Abstract
Homoaconitase enzymes catalyze hydrolyase reactions in the α-aminoadipate pathway for lysine biosynthesis or the 2-oxosuberate pathway for methanogenic coenzyme B biosynthesis. Despite the homology of this iron-sulfur protein to aconitase, previously studied homoaconitases catalyze only the hydration of cis-homoaconitate to form homoisocitrate rather than the complete isomerization of homocitrate to homoisocitrate. The MJ1003 and MJ1271 proteins from the methanogen Methanocaldococcus jannaschii formed the first homoaconitase shown to catalyze both the dehydration of (R)-homocitrate to form cis-homoaconitate, and its hydration is shown to produce homoisocitrate. This heterotetrameric enzyme also used the analogous longer chain substrates cis-(homo)2aconitate, cis-(homo)3aconitate, and cis-(homo)4aconitate, all with similar specificities. A combination of the homoaconitase with the M. jannaschii homoisocitrate dehydrogenase catalyzed all of the isomerization and oxidative decarboxylation reactions required to form 2-oxoadipate, 2-oxopimelate, and 2-oxosuberate, completing three iterations of the 2-oxoacid elongation pathway. Methanogenic archaeal homoaconitases and fungal homoaconitases evolved in parallel in the aconitase superfamily. The archaeal homoaconitases share a common ancestor with isopropylmalate isomerases, and both enzymes catalyzed the hydration of the minimal substrate maleate to form d-malate. The variation in substrate specificity among these enzymes correlated with the amino acid sequences of a flexible loop in the small subunits.
In the final reaction of methanogenesis, the nickel metalloenzyme methyl-coenzyme M (CoM)2 reductase catalyzes the attack of the coenzyme B (CoB) thiol on the methyl thioether of methyl-CoM, releasing methane and forming a heterodisulfide compound (CoM-S-S-CoB) (1). To reach the deeply buried active site of methyl-CoM reductase, CoB contains a long 7-mercaptoheptanoyl chain with an amide bond to phosphothreonine (2, 3).
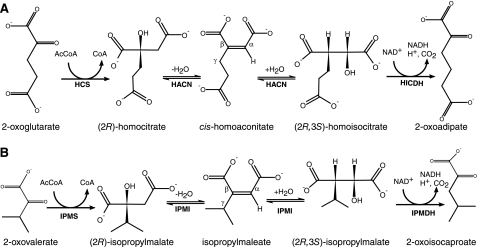
Methanogens make the 7-mercaptoheptanoate moiety of CoB from a 2-oxosuberate intermediate, which these cells produce using a series of 2-oxoacid elongation reactions (4). 2-Oxosuberate biosynthesis begins with 2-oxoglutarate from central metabolism (Fig. 1A). The homocitrate synthase enzyme (HCS) catalyzes the aldol-like addition of an acetyl group from acetyl-coenzyme A to 2-oxoglutarate to produce (R)-homocitrate. The homoaconitase enzyme (HACN) is proposed to catalyze the anti-elimination of water, forming the cis-homoaconitate intermediate, and its hydration is proposed to produce (2R,3S)-homoisocitrate. Finally, the NAD+-dependent homoisocitrate dehydrogenase enzyme (HICDH) oxidizes and decarboxylates the β-hydroxytricarboxylic acid to produce 2-oxoadipate. Analogous reactions are found in the citric acid (Krebs) cycle, the isopropylmalate pathway to leucine, the pyruvate pathway to 2-oxobutyrate, and the α-aminoadipate pathway (5). In the latter case, some bacteria and fungi use HCS, HACN, and HICDH enzymes to produce 2-oxoadipate for lysine biosynthesis (6). However, methanogens repeat this series of reactions two more times, using 2-oxoadipate to produce 2-oxopimelate and using 2-oxopimelate to produce 2-oxosuberate.
FIGURE 1.
Biosynthetic 2-oxoacid elongation pathways. A, three enzymes convert 2-oxoglutarate to 2-oxoadipate in the α-aminoadipate pathway and the biosynthesis of the CoB acyl chain. HCS (MJ0503) catalyzes the aldol-type addition of an acetyl group to the re-face of 2-oxoglutarate, producing (R)-homocitrate. Aconitase or methanogen HACN (MJ1003 and MJ1271) catalyzes the anti-elimination of the hydroxyl group and pro-R hydrogen from homocitrate forming the intermediate cis-homoaconitate. All of the HACN enzymes catalyze the anti-addition of water to produce (2R,3S)-homoisocitrate. Finally, HICDH (MJ1596) catalyzes an oxidative decarboxylation reaction forming 2-oxoadipate. Replacing 2-oxoglutarate with 2-oxoadipate produces 2-oxopimelate, and a third iteration of this pathway produces 2-oxosuberate. B, the analogous reactions in leucine biosynthesis in M. jannaschii use isopropylmalate synthase (IPMS; MJ1195), IPMI (MJ0499 and MJ1277), and isopropylmalate dehydrogenase (IPMDH; MJ0720). The citramalate synthase (MJ1392) replaces isopropylmalate synthase in isoleucine biosynthesis, using pyruvate to produce (R)-citramalate that is converted to 2-oxobutyrate by IPMI and isopropylmalate dehydrogenase (28). The conserved α and β groups and the variable γ groups of cis-homoaconitate and isopropylmaleate are labeled for reference.
The enzymes in the CoB pathway evolved from leucine biosynthetic proteins that catalyze analogous reactions in the isopropylmalate pathway to 2-oxoisocaproate (Fig. 1B). HCS was identified in the methanogen Methanocaldococcus jannaschii as a paralog of isopropylmalate synthase (7). Remarkably, this HCS was reported to convert 2-oxoglutarate and acetyl-CoA to trans-homoaconitate, which was hydrated to form (S)-homocitrate. But in analogous reactions, HCS produces (R)-homo2citrate from 2-oxoadipate and (R)-homo3citrate from 2-oxopimelate. Although different acetyl-CoA condensing enzymes are known to have different stereospecificities, each enzyme usually forms only one product (8). The yeast HCS produces only (R)-homocitrate (9). (R)-Homocitrate is also a key component of the nitrogenase iron-molybdenum cofactor found in many bacteria and some methanogens (10).
Subsequently the M. jannaschii HICDH was identified as a paralog of isopropylmalate dehydrogenase (11). This enzyme catalyzes the NAD+-dependent oxidation of threo-homoisocitrate, threo-(homo)2isocitrate, and threo-(homo)3isocitrate with similar specificity constants. Experiments using cell-free extract from Methanosarcina thermophila indicated that (–)-threo-homoisocitrate or (2R,3S)-homoisocitrate is the relevant diastereomer substrate for methanogen HICDH, similar to bacterial and yeast enzymes (7, 12).
The HACN enzyme in CoB biosynthesis has not yet been identified. It is predicted to differ from the previously characterized HACN enzymes that function in α-aminoadipate pathways for lysine biosynthesis in Saccharomyces cerevisiae (13), Aspergillus nidulans (14), and the bacterium Thermus thermophilus (15). Along with the homologous 2-methyl-cis-aconitate hydratase and dimethylmalate dehydratase proteins, those HACN proteins catalyze only half of an aconitase-like reaction (16, 17). The HACN enzymes catalyze the hydration of cis-homoaconitate to form homoisocitrate (18). No HACN has been shown to catalyze the dehydration of homocitrate to form cis-homoaconitate, but an aconitase enzyme was proposed to catalyze the dehydration of homocitrate in aerobic fungi and T. thermophilus. Methanogens have no aconitase, so their HACN must catalyze both half-reactions. Additionally, the methanogen HACN must use tricarboxylate substrates of varying chain length, whereas the HACN proteins from α-aminoadipate pathways need only recognize cis-homoaconitate.
Genomes from methanogens that produce CoB contain two copies of genes annotated as encoding large and small subunits of [4Fe-4S]-dependent isopropylmalate isomerases. The large subunit paralogs share more than 50% amino acid sequence identity with each other and more than 40% identity with the T. thermophilus HACN subunits. The methanogen small subunits are likewise more similar to each other than to bacterial or eukaryotic sequences. These genes are scattered around the chromosomes of most methanogens, with little contextual information about gene function. Therefore bioinformatic analysis cannot distinguish the functions of these proteins. We previously identified one combination of M. jannaschii proteins (MJ0499 and MJ1277) as the isopropylmalate/citramalate isomerase that is involved in leucine and isoleucine biosyntheses (19).
In this work, the MJ1003 and MJ1271 proteins from M. jannaschii formed an active homoaconitase enzyme. HACN activity evolved in parallel in the archaeal and fungal lineages from isopropylmalate isomerase or aconitase ancestors, respectively. Direct assays showed that this reconstituted ironsulfur enzyme catalyzed the hydration of cis-homoaconitate, cis-homo2aconitate, cis-homo3aconitate, and even cis-homo4aconitate with comparable specificity. cis-Aconitate and citraconate were not substrates, although the minimal substrate maleate was hydrated to produce d-malate. In a coupled reaction with HICDH, the homoaconitate analogs were converted to their corresponding 2-oxoacids, demonstrating that the homoisocitrate products are substrates for HICDH. In contrast to previously studied HACN proteins, the methanogen enzyme catalyzed both the dehydration of (R)-homocitrate to form cis-homoaconitate and the subsequent hydration reaction that forms homoisocitrate. HICDH oxidizes and decarboxylates homoisocitrate to make 2-oxoadipate. (S)-Homocitrate and trans-homoaconitate were inhibitors rather than substrates for HACN, so HACN cannot catalyze all of the predicted dehydratase reactions in the original pathway for CoB biosynthesis.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—(R)-Homocitrate and (S)-homocitrate were synthesized through the diastereoselective alkylation of malic acid. (2R,3S)-Homoisocitrate was synthesized from dimethyl d-malate (20). p-Toluenesulfonic acid was recrystallized from ethyl acetate.
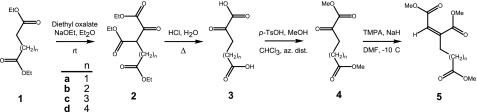
Synthesis of 2-Oxoacids and Homoaconitic Acids—Massoudi et al. (21) used the unstable t-butyl ester of 2-oxoglutarate in a Wittig-Horner reaction to preferentially produce cis-homoaconitate. We have used the stable methyl ester of 2-oxoglutarate 4a (Sigma) to produce homoaconitate trimethyl ester 5a (12:1 cis:trans) (Fig. 2) (7). trans-Homoaconitate trimethyl ester was separated from the cis-homoaconitate trimethyl ester by flash chromatography. The purified ester was saponified with sodium hydroxide at 50 °C and extracted from the acidified solution to produce cis-homoaconitic acid. The purity of the homoaconitate isomers was established by 1H NMR. The assignments of the vinylic proton resonances for each trimethyl ester isomer were confirmed by nuclear Overhauser effect difference spectroscopy. Mass spectra of these compounds were acquired at the University of Texas Mass Spectrometry facility, and 1H NMR and 13C NMR spectra were acquired at the University of Texas Nuclear Magnetic Resonance facility; these spectra are described in the supplemental materials.
FIGURE 2.
Chemical synthesis of cis-homoaconitate methyl esters with different chain lengths in the γ-moiety. The reaction conditions are described under “Experimental Procedures.”
2-Oxoadipic Acid—Diethyl glutarate (4.7 g, 25 mmol) was mixed with sodium ethoxide (2.7 g, 40 mmol) in 25 ml of dry diethyl ether at ambient temperature. Diethyl oxalate (4.4 g, 30 mmol) was added dropwise to the stirred solution at ambient temperature. The reaction was quenched with 15 ml of water after 8 h, and the pH was adjusted to neutral with HCl. The triethyl α-oxalyl glutarate 2b was extracted into diethyl ether and dried over sodium sulfate. The solvent was removed by evaporation under vacuum to afford a yellow oil in 80% yield. Compound 2b was stirred in 30 ml of 4 m HCl at 80 °C for 6 h to produce 2-oxoadipic acid 3b (22). The solution was dried by evaporation under vacuum, and 2-oxoadipic acid was crystallized from diethyl ether (34% yield).
2-Oxopimelic Acid—Compound 3c was prepared by the procedure described for 2-oxoadipic acid, using diethyl adipate instead of diethyl glutarate.
2-Oxosuberic Acid—Compound 3d was prepared by the procedure described for 2-oxoadipic acid, using diethyl pimelate instead of diethyl glutarate.
cis-(Homo)2aconitic Acid—Dimethyl 2-oxoadipate 4b was prepared by the azeotropic distillation of 2-oxoadipate (1.2 g, 7.2 mmol) in 20 ml of dry chloroform with 10 ml of dry methanol and p-toluenesulfonic acid (57 mg, 0.3 mmol) (23). The solvent was removed by evaporation under vacuum, and the product was filtered through a plug of silica (90% yield). Sodium hydride (0.3 g, 8.1 mmol) was dissolved in 10 ml of dry dimethylformamide at 5 °C. Trimethyl phosphonoacetate (Aldrich; 1.2 g, 6.5 mmol) dissolved in 3 ml of dimethylformamide was added dropwise at –5 °C with stirring for 30 min. Compound 4b (1.2 g, 6.5 mmol) in 3 ml of dimethylformamide was added dropwise, and the mixture was stirred for 1 h. The reaction was quenched with 50 ml of ice water and extracted with diethyl ether. The ether phase was dried with sodium sulfate and concentrated by evaporation under vacuum to produce trimethyl (homo)2aconitate 5b in 60% yield as a yellow oil (17:1 cis:trans mixture). The isomers were separated by flash chromatography on silica with pentane:ethyl acetate (90:10). Deprotection by saponification and purification of the free acids were carried out as described above to produce cis-(homo)2aconitic acid and trans-(homo)2aconitic acid as yellow oils. Solutions of acidic substrates were adjusted to pH 7 with KOH before enzymatic analysis.
cis-(Homo)3aconitic Acid—Trimethyl cis-(homo)3aconitate 5c was prepared from diethyl adipate 1c, as described for cis-(homo)2aconitate producing a 10:1 cis:trans mixture. cis-(Homo)4aconitic Acid—Trimethyl cis-(homo)4aconitic 5d acid was prepared from diethyl pimelate 1d, producing a 23:1 cis:trans mixture.
Cloning and Molecular Biology—Plasmid pT7-MJ1596 (encoding the protein with RefSeq accession number NP_248605.1) was a gift from Huimin Xu and Robert H. White (Virginia Tech) (11). DNA sequencing showed that this cloned gene contained two nucleotide substitutions (C229A and T378C) compared with the MJ1596 sequence reported by the genome sequencing project (24). The C229A transversion resulted in a Q77K amino acid substitution, whereas the second nucleotide difference was silent. Therefore we reamplified the MJ1596 gene from chromosomal DNA of M. jannaschii JAL-1 using PCR. DNA amplified using oligonucleotide primers 5MJ1596BN and 3MJ1596B (see supplemental material) was ligated between the NdeI and BamHI sites of vector pET-19b (Novagen) to create vector pDG382, and in the BamHI site of vector pET-43.1c (Novagen) to create vector pDG130. Deletion of the NdeI fragment from pDG130 created vector pDG131. The C229A substitution was also identified in the sequences of these plasmids (the polymorphic gene sequence was deposited into GenBank™ under accession number EU447775). Plasmids were propagated in Escherichia coli DH5α. Sequencing by the DNA facility at the Institute for Cellular and Molecular Biology (University of Texas, Austin) confirmed the cloned DNA sequences. Plasmids for expressing the M. jannaschii dehydratase subunits were described previously (19) and are summarized in the supplemental material.
Protein Expression and Purification—The MJ1003 and MJ1271 proteins were coexpressed in an E. coli BL21(DE3) (pDG141 pDG163) strain described previously (19). The cells were grown at 37 °C with shaking at 250 rpm until the culture reached an optical density at 600 nm of 0.6–0.8. Expression was then induced by the addition of 50 μm isopropyl-1-thio-β-d-galactopyranoside, and the culture was transferred to a 16 °C water bath and shaken continuously for 15–20 h. The MJ1003 and MJ1271 proteins were purified by heat treatment of the cell lysate, followed by anion exchange chromatography, dialysis, and concentration (19). The protein was further concentrated using a stirred ultrafiltration cell (Amicon) under N2 with a 10-kDa molecular mass cut-off filter (Pall). The untagged MJ1596 HICDH protein was expressed in an E. coli Arctic-Express (DE3)-RIL (pDG131) strain. The cells were grown in medium containing streptomycin (50 μgml–1), tetracycline (50 μgml–1), and gentamycin (50 μgml–1) at 16 °C with shaking at 250 rpm. HICDH protein expression, extraction, purification, and concentration were performed as described for the MJ1003/MJ1271 proteins.
Reconstitution of the Iron-Sulfur Center—In a typical reconstitution reaction, copurified MJ1003 and MJ1271 proteins (1 mg ml–1 apoenzyme) were mixed with Fe(NH4)2(SO4)2 and sodium sulfide as described previously (19). The reconstitution mixture was stirred under argon until maximum activity was observed (typically 3–4 h). Hydrolyase activity was monitored in a standard assay with 0.2 mm cis-homoaconitate as described below. A mock reconstitution control consisted of the above mixture without protein.
Determination of Iron-Sulfur Content—The holoenzyme was reconstituted using concentrated MJ1003/MJ1271 apoprotein (2.5 mg ml–1), and then it was desalted using a PD-10 column (GE Healthcare) equilibrated with an anoxic solution of 50 mm Tris/HCl (pH 8.0). Desalting was performed at room temperature in an anaerobic chamber (Coy Laboratory Products) with an atmosphere of N2:CO2:H2 (75:20:5 by volume). Fractions containing the desalted protein were combined, and aliquots were removed for iron and sulfide analyses. Total iron analysis was performed using the colorimetric bathophenanthrolinedisulfonate assay with a standard curve prepared from ferric chloride (25). The labile sulfide concentration was determined using the methylene blue assay with the standard addition of sodium sulfide (26). The total protein concentration was determined by the Bradford dye binding method (Thermo-Pierce) using bovine serum albumin as a standard. The standard errors calculated for each assay were propagated to estimate the error associated with each analysis.
Measurement of Hydrolyase Activities—Reactions (1 ml) were conducted in quartz semimicrocells with screw cap septa (Starna) containing 50 mm CHES-KOH (pH 9.0), 200 mm KCl, 20 μgml–1 holoenzyme, and various substrate concentrations. The sealed quartz cell containing buffer and substrate was degassed under argon for 10 min and equilibrated at 60 °C for 10 min. The reactions were initiated by the addition of holoenzyme, and the decrease in UV absorbance was monitored at 60 °C. Initial rates were measured from the linear portion of the reaction progress curve at 235 nm for cis-(homo)1aconitate (ε235 = 4580 m–1 cm–1), 235 nm for cis-(homo)2aconitate (ε235 = 4420 m–1 cm–1), 240 nm for cis-(homo)3aconitate (ε240 = 3180 m–1 cm–1), and 240 nm and 250 nm for cis-(homo)4aconitate (ε240 = 3140 m–1 cm–1, ε250 = 1370 m–1 cm–1). All of the molar absorption coefficients were determined in the reaction buffer at 60 °C. One unit of hydrolyase activity catalyzed the conversion of 1 μmol of substrate to product per min. Steady-state kinetic constants were estimated by nonlinear regression of initial rate data fit to the Michaelis-Menten-Henri equation (19). The inhibitors were screened in direct assays containing buffer, 20 μgml–1 HACN, and various concentrations of substrate analogs; these reactions were initiated by the anaerobic addition of 100 μm cis-homoaconitate, and hydrolyase activity was monitored at 240 nm. The stereochemistry of maleate hydration products was determined enzymatically using tartrate dehydrogenase and l-malate dehydrogenase enzymes (19).
Coupled Assay of Hydratase Activity—The hydration of unsaturated intermediates in the forward direction was measured by a coupled reaction with HICDH by monitoring the reduction of NAD+ to NADH at 340 nm (15). Reaction mixtures containing 50 mm TAPS-KOH (pH 8.5), 50 mm KCl, 5 mm MgCl2, 20 mm β-mercaptoethanol, 1 mm NAD+, and 30 μg ml–1 HICDH were degassed as described above, after which MJ1003/MJ1271 holoenzyme was added to a final concentration of 10 μgml–1. The reaction mixtures were equilibrated to 60 °C, and the reactions were initiated with 5 μl of substrate. The initial rates were measured using the linear portion of the reaction progress curve. One unit of enzymatic activity catalyzed the reduction of 1 μmol NAD+ min–1. The apparent kinetic constants were estimated as described above.
HPLC Analysis of Reaction Products—Standard reactions containing 10 mm cis-homoaconitate analogs and HACN holoenzyme (40 μgml–1) were incubated for 20 h at 60 °C. A portion of the reaction product was analyzed by reversed phase HPLC using a Synergi Hydro-RP column (Phenomenex; 250 by 4.6 mm) with a guard column (4 by 3 mm). Chromatography was performed at 35 °C with an isocratic elution with 20 mm potassium phosphate (pH 2.5) and 5% (v/v) acetonitrile in water at a flow rate of 1 ml min–1. Retention factors for standard compounds (t0 = 2.5 min) were 0.4 (homoisocitrate), 1.0 (homocitrate), 1.1 (cis-homoaconitate), and 2.2 (cis-homo2aconitate). Reaction products from incubations with cis-homo3aconitate and cis-homo4aconitate were analyzed using mobile phase containing buffer and 15% (v/v) acetonitrile in water. The retention factors were 1.5 (cis-homo3aconitate) and 3.5 (cis-homo4aconitate). A photodiode array detector was used to identify analytes; cis-homoaconitate analogs had absorption maxima near 215 nm, whereas hydroxyacid products absorbed maximally near 205 nm.
LC-MS Analysis of Enzyme Reaction Products—Standard reactions (1 ml) containing 250 μm homoaconitate (or analog), and MJ1003/MJ1271 holoenzyme were incubated at 60 °C for 20 h under argon. The reaction mixture was adjusted to pH 1 with HCl and extracted twice with ethyl acetate and once with diethyl ether. The combined organic phases were dried under N2 at 60 °C. The residue was dissolved in water and analyzed by LC-MS using a Thermo LTQ-XL instrument. The sample was applied to a reversed phase column (Macro Bullet 3 μm 200 Å Magic C18AQ; Michrom Bioresources; 25 × 4 to 1 mm tapered) and eluted with a gradient from 5 to 95% acetonitrile with 0.1% formic acid in water at a flow rate of 0.5 ml min–1. Electrospray ionization mass spectrometry was performed in the negative ion mode.
For the analysis of coupled reaction products, 1 mm NAD+ and HICDH were added to the reaction mixture. To form the methoxime derivatives of 2-oxoacid products, the pH was adjusted to 13–14 with sodium hydroxide and 0.25 ml of 10% O-methylhydroxylamine hydrochloride was added. The mixtures were incubated at room temperature for 1 h and then adjusted to pH 1 with HCl and saturated with NaCl. The acidic solution was extracted as described above. The concentrated sample was applied to a reversed phase column (Hypersil Gold; Thermo; 50 × 2.1 mm) and eluted with a gradient from 95 to 20% acetonitrile in aqueous 0.1% formic acid at a flow rate of 0.5 ml min–1. Electrospray ionization mass spectrometry was performed in the positive ion mode.
RESULTS
Hydrolyase Activity of the MJ1003/MJ1271 Proteins—The MJ1003 and MJ1271 proteins were previously coexpressed in E. coli, and the apoenzyme was purified to homogeneity by heating and ion exchange chromatography. The subunits associated to form a heterotetramer, similar to the homologous isopropylmalate isomerase complex (19). The MJ1003/MJ1271 apoprotein was mixed with iron(II) and sulfide to reconstitute an air-sensitive iron-sulfur cluster. The resulting holoenzyme contained 5.1 ± 0.2 iron equivalents per MJ1003/MJ1271 dimer and 5.1 ± 0.8 sulfide equivalents, characteristic of a single [4Fe-4S] cluster. Unlike the IPMI enzyme, the MJ1003/MJ1271 complex did not catalyze the hydration of citraconate or the dehydration of citramalate. To test whether this complex had homoaconitase activity, cis-homoaconitate was synthesized using the Wittig-Horner reaction (Fig. 2). The fully reconstituted MJ1003/MJ1271 holoenzyme catalyzed the hydration of cis-homoaconitate with a specific activity of 0.8 unit mg–1 (0.9 s–1) at 60 °C, measured in a continuous spectrophotometric assay. Enzymatic activity was observed between pH 8 and 10, with maximal activity at pH 9. No MgCl2 was required in this reaction, although activity was highest with 200 mm KCl (activity in the absence of KCl was only 30% of the activity with KCl). In contrast, the MJ1003 and MJ1277 (IPMI small subunit) proteins that also formed a heterotetramer demonstrated no activity with any tested substrate.
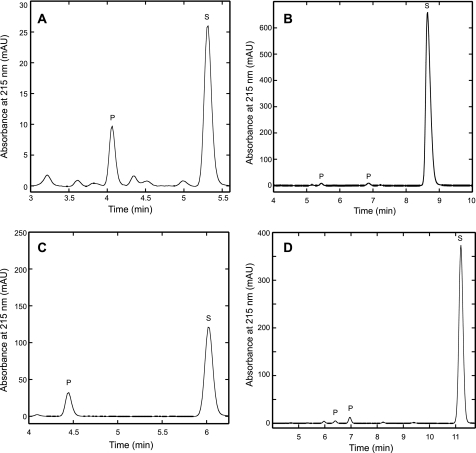
Reversed phase HPLC analysis of the MJ1003/MJ1271 reaction products showed a decrease in substrate concentration and the appearance of one new peak corresponding to homocitrate (Fig. 3A). No homoisocitrate was detected. LC-MS analysis of the reaction mixture identified peaks at 205 m/z corresponding to the [M-H]– ion of homocitrate or homoisocitrate and 187 m/z corresponding to the [M-H]– ion of homoaconitate. Neither apoenzyme nor a mock reconstitution mixture had hydrolyase activity, confirming that the iron-sulfur cluster in the holoenzyme is essential for activity.
FIGURE 3.
HACN catalyzed the hydration of cis-unsaturated tricarboxylic acids to produce hydroxyacids. Holoenzyme was incubated with cis-homoaconitate analogs for 20 h at 60 °C. The reaction products were separated by reversed phase HPLC with UV absorbance detection in milli-absorbance units (mAU) as described in the text. Substrate peaks (S) were identified by their retention times compared with external standards. Product peaks (P) had absorbance maxima near 205 nm, and they were not observed in control reactions. The unlabeled peaks were also observed in control reactions without enzyme, so they are considered to be contaminants. A, incubation with cis-homoaconitate (S) produced homocitrate (P) that was confirmed by coinjection with (R)-homocitrate. No homoisocitrate was detected in this experiment. B, incubation with cis-homo2aconitate produced two product peaks. C, incubation with cis-homo3aconitate produced a single product peak. D, incubation with cis-homo4aconitate produced two product peaks. The homoaconitate analogs have significantly higher molar absorptivities than their corresponding hydroxy-acids, which preclude direct comparisons of their concentrations by peak area integration.
2-Oxosuberate biosynthesis requires three rounds of 2-oxoacid elongation. Therefore the methanogen HACN was predicted to catalyze the isomerization of homocitrate as well as (homo)2citrate and (homo)3citrate. As expected, the MJ1003/MJ1271 holoenzyme catalyzed the hydration of cis-homoaconitate, cis-homo2aconitate, cis-homo3aconitate, and even the nonphysiological cis-homo4aconitate substrates with similar efficiency (Fig. 3). Surprisingly, no hydration activity was detected in a continuous assay when 0.8 mm cis-aconitate was used as a substrate (i.e. the rate was less than the limit of detection of 0.029 s–1). Nor was any dehydratase activity observed in reactions containing 5 mm citrate or threo-dl-isocitrate.
Steady-state kinetic parameters for HACN were calculated from the initial rates of the hydration reactions for all four substrates (Table 1). The specificity constants for the four substrates are similar, although the turnover number increases for longer γ-carboxylate chain substrates. The Michaelis constant is similar for all three physiologically relevant substrates but is higher for cis-homo4aconitate. Therefore the methanogen HACN does not significantly discriminate between substrates with 2–5 methylene groups in their γ-carboxylate chains.
TABLE 1.
Steady-state kinetic parameters for the direct assay of hydrolyase activity
Reactions containing substrate and buffer salts were preincubated at 60 °C before the addition of reconstituted MJ1003/MJ1271 protein to initiate the reaction. Hydrolase activity was determined by measuring the rate of decrease in UV absorbance caused by the unsaturated substrate, as described under “Experimental Procedures.”
| Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| μm | units mg-1 | s-1 | m-1 s-1 | |
| cis-Homo1aconitate | 22 ± 3 | 0.68 ± 0.03 | 0.75 | 3.4 × 104 |
| cis-Homo2aconitate | 30 ± 5 | 0.60 ± 0.03 | 0.66 | 2.2 × 104 |
| cis-Homo3aconitate | 36 ± 6 | 2.2 ± 0.09 | 2.5 | 6.8 × 104 |
| cis-Homo4aconitate | 175 ± 39 | 5.1 ± 0.3 | 5.6 | 3.2 × 104 |
| Maleate | 330 ± 50 | 5.5 ± 0.3 | 6.0 | 1.8 × 104 |
| (R)-Homocitratea | 1500 ± 200 | 0.59 ± 0.03 | 0.37 | 2.5 × 102 |
Dehydratase activity was determined by measuring the rate of increase in UV absorbance caused by homoaconitate formation.
Despite its preference for long γ-chain carboxylate substrates, HACN catalyzed the hydration of maleate, a minimal analog of homoaconitate with a hydrogen atom replacing the γ-chain. HACN reactions containing 5 mm maleate produced 5 mm d-malate or (R)-2-hydroxysuccinate and no l-malate (<10 μm), determined by enzymatic analysis (19). Therefore the stereochemistry of this hydrolyase reaction is the same as in the physiological reactions, suggesting a similar mode of binding to the active site. The enzyme's Km for maleate is 10-fold higher than its Km for physiological substrates (Table 1). However, the turnover number for maleate is high compared with turnovers for the tricarboxylate substrates.
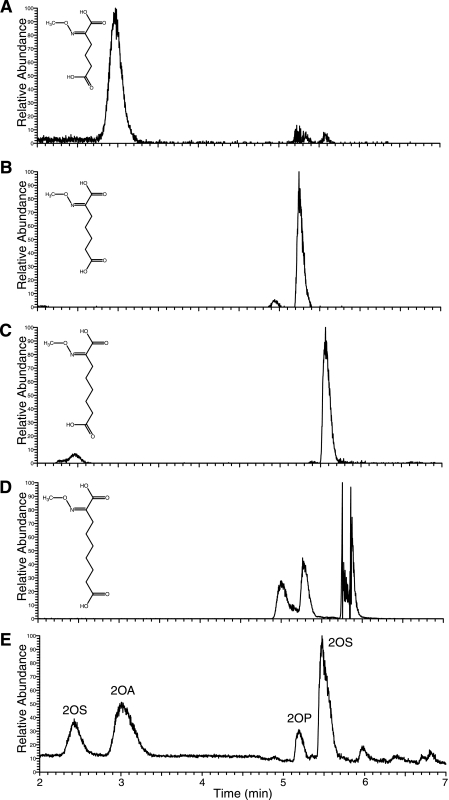
Coupled Activity of HACN with HICDH—The M. jannaschii HICDH enzyme was previously shown to catalyze the oxidative decarboxylation of (–)-threo-homoisocitrate, (–)-threo-homo2isocitrate, and (–)-threo-homo3isocitrate (11). To specifically measure the rate of the forward HACN hydration reaction (the conversion of cis-homoaconitate to homoisocitrate), coupled reactions were performed with HACN and excess HICDH monitoring the rate of NADH production. This coupled reaction had maximal activity with 5 mm MgCl2, 100 mm KCl, 20 mm 2-mercaptoethanol, and a pH of 8.5. The 2-oxoadipate product of these enzymes was identified as the methoxime derivative by LC-MS (Fig. 4). The Km for cis-homoaconitate in this coupled reaction is similar to that measured in the direct assay (Table 2). However, the turnover number increases 2-fold because of the removal of product by HICDH. Comparable results were obtained for cis-homo2aconitate and cis-homo3aconitate substrates, producing 2-oxopimelate and 2-oxosuberate, respectively (Fig. 4, B and C). The cis-homo4aconitate substrate was also efficiently hydrated to produce homo4isocitrate that HICDH oxidatively decarboxylated to form 2-oxoazelate (Fig. 4D). Although cis-homo4aconitate is probably not physiologically relevant, both the HACN and HICDH enzymes efficiently recognize tricarboxylate substrates with two to five methylene groups in their γ-chains.
FIGURE 4.
HACN and HICDH hydrated cis-homoaconitate analogs and catalyzed their oxidative decarboxylation to produce the corresponding 2-oxoacids. Methoxime derivatives extracted from the reaction products were analyzed by LC-MS. A–D show mass chromatograms for the MH+ ions of the methoxime-derivatized 2-oxoacids depicted in the inset. Similar plots were obtained for the sodium adducts of each acid. A, reactions with cis-homoaconitate produced 2-oxoadipate indicated by the m/z 190 peak. B, reactions containing cis-(homo)2aconitate produced 2-oxopimelate identified by the m/z 204 peak. C, reactions containing cis-(homo)3aconitate produced 2-oxosuberate identified by the m/z 218 peak. D, reactions containing cis-(homo)4aconitate produced 2-oxoazelate identified by the m/z 232 peak. This derivative formed several noncovalent complexes producing multiple peaks. E contains a chromatogram of 2-methoxime dicarboxylic acid standards showing the reconstructed total ion current. The standards included 2-oxoadipic acid (2OA), 2-oxopimelic acid (2OP), and 2-oxosuberic acid (2OS).
TABLE 2.
Kinetic parameters for the coupled assay of HACN hydrolyase activity with HICDH
Reactions containing 10 μg ml–1 HICDH, 30 μg ml–1 HACN holoenzyme, 1 mm NAD+, and buffer salts were preincubated at 60 °C before the addition of substrate to initiate the reaction. Hydrolase activity was determined by measuring the rate of increase in UV absorbance at 340 nm caused by the reduction of NAD+, as described under “Experimental Procedures.”
| Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| μm | units mg-1 | s-1 | m-1s-1 | |
| cis-Homo1aconitate | 25 ± 3 | 1.4 ± 0.04 | 1.5 | 5.9 × 104 |
| cis-Homo2aconitate | 20 ± 2 | 0.93 ± 0.02 | 1.0 | 5.0 × 104 |
| cis-Homo3aconitate | 46 ± 9 | 4.7 ± 0.3 | 5.1 | 1.1 × 105 |
| cis-Homo4aconitate | NDa | 5.8b | ND | ND |
ND, not determined.
Specific activity determined with 0.78 mm cis-homo4aconitate.
Dehydration Reaction and Stereospecificity of HACN—HPLC analysis showed that the primary product of homoaconitate hydration was homocitrate, indicating that the methanogen HACN catalyzed both half-reactions in the isomerization of homocitrate. However, the stereochemistry of homocitrate in this pathway was ambiguous. The partially purified M. jannaschii HCS was previously reported to produce (S)-homocitrate from 2-oxoglutarate and acetyl-CoA, but it produced (R)-(homo)2citrate from 2-oxoadipate (7). To test which homocitrate enantiomer is a substrate for HACN, we incubated each isomer with HACN and HICDH in coupled assays. (S)-Homocitrate was not a substrate for these enzymes (Fig. 5). However, (R)-homocitrate was isomerized and oxidatively decarboxylated with a specific activity of 0.3 unit mg–1 (corresponding to a rate of 0.3 s–1 for HACN). In continuous spectrophotometric assays monitoring the formation of homoaconitate, the HACN protein catalyzed the dehydration of (R)-homocitrate with a kcat of 0.37 s–1 (Table 1). The rate of dehydration is lower than the rate of cis-homoaconitate hydration, as observed for aconitase (27).
FIGURE 5.
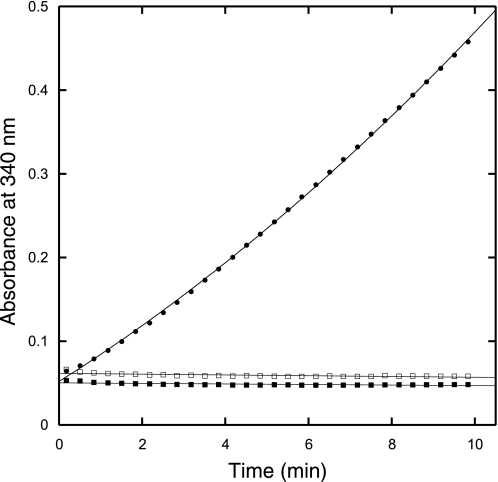
Homoaconitase and homoisocitrate dehydrogenase specifically converted (R)-homocitrate to 2-oxoadipate in a coupled reaction. The reaction progress curves show the NADH absorbance in reactions containing 0.5 mm (R)-homocitrate (solid circles), 0.5 mm (S)-homocitrate (open squares), or no substrate (solid squares). A control reaction without homoaconitase enzyme also showed no NADH production (data not shown).
Inhibitors of HACN—Product inhibition tests using mixtures of 100 μm cis-homoaconitate and 50 μm (R)-homocitrate reduced the HACN hydrolyase activity by 50%; no activity was observed in the presence of 100 μm (S)-homocitrate. Thus HACN specifically acts on (R)-homocitrate, whereas (S)-homocitrate inhibits the reaction. In reactions containing 100 μm cis-homoaconitate, hydrolyase activity was reduced 50% by the following analogs: 25 μm cis-aconitate, 100 μm trans-aconitate, and 400 μm trans-(homo)3aconitate. No inhibition was observed with citraconate (up to 200 μm).
DISCUSSION
Yeast, some bacteria, and many archaea use a homoaconitase enzyme in the α-aminoadipate pathway for lysine biosynthesis. By comparison with the homologous aconitase protein, the HACN enzyme is expected to catalyze the isomerization of homocitrate to homoisocitrate. Yet the previously studied HACNs catalyzed only the hydration of cis-homoaconitate, forming homoisocitrate. The M. jannaschii enzyme described here is the first HACN shown to catalyze both hydrolyase reactions. HPLC analysis showed that this enzyme preferentially converts homoaconitate to homocitrate. Direct assays demonstrated that (R)-homocitrate was dehydrated to form homoaconitate. Coupled spectrophotometric assays with HICDH showed that homoaconitate was also converted to homoisocitrate. The same system was used to show that (R)-homocitrate could be converted to 2-oxoadipate, confirming that the methanogen HACN catalyzes the complete isomerization of (R)-homocitrate to (2R,3S)-homoisocitrate.
The stereospecificity of HACN for (2R)-homocitrate and cis-homoaconitate analogs shows that it cannot catalyze all of the dehydratase reactions in CoB biosynthesis proposed previously (7). Either additional enzymes are required in that pathway, or 2-oxoacid elongation proceeds through a simpler, evolutionarily conserved pathway using only (R)-homocitrate analogs as shown in Fig. 1A (28).
The rate of M. jannaschii HACN-catalyzed cis-homoaconitate hydration is similar to the rate for the T. thermophilus HACN, which has a similar turnover (1.3 s–1) but a lower Km (8.2 μm) (15). Furthermore, the M. jannaschii HACN uses cis-homoaconitate analogs with two to five methylene groups in the γ-carboxylate chain, all with similar specificity constants. This broad substrate specificity is surprising given our inability to detect the hydration of cis-aconitate, which has a single methylene group. The M. jannaschii HACN also catalyzes the hydration of the minimal substrate maleate that has no γ-carboxylate chain. This reaction exclusively produces (R)-malate, suggesting that the binding modes for the maleate and homoaconitate substrates are the same. Neither maleate nor the longer chain analogs of cis-homoaconitate have been tested as substrates for the yeast or T. thermophilus HACN, so it is unclear whether this broad substrate specificity is unique to the methanogen HACN.
Compared with the rates of homoaconitate hydration catalyzed by the M. jannaschii HACN, the rate of (R)-homocitrate dehydration is slow. The same discrepancy was observed in the kinetic analysis of the M. jannaschii IPMI, which catalyzed the dehydration of (R)-citramalate and 2-isopropylmalate with kcat/Km values of 2 × 103 m–1 s–1, more than 2 orders of magnitude lower than the specificity constant for citraconate hydration (19). Future studies may determine the equilibrium constant for this reaction and distinguish the different modes of substrate binding to HACN.
Almost all of the information about the reaction mechanisms of IPMI and HACN proteins has been inferred from aconitase studies (29). Crystal structure models of mitochondrial aconitase with substrate analogs indicate that the protein undergoes long range conformational shifts upon substrate binding (30). The side chains of three residues, Gln72, Arg580, and Arg644 form hydrogen bonds with the γ-carboxylate of isocitrate. The Gln72 residue is replaced with His in both IPMI and HACN proteins. The Arg644 is universally conserved in the small subunit proteins of IPMI and HACN. Therefore these two residues cannot affect substrate specificity.
Aconitase domain 4 (corresponding to the small subunit proteins of IPMI and HACN) contains a flexible loop with Arg580. The sequence of this loop is highly variable and appears to be the best predictor of substrate specificity; archaeal IPMI subunits have a consensus sequence motif of YLV(I/Y/M), whereas archaeal HACN subunits have a consensus sequence of YLRT. The residues in the third and fourth positions of this motif are hydrophobic in IPMI proteins, which could accommodate the hydrophobic γ-chains of their substrates. In contrast the HACN subunits have basic arginine and polar threonine residues that could form an ion pair or hydrogen bond with the γ-carboxylate.
Most crenarchaea have only one HACN/IPMI homolog, even though these microbes apparently use both the isopropylmalate pathway and the α-aminoadipate pathway. Therefore a single isomerase functions in both pathways, suggesting that the ancestral protein could have recognized a broad pool of hydroxyacid substrates. The consensus sequence for the crenarchaeal flexible loop region is YL(K/V)Y, which contains a mixture of residues from both HACN and IPMI motifs. To identify substrate specificity determinants will require cocrystal structure models of HACN holoenzyme bound to a substrate analog. Future studies will also replace the amino acids in the HACN motif to change the substrate specificity of the enzyme.
The aconitase crystal structure models show that the same residues bind citrate, cis-aconitate, and isocitrate (29). This symmetry makes the inability of the yeast and bacterial HACN proteins to catalyze the dehydration of homocitrate puzzling. The yeast Lys4p HACN homologs have a highly conserved flexible loop consensus sequence of YTYQ, where the tyrosine or glutamine residues could form a hydrogen bond with the γ-carboxylate of cis-homoaconitate. The bacterial HACNs have a conserved loop sequence of YAP(Y/F), suggesting that steric effects or amide backbone interactions with the substrate may be more important than ion pair formation in these proteins. However, changes in the length or conformation of the loop could also affect binding, so additional experiments will be required to determine whether this region is fully modular and responsible for the HACN specificity.
The similarity among 2-oxoacid elongation reactions of the Krebs cycle, leucine biosynthesis, and the α-aminoadipate pathway has been recognized for at least 30 years, along with the proposal that the respective enzymes share common ancestors (5). The homocitrate (re)-synthase belongs to the aldolase superfamily. It is related to isopropylmalate synthase and the recently discovered citrate (re)-synthase but not to the common citrate (si)-synthase (31), demonstrating convergent evolution. Aconitase, HACN, and IPMI all belong to the same family of [Fe-S] dehydratases. We have shown that M. jannaschii HACN and IPMI have relatively broad substrate specificities. The isocitrate, homoisocitrate, and isopropylmalate dehydrogenases belong to another protein superfamily. We have also shown that M. jannaschii HICDH and isopropylmalate dehydrogenase recognize diverse dehydratase products with (2R,3S)-stereocenters. Therefore the invention of an acetyl-transferase with new substrate specificity probably spurred the evolution of each 2-oxoacid elongation pathway. This model is consistent with the parallel evolution of HACN activity several times within the dehydratase enzyme family (Fig. 6) and the conserved malease activity.
FIGURE 6.
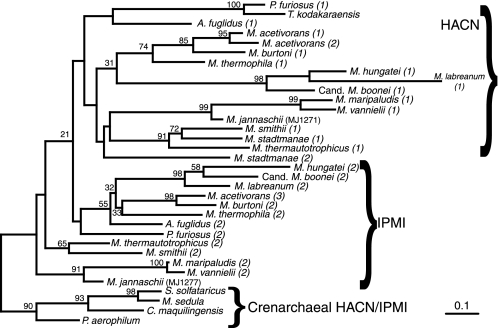
Phylogeny of small subunit IPMI and HACN proteins from archaea. The phylogeny was inferred by the protein maximum likelihood method and rooted using the crenarchaeal IPMI/HACN homologs as an out-group. The bootstrap values are shown near branches supported by a plurality of 100 trees inferred by the same method. Only the functions of the MJ1277 (IPMI) and MJ1271 (HACN) proteins have been experimentally confirmed, but related proteins predicted to catalyze each reaction are grouped by braces. The numbers in parentheses differentiate the IPMI and HACN paralogs in each species. The full organism names and sequence accession numbers are listed in the supplemental materials. The scale bar indicates 0.1 amino acid changes expected per site.
Supplementary Material
Acknowledgments
We thank Mehdi Moini and Mark Fountain for assistance with mass spectrometry and Philip Magnus and Ben Shoulders for helpful discussions.
This work was supported in part by National Science Foundation Grant MCB-0817903, Petroleum Research Foundation Grant 44382-G4, and Welch Foundation Grant F-1576. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EU447775.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and supplemental tables.
Footnotes
The abbreviations used are: CoM, coenzyme M; CoB, coenzyme B; HCS, homocitrate synthase; HACN, homoaconitase; HICDH, homoisocitrate dehydrogenase; IPMI, isopropylmalate isomerase; CHES, 2-(cyclohexylamino)ethanesulfonic acid; TAPS, tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid; HPLC, high pressure liquid chromatography; LC-MS, liquid chromatography-mass spectrometry.
References
- 1.Horng, Y.-C., Becker, D. F., and Ragsdale, S. W. (2001) Biochemistry 40 12875–12885 [DOI] [PubMed] [Google Scholar]
- 2.Noll, K. M., Rinehart, K. L., Jr., Tanner, R. S., and Wolfe, R. S. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ermler, U., Grabarse, W., Shima, S., Goubeaud, M., and Thauer, R. K. (1997) Science 278 1457–1462 [DOI] [PubMed] [Google Scholar]
- 4.White, R. H. (1989) Biochemistry 28 860–865 [Google Scholar]
- 5.Jensen, R. A. (1976) Annu. Rev. Microbiol. 30 409–425 [DOI] [PubMed] [Google Scholar]
- 6.Xu, H., Andi, B., Qian, J., West, A. H., and Cook, P. F. (2006) Cell Biochem. Biophys. 46 43–64 [DOI] [PubMed] [Google Scholar]
- 7.Howell, D. M., Harich, K., Xu, H., and White, R. H. (1998) Biochemistry 37 10108–10117 [DOI] [PubMed] [Google Scholar]
- 8.Bentley, R. (1970) in Molecular Asymmetry in Biology (Bentley, R., ed) pp. 90–163, Academic Press, New York
- 9.Thomas, U., Kalyanpur, M. G., and Stevens, C. M. (1966) Biochemistry 5 2513–2516 [DOI] [PubMed] [Google Scholar]
- 10.Curatti, L., Hernandez, J. A., Igarashi, R. Y., Soboh, B., Zhao, D., and Rubio, L. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 17626–17631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell, D. M., Graupner, M., Xu, H., and White, R. H. (2000) J. Bacteriol. 182 5013–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto, T., Miyazaki, K., and Eguchi, T. (2007) Bioorg. Med. Chem. 15 1346–1355 [DOI] [PubMed] [Google Scholar]
- 13.Strassman, M., and Ceci, L. N. (1966) J. Biol. Chem. 241 5401–5407 [PubMed] [Google Scholar]
- 14.Weidner, G., Steffan, B., and Brakhage, A. A. (1997) Mol. Gen. Genet. 255 237–247 [DOI] [PubMed] [Google Scholar]
- 15.Jia, Y., Tomita, T., Yamauchi, K., Nishiyama, M., and Palmer, D. R. J. (2006) Biochem. J. 396 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimek, T. L., and Escalante-Semerena, J. C. (2004) J. Bacteriol. 186 454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kollmann-Koch, A., and Eggerer, H. (1984) Hoppe-Seyler's Z. Physiol. Chem. 365 847–857 [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharjee, J. K. (1992) in The Evolution of Metabolic Function (Mortlock, R. P., ed) pp. 47–80, CRC Press, Boca Raton, FL
- 19.Drevland, R. M., Waheed, A., and Graham, D. E. (2007) J. Bacteriol. 189 4391–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, G., and Palmer, D. R. J. (2000) Tetrahedron Lett. 41 9209–9212 [Google Scholar]
- 21.Massoudi, H. H., Cantacuzene, D., Wakselman, C., and de la Tour, C. B. (1983) Synthesis 12 1010–1012 [Google Scholar]
- 22.Nelson, R. B., and Gribble, G. W. (1973) Org. Prep. Proc. Int. 5 55–58 [Google Scholar]
- 23.Clive, D. L. J., Coltart, D. M., and Zhou, Y. (1999) J. Org. Chem. 64 1447–1454 [DOI] [PubMed] [Google Scholar]
- 24.Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., Blake, J. A., FitzGerald, L. M., Clayton, R. A., Gocayne, J. D., Kerlavage, A. R., Dougherty, B. A., Tomb, J.-F., Adams, M. D., Reich, C. I., Overbeek, R., Kirkness, E. F., Weinstock, K. G., Merrick, J. M., Glodek, A., Scott, J. L., Geoghagen, N. S. M., Smith, H. O., Woese, C. R., and Venter, J. C. (1996) Science 273 1017–1140 [DOI] [PubMed] [Google Scholar]
- 25.Wu, S.-p., Wu, G., Surerus, K. K., and Cowan, J. A. (2002) Biochemistry 41 8876–8885 [DOI] [PubMed] [Google Scholar]
- 26.Beinert, H. (1983) Anal. Biochem. 131 373–378 [DOI] [PubMed] [Google Scholar]
- 27.Henson, C. P., and Cleland, W. W. (1967) J. Biol. Chem. 242 3833–3838 [PubMed] [Google Scholar]
- 28.Graham, D. E., and White, R. H. (2002) Nat. Prod. Rep. 19 133–147 [DOI] [PubMed] [Google Scholar]
- 29.Beinert, H., Kennedy, M. C., and Stout, C. D. (1996) Chem. Rev. 96 2335–2374 [DOI] [PubMed] [Google Scholar]
- 30.Lauble, H., Kennedy, M. C., Beinert, H., and Stout, C. D. (1992) Biochemistry 31 2735–2748 [DOI] [PubMed] [Google Scholar]
- 31.Li, F., Hagemeier, C. H., Seedorf, H., Gottschalk, G., and Thauer, R. K. (2007) J. Bacteriol. 189 4299–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.