brewster et al. 1993; Bilsland et al. 2004; Swaminathan et al. 2006). In other eukaryotes, there is also evidence for the involvement of SAPK homologs in translational control. Thus, activation of the mammalian SAPK p38 stimulates translation of tumor necrosis factor α mRNA (Kontoyiannis et al. 2001; Hitti et al. 2006). The fission yeast SAPK, Sty1, binds to translation factors (Asp et al. 2008), and sty1 mutants have defects in recovery of translation after stress (Dunand-Sauthier et al. 2005; Asp et al. 2008).
Messenger RNA levels are determined not only by transcription rates but also by degradation rates. Hence, regulation of degradation rate in response to different stimuli is a potentially important stress response mechanism. For example, mammalian gene transcripts encoding interleukins and containing A/U-rich elements (AREs), which are involved in inflammation, are stabilized following signaling from the p38 or JNK SAPK pathways (Winzen et al. 1999; Chen et al. 2000). Similarly, the mammalian Rck2 homolog MAPKAPK-2 regulates mRNA stability through phosphorylation of the ARE-binding protein tristetraproline (Hitti et al. 2006). The stability of large functional mRNA groups can be co-regulated via mRNA sequence elements, such as the iron responsive element (IRE) in yeast (Puig et al. 2005).
Genome-wide analysis of intrinsic mRNA stability has been performed in S. cerevisiae using microarrays (Wang et al. 2002; Grigull et al. 2004; Duttagupta et al. 2005; Puig et al. 2005). In this study, we have used microarrays to investigate global changes in transcript stability in response to mild (0.4 M) NaCl stress in yeast. We find that the changes in mRNA stability during the adaptation and recovery phases are of a magnitude that could account for a major fraction of the overall changes in mRNA steady-state levels. The stability changes are most pronounced in the latest (recovery) phase of the stress response. During this phase, previously induced mRNAs encoding proteins involved in stress survival undergo a distinct loss of stability. Conversely, mRNAs that are transcriptionally repressed in the early adaptation phase, principally encoding growth-related functions, e.g., protein translation, are markedly stabilized in the recovery phase. Thus, stability changes precede changes in steady-state mRNA levels. We also find that most of these changes are dependent on Hog1 and, in some cases, on Rck2.
RESULTS
Several gene categories are affected at the stability level upon osmotic stress
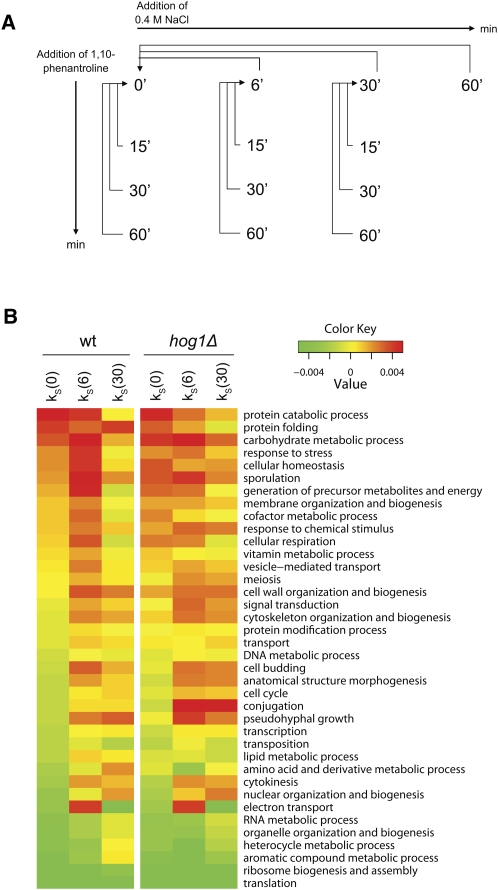
Transcript steady-state levels are determined not only by transcription rate but also by mRNA stability. The importance of transcriptional regulation in response to salt stress has been well studied. In order to investigate the importance of a regulation of mRNA stability in response to stress, we collected global data on mRNA stability and steady-state levels from unstressed cells and after mild hyperosmotic (0.4 M NaCl) shock (Fig. 1A). 1,10-Phenanthroline (Phen) blocks de novo synthesis of transcripts by inhibiting RNA polymerase II (Brown 1994). Hence, changes in transcript levels in Phen-treated cells may be assumed to be due exclusively to transcript degradation (Rodriguez-Gabriel et al. 2003; Grigull et al. 2004; Lackner et al. 2007). On the basis of mRNA levels measured at different time points after Phen treatment, relative slope coefficients representing stability were calculated (stability indices, k S) (see Materials and Methods). Positive and negative k S values indicate transcripts more stable and unstable than the average transcript. Positive differences in stability indices (Δk S) indicate stabilization (with an unknown scale since absolute half-lives were not measured), while negative differences indicate destabilization. Genome-wide stability indices (k S) were calculated in unstressed cells and after 6 and 30 min after NaCl exposure. The amplitudes of the Δk S indicate that the overall contribution of stress-induced changes in mRNA stability on steady-state levels is considerable (see Materials and Methods, section Modeling mRNA Stability).
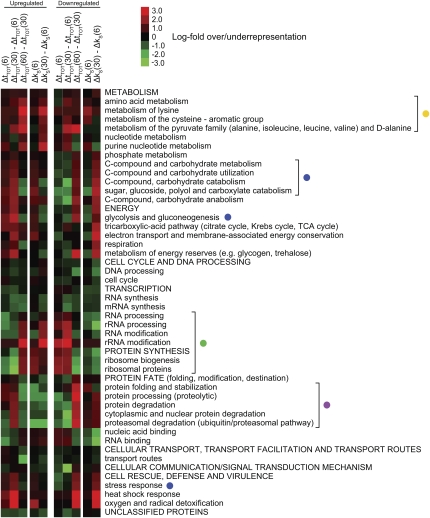
FIGURE 1.
Several functional transcript categories are regulated at the stability level after salt stress. (A) Study design. Transcript steady-state levels were investigated at 6, 30, and 60 min after 0.4 M NaCl shock. Stability was investigated at the time points 0, 6, and 30 min by the addition of Phen. Aliquots were harvested at 15, 30, and 60 min after Phen addition to monitor mRNA decay and stability indices (k S) were calculated. (B) mRNA stability in functional categories before and after stress. All 38 GO Slim broad functional categories (biological process) were ranked from most (top) to least stable (bottom) based on the mean k S of unstressed wt cells (left-most column). Unstressed wt and hog1Δ cells have similar mean k S for most categories. After 6 min of stress, categories that are initially stable tend to get more stabilized in the wild type, while initially unstable categories are further destabilized. After 30 min, the reverse is true, as stabilized categories are destabilized and vice versa. In the hog1Δ mutant, both steps in this response at the stability level are less pronounced.
Figure 1B shows the average stability indices (k S) for the 38 Gene Ontology (GO) Slim categories of biological processes in wild-type (wt) and hog1Δ cells before and during stress. In wt cells, the mean mRNA stability of several GO Slim categories increases after 6 min of stress, whereas after 30 min of stress almost all of these stabilized categories are destabilized to a level similar to or even below the original. These stabilized categories already tend to have a relatively high mean stability before stress. This includes processes such as “response to stress,” the “carbohydrate metabolic process,” and “cellular homeostasis.” On the other hand, transcripts belonging to categories that already are unstable in unstressed conditions, such as “ribosome biogenesis and assembly” (RiBi) (Jorgensen et al. 2004), and the “RNA metabolic process,” increase in stability in the later phase after stress. Before stress, wt and hog1Δ cells have very similar stability profiles (Fig. 1B). After stress, however, the mRNA stability response of the hog1Δ mutant clearly is weaker than that of the wt. Stability in the category “conjugation” stands out as misregulated in the hog1Δ mutant, indicating that the improper activation of the mating pheromone MAPK pathway observed in this mutant (O'Rourke and Herskowitz 1998) also extends to the mRNA stability regulation level.
Changes in mRNA stability precede changes in steady-state levels after hyperosmotic shock
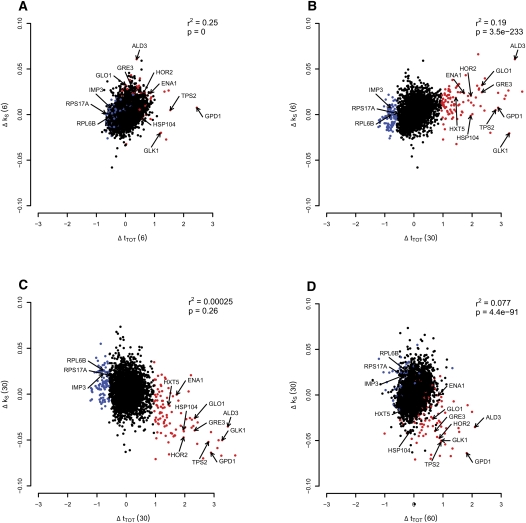
Since there was a sharp decrease in mean stability after 30 min of stress of virtually all initially stabilized categories, indicating a narrow time frame of regulation, the temporal relationship between salt-induced changes in transcript stability (Δk S) and changes in steady-state transcript levels (Δt TOT) were investigated (Fig. 2A–D). We marked the 100 most up-regulated genes at 30 min in red in Figure 2, A–D. This group is dominated by genes known to be functionally important in the salt response (GO overrepresentation, Fisher's exact test: stress response [P = 3 × 10−16], carbohydrate metabolic process [P = 6 × 10−6], and sodium ion transport [P = 0.0027]).
FIGURE 2.
Transcript regulation at the stability level precedes the regulation at the steady-state level. Scatter plots of changes in mRNA stability (Δk S) versus changes in steady-state levels (Δt TOT) at various times after stress. The 100 most highly induced genes after 30 min at the steady-state level [Δt TOT (30)] are colored in red, and the 100 most down-regulated genes at the same time point are colored blue. Ten well-known stress-responsive genes and three down-regulated genes required for protein synthesis are indicated by arrows (including the six genes verified by qPCR: HOR2, GRE3, GPD1, IMP3, RPS17A, and RPL6B). The alterations in steady-state levels and mRNA stability correlate positively when comparing stability after 6 min [Δk S (6)] with steady-state levels after 6 min [Δt TOT (6)] (A), as well as with steady-state levels after 30 min [Δt TOT (30)] (B). The change in stability after 30 min [Δk S (30)] was not globally correlated with the difference in steady-state levels after 30 min [Δt TOT (30)] (C), but a group of outliers consisting of the most induced genes at the steady-state level (marked in red) show an inverse relationship, indicating that these salt-responsive genes are now destabilized. After 60 min of stress, the steady-state levels are lower for the salt-responsive genes (D). For information about the correlation test, see Materials and Methods.
Interestingly, the relative contribution of changes in transcript stability to the overall changes in transcript levels showed drastic temporal variations following salt addition. After 6 min of salt adaptation, a strong positive global correlation between Δt TOT and Δk S was observed (Fig. 2A), suggesting that changes in transcript stability account for a large fraction of the total salt-induced changes in transcript levels. Stability changes after 6 min also correlated positively with steady-state changes after 30 min, indicating that early stabilization influences later steady-state level changes (Fig. 2B), which demonstrates an expected lag in the effects of stabilizing mRNAs.
In contrast to the findings above, no correlation between Δk S and Δt TOT was seen after 30 min of salt adaptation (Fig. 2C), nor when comparing Δk S (30) to Δt TOT (60) min (Fig. 2D). Interestingly, however, at 30 min a group of genes corresponding to the red-colored salt-induced genes (including well-known salt targets such as the glycerol dehydrogenase GPD1, the glycerol phosphatase HOR2, and the aldose reductase GRE3) are still induced on a steady-state level, but now display a strongly reduced stability. This suggests that regulation of steady-state levels has a temporal dependency to mRNA stability through the phases of shock, adaptation, and recovery, and that increased mRNA degradation underlies the decline from the peak of steady-state levels in the late recovery phase. The indicated genes would correspond to transcripts that are required at high levels for salt adaptation, but not for recovery from salt stress. Out of the 100 most steady-state-induced genes at 30 min, 81 have significantly (moderated t-test: P < 0.05) reduced transcript stability at the same time point, indicating that a reduction in stability is the most common fate among this group of mRNAs. Also, the changes in stability between 6 and 30 min correlate with the changes in steady-state levels between 30 and 60 min (Spearman rank correlation r 2 = 0.149, Spearman rank test P = 4 × 10−176) (data not shown), strongly indicating that late in the salt response, mRNA turnover is a major factor driving changes in mRNA levels.
The 100 most down-regulated genes at Δt TOT (30) are colored blue in Figure 2, A–D. This group includes ribosomal protein genes and the RiBi regulon encoding nucleolar proteins involved in ribosome biogenesis (GO overrepresentation, Fisher's exact test: translation [P = 3 × 10−3], ribosome biogenesis and assembly [P = 3 × 10−14]). These genes show an inverted, but less pronounced behavior compared to the up-regulated transcripts (Fig. 2, in red). Among these are the genes encoding the ribosomal proteins Rps17A and Rpl6B and the rRNA processor Imp3, for which the expression profiles were confirmed with quantitative PCR (qPCR) (Fig. 6C, see below).
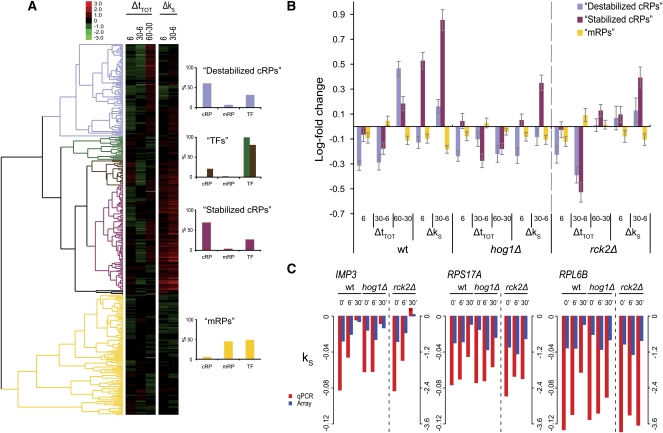
FIGURE 6.
Cluster analysis of the GO Slim functional category translation. (A) The genes in the GO Slim category Translation were hierarchically clustered according to transcript behavior in the wild type, and the Δk S values were multiplied by 30 as in Fig. 5 (genes with >1 missing value omitted). Five clusters were defined, highly enriched for cytoplasmic ribosomal proteins (cRPs), mitochondrial ribosomal proteins (mRPs), and translation factors (TFs), respectively, as shown in bar diagrams to the right. Note that the composition of clusters 2 and 3 are shown in the same diagram. (B) The mean log fold change at the steady-state and stability levels of the three RP-enriched clusters are plotted. The mean changes across the conditions were calculated for wt, hog1Δ, and rck2Δ cells. The mRPs cluster show only small changes during stress, while the cRPs are divided into two clusters, which differ in their regulation at the steady-state as well as the stability level. Especially the recuperation in the recovery phase is affected in both mutants to a similar extent. Error bars denote 95% confidence intervals. (C) The stability indices of two cRPs (RPS17A and RPL6B) were confirmed by qPCR, along with a gene in the RiBi group (IMP3). Spearman rank correlation between array and qPCR data: 0.9 (see Materials and Methods).
Hence, at 6 min, transcription initiation and transcript turnover are in phase, stabilizing and transcribing transcripts critical for adaptation, whereas at 30 min the same transcripts are no longer stabilized, while still at a high steady-state level. Transcript turnover is at this time already preparing the entry into the recovery phase, suggesting a turning point of the response between 6 and 30 min.
Hog1 influences both early and late changes in mRNA stability
It is well established that the major part of the transcriptional initiation response following osmostress is mediated via the HOG pathway and its key component Hog1 (Posas et al. 2000; Proft et al. 2001; de Nadal et al. 2004). To investigate to what extent Hog1 also controls the regulation of transcript degradation during osmoadaptation, we compared the pattern of changes in transcript stability following 0.4 M NaCl stress in wt and hog1Δ mutant cells.
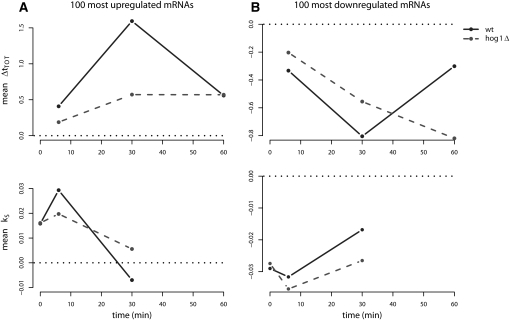
Confirming the function of Hog1 in transcriptional adaptation, we find that the average Δt TOT (30) of the 100 most induced salt-responsive genes is significantly (Student's t-test, P = 3 × 10−27) lower in hog1Δ than in wt cells (Fig. 3A, upper panel). Correspondingly, for the 100 most repressed genes, the average Δt TOT (30) is significantly higher in the hog1Δ mutant (Student's t-test, P = 6.9 × 10−11) (Fig. 3B, upper panel). Interestingly however, at 60 min the levels of these down-regulated genes have recuperated in the wt, while in the hog1Δ mutant, the levels keep declining until significantly lower than in the wt (Student's t-test, P = 5 × 10−25). Hog1 also has a clear effect on mRNA stability, especially with regard to the up-regulated genes. While similar in stability in unstressed cells, after 6 min of salt stress the top 100 salt-induced genes (Fig. 3A, lower panel) are stabilized in the wt but significantly less so in the hog1Δ mutant (Student's t-test, P = 2 × 10−4). Even more apparent is the difference in destabilization of these genes taking place between 6 and 30 min (Student's t-test, P = 1.6 × 10−7). As for the top 100 salt repressed genes, hog1Δ displays only a marginally enhanced destabilization after 6 min (Student's t-test, P = 0.037; Fig. 3B, lower panel), while the later stabilization is somewhat more affected (Student's t-test, P = 5.5 × 10−5). Hence, it is clear that Hog1 plays a critical role not only in regulating transcription initiation, but also transcript degradation rate in response to salt exposure, and most specifically so with regard to transcripts that are in high demand during the salt adaptation phase.
FIGURE 3.
Hog1 affects both steady-state levels and stability. Average stability index (k S) and steady-state log-fold changes (Δt TOT) of the 100 most up-regulated (A) and down-regulated (B) genes at the steady-state level at 30 min in the wild type against time in wt and hog1Δ cells. Zero on the y-axis is indicated by a reference line (dotted black). These groups of genes display a distinct temporal pattern where changes in stability precede changes at the steady-state level. Hog1 influences both the changes in stability and at the steady-state level for both groups. Two-sample t-tests were used to calculate P-values for the average differences between the strains in all panels (see Materials and Methods).
The functional roles of mRNAs constrain their temporal profiles with regard to stability and steady-state levels
The temporal profile of stability and steady-state levels led us to define the intervals between 0 and 6 min (the “shock” phase), 6 and 30 min (the “adaptation” phase), and 30 and 60 min (the “recovery” phase). In order to investigate the impact of changing mRNA turnover rates in different functional categories in more detail, we utilized the highly resolved GO tree of the MIPS functional catalog (http://mips.gsf.de/genre/proj/yeast/), and examined the 200 most affected genes in the different phases with respect to Δk S and Δt TOT (Fig. 4). All categories exhibiting a significant enrichment in any of the intervals were included to get an overview of how the different categories behaved during the stress response. The categories that display the turning-point behavior seen in Figure 3A include “C (carbon)-compound and carbohydrate metabolism,” “glycolysis and gluconeogenesis,” and “stress response” (Fig. 4, blue dots). The transcriptional induction of genes in these categories during salt stress is well established. Carbohydrate metabolism, glycolysis and gluconeogenesis are important both for the production of the osmolytes glycerol and trehalose, as well as for glycogen, and for regulation of the energy needs of the cell. Stress response includes genes involved in sodium transport, redox metabolism, and heat shock proteins, all of which are known to be up-regulated after salt stress, but also several genes included in the carbohydrate metabolism processes mentioned above. Protein degradation (Fig. 4, violet dot) and related categories display a turning-point behavior at the steady-state level, while there is an immediate decrease in stability during the shock phase, which is continued into the adaptation phase. A plausible explanation is that the initial increase in steady-state levels is accomplished through transcription without an accompanying increase in stability. As can be seen in Figure 1B, the initial mean stability for the corresponding GO Slim category (“protein catabolic process”) is very high, possibly negating a need for further stabilization in response to stress. An inverse behavior is seen for the categories involved in ribosome biogenesis (see below). Categories involved in amino acid metabolism (Fig. 4, yellow dot) show a tendency to be up-regulated in the recovery phase. It has previously been shown that salt stress causes starvation for amino acids through inhibition of their uptake, which in turn induces genes involved in their biosynthesis (Norbeck and Blomberg 1998; Pandey et al. 2007). The categories “DNA processing” and “cell cycle” are underrepresented throughout the response, indicating that those gene products are not in increased demand during the response. Categories involved in protein synthesis, such as rRNA processing, RiBi, and ribosomal proteins (Fig. 4, green dot) mirror the profiles seen in Figure 3B, although an early destabilization in the shock phase is not so apparent. Together, these observations imply that the temporal dependency of the steady-state levels to the stability regulation is a part of the response of several functional categories, first and foremost those involved in carbohydrate metabolism, response to stress, amino acid synthesis, protein degradation, and protein synthesis.
FIGURE 4.
Functional categories have different temporal regulation profiles at the steady-state and the stability levels. Enrichment analysis (using hypergeometrical distribution) of functional categories on the 200 most up-regulated (left panel) and down-regulated (right panel) transcripts in the different phases of the response: the shock phase (0–6 min), the adaptation phase (6–30 min), and the recovery phase (30–60 min) in the wt. In each panel, the first three lanes show changes in steady-state levels, and the last two lanes show changes in stability. Only the categories significantly over- or underrepresented in any of the conditions (P-value < 0.01, Bonferroni corrected) were included in the plot. Red indicates overrepresentation and green indicates underrepresentation. Functional categories emphasized in the text are bracketed and marked with colored dots.
A subgroup of stress response genes is responsible for the distinct turning-point behavior
Hierarchical clustering of all 5551 genes reveals a subgroup of 338 genes with a turning-point behavior similar to the one seen in Figure 3A (data not shown). This is the most striking subgroup, with the biggest response magnitude both with respect to the steady-state and stability levels. GO enrichments include most categories mentioned above, including the “trehalose metabolic process” (P = 9.6 × 10−7), catabolic process (P = 4.8 × 10−5), and response to stress (P = 0.0008).
To provide a higher resolution of the stress-responsive transcripts, the 430 genes of the GO Slim category response to stress, encompassing a wide diversity of functions, were hierarchically clustered according to their profiles during the different phases at the steady-state as well as the stability levels in the wt and shown in Figure 5A. A group of 113 genes are responsible for the “turning-point” profile, while the rest of the genes show only a small or no response. This cluster consists of two subgroups that mainly differ in that one “early stabilized” cluster is stabilized in the shock phase, while the other, “destabilized” cluster is destabilized in the shock phase. The early-stabilized cluster contains well-known osmotic stress response genes such as the methylglyoxal reductase GRE2, the glycerol phosphatase RHR2, the aldehyde dehydrogenase ALD3, the dihydroxyacetone kinase DAK1, and the trehalose phosphate phosphatase TPS2. The destabilized cluster contains genes less well known in this context, but includes several chaperone-coding genes, such as SSA1, HSP104, HSP78, and HSC82 (protein folding; GO enrichment, Fisher's exact test, P = 0.007).
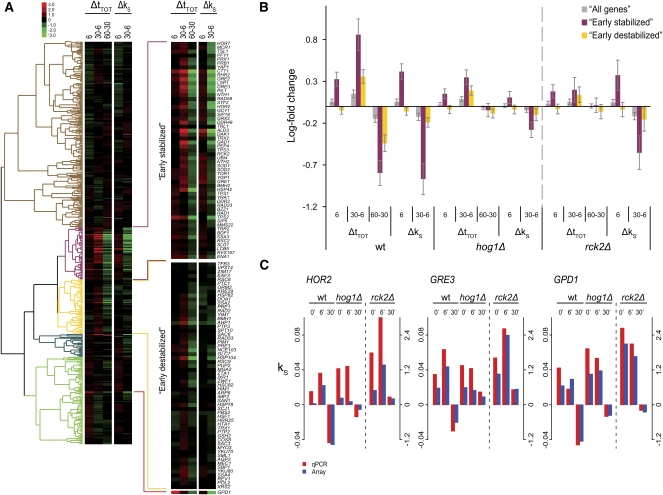
FIGURE 5.
Cluster analysis of the GO Slim category Response to stress. (A) The genes in the GO Slim category response to stress were hierarchically clustered (uncentered Pearson correlation metric) with respect to the behavior of the transcripts in the wt during the phases of salt stress response. Genes with more than one missing value were omitted. To obtain value ranges comparable between steady state and stability, the stability indices were multiplied by 30 to approximate the log-fold differences 30 min after transcription inhibition (see Materials and Methods). The two clusters (“Early stabilized” and “Early destabilized,” shown magnified to the right) include genes responsive to 0.4 M NaCl stress. GPD1 fell outside the two clusters because of its fast up-regulation at the steady-state level. (B) Mean changes in steady-state and stability levels were calculated for the two clusters defined in (A) across the strains (wt, hog1Δ, and rck2Δ) shows that Hog1 influences both steady-state levels and stability, especially for the early stabilized cluster, while Rck2 mainly influences steady-state levels without major effects on stability. Error bars denote 95% confidence intervals. (C) Stability indices were confirmed by qPCR for three genes (HOR2, GRE3, and GPD1). HOR2 and GRE3 are part of the early-stabilized cluster, while GPD1 fell outside the clusters. Spearman rank correlation between array and qPCR data: 0.9 (see Materials and Methods).
The mean steady-state and stability changes in all phases were calculated for the two clusters for wt and hog1Δ, as well as for rck2Δ (Fig. 5B). In the wt, the early stabilized cluster has a stronger response at all time points, especially on the stability level where it displays a very clear turning-point behavior between 6 and 30 min. A similar turning-point behavior was observed on the steady-state level between 30 and 60 min, again reflecting the temporal difference between stability and steady-state level regulation. Hog1 affects both levels of regulation for this cluster, while in rck2Δ mutants, effects are seen mainly on the steady-state level. Contrary to expectations, this links Rck2 to transcription rather than stability regulation. This is especially true regarding the shock and adaptation phases, as the destabilization during the recovery phase is only mildly affected in the rck2Δ mutant. The stability indices and steady-state levels of three genes involved in the stress response (HOR2, GRE3, and GPD1) were confirmed with qPCR (Fig. 5C). Agreement between the qPCR data and the microarray data is very good, and the turning-point behavior, which is severely hampered in the mutants for these three genes, is clearly seen. qPCR confirmation of the steady-state level changes is also well in agreement with microarray data (Supplemental Fig. S3), although the microarray data give consistently lower responses (“signal compression,” a well-known phenomenon) (see Materials and Methods).
In addition, the GO Slim categories “carbohydrate metabolism,” “amino acid metabolism,” and “proteasomal catabolism” were clustered (Supplemental Fig. S4A–C). All three categories contain groups of stress-responsive genes with an increase in steady-state levels along with a distinct destabilization during the adaptation phase (a turning-point behavior). The turning-point cluster from carbohydrate metabolism contains 28 genes, which are involved mainly in trehalose metabolism (trehalose metabolic process, Fisher's exact test, P = 1.35 × 10−16) and glucose transport (P = 3.55 × 10−5). This cluster shares 11 genes with the two clusters from response to stress of which nine are from the early stabilized cluster. The category amino acid metabolism mainly contains transcripts, which are weakly stabilized in the adaptation phase and up-regulated in the recovery phase (which is the behavior seen in Fig. 4), but a cluster of 33 genes instead has a turning-point profile. GO enrichment analysis of this group as compared to the rest of the category reveals that several of the genes in this group are involved in catabolic processes (catabolic process, P = 1.44 × 10−10) including the amino acid derivative catabolic process (P = 1.66 × 10−6), proline catabolic process (P = 4.91 × 10−5), and acetyl-CoA catabolic process (P = 6.8 × 10−5). A large cluster from protein catabolism (67 genes) is homogenous but does not have a prominent early stabilization. See Supplemental Table S5 for cluster gene lists.
Increased transcript stability contributes to restoration of ribosomal components in the recovery phase after NaCl exposure
Genes with ribosomal functions display quick and strong variations in transcript levels during the salt stress response phases (Gasch et al. 2000; Yale and Bohnert 2001). Our data show heterogeneity within this functional group with respect to changes in transcript stability (Δk S) and transcript steady-state levels (Δt TOT) concerning timing and strength of the shifts. A hierarchical clustering was performed of the GO Slim category “translation,” which contains 137 cytoplasmic (cRP) and 77 mitochondrial (mRP) ribosomal proteins along with slightly more than a hundred different translation factors and other proteins involved in the process of translation (all denoted TFs). The clustering resolved the translation genes into three large and two smaller groups with different expression profiles during the three phases of stress (Fig. 6A; see Supplemental Table S5 for lists of genes within each cluster). The mean Δt TOT and Δk S of the three large clusters across the different phases in wt, hog1Δ, and Δrck2 cells were calculated (Fig. 6B). The first cluster (“destabilized cRPs”) contained mainly cRPs and TFs and displayed an initial destabilization in the shock phase with a later increase in stability in the adaption phase. At the steady-state level, this cluster is repressed in the shock as well as in the adaption phase in the wt, while it is clearly up-regulated between 30 and 60 min, presumably reflecting the increase in stabilization. The cluster “stabilized cRPs” contains most of the remaining cRPs, along with a smaller proportion of TFs. This cluster is immediately stabilized in the wt, and even more so between 6 and 30 min, while the changes at the steady-state levels are less pronounced than for the destabilized cRPs cluster. Presumably this strong stabilization cluster corresponds to cRPs that are critical for the very early recovery from salt stress. The cluster “mRPs” contains about equal amounts mRPs and TFs but almost no cRPs. This cluster displays only marginal changes in stability and steady-state levels during the stress response, which is in line with earlier observations of mRP mRNA level responses to other stresses (Ihmels et al. 2002; Ihmels et al. 2005; Swaminathan et al. 2006).
The behavior of both cRP clusters was partially dependent on a functional HOG pathway. This is most apparent in the recovery phase where both the stabilization and the steady-state level induction are affected in both hog1Δ and rck2Δ, although the failure to up-regulate the steady-state level in the recovery phase is more apparent in the hog1Δ mutant. Hence, at least a part of the cRP mRNA regulation following NaCl exposure is dependent on a functional HOG pathway. To verify the expression profiles of cRPs following salt exposure, we quantified the decay rates of two cRP transcripts, RPS17A and RPL6B, independently and individually using qPCR. The relative trends were essentially confirmed, showing a marked stabilization of both transcripts at 30 min after salt exposure (Fig. 6C). Although these two genes belong to the destabilized cRP cluster, neither appeared destabilized in the shock phase either by arrays or qPCR. Instead, both displayed a very small stabilization. The qPCR data show that both these genes appear to be slightly more stable in the mutants in unstressed cells, but are destabilized during the shock phase in both hog1Δ and rck2Δ mutants. During the adaptation phase, however, they are not stabilized to the extent of the wt. IMP3, a gene belonging to the functional category ribosomal biogenesis and assembly (RiBi) was also included in the qPCR analysis. Genes belonging to RiBi behaved in the wt essentially like translation (named ribosomal proteins in the MIPS functional catalog) (Figs. 3, 4), with a strong stabilization in the adaptation phase, but IMP3 does not seem to be affected in a hog1Δ mutant, except that the initial stability appears to be higher, as is the case for the cRPs.
DISCUSSION
Stability regulation precedes changes in steady-state levels
We have found that almost all genes known to be induced by salt shock at the steady-state level are also regulated at the stability level, suggesting that regulation of mRNA stability is a general and integral part of the regulation of transcript levels, and that most stress-responsive mRNAs have a dynamic range of half-lives that can be modulated to fit the situation, in line with earlier studies of the response to diauxic shift (Garcia-Martinez et al. 2004). The changes in stability can be fast and dramatic, exemplified by such well-known salt stress induced genes as GPD1, GRE3, and HOR2, which are stabilized early on after stress (0–6 min, the shock phase) and then greatly destabilized between 6 and 30 min (the adaptation phase). In fact, the relative stability of many salt-induced genes drop from being among the most stable in the shock phase to being among the least stable in the entire transcriptome during the adaptation phase. The steady-state levels of these genes only start to fall after 30 min (the recovery phase), indicating that the regulation at the stability level precedes the changes at the steady-state level. Genes that are repressed at the steady-state level display an inverse pattern with a similar temporal difference in regulation. It might be argued that within the population of induced transcripts, those species transcribed early would age and become susceptible to later degradation. However, this would not provide a satisfactory explanation for the behavior of repressed transcripts, which instead become stabilized late in the response. A recent study (Shalem et al. 2008) where fast destabilization of induced mRNAs was also observed, led to the seemingly paradoxical conclusion that changes in transcript abundance and stability in yeast cells were counterdirectional at a particular time point in two stress conditions. We believe that this paradox is resolved by considering the delay that occurs between a change in stability and resulting change in steady-state level for an mRNA: A stability change at an earlier time point is homodirectional with a change in abundance at a later time, as seen in our work.
It is important to emphasize, however, that although the phenomenon of a contribution from stability changes to the stress-induced increases or decreases in steady-state level is wide-spread, it does not apply to all stress-regulated mRNAs. This is noticeable for certain functional groups: among the stress-induced genes, there is no stability increase in the early destabilized cluster (Fig. 5). Similarly, for the cluster enriched for gene products with a role in protein catabolism, there is no mRNA stabilization despite a marked increase in steady-state mRNA levels (Supplemental Fig. S4).
Functional characteristics of the mRNA turnover response
In response to salt stress, the cell up-regulates the expression of genes involved in processes such as glycerol and trehalose metabolism, glycolysis and ion homeostasis, in order to relieve the stress. mRNAs encoding proteins involved in all these processes display the behavior seen for the above-mentioned genes, GPD1, HOR2, and GRE3, suggesting that the processes that are induced have an as well-defined and common regulation at the mRNA stability level as they do at the level of transcription initiation. Additionally, groups of genes with a role in amino acid metabolism and protein catabolism show a similar behavior. The pattern of an increase in steady-state levels and a decrease in stability during the adaptation phase followed by a decrease in steady-state levels in the recovery phase is common to all these genes, but there are differences in the stability regulation during the shock phase. This could reflect a difference in the timing or the intensity of transcriptional activity during the shock phase.
The production of osmolytes potentially involves several parts of the cellular metabolism; aside from the direct production of glycerol and trehalose, sugar transport and glycolysis, and thus, energy metabolism is affected since the metabolites involved are in constant flux. This should also affect amino acid synthesis, which shares much of the same metabolites, and this may be the explanation why amino acid catabolic processes are transiently up-regulated in response to stress. The slow up-regulation and stabilization of the majority of the mRNAs annotated to amino acid synthesis might reflect a recovery process, or it could reflect the amino acid starvation induced by hyperosmotic stress caused by an impaired ability to import amino acids (Norbeck and Blomberg 1998; Pandey et al. 2007). A group of mRNAs involved in protein catabolism is transiently up-regulated, but not stabilized in response to stress. This could be an effort to remove damaged or unwanted proteins, or a response to amino acid starvation, elevating recycling of protein. The early destabilization might indicate that changes in transcription rate are more important than changes in stability for this group. It should be noted that this group of genes have a very high initial stability (before stress), which might mean that transcripts can accumulate even though the stability declines.
In response to osmotic stress, the cell down-regulates the steady-state levels of transcripts involved in protein synthesis, cRP, and RiBi mRNAs. These transcripts are stabilized during the adaptation phase and their steady-state levels increase during the recovery phase. The cRPs follow a common trend, but differ considerably with respect to the amount and timing of stabilization. The fact that heterogeneity exists among the cRPs indicates that functional differences exist within this group, where a specific subgroup could be required early in recovery.
Generally, categories that are up-regulated during stress such as response to stress and carbohydrate metabolism tend to have a high mean stability already before stress. We find that categories with a high relative stability in the unstressed condition (most “stress genes”) tend to get further stabilized in the shock phase, while categories with a low stability before stress (most growth-related genes including cRP and RiBi) tend to get destabilized.
A recent paper (Molina-Navarro et al. 2008) estimated the regulation of transcriptional rate during oxidative stress in S. cerevisiae (“genomic run-on”), and from this inferred the regulation at the stability level. The authors demonstrated that regulation of mRNA stability was a wide-spread feature of the response also to that stress condition. The results concerning genes involved in stress response and ribosome biogenesis were well in agreement with our direct measurements, although in our case the switch from adaptation to recovery (the turning point) is more dramatic. This could be because of different stress conditions, of time frames, or as a consequence of the methodology used. Block of RNA polymerase II, as in this paper, provides direct estimates of mRNA degradation rates, while genomic run-on only provides an indirect measure of transcript turnover. The authors express concern that the combination of Phen with stress would not be feasible. However, the apparent specificity of the regulation of stability in response to stress in our experiments, and the elimination from our analysis of genes previously shown to be Phen dependent (Grigull et al. 2004), give confidence that the use of Phen as a transcription inhibitor allows for a sensitive and convenient method of measuring mRNA stability, also in the presence of stress.
Hog1 dependency
Previously it has been shown that the osmostress-induced MAPK Hog1 affects mRNA levels through regulation of transcription. We have shown that Hog1 also regulates the stability of almost all transcripts that are up-regulated at the steady-state level after 30 min, both during the shock and adaptation phases. Hog1 also influences stability of the down-regulated transcripts, although to a lesser degree. The MAPKAP kinase Rck2 was suspected to be involved in stability regulation since the mammalian homolog MAPKAPK-2 is involved in the regulation of interleukin mRNA stability. Surprisingly however, Rck2 seems to influence steady-state levels of stress-responsive transcripts, with only minor effects on stability. This suggests a role for Rck2 in transcriptional regulation rather that stability regulation. It is noteworthy that the transcriptional response was abolished in rck2Δ mutants after hyperosmotic shock (this work), but not in oxidative stress (Swaminathan et al. 2006), indicating that the requirement for Rck2 is specific for the hyperosmotic transcriptional response.
Future perspectives
Degradation of mRNA is accomplished through several different mechanisms and regulated through mRNA binding proteins such as the poly A-binding proteins Pab1 and Pub1 and the RNA-binding proteins Puf1-5. mRNA degradation is also tightly coupled to translation, acting as a regulatory mechanism for mRNAs that are not properly translated. Since mammalian p38 acts on mRNA stability through RNA binding proteins like TTP and Hur, it is conceivable that this applies also to Hog1. Transcripts binding to both Pub1 and Puf1-5 have been investigated (Gerber et al. 2004; Duttagupta et al. 2005), but we have not found any obvious connection to our data, except that RiBi mRNAs are bound by both Puf4 and Pub1. If a primary RNA binding protein is the Hog1-targeted effector of mRNA stability regulation, it might be feasible to investigate sequences of, first and foremost, the untranslated regions of the mRNAs affected during stress. Another possibility is that Hog1 primarily affects the translation of the stress-responsive mRNAs and the stability only as a consequence of that. MAP kinases have been shown to regulate translation through phosphorylation of the initiation factor eIF2α and the elongation factor EF2, and might in this manner influence stability. A study of translational regulation of mRNAs following hyperosmotic shock in S. cerevisiae has recently been performed (Melamed et al. 2008). However, a preliminary comparison of that data set with ours shows a low degree of overlap in the response profile of steady-state total mRNA (not shown). This indicates that the conditions studied in their work (1 h after exposure to high salinity at 1 M NaCl) are too different from ours to allow a detailed analysis of this issue, especially in view of the strict time dependence revealed in this work.
Studies of mRNA stability provide insight into yet another layer of the complex regulatory network between gene and protein production. Together with investigations of transcription activity and translation, a picture can be shaped of the intricate and interconnected pattern of processes helping the cell to regulate its protein activity in its interaction with the environment.
CONCLUSIONS
Changes in mRNA stability are known to be an important regulatory mechanism in yeast, as well as in mammals. Here we performed global measurements of mRNA stability changes in the course of the response to hyperosmotic stress in S. cerevisiae. We show that regulation of stability makes general and substantial contributions to the changes in the steady-state level during transient stress. For certain stress-regulated functional categories, changes in mRNA stability are prominent, whereas for others, steady-state levels are determined solely by transcription rate. The changes at the stability level of stress-induced genes precede and anticipate the changes at the steady-state level, and display a distinct turning point before the peak of the steady-state levels. Finally, the MAP kinase Hog1 is required for most early as well as late changes in stability.
MATERIALS AND METHODS
Strains and growth conditions
S. cerevisiae strains used in the study were from the W303-1A background (wt genotypes: MAT a ura3-1, ade2-1, leu2-3,112, trp1-1, his3-11,15; mutant genotypes: W303-1A, hog1∷KanMX; W303-1A, rck2∷KanMX), and stored in 20% glycerol at −80°C. Cells were precultivated for 24 h in rich medium (YPAD, 1.5% peptone, 1% yeast extract, 2% glucose), inoculated to A595 nm = 0.05–0.1 in fresh medium (as above) and cultivated for 6–8 h to A 595 nm = 0.7, whereupon NaCl was added to a final concentration of 0.4 M. Ten microliter samples were harvested by centrifugation before (0 min) as well as after 6, 30, and 60 min of NaCl exposure. Samples were divided in two; one of these was washed (twice) and RNA was isolated as below, in the other, transcription was stopped by the addition of the transcriptional inhibitor Phen (Sigma-Aldrich) to a final concentration of 100 ng/mL. No transcriptional inhibition was performed on samples collected 60 min post-NaCl shock. From transcriptionally inhibited samples, 2.5 mL aliquots were harvested by centrifugation and washing (twice) at 5, 15, 30, and 60 min (the 5 min time point was not included in the subsequent analysis due to the high variability of the measurements). For rck2Δ mutants, aliquots were harvested only 30 min after Phen addition. Cells were disrupted and RNA extracted using the RiboPure-yeast kit and instructions from the supplier (Applied Biosystems/Ambion).
Microarray hybridization
Labeling of samples and array hybridization were performed essentially as previously described (Bilsland et al. 2007). Briefly, cDNA was synthesized from 15 to 20 μg of RNA using Superscript III reverse transcriptase and an 1:1 mixture of random primers and anchored oligo dT (all from Invitrogen). Samples were labeled with Cy5-dUTP (reference) or Cy3-dUTP (experiment sample) (GE Healthcare). Post-labeling, RNA was hydrolyzed with NaOH and labeled cDNA probes were purified using a CyScribe GFX purification kit (GE Healthcare). Probes were dissolved in DIG Easy hybridization buffer (Roche) and hybridized onto Yeast 6.4K microarray slides (Microarray center, University Health Network) overnight at 42°C. After washing, slides were scanned in a VersArray Chipreader (Bio-Rad).
For quantification of steady-state transcript levels, RNA from NaCl-stressed (experiment) samples were paired with RNA from 0 min, unstressed (reference) samples. For quantification of transcript stabilities, RNA from each Phen time point (experiment) sample was paired with a matching 0 min (before Phen addition) sample (reference).
Microarray data processing
Array spot intensities were quantified using ImaGene v. 6.0.1 (BioDiscovery). Spots flagged as empty, poor, or negative by the ImaGene software were removed. Data analysis was performed using the open source statistical software R, and the LIMMA array analysis package, which is available at the Bioconductor repository (http://www.bioconductor.org/). A loess smoother was applied to remove intensity dependent trends. For each gene on each array, the log(2)-fold change (M-value) was calculated and retained for downstream analysis. To ensure comparability within groups of arrays, scale normalization was applied to create the same median-absolute-deviation (MAD) across arrays within a group. The normalization groups were (1) all arrays measuring non-Phen treated pools (steady-state levels); (2) arrays measuring pools within the same Phen time point across strains and stress conditions (i.e., three groups comprising arrays hybridized with mRNA from cells treated with Phen after 15, 30, and 60 min, respectively). Annotations for all open reading frames (ORFs) were taken from the Saccharomyces Genome Database (www.yeastgenome.org/; February 2008). All dubious and deleted ORFs were excluded (Supplemental Table S1). In addition, a set of 31 Phen-induced genes previously identified by Grigull et al. (2004) were removed in order to avoid inhibitor specific effects (Supplemental Table S2). Five thousand five hundred and fifty-one genes were retained.
Modeling mRNA stability
To model transcript stability, a simple exponential decay model was adopted, implying that a regression line through the origin can be fit to the log(2) scale M-values for individual mRNAs using the different time points (15, 30, and 60 min after Phen addition). Positive and negative values of the slope of the regression line (denoted k S and referred to as the stability index) indicate transcripts more stable and unstable than an average transcript. The stability indices range from −0.135 to 0.138 for the least and most stable transcripts, respectively, with median negative and positive values of −0.0045 and 0.0047.
Comparisons of different transcriptional inhibitors in yeast using microarrays have been published previously (Grigull et al. 2004). From this data set, arrays hybridized with Phen-treated pools of mRNA after 12, 30, and 75 min were downloaded (experimental design similar to the design in this study). All arrays were normalized in the same way as described above, and stability indices (k S) were calculated (only wild-type and unstressed conditions). Mean values for k S within 38 GO Slim biological process categories (see Functional Annotations) were found to be similar in the two studies (Supplemental Fig. S1; r 2 = 0.9).
For the rck2Δ mutant, samples were only collected after 30 min of Phen treatment, and a reduced decay model was therefore adapted. With the reduced model, stability is indicated by the log(2)-fold changes (M-values) as compared to the corresponding zero time point in all stress conditions. The reduced model was also adapted for the wild-type and hog1Δ strains for validation purposes. The full and reduced models in both strains were in good agreement (Supplemental Fig. S2).
Assessing the impact of a stability change after, for example, 30 min can be achieved by multiplying Δk S with 30. The magnitude of this entity represents a log(2)-fold change contributable to the alteration in stability for a given gene. For example, in Figure 3A (upper and lower panels), the change in stability from 6 to 30 min of stress in the wt contributes to a depletion of half of the transcripts for an average gene at the 60 min time point, as observed in the steady-state levels. This example shows that that the magnitude of fold changes induced by mRNA stability regulation is approximately at par with the changes at the steady-state level. Although the aim of this study was to compare relative changes and not to calculate absolute half-lives, the spread of our stability indices suggests that the average half-life would be less than 13.3 min in the wild type in the unstressed condition [max k S(0) for wt: 0.074], which is lower than the 23 min reported by Wang et al. (2002). We confirmed the expression levels and calculated stability indices for eight genes using qPCR. The trends were essentially confirmed although changes in k S values calculated from qPCR data were of higher amplitude than from the array data. Such signal compression is commonly seen when comparing DNA arrays to quantitative methods such as qPCR (Canales et al. 2006; Shi et al. 2006; Arikawa et al. 2008). The signal compression was also apparent at the steady-state level. This complicates the estimation of half-lives, indicating that the spread is, in fact, larger than suggested from the microarrays. Therefore, we use comparisons between stability indices for our investigations of the effect of NaCl stress on mRNA stability instead of absolute half-lives.
Statistical analysis
Differences between estimated coefficients were modeled and ranked using the moderated t-statistic (Smyth 2004) in both decay models and between estimated coefficients in the steady-state level data. Gene-specific variances were estimated using all available experiments.
Correlations between data sets were modeled and tested with the Spearman rank correlation test. The r 2-value, the coefficient of determination, is a measure of explanatory power of the chosen predictive variable.
Differences between mean values for groups of genes were tested with the Welch two-sample t-test.
Gene clustering
Clustering was performed using Cluster 3.0 (Stanford University) and an uncentered Pearson correlation metric (average linkage mapping) and visualized using JavaTreeview (University of Tokyo).
Comparisons with previously published data at the steady-state level
Comparisons were made with previously published data on steady-state level changes of mRNA. Lists of osmoregulated genes from five papers were compared (Posas et al. 2000; Rep et al. 2000; Causton et al. 2001; Yale and Bohnert 2001; Krantz et al. 2004), and the 73 genes that were found to be up-regulated in at least three of the papers were selected as “common osmoregulated genes.” Sixty-six of these were found on our arrays and compared to our data (Supplemental Table S3).
Quantitative RT-PCR data generation, processing, and analysis
Quantitative RT-PCR was performed to confirm results from the microarray data both on steady-state levels and stability for eight genes: RIB5 (YBR256C), ECM31 (YBR176W), IMP3 (YHR148W), RPS17A (YML024W), RPL6B (YLR448W), HOR2 (YER062C), GRE3 (YHR104W), and GPD1 (YDL022W). Steady-state levels (t TOT) for all genes were examined 0, 6, 30, and 60 min after the addition of 0.4 M NaCl in wild type and the hog1Δ and rck2Δ mutants (Supplemental Fig. S3). All genes were also examined on the stability level with the same design as in the microarray experiment (Figs. 5C, 6C). cDNA synthesis was performed using 1.1–4.5 μg of total RNA. Random primers and Superscript III reverse transcriptase (Invitrogen) were used. Primers were designed using the Primer express 2.0 software (Applied Biosystems) using standard settings. RT-PCR reaction was performed using SYBR Green detection in the Göteborg genomics core facility (Swegene) in an ABI PRISM 7900HT Sequence Detection system (Applied Biosystems). Two biological and three technical replicates were used for each sample.
Both RIB5 and ECM31 were included as potential reference genes, and since RIB5 proved to be stable over all conditions with the smallest variance, all signals were normalized against this gene. For each Phen time series, ratios were computed to the corresponding Phen untreated pool (e.g., unstressed wt cells harvested after 15 min of Phen against unstressed wt cells without Phen treatment). Both the reduced and full decay models were adapted to the data as in the microarray experiment. For the steady-state level data, expression data were modeled similarly as in the microarray experiment in order to make direct comparisons. Spearman rank correlation between array and qPCR data: 0.9 using data for rck2Δ and transformed stability index data for wt and hog1Δ. Fitting a regression line to the measurements with the qPCR data as the independent variable will produce a small intercept, indicating comparable normalization procedures between the two sets.
Data availability at ArrayExpress
The microarray data from this study are available at the ArrayExpress repository with accession number XY-123 (E-TABM-622).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by a grant from the Swedish Research Council (2007-5460) to P.S. C.M. was supported by the National Graduate School of Genomics and Bioinformatics. A.J. and O.N. received support from the Swedish Research Council, the Gothenburg Stochastic Centre, Gothenburg Mathematical Modeling Centre, and the Swedish Foundation for Strategic Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1403509.
REFERENCES
- Albertyn J., Hohmann S., Thevelein J.M., Prior B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikawa E., Sun Y., Wang J., Zhou Q., Ning B., Dial S.L., Guo L., Yang J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genomics. 2008;9:328. doi: 10.1186/1471-2164-9-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp E., Nilsson D., Sunnerhagen P. Fission yeast mitogen-activated protein kinase Sty1 interacts with translation factors. Eukaryot. Cell. 2008;7:328–338. doi: 10.1128/EC.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland E., Molin C., Swaminathan S., Ramne A., Sunnerhagen P. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 2004;53:1743–1756. doi: 10.1111/j.1365-2958.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- Bilsland E., Hult M., Bell S.D., Sunnerhagen P., Downs J.A. The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair (Amst.) 2007;6:1471–1484. doi: 10.1016/j.dnarep.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan E., Ariño J., Saito H., Sunnerhagen P., Posas F. Rck2 kinase is a substrate for the osmotic stress activated mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster J.L., de Valoir T., Dwyer N.D., Winter E., Gustin M.C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Brown A.J.P. Measurement of mRNA stability. In: Johnston J.R., editor. Molecular genetics of yeast: A practical approach. Oxford University Press; Oxford, UK: 1994. pp. 147–160. [Google Scholar]
- Canales R.D., Luo Y., Willey J.C., Austermiller B., Barbacioru C.C., Boysen C., Hunkapiller K., Jensen R.V., Knight C.R., Lee K.Y., et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G., Lee T.I., True H.L., Lander E.S., Young R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Andersen J.S., Gaietta G., Jurchott K., Royer H.D., Mann M., Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes & Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- Clotet J., Escote X., Adrover M.A., Yaakov G., Garí E., Aldea M., de Nadal E., Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Casadome L., Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Zapater M., Alepuz P.M., Sumoy L., Mas G., Posas F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier I., Walker C.A., Narasimhan J., Pearce A.K., Wek R.C., Humphrey T.C. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot. Cell. 2005;4:1785–1793. doi: 10.1128/EC.4.11.1785-1793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta R., Tian B., Wilusz C.J., Khounh D.T., Soteropoulos P., Ouyang M., Dougherty J.P., Peltz S.W. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 2005;25:5499–5513. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escote X., Zapater M., Clotet J., Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J., Aranda A., Pérez-Ortín J.E. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M.D., Hughes T.R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti E., Iakovleva T., Brook M., Deppenmeier S., Gruber A.D., Radzioch D., Clark A.R., Blackshear P.J., Kotlyarov A., Gaestel M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J., Friedlander G., Bergmann S., Sarig O., Ziv Y., Barkai N. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Ihmels J., Bergmann S., Gerami-Nejad M., Yanai I., McClellan M., Berman J., Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes & Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D., Kotlyarov A., Carballo E., Alexopoulou L., Blackshear P.J., Gaestel M., Davis R., Flavell R., Kollias G. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 2001;20:3760–3770. doi: 10.1093/emboj/20.14.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz M., Nordlander B., Valadi H., Johansson M., Gustafsson L., Hohmann S. Anaerobicity prepares Saccharomyces cerevisiae cells for faster adaptation to osmotic shock. Eukaryot. Cell. 2004;3:1381–1390. doi: 10.1128/EC.3.6.1381-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner D.H., Beilharz T.H., Marguerat S., Mata J., Watt S., Schubert F., Preiss T., Bähler J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACQ Consortium. The MicroArray Quality Control (MAQC) Project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S.M., Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Mager W.H., Varela J.C. Osmostress response of the yeast Saccharomyces . Mol. Microbiol. 1993;10:253–258. [PubMed] [Google Scholar]
- Marquez J.A., Serrano R. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- Melamed D., Pnueli L., Arava Y. Yeast translational response to high salinity: Global analysis reveals regulation at multiple levels. RNA. 2008;14:1337–1351. doi: 10.1261/rna.864908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Navarro M.M., Castells-Roca L., Bellí G., García-Martínez J., Marín-Navarro J., Moreno J., Pérez-Ortín J.E., Herrero E. Comprehensive transcriptional analysis of the oxidative response in yeast. J. Biol. Chem. 2008;283:17908–17918. doi: 10.1074/jbc.M800295200. [DOI] [PubMed] [Google Scholar]
- Norbeck J., Blomberg A. Amino acid uptake is strongly affected during exponential growth of Saccharomyces cerevisiae in 0.7 M NaCl medium. FEMS Microbiol. Lett. 1998;158:121–126. doi: 10.1111/j.1574-6968.1998.tb12810.x. [DOI] [PubMed] [Google Scholar]
- O'Rourke S.M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae . Genes & Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G., Yoshikawa K., Hirasawa T., Nagahisa K., Katakura Y., Furusawa C., Shimizu H., Shioya S. Extracting the hidden features in saline osmotic tolerance in Saccharomyces cerevisiae from DNA microarray data using the self-organizing map: Biosynthesis of amino acids. Appl. Microbiol. Biotechnol. 2007;75:415–426. doi: 10.1007/s00253-007-0837-8. [DOI] [PubMed] [Google Scholar]
- Posas F., Chambers J.R., Heyman J.A., Hoeffler J.P., de Nadal E., Ariño J. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Proft M., Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell. 2004;118:351–361. doi: 10.1016/j.cell.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Proft M., Pascual-Ahuir A., de Nadal E., Ariño J., Serrano R., Posas F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Askeland E., Thiele D.J. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Rep M., Reiser V., Gartner U., Thevelein J.M., Hohmann S., Ammerer G., Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Krantz M., Thevelein J.M., Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gabriel M.A., Burns G., McDonald W.H., Martin V., Yates J.R., 3rd, Bähler J., Russell P. RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 2003;22:6256–6266. doi: 10.1093/emboj/cdg597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller C., Brewster J.L., Alexander M.R., Gustin M.C., Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Dahan O., Levo M., Martinez M.R., Furman I., Segal E., Pilpel Y. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 2008;4:223. doi: 10.1037/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004 doi: 10.2202/1544-6115.1027. 3: Article 3. http://www.bepress.com/sagmb/vol3/iss1/art3. [DOI] [PubMed]
- Swaminathan S., Masek T., Molin C., Pospisek M., Sunnerhagen P. Rck2 is required for reprogramming of ribosomes during oxidative stress. Mol. Biol. Cell. 2006;17:1472–1482. doi: 10.1091/mbc.E05-07-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Reiser V., Ruis H., Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc. Natl. Acad. Sci. 2001;98:5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu C.L., Storey J.D., Tibshirani R.J., Herschlag D., Brown P.O. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Winzen R., Kracht M., Ritter B., Wilhelm A., Chen C.Y., Shyu A.B., Müller M., Gaestel M., Resch K., Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale J., Bohnert H.J. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 2001;276:15996–16007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microarray data from this study are available at the ArrayExpress repository with accession number XY-123 (E-TABM-622).