Abstract
The human pathogen Helicobacter pylori influences cell adhesion, proliferation, and apoptosis and is involved in gastric adenocarcinoma formation. In our study we analyzed the impact of H. pylori infection on the regulation of β-catenin, which plays a central role in both cell adhesion and tumorigenesis. Infection of Madin-Darby canine kidney cells with H. pylori led to suppression of Ser/Thr phosphorylation and ubiquitin-dependent degradation of β-catenin and to up-regulation of lymphoid enhancer-binding factor/T cell factor (LEF/TCF)-dependent transcription. The impaired Ser/Thr phosphorylation of β-catenin was accompanied by an increase of glycogen synthase kinase 3β phosphorylation. Inhibition of Akt kinase, an up-stream regulator of glycogen synthase kinase 3, by a specific inhibitor Akti-1/2 or depletion of Akt with siRNA restored Ser/Thr phosphorylation of β-catenin. We conclude that glycogen synthase kinase 3β activity exerts an important role in β-catenin regulation and LEF/TCF transactivation in H. pylori-infected Madin-Darby canine kidney cells.
The highly adapted pathogen Helicobacter pylori persists life-long within the human stomach. Under certain host and environmental conditions H. pylori induces chronic inflammation of the gastric mucosa and contributes to the development of peptic ulcer, gastric malignancies, and mucosa-associated lymphoid tissue lymphomas (1). Bacteria-host interaction and disease progression depend on different bacterial factors, among them VacA, Fld, BabA2, SabA, and CagA (2-5). Gastric carcinogenesis was shown to be associated with alterations in E-cadherin/catenin signaling (6, 7), but the role of β-catenin in H. pylori infection was not clarified so far (8-10).
β-Catenin is a multifunctional protein that (i) participates in cell adhesion by bridging E-cadherin to α-catenin and (ii) mediates transcriptional regulation by forming a complex with LEF/TCF2 transcription factors (11, 12). β-Catenin redistribution in the cell is tightly regulated and, as was proposed by Gottardi and Gumbiner (13), could be dictated by the existence of distinct molecular forms of β-catenin with different affinity to E-cadherin and TCFs. The level of free β-catenin in the cytoplasm is controlled by ubiquitinylation and degradation in the 26 S proteasome (14). The multiprotein destruction complex involves axin and adenomatous polyposis coli (APC) scaffolds which bind β-catenin to facilitate its phosphorylation by casein kinase 1 at Ser-45 and by GSK3β at Ser-33, Ser-37, and Thr-41 (15). Ubiquitinylation and degradation of N-terminal-phosphorylated β-catenin is mediated by the cullin-RING ubiquitin ligase Skp1-Cul1-F-box (SCFβ-TrCP) (16, 17). If some components of the degradation complex are mutated or if wingless-type (Wnt) ligands bind to Frizzled membrane receptors and interfere with the multi-protein destruction complex, non-phosphorylated β-catenin accumulates and co-activates TCF-dependent expression of target genes, among them c-myc, c-jun, axin2, twin, ubx, MMP7, and cyclin D1 (18-20). The fact that H. pylori infection leads to up-regulation of Wnt10A and activates β-catenin target genes (21-23) contributes to the hypothesis that H. pylori induces activation of the β-catenin-TCF pathway during human gastric cancer development. The precise mechanism by which H. pylori could influence β-catenin nuclear activity remains obscure. Disruption of adherens junctions by H. pylori (24) could promote β-catenin translocation into the nucleus, as was recently shown using cultured gastric cancer cells MKN-28 and MKN-45 (25).
In this work we investigated β-catenin regulation in H. pylori-infected epithelial cells. Herein, we report that 1) H. pylori suppresses GSK3β activity in a CagA-independent manner, 2) H. pylori-induced suppression of GSK3β leads to inhibition of Ser/Thr phosphorylation, ubiquitinylation, and degradation of β-catenin, and 3) H. pylori stimulates β-catenin-dependent LEF/TCF transactivation activity and causes up-regulation of cyclin D1.
EXPERIMENTAL PROCEDURES
Materials—Antibodies used in the work were phospho-β-catenin (Ser-33/Ser-37/Thr-41), phospho-GSK3α/β, histone H3, Akt, and phospho-Akt (Cell Signaling Technology Inc.), β-catenin, GSK3β, and occludin (BD Biosciences), ubiquitin (Berkeley Antibody Co., Inc.), actin and phospho-tyrosine (PY99) (Santa Cruz Biotechnology), cyclin D1 (United States Biological/Biomol), glyceraldehyde-3-phosphate dehydrogenase (Chemicon International), CagA (26). MG132, Akti-1/2 (Akt inhibitor VIII), LY294002, AG1478, AG828, and U73122 were purchased from Calbiochem. LiCl and DMSO were purchased from Sigma. PHA-665752 was kindly provided (Pfizer Global Pharmaceuticals).
Cell Culture and Bacteria—MDCK cell line (European Collection of Cell Cultures) and AGS cells were cultured in RPMI 1640 medium (PAA Laboratories) supplemented with 10% fetal calf serum and penicillin/streptomycin in a humid atmosphere at 37 °C with 5% CO2. For infection with H. pylori, MDCK cells (2 × 104 cells/35-mm dish) were grown in complete medium to reach 70% of confluency. 16 h before infection the complete medium was replaced by fresh RPMI 1640 supplemented with 0.5% fetal calf serum.
The H. pylori strain P1 wild type (wt) and isogenic mutants cagA and virB7 (26, 27) were cultured for 48-72 h as described (28) and added to the epithelial cells at a multiplicity of infection of 100. Heat-inactivated H. pylori was prepared by incubating bacteria suspension at 65 °C for 15 min.
Transfection and Reporter Gene Assay—pSP65/SRα-CagA-HA plasmid encoding CagA was kindly provided by M. Hatakeyama (Sapporo, Japan). MDCK cells (2 × 104 cells/35-mm dish) were transfected with 0.1 μg of DNA (DNA/Effectene transfection reagent (Qiagen) ratio was 1:10). 48 h later the cells were subjected either to subcellular fraction separation or to cell lysate preparation.
MDCK cells (2 × 104 cells/35 mm dish) were transfected with 0.1 μg of siRNA against human Akt1/2 (100% sequence homology to canine Akt2 mRNA) or with control (non-targeting) siRNA (Santa Cruz Biotechnology) using siLentFect™ Lipid Reagent (Bio-Rad). At 24 h of transfection, cells were infected with H. pylori for 24 h.
For luciferase reporter assay, MDCK cells were seeded into 24-well plates at a density of 0.6 × 104 cells per well 1 day before transfection. The cells were transfected with 0.1 μg of Firefly TOP- or FOPflash (Biomol) and 0.01 μg of Renilla (Promega) luciferase plasmids using Effectene transfection reagent. Luciferase activity was estimated using the Dual Luciferase Reporter Assay System (Promega) at Lumat LB 9507 luminometer (Berthold Technologies). All samples were assayed in duplicate.
RNA Isolation and Reverse Transcription-PCR—Total RNA was extracted by TRIzol reagent (Invitrogen). cDNA was synthesized from 1 μg of RNA using random hexamer primers and a RevertAid™ First Strand cDNA Synthesis kit (Fermentas). 1/20 of cDNA mixture was combined with primers (0.25 μm), fluorescein (1:105; Bio-Rad) and components of the SensiMix DNA kit (Quantace) according to the manufacture's instructions and amplified in the iCycler (Bio-Rad). The following iCycler protocol was used: denaturation (95 °C for 10 min), amplification and quantification (40 cycles: 95 °C for 15 s, 60 °C for 30 s, 72 °C for 60 s), melting curve program (72-95 °C; 0.5 °C/s), and cooling to 22 °C. Primers 5′-TACTGAGCCTGCCATCTGTGC-3′ (forward) and 5′-CCTCCACAAACTGCTGCTGTG-3′ (reverse) and primers 5′-AAGATGACTCAGATCATGTTCGAG-3′ (forward) and 5′-AGGGGCGATGATCTTGATCTTCAT-3′ (reverse) were used for amplification of β-catenin and β-actin fragment, respectively. Serial dilutions of the dipeptidyl peptidase IV gene cloned into a pCRR2.1-TOPO vector and primers 5′-GATGCTACAGCTGACAGTCGC-3′ (forward) and 5′-TGGTGACCATGTGACCCACTG-3′(reverse) served for generation of a calibration curve.
Preparation of Cell Lysates, Fractions, and Immunoprecipitation—Whole cell extracts were prepared with modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm EDTA, 1% Triton X-100, 10% glycerol, 10 mm K2HPO4, 0.5% Nonidet P40, 1× protease inhibitor mixture (Roche Applied Science), 1 mm Na3VO4, 1 mm Na2MoO4, 20 mm NaF, 0.1 mm PMSF, 20 mm β-glycerol-2-phosphate, 10 mm Na4P2O7). Aliquots of the lysates were boiled with sample buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 100 mm dithiothreitol, 0.1% bromphenol blue) for 5 min. Subcellular fractions of MDCK were prepared with ProteoExtract kit (Calbiochem) according to the manufacturer's instructions. For immunoprecipitation, the lysates were incubated with appropriate antibodies for 4 h to overnight at 4 °C. The immune complexes were trapped on protein G-Sepharose (Amersham Biosciences), washed, boiled in the sample buffer, and subjected to Western blotting.
Pulse-Chase Assay—Cells were washed and incubated in the pulse medium (methionine/cysteine-free RPMI 1640 (Sigma), 2 mm l-glutamine (Sigma), 0.5% dialyzed fetal bovine serum (Invitrogen)) for 1 h. The cells were labeled with [35S]methionine/cysteine using the Amersham Biosciences Redivue Pro-Mix Cell Labeling Mix, 0.05 mCi (with respect to l-[35S]methionine) per 1 ml of pulse medium for 1 h. Labeled cells were washed twice with pulse medium and chased in complete RPMI 1640, 0.5% fetal calf serum for 2-8 h, after which β-catenin was immunoprecipitated from the cell lysates. Immunocomplexes were separated by 8% SDS-PAGE; the gels were incubated in Amplify Fluorographic Reagent (Amersham Biosciences), dried, and exposed to film. A part of the samples was subjected to Western blotting.
Western Blotting—The proteins were separated by SDS-PAGE, electrotransferred to Immobilon-P transfer polyvinylidene fluoride membrane (Millipore), and stained with specific antibodies as described (29). Immunoreactivity was detected using the enhanced chemiluminescence detection kit ECL™ (Amersham Biosciences). The protein bands on x-ray films were scanned by using VersaDoc Imaging System (Bio-Rad) and analyzed using Quantity One software (Bio-Rad). By applying several software functions, the Quantity One software allows subtraction of the background from the images. The data from three independent experiments were used for the calculations of the results.
Immunofluorescence—Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, permeabilized, and blocked in PBS, 0.1% Triton X-100, 5% goat serum (Sigma) for 30 min on ice. Then the cells were incubated with the first antibody for 2 h at room temperature (RT), washed, and exposed for indocarbocyanine-conjugated secondary antibody (Dianova) for 1 h at RT. Slides were analyzed by Leica DMRE 7 fluorescence microscope.
Statistical Analysis—The statistical analysis of the results was done using Student's t test. p < 0.05 was considered significant. Data are expressed as the mean ± S.E.
RESULTS
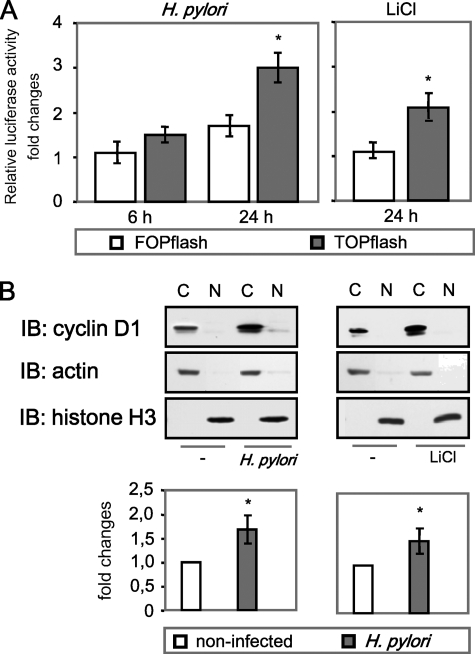
H. pylori Induces LEF/TCF Transactivation and Regulates Cyclin D1 Expression—To investigate whether H. pylori controls LEF/TCF activity, non-tumor epithelial MDCK cells were transfected with luciferase reporter plasmid containing either three copies of wild type (TOPflash) or mutated (FOPflash) LEF/TCF binding sites and infected with H. pylori. At different periods of time, cells were collected, and luciferase activity was measured. The firefly luciferase activity was normalized relative to renilla luciferase activity. As shown in Fig. 1A, H. pylori regulates luciferase activity within 24 h post-infection (1.7-fold compared with FOPflash). LiCl, a known activator of β-catenin signaling (30), was used as a positive control. LEF/TCF transcription factors are known to bind to cyclin D1 promoter and regulate gene expression (20). Thus, we tested the ability of the bacteria to regulate cyclin D1. We found an approximate 2.1 and 1.9-fold increase of cyclin D1 mRNA in MDCK cells infected with H. pylori for 10 and 24 h, respectively (data not shown). To investigate protein levels of cyclin D1, cytosolic and nuclear fractions of MDCK cells were subjected to Western blot analysis. As shown in Fig. 1B, infection with H. pylori led to an increase of cyclin D1 in the cytosol of the cells. A similar effect on cyclin D1 expression was achieved in MDCK cells by treatment with LiCl (10 μm, 24 h). Thus, H. pylori stimulates β-catenin-dependent LEF/TCF transactivation activity and causes an increase of cyclin D1 level in MDCK cells.
FIGURE 1.
H. pylori induces LEF/TCF transactivation and regulates cyclin D1 expression. A, cells were transiently transfected with either TOPflash or FOPflash and cotransfected with renilla luciferase plasmid. 24 h later the cells were infected with H. pylori wt or treated with LiCl and lysed, and firefly/renilla luciferase activity was measured. Graphs represent the mean -fold changes ±S.E. of three separate experiments performed in duplicate. *, p < 0.05, relative to activity in FOPflash transfected cells. B, cells were infected with H. pylori or stimulated with LiCl for 24 h. Subcellular fractions were separated by using the ProteoExtract kit, and Western blot (IB) analysis was performed using specific antibodies as indicated. The graph represents the mean -fold changes of band intensity from three separate experiments ±S.E. with the value for the control as 1 arbitrarily. Actin and histone H3 were immunodetected to show appropriate separation of cellular fractions as well as equal protein amounts in the cytosolic and nuclear fractions, respectively. C, cytosol; N, nuclear fraction. *, p < 0.05, relative to non-infected cells.
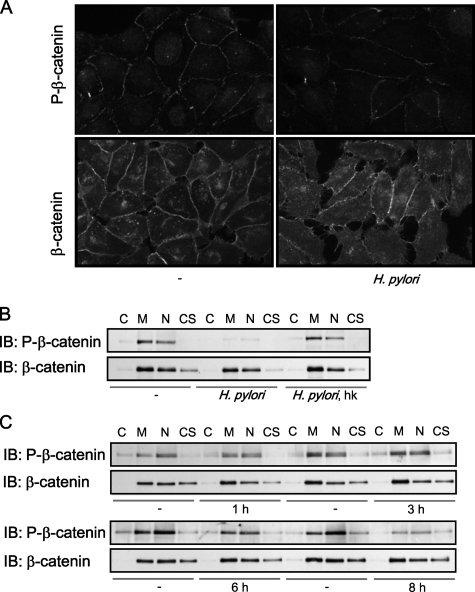
H. pylori Inhibits Ser/Thr Phosphorylation of β-Catenin—Studying phosphorylation status and localization of β-catenin in H. pylori-infected cells, fluorescence microscopy revealed that Ser-33/Ser-37/Thr-41-phosphorylated β-catenin was localized at cell-cell contacts and in nuclei of non-stimulated cells. Total β-catenin was detected at cell-cell contacts (Fig. 2A). Ser/Thr-phosphorylated β-catenin was prominently removed from adherens junctions, and cytosolic staining of β-catenin increased at 24 h post-infection (Fig. 2A). We failed to see any changes of β-catenin amount in H. pylori-infected cells in comparison to non-infected MDCK cells in a Western blot analysis of subcellular fractions (Fig. 2B). We observed a prominent decrease of Ser/Thr-phosphorylated β-catenin in membranes/organelles and nuclear fractions of cells infected with H. pylori for 24 h (Fig. 2B). Heat-inactivated H. pylori had no effect on β-catenin phosphorylation (Fig. 2B). Thus, physical interaction of living bacteria with the host cell is requested for β-catenin regulation. Decrease of β-catenin phosphorylation in H. pylori-infected MDCK cells was detected at 6 h and more prominently at 8 h post-infection (Fig. 2C). Treatment of MDCK cells with LiCl provoked similar suppression of Ser/Thr phosphorylation of β-catenin (supplemental Fig. S1).
FIGURE 2.
H. pylori inhibits Ser/Thr phosphorylation of β-catenin. A, cells were stimulated with H. pylori for 24 h and analyzed by immunofluorescence using antibodies against Ser-33/Ser-37/Thr-41-phospho-β-catenin or β-catenin. B, cells were infected with H. pylori for 24 h. Subcellular fractions were separated by using ProteoExtract kit, and Western blot (IB) analysis was performed using antibodies as indicated. C, cytosol; M, membrane/organelles fraction; N, nuclear fraction; CS, cytoskeleton; hk, heat-killed H. pylori. C, time kinetics.
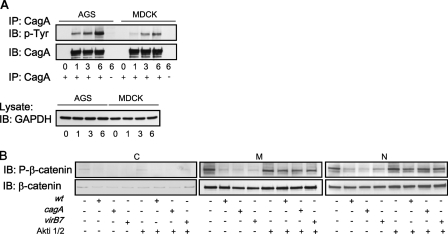
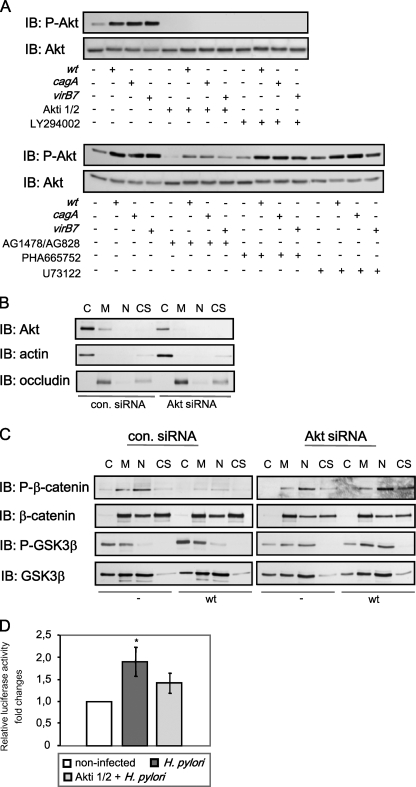
Regulation of β-Catenin Phosphorylation in H. pylori Infection is CagA-independent—The H. pylori type 4 secretion system (T4SS) serves for delivery of CagA and other yet unknown bacterial effectors into epithelial cells (31). CagA is effectively translocated into MDCK cells, similar to gastric AGS cells, where it becomes phosphorylated at tyrosine residues (Fig. 3A). To investigate the role of CagA and the functional T4SS in H. pylori-induced changes of β-catenin signaling, we infected MDCK cells with H. pylori P1 isogenic mutants lacking cagA or virB7 for 24 h. Compared with the H. pylori wt, cagA and virB7 mutants also inhibited β-catenin phosphorylation (Fig. 3B).
FIGURE 3.
Regulation of β-catenin in H. pylori infection is CagA-independent. A, H. pylori CagA is translocated into and phosphorylated at tyrosine residues in both AGS and MDCK cells. Cells were infected with H. pylori for the indicated periods of time, lysed, and incubated with anti-CagA antibody. Western blot (IB) analysis of immunoprecipitates (IP) was performed using anti-phospho-Tyr-99 antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was immunodetected to show equal protein amounts in the samples subjected to immunoprecipitation. B, MDCK cells were infected with H. pylori wt or cagA or with virB7 mutants for 24 h in the absence or presence of Akti-1/2. Subcellular fractions were separated by using the ProteoExtract kit, and Western blot analysis was performed using antibodies indicated. C, cytosol; M, membrane/organelles fraction; N, nuclear fraction.
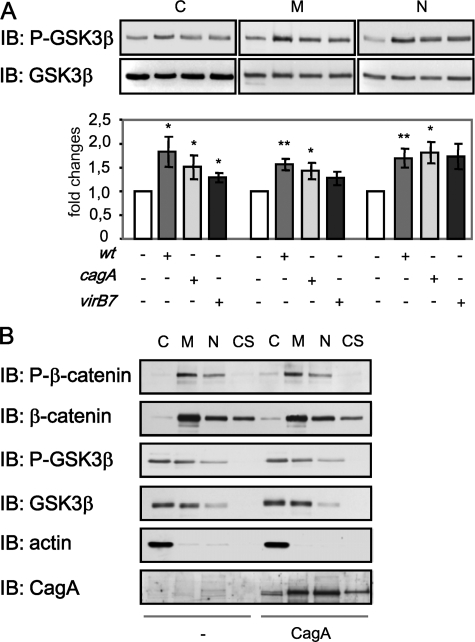
H. pylori Suppresses Ser/Thr Phosphorylation of β-Catenin in a GSK3β-dependent Manner—Ser-33, Ser-37, and Thr-41 of β-catenin are known target sites for GSK3β which phosphorylates and thereby marks the protein for ubiquitinylation and subsequent proteasomal degradation (32). Thus, decrease of Ser/Thr phosphorylation in the β-catenin protein can be caused either by GSK3β inhibition or by degradation of β-catenin. Because the total amount of β-catenin in infected cells was not affected (Figs. 2B and 3B), we supposed that GSK3β inhibition could be responsible for the suppression of β-catenin phosphorylation. GSK3β activity is negatively regulated through its phosphorylation at Ser-9 by Akt kinase (33). Western blot analysis of different cellular fractions revealed an induction of GSK3β Ser-9 phosphorylation by the wt strain of H. pylori and slightly less prominently by cagA and virB7 mutant strains (Fig. 4A). GSK3β was detected in nuclear fractions; this could be a consequence of partial serum deprivation during the experiments, as has been described (34). To investigate whether CagA is involved in the regulation of β-catenin phosphorylation, we transiently transfected MDCK cells with a plasmid encoding CagA. As shown in Fig. 4B, neither GSK3β nor β-catenin phosphorylations were affected in MDCK cells transfected with the plasmid encoding CagA. Therefore, H. pylori inhibits GSK3β in a CagA-independent manner, which leads to inhibition of Ser/Thr phosphorylation of β-catenin in infected cells.
FIGURE 4.
H. pylori induces GSK3β phosphorylation. A, MDCK cells were infected with H. pylori wt or cagA and virB7 mutants for 24 h, and subcellular fractions were separated by using a ProteoExtract kit. Western blot (IB) analysis was performed using anti-Ser-9-phospho-GSK3β antibody. Graphs represent the mean -fold changes of band intensity from three separate experiments ±S.E. with the value for the control as 1 arbitrarily. *, p < 0.05; **, p < 0.01, relative to non-infected cells. B, MDCK cells were transiently transfected with a plasmid encoding for CagA for 48 h. Whole cell lysates were prepared, and subcellular fractions were separated by using ProteoExtract kit. Western blot analysis was performed using antibodies as indicated. C, cytosol; M, membrane/organelles fraction; N, nuclear fraction; CS, cytoskeleton.
GSK3β is known to be regulated upon Akt kinase (35). To investigate up-stream regulators and to confirm that GSK3β plays a role in H. pylori-induced suppression of β-catenin phosphorylation, we infected cells in the presence of Akti-1/2, an allosteric inhibitor of Akt1 and Akt2 isozymes. This substance promotes the formation of an inactive conformation of Akt in a pleckstrin homology-dependent manner and selectively inhibits phosphorylation and kinase activity (36). The inhibitor does not exhibit any effect on protein kinases A and C even at concentration as high as 50 μm. In MDCK cells, Akti-1/2 (2 μm) blocked effectively Ser-473 phosphorylation of Akt kinase induced by H. pylori (Fig. 5A) and prevented bacteria-induced suppression of Ser/Thr phosphorylation of β-catenin (Fig. 3B). Viability of H. pylori was not affected by inhibitor treatment (data not shown). Stimulation of Akt kinase activity by H. pylori has been also observed in AGS cells (28). AGS cells infected with H. pylori also exert suppression of β-catenin phosphorylation, and Akti-1/2 was able to restore β-catenin phosphorylation (data not shown).
FIGURE 5.
H. pylori suppresses Ser/Thr phosphorylation of β-catenin in a GSK3β-dependent manner. A, MDCK cells were infected with H. pylori wt or cagA and virB7 mutants for 6 h in presence or absence of selective inhibitors as indicated. Whole cell lysates were prepared, and Western blot (IB) analysis was performed using antibodies indicated. B, siRNA specifically impairs Akt level in MDCK cells; the cells were transiently transfected with control siRNA or siRNA against Akt. At 48 h post-transfection subcellular fractions were separated by using the ProteoExtract kit, and Western blot analysis was performed using the antibodies indicated. C, cytosol; M, membrane/organelles fraction; N, nuclear fraction; CS, cytoskeleton. C, MDCK cells were transiently transfected with control siRNA or siRNA against Akt kinase and infected with H. pylori wt for 24 h. Subcellular fractions were separated and analyzed as described above in B. D, cells were transfected and treated with H. pylori wt for 24 h as described in Fig. 1A in the presence or absence of Akti-1/2. *, p < 0.05, relative to non-infected cells.
To unravel putative upstream components that could direct Akt activation, we pretreated the cells before infection with the following inhibitors: LY294002 (20 μm), an inhibitor of phosphoinositide 3-kinase (PI3K); AG1478/AG828 (5 μm), an inhibitor of epidermal growth factor (EGF) and Her2/Neu receptors; PHA-665752 (0.2 μm), an inhibitor of c-Met receptor; U73122 (5 μm), an inhibitor of phospholipase C (Fig. 5A).
Pretreatment of the cells with LY294002 (Fig. 5A) completely inhibited Akt phosphorylation induced by H. pylori P1 wt and isogenic mutants. Thus, we suggest that H. pylori induces T4SS-independent PI3K activity leading to Akt activation. PI3K activity can be induced by EGF receptor (EGFR) or c-Met, which become active in H. pylori infection (37, 28). Inhibition of EGFR activity significantly reduced H. pylori-induced phosphorylation of Akt, whereas inhibition of c-Met had no effect on phosphorylation of Akt kinase (Fig. 5A). EGFR activation in gastric epithelial cells was shown to be stimulated by H. pylori in a T4SS-independent manner (37). Thus, our results allow us to suppose that EGFR activation could contribute to Akt phosphorylation induced by H. pylori in a T4SS-independent manner. Inhibition of PLC had no effect on Akt phosphorylation in H. pylori-infected cells (Fig. 5A).
To confirm the data obtained by using Akt kinase inhibitor, Akt depletion with siRNA was performed. At 48 h of transfection, the amount of Akt in MDCK cell lysates was significantly decreased (Fig. 5B). As shown in Fig. 5C, anti-Akt siRNA prevented inhibitory GSK3β phosphorylation, leading to β-catenin phosphorylation in MDCK cells infected with H. pylori. Therefore, Akt kinase, an upstream modulator of GSK3β, is causally related to regulation of β-catenin signaling in H. pylori-infected cells.
Interestingly, besides restoration of the β-catenin phosphorylation, Akti-1/2 inhibited LEF/TCF transactivation in H. pylori-infected cells (Fig. 5D). These data indicate the important role of Akt/GSK3β and Ser/Thr phosphorylation of β-catenin in the regulation of β-catenin nuclear activity during H. pylori infection.
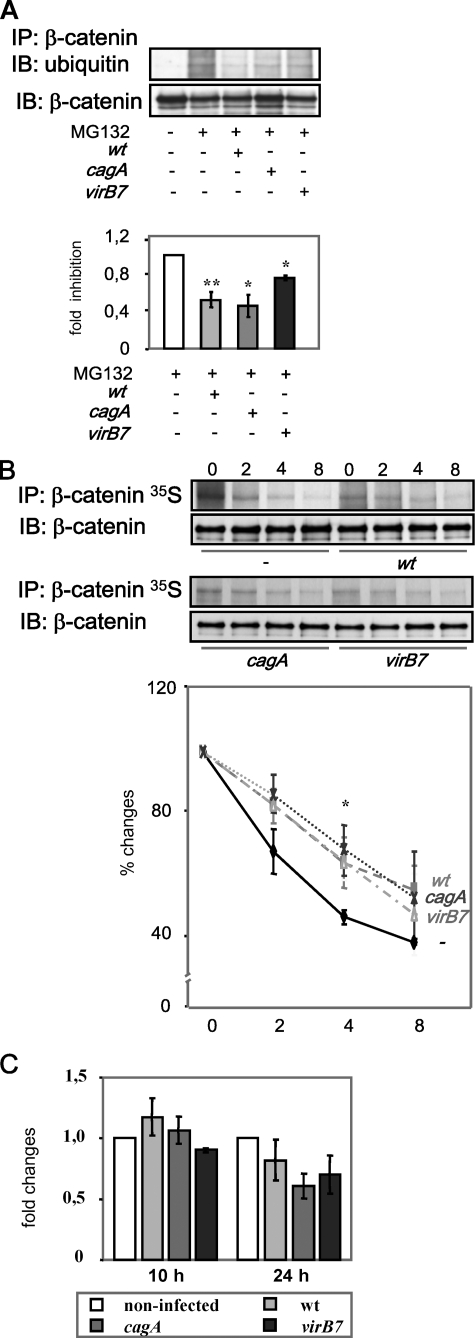
H. pylori Affects Ubiquitinylation of β-Catenin—Because Ser/Thr phosphorylation of β-catenin has been shown to be essential for the protein ubiquitinylation (38), we investigated whether H. pylori affects this process. MDCK cells were infected in the presence of the 26 S proteasome inhibitor MG132 (20 μm), and the lysates were subjected to immunoprecipitation with a β-catenin-specific antibody. Western blot analysis revealed a reduction of the ubiquitin level in β-catenin immunoprecipitates from MDCK cells infected by H. pylori, with T4SS mutant being less effective (Fig. 6A). The same result was obtained by using N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (25 μm), another inhibitor of proteasomal degradation (data not shown). Ubiquitinylation is known to be a prerequisite for β-catenin degradation. Thus, we examined further stability of β-catenin by pulse-chase analysis after 10 h of co-culture with H. pylori. Infection of cells with wt or cagA and virB7 mutants led to delayed β-catenin degradation within the periods of time analyzed (Fig. 6B). Therefore, H. pylori regulates β-catenin signaling by suppression of β-catenin ubiquitinylation and degradation in MDCK cells, and this effect is CagA-independent.
FIGURE 6.
H. pylori affects ubiquitinylation of β-catenin. A, MDCK cells were infected with H. pylori wt or cagA and virB7 mutants for 8 h in presence of MG132 and lysed, and the lysates were incubated with anti-β-catenin antibody. Western blot (IB) analysis of immunoprecipitates (IP) was performed using specific antibodies as indicated. B, MDCK cells were infected for 10 h, pulse-labeled, and then chased with nonradioactive medium for next 2-8 h. 35S incorporation was detected in β-catenin immunoprecipitates by radioautography. The graph represents the mean relative band intensity from 3 separate experiments ±S.E. with the value for appropriate control as 100%. *, p < 0.05, **, p < 0.01, relative to non-infected cells. C, H. pylori does not influence β-catenin transcription. MDCK cells were infected with H. pylori wt or cagA and virB7 for the times indicated. Total RNA was isolated and subjected to quantitative reverse transcription-PCR. β-Actin was used as a housekeeping gene for normalization of β-catenin product. The graph represents the mean -fold changes of relative amount of the β-catenin PCR product from at least three separate experiments ±S.E. with the value for control as 1 arbitrarily.
Interestingly, incorporation rate of [35S]methionine/cysteine during the pulse period was decreased in infected MDCK cells. We failed to observe significant changes in β-catenin gene expression at 10 and 24 h post-infection (Fig. 6C). Thus, the decreased incorporation could also reflect negative effect of H. pylori on β-catenin translation.
DISCUSSION
β-Catenin has been studied intensively in relation to the colorectal, breast, and prostate cancers (39-41). It becomes evident that alterations of expression and intracellular redistribution of β-catenin may be important for gastric cancer development as well (6, 7, 42). Up-to-date publications (9, 25) have described on the basis of results obtained by using LEF/TCF- or cyclin D1-luciferase reporters that infection with H. pylori stimulates nuclear activity of β-catenin. But the precise intracellular mechanism leading to β-catenin-dependent LEF/TCF transactivation in infected cells remains still unclear. Our work represents the first investigation which analyzes the regulation of β-catenin directed by upstream components in H. pylori-infected MDCK cells. We used MDCK cells, which express E-cadherin, possess no mutations in β-catenin, E-cadherin, GSK3β, or APC, and have no basal up-regulation of LEF/TCF, in contrast to AGS, MKN28, and MKN45 cell lines (43, 44). In accordance with the data obtained by other groups (9, 25), we observed LEF/TCF transactivation as well as up-regulation of LEF/TCF target gene cyclin D1 in MDCK cells after infection with H. pylori. This also reflects the in vivo situation; up-regulation of cyclin D1 has been reported to take place in gastric tissue of H. pylori-infected patients (22, 45). In contrast to data reported by Franco et al. (9), we failed to observe any nuclear accumulation of β-catenin in MDCK cells within 24 h of infection with H. pylori, which could be explained by the use of different cell types and bacterial strains. We found that despite that the amount of endogenous β-catenin was not changed, a quota of Ser-33-, Ser-37-, and Thr-41-phosphorylated β-catenin was decreased in infected cells.
Furthermore, we suppose that a balance between phosphorylated/unphosphorylated β-catenin could play an important role in TCF-dependent regulation of transcription. Ser/Thr-phosphorylated β-catenin itself does not influence LEF/TCF transcriptional activity (46). It has been described (47, 48) that an unphosphorylated form of β-catenin is able to promote transcription, but how this form is produced, kept, and regulated and regulates the TCF activity in H. pylori infection is not clear.
Ser-33, Ser-37, and Thr-41 of β-catenin are GSK3β target residues. GSK3β phosphorylates a range of enzymes, transcription factors, and structural proteins including these related to β-catenin signaling, e.g. E-cadherin, APC, cyclin D1 (35, 49-51). GSK3 is also a key player in Akt signaling which is regulated by H. pylori in gastric epithelial cells (28). To investigate the role of GSK3β in β-catenin regulation, we treated the cells with specific Akt kinase inhibitor Akti-1/2 (to activate GSK3β). The inhibitor impaired both an H. pylori-induced increase of GSK3 phosphorylation and decrease of Ser/Thr phosphorylation of β-catenin. These data were confirmed by using siRNA against Akt kinase. Therefore, H. pylori-stimulated reduction of β-catenin Ser/Thr phosphorylation involves Akt kinase and GSK3β in MDCK cells. Furthermore, these data suggest that casein kinase 1, whose phosphorylation of β-catenin at Ser-45 is obligatory for subsequent GSK3β-induced phosphorylation (52), is not impaired during infection. Importantly, Akti-1/2 inhibited H. pylori-induced LEF/TCF-dependent transactivation and cyclin D1 up-regulation. This provides evidence for a causal link between the decrease of β-catenin phosphorylation and up-regulation of LEF/TCF transactivation.
Akt kinase is a central player in the signaling pathways activated by growth factors and is thought to contribute to a broad range of cellular functions. Our data suggest that H. pylori induces T4SS-independent PI3K activity and Akt activation. PI3K phosphorylates phosphatidylinositol 4,5-diphosphate leading to the generation of phosphatidylinositol 3,4,5-triphosphate. Phosphatidylinositol 3,4,5-triphosphate acts as a lipid second messenger essential for the translocation of Akt to the plasma membrane and induces activation of phosphoinositide-dependent kinase-1, which phosphorylates Akt.
PI3K activity can be induced by growth factor receptors, e.g. EGFR or c-Met, and it has been shown that these receptors become active in H. pylori infection (37, 28). Interestingly, pretreatment of cells with pharmacological inhibitors against these tyrosine kinase receptors had a different outcome in regard of their impact on the phosphorylation of Akt kinase. Inhibition of the EGFR activity significantly reduced H. pylori-induced phosphorylation of Akt, whereas the selective c-Met inhibitor PHA-665752 did not block the phosphorylation of Akt kinase. EGFR in gastric epithelial cells was shown to be stimulated in a H. pylori T4SS-independent manner (37). In addition, downstream effectors of EGFR like ERK and MEK1 are both regulated in a T4SS-independent manner in H. pylori-infected cells (53). Thus, our results allow us to suppose that EGFR activation could contribute to Akt phosphorylation induced by H. pylori in a T4SS-independent manner.
Ser/Thr phosphorylation of β-catenin is known to be a prerequisite for ubiquitinylation and degradation of the protein. For the first time we show that H. pylori inhibits β-catenin ubiquitinylation. Using 35S-labeled amino acids, we performed pulse-chase experiments to confirm that impairment of β-catenin ubiquitinylation correlates to delayed metabolic turnover of the protein in infected cells. Therefore, our study demonstrates that GSK3 inhibition leads to suppression of phosphorylation/ubiquitinylation of β-catenin.
Decrease of β-catenin ubiquitinylation could be achieved not only through regulation of Ser/Thr phosphorylation but also through an increased deubiquitinylation by deubiquitinylating enzymes, for example by fat facets in mouse (FAM) (54). The role of H. pylori in the control of the deubiquitinylation machinery would be an interesting aim of future investigations.
Our experimental work indicates no obvious changes of β-catenin levels in subcellular fractions in MDCK cells infected for 24 h with H. pylori. The study by Weydig et al. (55) supports this observation. Herein, the authors observed no changes in the total amount of β-catenin in prolonged H. pylori infection. One explanation of why we failed to see the changes in the β-catenin levels might be that H. pylori impairs protein synthesis in infected cells. This hypothesis is based on our observation that the incorporation rate of [35S]methionine/cysteine in the β-catenin protein during the pulse period was decreased in infected cells (time 0, Fig. 6B). Thus, it would be potentially interesting to address this mechanism in the future in more detail, taking in account that GSK3β also is involved in the regulation of translation through eIF2B (56). Thus, regulation of β-catenin activity is highly complex and needs further detailed studies, e.g. about kinetics, stability, and de novo synthesis of β-catenin.
H. pylori is known to manipulate the host signaling by injecting pathogenic factors such as CagA into the target cell through the T4SS (31). Several studies have shown that nuclear β-catenin localization was affected in a CagA-dependent manner in different MKN gastric tumor cell lines infected with the NCTC11637 strain (24, 25, 57). Other investigations about β-catenin localization in MKN28 and AGS cell lines were performed without the use of isogenic H. pylori mutant strains (9, 58). It is worth mentioning that the published data about β-catenin localization in gastric epithelium harvested from patients clearly show that nuclear β-catenin also appears in gastric epithelium colonized with cag- H. pylori (9). Thus, other virulence factors could be relevant for β-catenin regulation; in particular, the H. pylori adhesin OipA (58).
In contrast to the studies mentioned above, an investigation using MCF-7 cells infected with H. pylori P12 strain clearly state a predominant cytoplasmic localization of β-catenin. In addition, Weydig et al. (55) provide experimental proof that the regulation of β-catenin is CagA-independent. Within this context, published work suggests that overexpression of CagA impairs the complex formation between the adherens junction protein E-cadherin and β-catenin (25), whereas another study shows that the E-cadherin-β-catenin complex was only slightly affected in a CagA-independent manner in H. pylori-infected cells (55). Furthermore, it has been shown that CagA is dispensable for the disruption of adherens junctions or loss of adhesion (55, 59).
However, the published data about the role of CagA in the regulation of adherens junctions and β-catenin are rather controversial. In addition, the application of overexpressed CagA protein and the use of different tumor cell lines with constitutively deregulated β-catenin signaling as well as use of different H. pylori strains contribute tremendously to the inconsistency of the published results.
The host molecular targets of CagA include Crk, SHP-2, c-Met, Csk, and focal adhesion kinase (24, 28, 60). A functionally active T4SS but not CagA is required for activation of the IκB kinase complex, c-Jun NH2-terminal kinase (JNK), p38, and Rac1/Cdc42 (3). The activation of MEK/ERK and upstream activators B-Raf/Rap1 (53) is T4SS-independent. Our data demonstrate that T4SS-independent effects include also suppression of GSK3β activity, decrease of β-catenin Ser/Thr phosphorylation, and delay of ubiquitinylation/degradation of β-catenin. Thus, alterations of Ser/Thr phosphorylation of β-catenin could play an important regulatory role in LEF/TCF transactivation induced by H. pylori.
Supplementary Material
Acknowledgments
We thank R. Hartig for fluorescent microscopy assistance, C. Wolke for help in reverse transcription-PCR, M. Hatakeyama for providing CagA-encoding plasmid, and Pfizer Global Pharmaceuticals for PHA-665752.
This work was supported in part by the Bundesministerium für Bildung und Forschung Grant 01ZZ0407. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: APC, axin and adenomatous polyposis coli; GSK, glycogen synthase kinase; MDCK, Madin-Darby canine kidney; T4SS, type 4 secretion system; ERK, extracellular signal-regulated kinase; wt, wild type; siRNA, small interfering RNA; EGF, epidermal growth factor; EGFR, EGF receptor; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; LEF/TCF, lymphoid enhancer-binding factor/T cell factor.
References
- 1.van Amsterdam, K., van Vliet, A. H., Kusters, J. G., and van der Ende, A. (2006) FEMS Microbiol. Rev. 30 131-156 [DOI] [PubMed] [Google Scholar]
- 2.Stoikov, C., Safari, R., Cai, X., Hasyagar, C., and Houghton, J. (2004) Gene (Amst.) 341 1-17 [DOI] [PubMed] [Google Scholar]
- 3.Naumann, M., and Crabtree, J. E. (2004) Trends Microbiol. 2 29-36 [DOI] [PubMed] [Google Scholar]
- 4.Penta, R., De Falco, M., Iaquinto, G., and De Luca, A. (2005) J Exp. Clin. Cancer Res. 24 337-345 [PubMed] [Google Scholar]
- 5.Yamaoka, Y., Ojo, O., Fujimoto, S., Odenbreit, S., Haas, R., Gutierrez, O., El-Zimaity, H. M., Reddy, R., Arnqvist, A., and Graham, D. Y. (2005) Gut 55 775-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nabais, S., Machado, J. C., Lopes, C., Seruca, R., Carneiro, F., and Sobrinho-Simoes, M. (2003) Int. J. Surg. Pathol. 11 1-9 [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. Y., Wang, Z. C., Li, H., Cheng, X. X., Sun, Y., Wang, X. W., Wu, M. L., and Liu, J. (2005) Hum. Pathol. 36 1294-1301 [DOI] [PubMed] [Google Scholar]
- 8.Shun, C. T., Wu, M. S., Lin, M. T., Chang, M. C., Lin, J. T., and Chuang, S. M. (2001) Oncology 60 339-345 [DOI] [PubMed] [Google Scholar]
- 9.Franco, A. T., Israel, D. A., Washington, M. K., Krishna, U., Fox, J. G., Rogers, A. B., Neish, A. S., Collier-Hyams, L., Perez-Perez, G., Hatakeyama, M., Whitehead, R., Gaus, K., O'Brien, D. P., Romero-Gallo, J., and Peek, R. M., Jr. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10646-10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romiti, A., Zullo, A., Borrini, F., Sarcina, I., Hassan, C., Winn, S., Tomao, S., Vecchione, A., Morini, S., and Mingazzini, P. (2005) World J. Gastroenterol. 11 4400-4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lilien, J., Balsamo, J., Arregui, C., and Xu, X. (2002) Dev. Dyn. 224 18-29 [DOI] [PubMed] [Google Scholar]
- 12.Nelson, W. J., and Nusse, R. (2004) Science 303 1483-1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottardi, C. J., and Gumbiner, B. M. (2004) J. Cell Biol. 167 339-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aberle, H., Bauer, A., Stappert, J., Kispert, A., and Kemler, R. (1997) EMBO J. 16 3797-3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peifer, M, and Polakis, P. (2000) Science 287 1606-1609 [DOI] [PubMed] [Google Scholar]
- 16.Hart, M., Concordet, J. P., Lassot, I., Albert, I., del los Santos, R., Durand, H., Perret, C., Rubinfeld, B., Margottin, F., Benarous, R., and Polakis, P. (1999) Curr. Biol. 9 207-210 [DOI] [PubMed] [Google Scholar]
- 17.Latres, E., Chiaur, D. S., and Pagano, M. (1999) Oncogene 18 849-854 [DOI] [PubMed] [Google Scholar]
- 18.Barker, N., Morin, P. J., and Clevers, H. (2000) Adv. Cancer Res. 77 1-24 [DOI] [PubMed] [Google Scholar]
- 19.Karim, R., Tse, G., Putti, T., Scolyer, R., and Lee, S. (2004) Pathology 36 120-128 [DOI] [PubMed] [Google Scholar]
- 20.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R., and Ben-Ze'ev, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5522-5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirikoshi, H., Sekihara, H., and Katoh, M. (2001) Int. J. Oncol. 19 533-536 [PubMed] [Google Scholar]
- 22.Sepulveda, A. R., Tao, H., Carloni, E., Sepulveda, J., Graham, D. Y., and Peterson, L. E. (2002) Aliment. Pharmacol. Ther. 16 145-157 [DOI] [PubMed] [Google Scholar]
- 23.Yang, Y., Deng, C. S., Peng, J. Z., Wong, B. C., Lam, S. K., and Xia, H. H. (2003) Mol. Pathol. 56 19-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki, M., Mimuro, H., Suzuki, T., Park, M., Yamamoto, T., and Sasakawa, C. (2005) J. Exp. Med. 202 1235-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata-Kamiya, N., Kurashima, Y., Teishikata, Y., Yamahashi, Y., Saito, Y., Higashi, H., Aburatani, H., Akiyama, T., Peek, R. M., Jr., Azuma, T., and Hatakeyama, M. (2007) Oncogene 26 4617-4626 [DOI] [PubMed] [Google Scholar]
- 26.Foryst-Ludwig, A., Neumann, M., Schneider-Brachert, W., and Naumann M. (2004) Biochem. Biophys. Res. Commun. 316 1065-1072 [DOI] [PubMed] [Google Scholar]
- 27.Schmitt, W., and Haas, R. (1994) Mol. Microbiol. 12 307-319 [DOI] [PubMed] [Google Scholar]
- 28.Churin, Y., Al-Ghoul, L., Kepp, O., Meyer, T. F., Birchmeier, W., and Naumann, M. (2003) J. Cell Biol. 161 249-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokolova, O., Heppel, N., Jägerhuber, R., Kim, K. S., Frosch, M., Eigenthaler, M., and Schubert-Unkmeir, A. (2004) Cell. Microbiol. 6 1153-1166 [DOI] [PubMed] [Google Scholar]
- 30.Stambolic, V., Ruel, L., and Woodgett, J. R. (1996) Curr. Biol. 6 1664-1668 [DOI] [PubMed] [Google Scholar]
- 31.Naumann, M. (2005) Int. J. Med. Microbiol. 295 335-341 [DOI] [PubMed] [Google Scholar]
- 32.Behrens, J. (2000) Ann. N. Y. Acad. Sci. 910 21-33 [DOI] [PubMed] [Google Scholar]
- 33.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995) Nature 378 785-789 [DOI] [PubMed] [Google Scholar]
- 34.Bijur, G. N., and Jope, R. S. (2001) J. Biol. Chem. 276 37436-37442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doble, B. W., and Woodgett, J. R. (2003) J. Cell Sci. 116 1175-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnett, S. F., DeFeo-Jones, D., Fu, S., Hancock, P. J., Haskell, K. M., Jones, R. E., Kahana, J. A., Kral, A. M., Leander, K., Lee, L. L., Malinowski, J., McAvoy, E. M., Nahas, D. D., Robinson, R. G., and Huber, H. E. (2005) Biochem. J. 385 399-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallasch, C., Crabtree, J. E., Bevec, D., Robinson, P. A., Wagner, H., and Ullrich, A. (2002) Biochem. Biophys. Res. Commun. 295 695-701 [DOI] [PubMed] [Google Scholar]
- 38.Orford, K., Crockett, C., Jensen, J. P., Weissman A. M, and Byers, S. W. (1997) J. Biol. Chem. 272 24735-24738 [DOI] [PubMed] [Google Scholar]
- 39.Chesire, D. R., and Isaacs, W. B. (2003) Endocr. Relat. Cancer 10 537-560 [DOI] [PubMed] [Google Scholar]
- 40.Brabletz, T., Hlubek, F., Spaderna, S., Schmalhofer, O., Hiendlmeyer, E., Jung, A., and Kirchner, T. (2005) Cells Tissues Organs. 179 56-65 [DOI] [PubMed] [Google Scholar]
- 41.Cowin, P., Rowlands, T. M., and Hatsell S. J. (2005) Curr. Opin. Cell Biol. 17 499-508 [DOI] [PubMed] [Google Scholar]
- 42.Lynch, H. T., Grady, W., Suriano, G., and Huntsman, D. (2005) J. Surg. Oncol. 90 114-133 [DOI] [PubMed] [Google Scholar]
- 43.Yokozaki, H. (2000) Pathol. Int. 50 767-777 [DOI] [PubMed] [Google Scholar]
- 44.Ikenoue, T., Ijichi, H., Kato, N., Kanai, F., Masaki, T., Rengifo, W., Okamoto, M., Matsumura, M, Kawabe, T., Shiratori, Y., and Omata, M. (2002) Jpn. J. Cancer Res. 93 1213-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polat, A., Cinel, L., Dusmez, D., Aydin, O., and Egilmez, R. (2002) Neoplasma (Bratisl.) 49 95-100 [PubMed] [Google Scholar]
- 46.Sadot, E., Conacci-Sorrell, M., Zhurinsky, J., Shnizer, D., Lando, Z., Zharhary, D., Kam, Z., Ben-Ze'ev, A., and Geiger, B. (2002) J. Cell Sci. 115 2771-2780 [DOI] [PubMed] [Google Scholar]
- 47.Staal, F. J., Noort, M., Strous, G. J., and Clevers, H. C. (2002) EMBO Rep. 3 63-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendriksen, J., Jansen, M., Brown, C. M., van der Velde, H., van Ham, M., Galjart, N., Offerhaus, G. J., Fagotto, F., and Fornerod, M. (2008) J. Cell Sci. 121 1793-1802 [DOI] [PubMed] [Google Scholar]
- 49.Rubinfeld, B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S., and Polakis, P. (1996) Science 272 1023-1026 [DOI] [PubMed] [Google Scholar]
- 50.Huber, A. H., and Weis, W. I. (2001) Cell 105 391-402 [DOI] [PubMed] [Google Scholar]
- 51.Diehl, J. A. (2002) Cancer Biol. Ther. 1 226-231 [DOI] [PubMed] [Google Scholar]
- 52.Liu, Ch., Li, Y., Semenov, M., Han, C., Baeg, G. H., Tan, Y., Zhang, Z., Lin, X., and Xe, X. (2002) Cell 22 837-847 [DOI] [PubMed] [Google Scholar]
- 53.Wessler, S., Rapp, U. R., Wiedenmann, B., Meyer, T. F., Schoneberg, T., Hocker, M., and Naumann, M. (2002) FASEB J. 16 417-419 [DOI] [PubMed] [Google Scholar]
- 54.Taya, S., Yamamoto, T., Kanai-Azuma, M., Wood, S. A., and Kaibuchi, K. (1999) Genes Cells 4 757-767 [DOI] [PubMed] [Google Scholar]
- 55.Weydig, C., Starzinski-Powitz, A., Carra, G., Löwer, J., and Wessler S. (2007) Exp. Cell Res. 313 3459-3471 [DOI] [PubMed] [Google Scholar]
- 56.Pap, M., and Cooper, G. M. (2002) Mol. Cell. Biol. 22 578-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurashima, Y., Murata-Kamiya, N., Kikuchi, K., Higashi, H., Azuma, T., Kondo, S., and Hatakeyama, M. (2008) Int. J. Cancer 22 823-831 [DOI] [PubMed] [Google Scholar]
- 58.Franco, A. T., Johnston, E., Krishna, U., Yamaoka, Y., Israel, D. A., Nagy, T. A., Wroblewski, L. E., Piazuelo, M. B., Correa, P., and Peek, R. M., Jr. (2008) Cancer Res. 68 379-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bebb, J. R., Leach, L., Zaitoun, A., Hand, N., Letley, D. P., Thomas, R., and Atherton, J. C. (2006) J. Clin. Pathol. 59 1261-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsutsumi, R., Takahashi, A., Azuma, T., Higashi, H., and Hatakeyama, M. (2006) Mol. Cell. Biol. 26 261-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.