Abstract
S100A6 is induced in myocardium post-infarction in vivo and in response to growth factors and inflammatory cytokines in vitro. Forced expression of S100A6 in cardiomyocytes inhibits regulation of cardiac specific gene expression in response to trophic stimulation. To define regulation and function of S100A6, we characterized the human S100A6 promoter and mapped upstream regulatory elements in rat neonatal cardiac myocytes, fibroblasts, and vascular smooth muscle cells and defined a functional role for S100A6 in tumor necrosis factor-α-induced myocyte apoptosis. The functional S100A6 promoter was localized to region -167/+134 containing 167 upstream base pairs. The S100A6 promoter is regulated by positive (-361/-167 and -588/-361) and negative (-1371/-1194) elements. Tumor necrosis factor-α induced the maximal S100A6 promoter and transcription factor NF-κB (p65 subunit). Electrophoretic mobility shift showed that tumor necrosis factor-α induced p65 binding to a potential NF-κB-binding site at -460/-451. Chromatin immunoprecipitation analysis revealed p65 is recruited to the S100A6 promoter upon tumor necrosis factor-α stimulation. The NF-κB inhibitor caffeic acid phenethyl ester and mutation of the NF-κB-binding site inhibited S100A6 promoter activation by tumor necrosis factor-α. Tumor necrosis factor-α induced cardiac myocyte apoptosis. Specific inhibition of S100A6 using a small interfering RNA directed against S100A6 potentiated tumor necrosis factor-α-induced myocyte apoptosis, whereas overexpression of S100A6 by gene transfer prevented tumor necrosis factor-α-induced myocyte apoptosis by interfering with p53 phosphorylation. These results demonstrate that S100A6 is induced by tumor necrosis factor-α via an NF-κB-dependent mechanism, serving a role in homeostasis to limit tumor necrosis factor-α-induced apoptosis by regulating p53 phosphorylation.
S100A6 (calcyclin), a 10-kDa protein belonging to the S100 subfamily of EF-hand, calcium-binding proteins (1–3), is expressed in diverse tissues, including smooth muscle, brain, lung, kidney, and heart (4–5), and has been linked with cell cycle progression (6–8), cytoskeleton rearrangement (9), exocytosis (10), and cell survival (11). Stimuli, including growth factors (12), inflammatory cytokines (13), vasopressin (14), phorbol esters (15), and cadmium ions (16), have been reported to induce an increase in S100A6 mRNA and/or protein levels.
We have previously reported that expression of S100A6 is increased in rat heart following experimental myocardial infarction and in cultured neonatal rat myocytes by treatment with trophic agonists known to stimulate myocardial hypertrophy (17). Furthermore, in co-transfection experiments, S100A6 was shown to block activation of the fetal gene program, e.g. skeletal α-actin and β-myosin heavy chain by trophic signals, and down-regulate normal expression of adult α-myosin heavy chain in keeping with its role in cell cycle progression in proliferating cell lines (17).
The signaling mechanisms producing increased expression of S100A6 in normal and diseased cells have not been defined, and experimental data concerning regulation of S100A6 gene transcription is limited. The promoter region of the S100A6 gene has been sequenced to residues 3000 bp upstream of the cap site, and deletion analysis has identified a basic promoter essential for transcription spanning 167 bp upstream of transcription initiation (12, 18). The presence of an NF-κB-binding site at position -460/-451 points to the potential role of inflammatory processes in activation of this gene (13).
TNF-α2 is a pro-inflammatory cytokine whose serum levels are elevated in many human cardiac related pathogenic conditions, including heart failure (19). In cardiac myocytes, TNF-α can induce apoptosis through death domain pathways initiated by the stimulation of TNFR1 (20). Besides the activation of proapoptotic pathways, the trimerization of TNFR1 by TNF-α also activates NF-κB, which may induce the expression of survival genes and counteract the cell death pathways (20). Currently, the NF-κB-induced survival genes remain to be fully elucidated.
In this study we extend our studies to an investigation of the following: 1) transcriptional regulation of the S100A6 promoter in myocardial cells and its response to the inflammatory cytokine, TNF-α, and 2) the functional consequences of S100A6 induction in the setting of TNF-α-induced myocyte apoptosis. We report that transcriptional regulation of the S100A6 promoter is complex and involves common and cell-specific positive and negative upstream regulatory elements. Under basal conditions, S100A6 is repressed, and TNF-α signaling via NF-κB activates S100A6. Induction of S100A6 functions to limit TNF-α-induced myocyte apoptosis by associating with p53 and inhibiting p53 phosphorylation.
EXPERIMENTAL PROCEDURES
Materials—A luciferase reporter gene system was used to study the relative ability of different 5′ DNA regions of the human S100A6 gene to promote transcription in transient transfection assays, as described previously (21) (gift from W. Lesniak, Nencki Institute of Experimental Biology, Warsaw, Poland). Mutations to the NF-κB(-460/-451) site of the full promoter (-3000/+134) were introduced by the QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the directions provided with the following primers (lowercase letters indicate the mutations), 5′-TAGGAGATAGGGGctGggGCCAG-3, 5′-AGGGGctGggGCCAGTGGTGTAA-3′ (13). The cis-reporter plasmid pNF-κB-LUX contained a luciferase cDNA under a regular TATA box and an enhancer element with five NF-κB-binding sites (22). The expression plasmids were obtained from the following sources: S100A6 expression vector encoding the complete 90-amino acid sequence of human S100A6 has been described (17); Sra-Δ protein kinase C (PKC)-β (a partially deleted constitutively active PKC-β cDNA) was from P.C. Simpson (23); pXJ40 (transcription enhancer factor (TEF))-1 and pXJ40-related (R)TEF-1 (containing human TEF-1 and RTEF-1 cDNA, respectively) were from A. F. R. Stewart (24); p65 expression vector was from R. I. Tapping (25); p53 wild type, p53His-175 mutant form of the p53 gene acting as a dominant negative was from S. Benchimol (26); and Rous sarcoma virus (RSV)-chloramphenicol acetyltransferase (CAT) (containing a CAT cDNA) was from Stratagene (La Jolla, CA). The S100A6 siRNA duplex containing an S100A6 sense (5′-rCrcUUrCUrCrGUrGrGrCUrAUrCUUrCTT-3′) and S100A6 antisense (5′-rGrArGrAUrArGrCrCrArCrGrArGrArArGrGTT-3′) was obtained from Sigma Proligo (The Woodlands, TX).
Cell Culture and Transfection—Low density (∼150 cells/mm2) non-contractile neonatal cardiac myocytes and non-myocytes were isolated from ventricles of 2-day-old Sprague-Dawley rats as described previously (17, 27). (All animal experiments conformed with protocols approved by the host institution.) In our culture system, contaminating non-myocytes constitute 5% of the total cell population (17, 27). Rat aortic smooth muscle cells (SMCs) were isolated from 2-day-old Sprague-Dawley rats as described previously (28). Confluent fibroblast and SMCs were used between passages 2 and 6. Freshly isolated myocytes were plated in media containing 5% fetal bovine serum for 24 h before use. Transfection was carried out with FuGENE 6 reagent according to the manufacturer's instructions (Roche Applied Science), using specific quantities of the following plasmids: S100A6 promoter-luciferase constructs and NF-κB-LUX (5 μg); SRα-ΔPKC-β, pXJ40-TEF-1, pXJ40-RTEF-1, S100A6 (0.1 μg), p65 (0.1 μg), wild type p53 (0.1 μg), mutant p53 (0.1 μg)m and S100A6 siRNA (1 μg). RSV-CAT (0.1 μg) was included as an internal control for the transfection efficiency. All cell types were maintained in medium supplemented with 5% fetal bovine serum for 18 h following transfection, prior to transfer to serum-free medium and treatment with norepinephrine (NE, 20 μm), phenylephrine (PE, μm), phorbol 12-myristate 13-acetate (PMA, 10 nm), porcine platelet-derived growth factor (PDGF, 5 ng/ml), recombinant human transforming growth factor (TGF) β1 (5 ng/ml), recombinant bovine TNF-α (5 ng/ml) (PDGF, TGFβ1, and TNF-α all from R & D Systems, Minneapolis, MN), 5% fetal bovine serum or vehicle diluent (100 μm ascorbic acid for NE and PE; phosphate-buffered saline, 0.1% bovine serum albumin for PDGF, TGFβ, and TNF-α) for 48 h. Cells were preincubated for 2 h with the NF-κB inhibitor caffeic acid phenethyl ester (13) (CAPE) (50 μg/ml) or vehicle (0.01% dimethyl sulfoxide). The cell lysates were assayed for luciferase activity, as described previously (29), and for CAT activity using the FAST CAT (deoxy) chloramphenicol acetyltransferase assay kit according to the manufacturer's instructions (Molecular Probes, Eugene, OR).
Western Blotting and Immunoprecipitation—Immunoprecipitation and Western blotting were performed as described previously (17). Blots were incubated with either goat anti-S100A6 antibody (1:1000), goat anti-phospho-p53 (Ser-15) (1:1000) (Cell Signaling, Danvers, MA), rabbit anti-p53 (1:1000) (R & D Systems, Minneapolis, MN), or mouse anti-β-actin (Sigma) (1:1000) followed by peroxidase-conjugated secondary antibody, either chicken anti-goat IgG, sheep anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:1000 dilution, or sheep anti-mouse IgG (Amersham Biosciences) at a 1:2500 dilution.
Electronic Mobility Shift Assays (EMSA)—Cardiac myocytes were treated for 1 h with either vehicle or TNF-α (5 ng/ml). Nuclear protein was extracted from whole cells using NE_PER nuclear and cytoplasmic extraction reagents (Pierce). EMSA was performed using a LightShift chemiluminescent EMSA kit (Pierce). The sequences (13) of the double-stranded oligonucleotides used for EMSA were as follows: NF-κB(-466/-445) 5′-GATAGGGGGAGTAGCCAGTGGT-3′; mutated (lowercase letters indicate the mutations introduced) NF-κB(-466/-445) 5′-GATAGGGGctGggGCCAGTGGT-3′.
Chromatin Immunoprecipitation Assays (ChIP)—Cardiac myocytes were transfected with either vector or the (-588/+134) human S100A6 promoter construct (containing the NF-κB-binding site at position -460/-451) and treated with vehicle or TNF-α (5 ng/ml) for 1 h. ChIP assays were performed according to the manufacturer's instructions using a kit from Upstate Biotechnology/Millipore (Temecula, CA). The sonicated chromatin fraction was immunoprecipitated with an equal amount of either rabbit IgG or anti-NF-κB (p65) antibody (Santa Cruz Biotechnology). The primer sequences used for PCR were as follows: human, forward 5′-AGCCCTGTAAGCACCATTGG-3′ and reverse 5′-CGTCTTCCATGGTGGCTTTACC-3′ (25); rat, forward 5′-TGGGAGCCCAGGACCCTAACC-3′ and reverse 5′-GCTGGGCTCCCCCATCCCGT-3′.
Apoptosis and Caspase-3 Activation—Neonatal rat cardiac myocytes were analyzed for apoptosis by terminal deoxynucleotidyl transfer-mediated end labeling of fragmented nuclei (TUNEL assay) using the ApopTag in situ apoptosis detection kit (Chemicon, Temecula, CA). Individual nuclei were visualized at ×400 using 4′-6-diamidino-2-phenylindole (Chemicon, Temecula, CA). To identify cells of cardiac origin, sections were double-stained with MF 20, a monoclonal anti-myosin antibody (working dilution 1:50) (Developmental Studies Hybridoma Bank, University of Iowa). Apoptosis was also assessed using FITC annexin V (BD Biosciences) and propidium iodide staining of cells and flow cytometry. Caspase-3 activity was measured from cell lysates using EnzChek Caspase-3 assay kit (Invitrogen).
Statistical Analysis—Treated/control ratios were tested for deviation from unity by calculation of confidence limits. Mean values were compared by one-way analysis of variance followed by either Dunnett's or Bonferroni's post hoc tests when appropriate. Probability values of p < 0.05 were accepted as statistically significant.
RESULTS
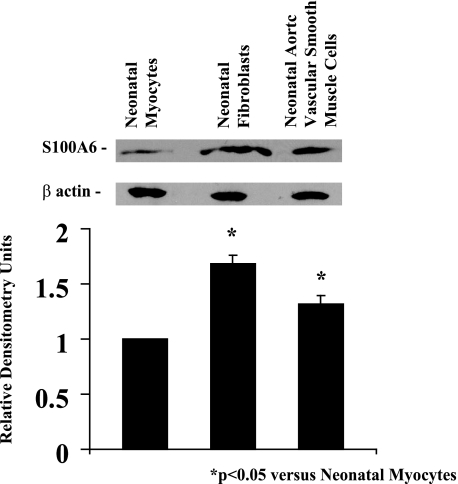
Cardiac Myocytes, Fibroblasts, and Aortic SMCs Differentially Express S100A6 under Basal Conditions—Western blot analysis detected S100A6 protein in cardiac myocytes, to a greater extent in SMCs, and most abundantly in fibroblasts (Fig. 1), a similar pattern to mRNA levels described previously (8, 17). Densitometry ratios (S100A6/β-actin), shown relative to cardiac myocytes, were significantly (p < 0.05) increased in SMCs and fibroblasts.
FIGURE 1.
Western blot analysis of S100A6 and β-actin proteins in rat neonatal cultured myocytes, fibroblasts, and aortic smooth muscle cells. Protein extracts from each treatment group were analyzed by Western blotting using goat anti-S100A6 and mouse anti-β-actin antibodies. Respective relative densitometry units are displayed below blots. *, p < 0.05 versus neonatal myocytes. n = 3.
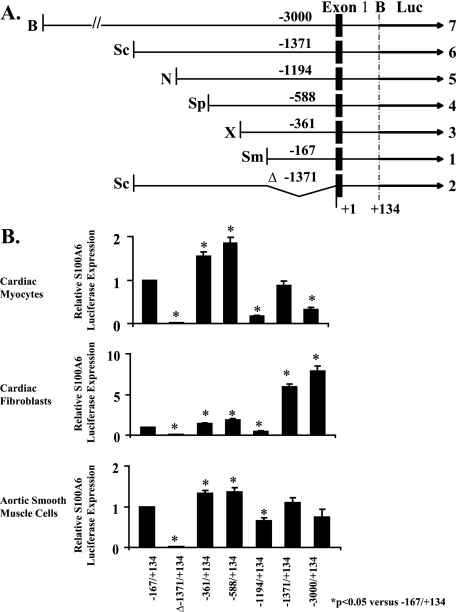
Transcriptional Regulation of the S100A6 Promoter in Cardiac Myocytes, Fibroblasts, and Aortic SMCs—Cardiac myocytes, fibroblasts, and aortic smooth muscle cells were transfected with S100A6 promoter/luciferase constructs to localize cis-acting elements within the S100A6 promoter, which may direct S100A6 expression in a cell-specific manner. Fig. 2A is a schematic representation of the constructs used. All cell types transfected with plasmids containing the reporter gene under the control of the S100A6 gene promoter fragments were able to express luciferase (Fig. 2B). The construct -167/+134 spanning 167 bp upstream of the transcription initiation site, 68 bp of exon 1 and 66 bp of intron 1, the shortest S100A6 promoter fragment assayed, was active in all cell types and identified as the basic promoter. The basic promoter contains several transcription elements, including a TATA box, three GC boxes, and an enhancer-like structure (30). Absence of the basic promoter from the cloned construct Δ-1371/+134 abolished transcription. When the S100A6 promoter element was extended upstream with 194 bp (-361/+134), luciferase activity was up-regulated 1.3–1.8-fold in all cells suggesting a positive regulatory element. Further extension upstream with 227 bp (-588/+134) had no effect on luciferase activity. Further extension with 606 bp (-1194/+134) down-regulated luciferase activity ∼2–5-fold in all cells suggesting a negative regulatory element. Further extensions upstream showed cell-specific transcriptional regulation of the S100A6 promoter. Whereas upstream extensions of 177 bp (-1371/+134) and 1629 bp to the full promoter (-3000/+134) maintained the inhibitory response on luciferase activity in cardiac myocytes and to a lesser extent in SMCs, similar upstream extensions in fibroblasts up-regulated luciferase activity ∼6–8-fold implicating the presence of highly positive regulatory element(s). These results suggest that in the upstream sequence 3000 bp 5′ from the transcription initiation site, there is sequence information that drives S100A6 transcription according to the basal physiological pattern.
FIGURE 2.
Human S100A6 promoter-luciferase reporter constructs. A, using the full-length pBS construct, shorter fragments of the human S100A6 gene (solid boxes were prepared by restriction digests to generate deletion constructs). Restriction enzymes are abbreviated as follows: B, BamHI; Sc, ScaI; N, NcoI; Sp, SphI; X, XhoI; Sm, SmaI. The black arrows (luciferase (Luc)) represent the coding region of firefly luciferase. B, neonatal rat cardiac myocytes, cardiac fibroblasts, and aortic smooth muscle cells were co-transfected with the appropriate S100A6 promoter-luciferase construct and RSV-CAT. The bars shown are the mean ± S.E. of luciferase expression (normalized to the RSV-CAT activity) relative to pGBS-167 in the same cell type, which has been assigned the value of 1. *, p < 0.05 relative to pGBS-167. n = 6.
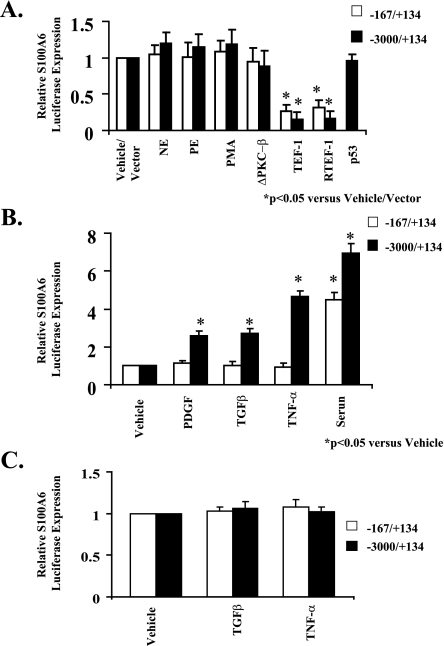
Trophic Stimuli Induce the Maximal S100A6 Promoter in Cardiac Myocytes—In response to α1-adrenergic PKC signaling, transcription from basic (-167/+134) and maximal (-3000/+134) promoters was not stimulated by the α1-agonists NE and PE, the phorbol ester PMA, or co-transfection with either PKCΔ-β, a constitutively active mutant of PKC-β, or wild type p53 (Fig. 3A). However, co-transfection with transcription enhancer factor (TEF)-1 or RTEF-1, which are distal components of the PKC signal pathway, similarly down-regulated transcription from basic (-167/+134) and maximal (-3000/+134) promoters implicating the presence of an MCAT element(s) located in the basic promoter. Whereas the basic S100A6 promoter (-167/+134) is not responsive to PDGF, TGFβ, and TNF-α, the maximal promoter (-3000/+134) is activated ∼2.5–5-fold by these stimuli suggesting the presence of upstream growth-responsive element(s) (Fig. 3B). In cultured neonatal rat cardiac fibroblasts, neither TGFβ nor TNF-α induced transcription from the basic (-167/+134) or the maximal (-3000/+134) promoters (Fig. 3C).
FIGURE 3.
Selective stimulation of the S100A6 promoter by trophic stimuli. A, myocyte cultures were co-transfected with the human S100A6 promoter-luciferase reporter constructs and treated for 48 h with either norepinephrine (NE, 20 μm), phenylephrine (PE, 20 μm), PMA (10 nm), or co-transfected with the expression plasmids ΔPKC-β, TEF-1, RTEF-1, or p53. B, myocyte cultures were co-transfected with the human S100A6 promoter-luciferase reporter constructs and treated for 48 h with either PDGF (5 ng/ml), TGFβ (5 ng/ml), TNF-α (5 ng/ml), or 5% serum. C, non-myocyte (fibroblast) cultures were co-transfected with the human S100A6 promoter-luciferase reporter constructs and treated for 48 h with either TGFβ (5 ng/ml) or TNF-α (5 ng/ml). Bars shown are mean ± S.E. of luciferase expression (normalized to the RSV-CAT activity) relative to vehicle treatment of each deletion fragment (assigned the value of 1). *, p < 0.05 versus vehicle/vector. n = 6.
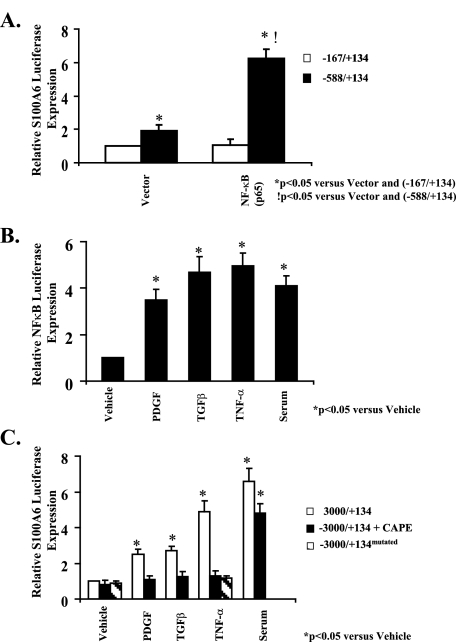
Induction of the Maximal S100A6 Promoter Is Dependent on NF-κB Signaling—We first confirmed that an NF-κB-binding site was present in the S100A6 promoter as described previously (13). Treatment of cardiac myocytes transfected with the -588/+134 S100A6 promoter construct containing the NF-κB-binding site with an NF-κB (p65 subunit) expression vector activated the promoter ∼3-fold (Fig. 4A). Because an NF-κB-binding site is present in the S100A6 promoter as described above, we tested the contribution of NF-κB to induction of the S100A6 promoter by peptide growth factors. Treatment of cardiac myocytes transfected with an NF-κB promoter luciferase construct with either PDGF, TGFβ, TNF-α or 5% serum induced a 3.5–5-fold increase in NF-κB-LUX activity (Fig. 4B). Pretreatment of cardiac myocytes with CAPE, a specific inhibitor of NF-κB activation (13), inhibited maximal (-3000/+134) promoter activation by PDGF, TGFβ, and TNF-α but not serum. Likewise, point mutagenesis of the NF-κB-binding site inhibited TNF-α activation of the maximal promoter (-3000/+134) suggesting that NF-κB signaling is involved in activation of the maximal S100A6 promoter in response to TNF-α (Fig. 4C).
FIGURE 4.
NF-κB activation is required for TNF-α induction of the human S100A6 promoter. A, myocyte cultures were co-transfected with either the -167/+134 or -588/+134 human S100A6 promoter-luciferase reporter constructs in combination with the p65 expression plasmid or empty vector as indicated. B, myocyte cultures were co-transfected with the human NF-κB promoter-luciferase reporter constructs and treated for 48 h with either PDGF (5 ng/ml), TGFβ (5 ng/ml), TNF-α (5 ng/ml), 5% serum, or vehicle. C, myocyte cultures were co-transfected with either the -3000/+134 or -3000/+134mutated (containing point mutations of the NF-κB-binding site) human S100A6 promoter-luciferase reporter constructs as indicated, and the -3000/+134 transfected cells were pretreated with the NF-κB inhibitor CAPE (50 μg/ml) or vehicle for 2 h and then treated for 48 h with either PDGF (5 ng/ml), TGFβ (5 ng/ml), TNF-α (5 ng/ml), or 5% serum. Bars shown are mean ± S.E. of luciferase expression (normalized to the RSV-CAT activity) relative to -167/+134 with vector alone (A) or vehicle control (B and C) (assigned the value of 1). *, p < 0.05 versus vehicle or vector.!, p < 0.05 versus -588/+134 and vector alone. n = 6.
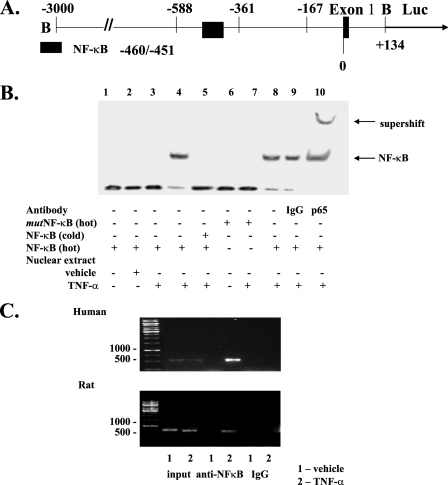
DNA Binding Activity of NF-κB Was Associated with S100A6 Promoter Activity—To study whether the NF-κB site at -460/-451 of the S100A6 promoter (Fig. 5A) is a functional site for nuclear proteins, EMSA was performed using nuclear extracts from cardiac myocytes stimulated with TNF-α. Treatment with TNF-α induced DNA binding activity of the nuclear extracts to a -466/-455 oligonucleotide (but not an NF-κB mutant oligonucleotide), including the putative NF-κB-binding site of the S100A6 promoter, and this binding was eliminated by addition of cold NF-κB oligonucleotide (Fig. 5B). Furthermore, addition of an antibody specific for the p65 subunit of NF-κB (but not IgG) induced a supershift (Fig. 5B). These results indicate that NF-κB is able to specifically interact with the predicted binding site in the S100A6 promoter.
FIGURE 5.
NF-κB binds to its putative site in the S100A6 promoter in vitro and in vivo in response to TNF-α. A, schematic representation of the -3000/+134 S100A6 promoter fragment containing the first exon. The location of the restriction sites (B, BamHI) used to generate the fragment and location of the NF-κB site are shown. B, electromobility shift assay using nuclear extracts from myocyte cultures treated with TNF-α (5 ng/ml) or vehicle as indicated. The binding of NF-κB was confirmed by point mutations (mut) in NF-κB oligonucleotides, addition of 100-fold excess cold competitor oligonucleotides, and by supershift with an antibody to NF-κB p65 as indicated. C, myocyte cultures were co-transfected with the -588/+134 human S100A6 promoter-luciferase reporter construct as indicated and treated with TNF-α (5 ng/ml) or vehicle for 1 h then harvested and then subjected to chromatin immunoprecipitation analysis using NF-κB p65 antibody or a nonspecific anti-rabbit IgG. The isolated DNA fragments were amplified by PCR using both human (C, top) and rat (C, bottom) S100A6-specific primers. Input, nonprecipitated chromatin.
NF-κB Is Recruited to Its Binding Site in the S100A6 Promoter—To measure the in vivo affinity of NF-κB for its site in the S100A6 promoter, cardiac myocytes were transfected with the -588/+134 S100A6 promoter reporter construct and subjected to ChIP analysis to monitor NF-κB recruitment. The primer sets used for this experiment were designed to amplify S100A6 DNA from either the reporter plasmid or genomic DNA. Fig. 5C shows that p65 was recruited to the reporter plasmid (Fig. 5C, top) and the endogenous gene (Fig. 5C, bottom) 1 h after stimulation with TNF-α. These data indicate that NF-κB is recruited to the S100A6 promoter, and the binding site in question is required for this recruitment.
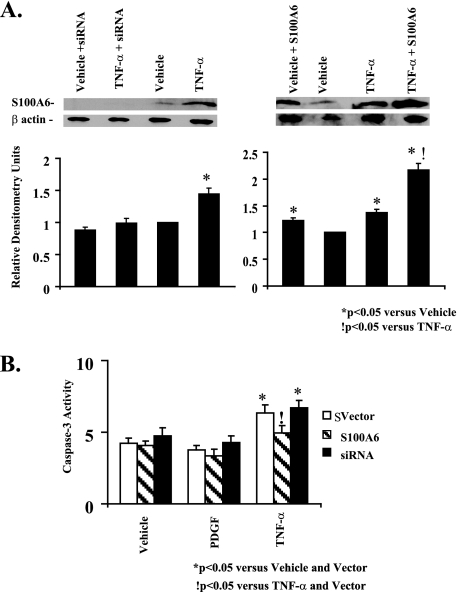
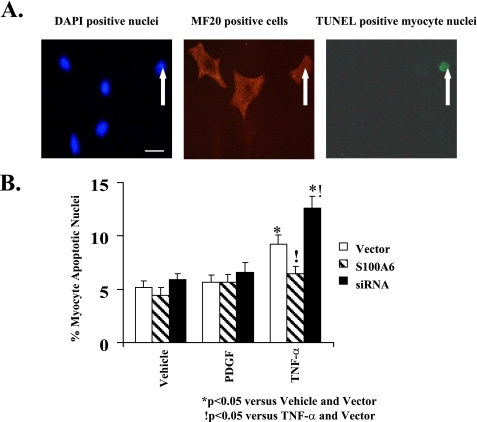
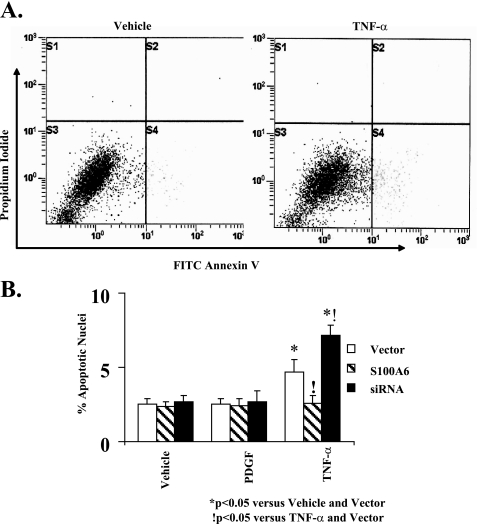
Up-regulation of S100A6 Limits TNF-α-induced Myocyte Apoptosis—We first showed that TNF-α stimulation of cardiac myocytes increases S100A6 protein (Fig. 6A) in keeping with transcriptional activation of the maximal S100A6 promoter (Fig. 3B). Densitometry ratios (S100A6/β-Actin), shown relative to vehicle treatment, were significantly (p < 0.05) increased in response to TNF-α (Fig. 6A). In cardiac myocytes, TNF-α stimulation is associated with activation of both anti-apoptotic (NF-κB signaling) and pro-apoptotic (caspase activation) pathways (20). To assess the net effect of TNF-α stimulation on cultured cardiomyocyte apoptosis, we employed a multiplicity of techniques, including TUNEL staining, FITC annexin V flow cytometry, and caspase-3 activation. Basal caspase-3 activity and cardiac myocyte apoptosis were seen in vehicle-treated myocytes (Figs. 6B, 7, and 8). Stimulation with TNF-α increased caspase-3 activity and cardiac myocyte apoptosis (Figs. 6B, 7, and 8). The overall frequency of apoptotic myocytes by TUNEL assay was somewhat higher than that detected by flow cytometry. The differential detection of apoptosis by flow cytometry and by TUNEL may be due to different sensitivities of the two approaches, e.g. the TUNEL assay may be more nonspecific detecting cells undergoing DNA repair, necrotic as well as apoptotic cells, whereas flow cytometry may be less sensitive detecting only cells in the early apoptotic phase (i.e. FITC annexin V-positive and propidium iodide-negative) (31). Other considerations include differences in the method of preparation. The TUNEL assay requires paraformaldehyde fixation of the cells, whereas annexin V flow cytometry does not. Because TNF-α increases S100A6 protein (Fig. 6A), we tested the contribution of S100A6 in TNF-α-induced cardiac myocyte apoptosis. Overexpression of S100A6 by gene transfer (Fig. 6A) inhibited TNF-α-induced caspase-3 activation and apoptosis (Figs. 6B, 7B, and 8B), whereas knocking down S100A6 using an siRNA duplex directed against S100A6 (Fig. 6A) further increased TNF-α-induced apoptosis (Fig. 7B and Fig. 8B) despite a minimal impact on caspase-3 activity (Fig. 6B). Densitometry ratios (S100A6/β-Actin), shown relative to TNF-α treatment, were significantly (p < 0.05) increased in response to TNF-α in combination with S100A6 overexpression (Fig. 6A). The above data suggest that up-regulation of S100A6 is a key mechanism in limiting TNF-α-induced cardiac myocyte apoptosis.
FIGURE 6.
TNF-α induction of S100A6 in cardiac myocytes. A, representative Western blots of S100A6 andβ-actin in myocyte cultures transfected with either vector alone or the human S100A6 expression plasmid or an siRNA duplex against S100A6 and treated for 48 h with either vehicle or TNF-α (5 ng/ml). Respective densitometry units relative to vehicle control are displayed below blots. *, p < 0.05 versus vehicle and vector.!, p < 0.05 versus TNF-α and vector. n = 3. B, myocyte cultures were transfected with either a human S100A6 expression plasmid or an siRNA duplex against S100A6 or empty vector and treated for 48 h with either PDGF (5 ng/ml), TNF-α (5 ng/ml), or vehicle. Myocytes were assessed for caspase-3 activity by the EnzChek caspase-3 assay kit. The results are the mean ± S.E. of 6–8 different experiments. *, p < 0.05 versus vehicle and/or vector.!, p < 0.05 versus TNF-α and vector alone.
FIGURE 7.
S100A6 modulation of TNF-α-induced myocyte apoptosis as assessed by TUNEL staining. A, representative photomicrographs (×400) of 4′,6-diamidino-2-phenylindole (DAPI)-nuclei (left) (bar = 10 μm), MF20-stained cardiac myocytes (middle), and TUNEL-positive (right) cardiac myocyte(s) treated with TNF-α (5 ng/ml). White arrow indicates myocyte apoptotic nucleus as visualized by TUNEL staining. B, myocyte cultures were transfected with either an S100A6 expression plasmid or an siRNA duplex against S100A6 and treated for 48 h with either PDGF (5 ng/ml), TNF-α (5 ng/ml), or vehicle. TUNEL-positive myocyte nuclei were counted and expressed as percentage of total myocyte nuclei. The results are the mean ± S.E. from random fields in blinded experiments. A minimum of 10 high power fields were scored per experiment of six different experiments. *, p < 0.05 versus vehicle/vector.!, p < 0.05 versus TNF-α/vector.
FIGURE 8.
S100A6 modulation of TNF-α-induced myocyte apoptosis as defined by flow cytometry. A, representative graphs showing the proportion of apoptotic myocytes (FITC-annexin V positive/propidium iodide negative) indicated in the lower right quadrant following treatment with either vehicle (left graph) or TNF-α (5 ng/ml) (right graph). B, myocyte cultures were transfected with either a human S100A6 expression plasmid or an siRNA duplex against S100A6 or empty vector and treated for 48 h with either PDGF (5 ng/ml), TNF-α (5 ng/ml), or vehicle. Myocytes were assessed for apoptosis by FITC annexin V/propidium iodide staining and flow cytometry. The results are the mean ± S.E. from 6 to 8 different experiments. *, p < 0.05 versus vehicle/vector. !, p < 0.05 versus TNF-α and vector.
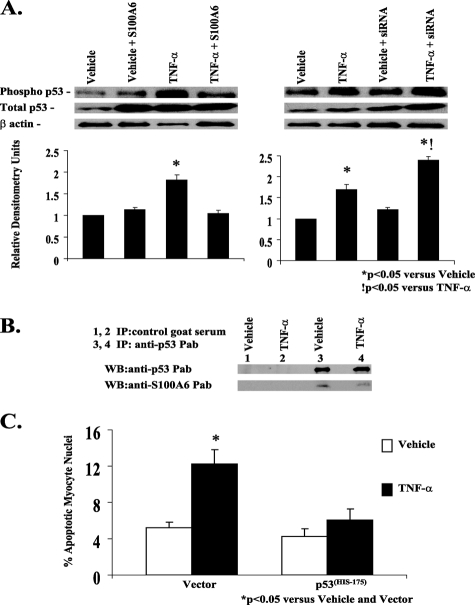
S100A6 Associates and Interferes with p53 Phosphorylation—The tumor suppressor protein p53 is activated in response to trophic stimuli and can induce apoptosis in a variety of cell types through activation of death-promoting gene targets (32). Stimulation of cardiac myocytes with TNF-α increased phosphorylation of p53 (Fig. 9A). Because binding of S100 proteins (e.g. S100B, S100A4, and S100A2) to p53 have been shown to inhibit its phosphorylation and consequently its activity by preventing p53 tetramerization (33–35), we tested whether S100A6 associates with p53 and interferes with its function. Overexpression of S100A6 inhibited TNF-α-induced phosphorylation of p53 (Fig. 9A), whereas knocking down S100A6 using an siRNA duplex further augmented TNF-α-induced phosphorylation of p53 (Fig. 9A). Densitometry ratios (phospho-p53/total p53), shown relative to vehicle treatment, were significantly (p < 0.05) increased in response to TNF-α alone and further augmented with an S100A6 siRNA (Fig. 9A). A co-immunoprecipitation strategy demonstrated heterodimerization of S100A6 and p53 in cardiomyocytes (Fig. 9B). Despite heterodimerization of S100A6 and p53, wild type p53 was not able to activate the S100A6 promoter (Fig. 3A). To further study the functional role of p53 in TNF-α-induced myocyte apoptosis, we transfected dominant negative p53 into cardiac myocyte cultures. Overexpression of dominant negative p53 inhibited TNF-α-induced myocyte apoptosis implicating p53 as a key mediator in TNF-α induced myocyte apoptosis (Fig. 9C).
FIGURE 9.
S100A6 modulation of TNF-α-induced myocyte apoptosis requires p53. A, representative Western blot of phospho-p53, total p53, and β-actin in myocyte cultures transfected with either vector alone, human S100A6 expression plasmid, or an siRNA duplex against S100A6 and treated for 48 h with either vehicle or TNF-α (5 ng/ml). Respective relative densitometry units are displayed below blots. *, p < 0.05 versus vehicle and vector.!, p < 0.05 versus TNF-α and vector. n = 3. B, co-immunoprecipitation of p53 and S100A6 in TNF-α-treated myocyte cultures. Aliquots of lysates of myocyte cultures treated with either vehicle or TNF-α were incubated with control goat serum (lanes 1 and 2) or anti-p53 polyclonal antibody (lanes 3 and 4), followed by incubation with protein A-Sepharose. The immune complexes were dissociated and analyzed by Western blotting with anti-S100A6 or anti-p53 antibodies as indicated in each blot. C, myocyte cultures were co-transfected with a mutant p53His175 plasmid acting as a dominant negative and treated for 48 h with either TNF-α (5 ng/ml), 5% serum, or vehicle. The results are the mean ± S.E. of six different experiments. *, p < 0.05 versus vehicle/vector.
DISCUSSION
S100A6 has been reported to be present in the heart of several species, including man, rat, mouse, and chicken, with the highest levels relative to other tissues reported for human heart (5). We reported the up-regulation of S100A6 in rat heart following experimental myocardial infarction and in response to trophic stimuli, and unlike the selective blockade of catecholamine-induced fetal gene expression by S100B, forced expression of S100A6 by gene transfer prevents induction of the skeletal α-actin and β-myosin heavy chain by multiple trophic signals and down-regulates the expression of the adult α-myosin heavy chain (17). We presently extend these observations by showing that S100A6 is part of a pro-survival pathway activated by TNF-α where it serves to limit TNF-α-induced myocyte apoptosis. As such, S100A6 regulates a spectrum of growth and differentiation properties in cardiac muscle, in keeping with its role in cell cycle progression in proliferating cell lineages.
Regulation of S100A6 in a variety of cell lineages is transcriptional (12, 18, 21, 30). We now provide the first evidence that expression in cardiovascular cells is also transcriptional. In cardiac myocytes, cardiac derived fibroblasts, and aortic smooth muscle cells, we defined a basic promoter essential for transcription spanning 167 bp upstream of the transcription initiation site, and we identified common positive (located in the region of -588/-162) and negative (located in the region -1194/-588) regulatory elements acting uniformly in the three different cell types as described previously in other cell lineages (12, 18, 21, 30). Upstream of the negative regulatory element, the region -1194/-3000 demonstrated cell-specific cis-acting elements. Cardiac fibroblasts, a proliferative cell lineage, exhibited enhanced transcriptional activation, whereas transcriptional repression occurred in terminally differentiated cardiac myocytes presumably in response to the binding of cell-specific transcription factors (18). Maximal promoter activity in SMCs, also a proliferative lineage, was similar to the minimal promoter. These results suggest that within the upstream region of the S100A6 promoter, there is sequence information that drives transcription according to the physiological pattern, i.e. relatively lower expression in cardiac myocytes versus fibroblasts.
Because S100A6 is a growth-regulated gene inducible by growth factors (12, 15, 17, 36), we set out to identify the sequences in the 5′-flanking region that are regulated by trophic stimuli. In cardiac fibroblasts, despite high basal S100A6 expression, trophic stimuli did not induce either the minimal or maximal S100A6 promoters. The role of S100A6 in cardiac fibroblasts in modulating cardiac pathophysiology is amenable to future investigation. In cardiac myocytes, because we have previously shown that the S100B promoter was selectively responsive to α1-adrenergic signaling, we used the basic and maximal S100A6 promoters to study induction of S100A6 by α1-adrenergic agonists and downstream components of α1-adrenergic signaling, the PKC pathway, and transcription factors TEF-1 and RTEF1 (37). The α1-adrenergic agonists NE, PE, the phorbol ester PMA, an activator of PKC, and a constitutively active mutant of PKC-β, ΔPKC-β did not activate the S100A6 promoters. Although the S100A6 promoters were unresponsive to α1-adrenergic stimulation, the transcription factors TEF-1 and RTEF-1 transrepressed the basic and maximal S100A6 promoters consistent with the presence of an atypical M-CAT sequence (the binding site for TEF-1/RTEF-1) in the basic promoter (30). The inhibition of the S100A6 promoter was not seen with 10- and 100-fold lower concentrations of TEF-1 and RTEF-1 (data not shown). The inhibitory effect of TEF-1 is comparable with the response of the S100B promoter and to the response of the skACT promoter and may result from TEF-1 overabundance sequestering or “squelching” a limiting factor (24, 37). The transrepression of the S100A6 promoter by TEF-1 and RTEF-1 may also be involved in the low level expression of this gene in cardiac myocytes.
Serum induced both the basic and maximal promoters equally suggesting that the serum-inducible sequences must be located in the basic promoter. The basic promoter includes a TATA homology, GC boxes, and an enhancer-like sequence (12). These sequences do not contain appreciable similarity to known serum-inducible sequences, and the closest similarity is in the enhancer-like structure, and removal of this sequence does not abolish the response to serum (12). Although serum stimulation activated both the basic and maximal promoters, PDGF, TGFβ, and TNF-α required the maximal promoter for their stimulatory action. Therefore, the peptide growth factor and cytokine-responsive sequences must be located upstream of the basic promoter fragment. Reported sequences upstream of the -167/+134 include an NF-κB-binding site at position -460/-451 (13), an antioxidant response element located at position -290/-281 (16) and two canonical E-boxes located at position -283/-278 that bind the ubiquitous upstream stimulatory factors 1 and 2 (38). Of interest, the NF-κB-binding site has been implicated in S100A6 transcriptional regulation by TNF-α in a liver cell line (13).
The NF-κB family of transcription factors consists of homo- or heterodimeric subunits of the Re1 family, including p65, p50, p52, c-Re1, and Re1-B (39). In unstimulated cells, most of the NF-κB is inactive and localized in the cytoplasm in complex with an inhibitory protein, IκB (39). Upon stimulation by a variety of agents, IκBα becomes phosphorylated and subsequently translocates to the nucleus and activates transcription of specific target genes (39). In this study, we first showed that NF-κB (p65 subunit) is able to activate the maximal S100A6 promoter implicating an active NF-κB-binding site in this promoter. We then demonstrated that PDGF, TGFβ, and TNF-α all activate NF-κB, and by using EMSA and chromatin immunoprecipitation analysis, we determined that a consensus NF-κB-binding site (-460/-451) in the maximal promoter of the human and rat S100A6 binds NF-κB in vitro and in vivo in response to TNF-α. Pretreatment with CAPE, a potent and specific inhibitor of NF-κB or point mutations of the NF-κB-binding site of the maximal promoter, blocked S100A6 promoter activation by these ligands. The above data suggest that NF-κB activation and subsequent binding to an NF-κB-binding site correlates with the induction of S100A6 promoter activity by PDGF, TGFβ, and TNF-α.
As cardiomyocytes are differentiated cells with limited capacity for cell division, maintenance of this cell population via prevention of apoptosis is of critical importance. The cellular decision to undergo apoptosis is governed by the integration of survival and death signals. Cell fate is ultimately determined by the balance of these antagonizing signals. It has been shown that cardiac myocyte apoptosis occurs in experimental models of ischemia/reperfusion injury and, importantly, that apoptosis is a feature of human ischemic myocardial damage that determines myocardial infarct size (40–42). In this study, we demonstrate that PDGF does not induce myocyte apoptosis, whereas TNF-α increases caspase-3 activity, increases the phosphorylation of the tumor suppressor gene p53, and induces myocyte apoptosis as described previously (20, 41–44). The observation that TNF-α is a trigger for apoptosis in the heart suggests that the cardiotoxicity of TNF-α observed in a variety of clinical conditions, including heart disease, may be due to TNF-α-induced cardiac myocyte apoptosis (40–42, 45). The dose of TNF-α capable of producing apoptosis in our cultured rat cardiac myocytes is within the range found in the serum of patients experiencing severe acute myocardial infarction (41), suggesting that TNF-α may contribute to cell death by apoptosis during ischemia. TNF-α induces cardiac myocyte apoptosis in vivo and in vitro through multiple death domain pathways initiated by the trimerization of TNFR1 culminating in the activation of caspases and myocyte apoptosis (20, 40–45). TNF-α may also induce cardiac myocyte apoptosis via induction of inducible nitric-oxide synthase and nitric oxide resulting in myocyte apoptosis (44). Besides activation of these multiple pro-apoptotic pathways, TNF-α also activates NF-κB (present study and see Refs. 20, 43). NF-κB signaling is involved in the pro-survival response of cardiomyocytes to other cytokines (20, 42). In fact, a large number of anti-apoptotic gene products are directly regulated by NF-κB (46). These include cellular inhibitors of apoptosis, anti-apoptotic Bcl-2 family members (A1, Bcl-xl), and the FLICE inhibitory protein (46). Many of these proteins can themselves directly influence the NF-κB pathway or block the apoptosis signaling cascade. We provide the first direct evidence that S100A6 is one such antiapoptotic gene product activated by TNF-α signaling. First, we showed that the induction of NF-κB in cardiac myocytes by TNF-α activated S100A6 at the transcriptional level. Second, overexpression of S100A6 by gene transfer inhibited myocyte apoptosis in response to TNF-α. Third, knocking down S100A6 expression using siRNA directed against S100A6 potentiated TNF-α-induced myocyte apoptosis. Taken together, the above data indicate that the same TNF-α pathway that initiates and sustains the apoptotic response in cardiac myocytes and is subject to negative modulation by S100A6 also induces S100A6. In agreement with our observation that S100A6 is an anti-apoptotic gene, in a cholesteatoma cell line, high levels of S100A6 were associated with the lowest levels of apoptosis (11). However, several recent publications have shown that exogenously administered S100A6 at micromolar concentrations induces glioblastoma cell apoptosis by interacting with the receptor for advanced glycation end products (47), and exogenously introduced S100A6 to a human liver cell line (Hep3B and HepG2) potentiated apoptotic response of the calcium ionophore A23187 (48). These data demonstrate that the differential effects of S100A6 on apoptosis may be dependent on the specific cell type, the concentration of S100A6, and the preferential activation of one distinct signaling pathway as has been suggested for S100B (47, 49). We also provide evidence as to the mechanism by which S100A6 modulates the TNF-α-induced apoptotic response. We demonstrate in co-immunoprecipitation experiments an association of S100A6 and p53. The effect of this association appears to be interference of p53 function as overexpression of S100A6 reduced phosphorylation of p53 by TNF-α, and inhibition of S100A6 by an siRNA restored and further potentiated the TNF-α-induced phosphorylation of p53. In this context, overexpression of wild type p53 does not activate S100A6 transcription. Taken together the evidence suggests that S100A6 associates with and inhibits the phosphorylation of p53 thereby limiting TNF-α-induced apoptosis. In keeping with a key role of p53 in this pathway, overexpression of mutant p53 acting as a dominant negative inhibited TNF-α-induced apoptosis. It is important to note that the induction of S100A6 by TNF-α is not sufficient to completely inhibit TNF-α-induced myocyte apoptosis. It is possible that the activation of the pro-apoptotic pathway(s) by TNF-α may simply overwhelm the pro-survival pathway(s) of which S100A6 is a component and tips the balance toward cell death.
Although the results of this study are restricted to cultured neonatal cardiomyocytes, they shed some light on S100A6 function in the pathological heart. The demonstration that TNF-α signaling gives rise to one or more cytoprotective signals that prevent and/or delay the development of cardiac myocyte apoptosis after ischemic injury (40) together with our observation that S100A6 expression is increased in the periinfarct zone of the rat heart post-infarction (17) suggest that the cytoprotective effects of TNF-α after ischemic injury may be mediated in part by S100A6. This possibility can be tested experimentally in vivo using animal models of S100A6 overexpression/inhibition. We are in the process of generating a cardiac tissue-specific and regulatable S100A6 transgenic mouse.
In conclusion, the transcriptional regulation of the S100A6 promoter is complex involving both common and cell selective positive and negative regulatory elements that modulate promoter activity by multiple stimuli. S100A6 is a component of the pro-survival pathway activated by TNF-α and provides a homeostatic mechanism to limit TNF-α-induced myocyte apoptosis by interfering with p53 phosphorylation. Additional delineation of this pathway may help elucidate mechanisms simultaneously modulating cardiac myocyte survival and inflammatory signaling while laying a foundation for novel approaches to intervention in related cardiac conditions.
This work was supported by the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: bp, base pairs; TNF, tumor necrosis factor; TEF, transcription enhancer factor; CAT, chloramphenicol acetyltransferase; RSV, Rous sarcoma virus; SMCs, smooth muscle cells; PKC, protein kinase C; NE, norepinephrine; PE, phenylephrine; TGF, transforming growth factor; PDGF, platelet derived growth factor; CAPE, caffeic acid phenethyl ester; EMSA, electronic mobility shift assay; ChIP, chromatin immunoprecipitation; TUNEL, terminal deoxynucleotidyl transfer-mediated end-labeling; FITC, fluorescein isothiocyanate.
References
- 1.Kligman, D., and Hilt, D. C. (1988) Trends Biochem. Sci. 13 437-443 [DOI] [PubMed] [Google Scholar]
- 2.Zimmer, D. B., Cornwall, E. H., Landar, A., and Song, W. (1995) Brain Res. Bull. 37 417-429 [DOI] [PubMed] [Google Scholar]
- 3.Schafer, B. W., and Heizmann, C. W. (1996) Trends Biochem. Sci. 21 134-140 [DOI] [PubMed] [Google Scholar]
- 4.Kuznicki, J., Filipek, A., Heimann, P., Kaczmarek, L., and Kaminska, B. (1989) FEBS Lett. 254 141-144 [DOI] [PubMed] [Google Scholar]
- 5.Engelkamp, D., Schafer, B. W., Erne, P., and Heizmann, C. W. (1992) Biochemistry 31 10258-10264 [DOI] [PubMed] [Google Scholar]
- 6.Hyland, J. K., Hirschhorn, R. R., Aller, P., Gibson, C. W., and Baserga, R. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 400-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabretta, B., Venturelli, D., Kaczmarek, L., Narni, F., Talpaz, M., Anderson, B., Beran, M., and Baserga, R. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 1495-1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuznicki, J., Kordowska, J., Puzianowska, M., and Wozniewicz, B. M. (1992) Exp. Cell Res. 200 425-430 [DOI] [PubMed] [Google Scholar]
- 9.Breen, E. C., and Tang, K. (2003) J. Cell. Biochem. 88 848-854 [DOI] [PubMed] [Google Scholar]
- 10.Timmons, P. M., Chan, C. T., Rigby, P. W., and Poirier, F. (1993) Cell Sci. 104 187-196 [DOI] [PubMed] [Google Scholar]
- 11.Choufani, G., Mahillon, V., Decaestecker, C., Lequeux, T., Danguy, A., Salmon, I., Gabius, H. J., Hassid, S., and Kiss, R. (1999) Laryngoscope 109 1825-1831 [DOI] [PubMed] [Google Scholar]
- 12.Ghezzo, F., Lauret, E., and Baserga, R. (1988) J. Biol. Chem. 263 4758-4763 [PubMed] [Google Scholar]
- 13.Joo, J. H., Kim, J. W., Lee, Y., Yoon, S. Y., Kim, J. H., Paik, S. G., and Choe, I. S. (2003) Biochem. Biophys. Res. Commun. 307 274-280 [DOI] [PubMed] [Google Scholar]
- 14.Courtois-Coutry, N., LeMoelic, C., Boulkroun, S., Fay, M., Cluzeaud, F., Escoubet, B., Farman, N., and Blot-Chabaud, M. (2002) J. Biol. Chem. 277 25728-25734 [DOI] [PubMed] [Google Scholar]
- 15.Gong, Y., Alkhalaf, B., Molmar, P., Murphy, L. C., and Murphy, L. J. (1992) Cell Growth & Differ. 3 847-853 [PubMed] [Google Scholar]
- 16.Lesniak, W., Szczepanska, A., and Kuznicki, J. (2005) Biochim. Biophys. Acta 1744 29-37 [DOI] [PubMed] [Google Scholar]
- 17.Tsoporis, J. N., Marks, A., Haddad, A., O'Hanlon, D., Jolly, S., and Parker, T. G. (2005) Exp. Cell Res. 303 471-481 [DOI] [PubMed] [Google Scholar]
- 18.Lesniak, W., Jezierska, A., and Kuznicki, J. (2000) Biochim. Biophys. Acta 1517 73-81 [DOI] [PubMed] [Google Scholar]
- 19.Satoh, M., Nakamura, M., Saitoh, H., Satoh, H., Maesawa, C., Segawa, I., Tashiro, A., and Hiramori, K. (1999) Circulation 99 3260-3265 [DOI] [PubMed] [Google Scholar]
- 20.Kubota, T., Miyagishima, M., Frye, C. S., Alber, S. M., Bounoutas, G. S., Kadokami, T., Watkins, S. C., McTiernan, C. F., and Feldman, A. M. (2001) J. Mol. Cell. Cardiol. 33 1331-1344 [DOI] [PubMed] [Google Scholar]
- 21.Lesniak, W., Swart, G. W. M., Bloemers, H. P. J., and Kuznicki, J. (2000) Acta Neurobiol. Exp. 60 569-575 [DOI] [PubMed] [Google Scholar]
- 22.Huttunen, H. J., Fages, C., and Rauvala, H. (1999) J. Biol. Chem. 274 19919-19924 [DOI] [PubMed] [Google Scholar]
- 23.Kariya, K., Karns, L. R., and Simpson, P. C. (1991) J. Biol. Chem. 266 10023-10026 [PubMed] [Google Scholar]
- 24.Stewart, A. F. R., Suzow, J., Kubota, T., Ueyama, T., and Chen, H. H. (1998) Circ. Res. 83 43-49 [DOI] [PubMed] [Google Scholar]
- 25.Johnson, C. M., and Tapping, R. I. (2007) J. Biol. Chem. 282 31197-31205 [DOI] [PubMed] [Google Scholar]
- 26.Johnson, P., Chung, S., and Benchimol, S. (1993) Mol. Cell. Biol. 13 1456-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long, C. C., Henrich, C. J., and Simpson, P. C. (1991) Cell Regul. 2 1081-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feener, E. P., Northrup, J. M., Aiello, L. P., and King, G. L. (1995) J. Clin. Investig. 95 1353-1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Wet, J. R., Wood, K. V., De Luca, M., Helinski, D. R., and Subramani, S. (1987) Mol. Cell. Biol. 7 725-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Groningen, J. J., Weterman, M. A., Swart, G. W. M., and Bloemers, H. P. J. (1995) Biochem. Biophys. Res. Commun. 213 1122-1131 [DOI] [PubMed] [Google Scholar]
- 31.Kanoh, M., Takemura, G., Misao, J., Hayakawa, Y., Aoyoma, T., Nishigaki, K., Noda, T., Fujiwara, T., and Fujiwara, H. (2000) Circulation 99 2757-2764 [DOI] [PubMed] [Google Scholar]
- 32.Canman, C. E., Gilmer, T. M., Coutts, S. B., and Kastan, M. B. (1995) Genes Dev. 9 600-611 [DOI] [PubMed] [Google Scholar]
- 33.Grigorian, M., Andresen, S., Tulchinsky, E., Kriajeska, M., Carlberg, C., Kruse, C., Cohnm, M., Ambartsumian, N., Christensen, A., Selivanova, G., and Lukanidin, E. (2001) J. Biol. Chem. 276 22699-22708 [DOI] [PubMed] [Google Scholar]
- 34.Lin, J., Blake, M., Tang, C., Zimmer, D., Rustandi, R. R., Weber, D. J., and Carrier, F. (2001) J. Biol. Chem. 276 35037-35041 [DOI] [PubMed] [Google Scholar]
- 35.Mueller, A., Schafer, B. W., Ferrari, S., Weibel, M., Makek, M., Hochli, M., and Heizmann, C. W. (2005) J. Biol. Chem. 280 29186-29193 [DOI] [PubMed] [Google Scholar]
- 36.Hirschhorn, R. R., Aller, P., Yuan, Z. A., Gibson, C. W., and Baserga, R. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 6004-6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsoporis, J. N., Marks, A., Van Eldyk, L. J., O'Hanlon, D., and Parker, T. G. (2003) Am. J. Physiol. 284 H193-H203 [DOI] [PubMed] [Google Scholar]
- 38.Lesniak, W., and Kuznicki, J. (2006) J. Cell. Biochem. 97 1017-1024 [DOI] [PubMed] [Google Scholar]
- 39.Baeuerle, P. A. (1991) Biochim. Biophys. Acta 1072 63-80 [DOI] [PubMed] [Google Scholar]
- 40.Hsieh, P. C., Davies, M. E., Gannon, J., MacGillivray, C., and Lee, R. T. (2007) J. Clin. Investig. 116 237-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krown, K. A., Page, T., Nguyan, C., Gutierrez, V., Comstock, K. L., Glembotski, C. C., Quintana, P. J. E., and Sabbadini, R. A. (1996) J. Clin. Investig. 98 2854-2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurrelmeyer, K. M., Michael, L. H., Baumgarten, G., Taffet, G. E., Peschon, J. J., Sivasubramanian, N., Entman, M. L., and Mann, D. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5456-5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergmann, M. W., Loser, P., Dietz, R., and Harsdorf, R. V. (2001) J. Mol. Cell. Cardiol. 33 1223-1232 [DOI] [PubMed] [Google Scholar]
- 44.Song, W., Lu, X., and Feng, Q. (2000) Cardiovasc. Res. 45 595-602 [DOI] [PubMed] [Google Scholar]
- 45.Sun, M., Chen, M., Dawood, F., Zurawska, U., Li, J. Y., Parker, T. G., Kassiri, Z., Kirshenbaum, L. A., Arnold, M., Khokla, R., and Liu, P. P. (2007) Circulation 115 1398-1407 [DOI] [PubMed] [Google Scholar]
- 46.Purcell, N. H., and Molkentin, J. D. (2003) Circulation 108 638-640 [DOI] [PubMed] [Google Scholar]
- 47.Leclerc, E., Fritz, G., Weibel, M., Heizmann, C. W., and Galichet, A. (2007) J. Biol. Chem. 282 31317-31331 [DOI] [PubMed] [Google Scholar]
- 48.Joo, J. H., Yoon, S. W., Kim, J. H., Paik, S.-P., Min, S. R., Lim, J.-S., Choe, I. S., Choi, I., and Kim, J. W. (2008) J. Cell. Biochem. 103 1183-1197 [DOI] [PubMed] [Google Scholar]
- 49.Huttunen, H. J., Kuja-Panula, J., Sorci, G., Agneletti, A. L., Donato, R., and Rauvala, H. (2000) J. Biol. Chem. 275 40096-40105 [DOI] [PubMed] [Google Scholar]