Abstract
The replication fork helicase in eukaryotes is a large complex that is composed of Mcm2-7, Cdc45, and GINS. The Mcm2-7 proteins form a heterohexameric ring that hydrolyzes ATP and provide the motor function for this unwinding complex. A comprehensive study of how individual Mcm subunit biochemical activities relate to unwinding function has not been accomplished. We studied the mechanism of the Mcm4-Mcm6-Mcm7 complex, a useful model system because this complex has helicase activity in vitro. We separately purified each of three Mcm subunits until they were each nuclease-free, and we then examined the biochemical properties of different combinations of Mcm subunits. We found that Mcm4 and Mcm7 form an active unwinding assembly. The addition of Mcm6 to Mcm4/Mcm7 results in the formation of an active Mcm4/Mcm6/Mcm7 helicase assembly. The Mcm4-Mcm7 complex forms a ringed-shaped hexamer that unwinds DNA with 3′ to 5′ polarity by a steric exclusion mechanism, similar to Mcm4/Mcm6/Mcm7. The Mcm4-Mcm7 complex has a high level of ATPase activity that is further stimulated by DNA. The ability of different Mcm mixtures to form rings or exhibit DNA stimulation of ATPase activity correlates with the ability of these complexes to unwind DNA. The Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 assemblies can open to load onto circular DNA to initiate unwinding. We conclude that the Mcm subunits are surprisingly flexible and dynamic in their ability to interact with one another to form active unwinding complexes.
The replication fork helicase unwinds parental duplex DNA to provide single-stranded DNA (ssDNA)3 substrates for the replicative polymerases (1, 2). Unwinding of the replication fork in eukaryotes is powered by six minichromosome maintenance proteins (Mcm2-7) acting in concert with Cdc45 and the GINS complex (3-8). There are an estimated 30,000 Mcm molecules in Saccharomyces cerevisiae, in vast excess of what is required to license origins of DNA replication (9-13). Thus, it is widely believed that Mcm proteins participate in functions other than DNA replication. There is evidence that Mcm7 acts as a transcription factor (14), and Mcm5 is essential for Stat-1-mediated transcriptional activation (15). There is also evidence that Mcm proteins are involved in chromatin remodeling (16-21), and Mcms interact directly with histone H3 (16, 22). Mcms are also important to maintain genome stability (23, 24), and it has been proposed that dormant origins licensed by Mcms are required to survive replication stress (25).
During purification of Mcm proteins from yeast, mouse, or Xenopus, Mcm4, Mcm6, and Mcm7 are most tightly associated (22, 26-32). The Mcm4-Mcm6-Mcm7 complex has helicase activity in vitro, as demonstrated by the ability of recombinant Mcm4/Mcm6/Mcm7 to unwind a duplex DNA substrate that resembles a replication fork (29, 33-36). The unwinding activity of Mcm4/Mcm6/Mcm7 is functionally conserved throughout eukaryotes, as Mcm4-Mcm6-Mcm7 complexes from budding yeast, fission yeast, Drosophila, Xenopus, and mouse exhibit helicase activity (29, 33-35, 37).
Investigations of ring-shaped helicases have been primarily focused on homohexameric assemblies (1, 2, 38). The eukaryotic Mcms are somewhat unusual in this regard, because they form heterohexamers. It is currently not known how the different eukaryotic Mcm subunits function in coordination to unwind DNA, and a comprehensive study of how individual Mcm subunit biochemical activities relate to unwinding activity has not been accomplished.
Study of the eukaryotic Mcms may reveal insight into how specific components of a hexameric helicase contribute to form an active unwinding assembly. The Mcm4-Mcm6-Mcm7 complex is a useful model system in this regard, because the complex purified from recombinant proteins is active as a helicase in vitro. In the past, Mcm4-Mcm6-Mcm7 was purified as a pre-assembled complex from partially purified subunits, because Mcm4, Mcm6, and Mcm7 subunits were not pure enough to allow for direct study with DNA (33, 36, 39, 40). In this study, the production of highly purified, nuclease-free individual Mcm subunits is used to study how each Mcm subunit contributes to DNA unwinding. We find that the combination of individually purified Mcm4 and Mcm7 subunits results in a complex that can unwind DNA. Incubation of Mcm6 with Mcm4/Mcm7 results in the formation of an active Mcm4/Mcm6/Mcm7 helicase assembly. Mcm4/Mcm7 forms ring-shaped particles that are likely to be hexameric, and Mcm4/Mcm7 unwinds DNA with 3′ to 5′ polarity in a manner that is consistent with a steric exclusion model. The Mcm4/Mcm7 and the Mcm4/Mcm6/Mcm7 rings spontaneously open to load onto circular ssDNA and unwind an annealed oligonucleotide. The ability of different Mcm mixtures to form ring shapes and exhibit DNA stimulation of ATPase activity correlates well with the ability of these Mcm mixtures to unwind DNA. We conclude that Mcm subunits are surprisingly flexible and dynamic in their ability to form active unwinding complexes.
EXPERIMENTAL PROCEDURES
Cloning and Purification of Mcm4, Mcm6, and Mcm7—The full-length open reading frame of S. cerevisiae Mcm4 was cloned into the NdeI/BamHI sites of the pET-28 vector (EMD Biosciences). The Mcm4-pET-28 vector was transformed into Escherichia coli BL-21 DE3 Codon Plus cells (Stratagene), and a colony from the transformation was inoculated into 60 liters of LB containing 30 μg/ml kanamycin. When the cells reached an A600 of 0.6, 0.7 mm isopropyl 1-thio-β-d-galactopyranoside was added, and the temperature was lowered to 12 °C. 16 h later, cells were harvested and lysed by French press in 500 ml of solution containing 10% sucrose, 3.5 g of spermidine, 500 mm NaCl, and 50 mm Tris-HCl, pH 8.0. The lysate was applied to a 40-ml chelating Sepharose fast flow resin (GE Healthcare) precharged with nickel sulfate. The column was washed with solution containing 500 mm NaCl, 50 mm Tris-HCl, pH 8.0, and 50 mm imidazole. The Mcm4 protein was eluted in solution containing 10% glycerol, 500 mm NaCl, 50 mm Tris-HCl, pH 8.0, and 250 mm imidazole, dialyzed into Buffer H (10% glycerol, 20 mm Hepes, pH 7.5, 0.1 mm EDTA, and 1 mm DTT) containing 50 mm NaCl, and applied to a 30-ml SP-Sepharose column. The SP column was washed with Buffer H containing 100 mm NaCl, and Mcm4 was eluted in a 300-ml linear gradient from Buffer H containing 100 mm NaCl to Buffer H containing 500 mm NaCl. Peak fractions containing Mcm4 were combined and precipitated with 0.3 g/ml ammonium sulfate, and the pellet was resuspended in 22 ml of Buffer H + 500 mm NaCl. 5 ml of this sample containing 22.5 mg of protein was applied to a 124-ml Superdex 200 column (GE Healthcare) that was pre-equilibrated in Buffer H + 500 mm NaCl. Peak fractions containing Mcm4 were pooled and dialyzed against Buffer A (10% glycerol, 20 mm Tris-HCl, pH 7.5, 0.1 mm EDTA, and 1 mm DTT) containing 50 mm NaCl and 10 mm magnesium chloride. The sample was applied to an ssDNA-cellulose resin, and the flow-through containing Mcm4 was flash-frozen.
Purification of Mcm6 was identical to the procedure followed previously (27), except the Mcm6 sample was further purified. The Mcm6 sample was subjected to preparative size-exclusion chromatography with a 124-ml Superdex 200 column (GE Healthcare) that was pre-equilibrated in Buffer A + 500 mm NaCl. Peak fractions containing Mcm6 were pooled and dialyzed against Buffer A containing 50 mm NaCl and 10 mm magnesium chloride. The sample was then applied to an ssDNA-cellulose resin, and the flow-through containing Mcm6 was flash-frozen.
Purification of Mcm7 was identical to the procedure followed previously (35), except the Mcm7 sample was further purified. The Mcm7 sample was subjected to preparative size-exclusion chromatography with a 124-ml Superdex 200 column that was pre-equilibrated in Buffer A + 500 mm NaCl. Peak fractions containing Mcm7 were pooled and dialyzed against Buffer A containing 50 mm NaCl and 10 mm magnesium chloride. The sample was then applied to an ssDNA-cellulose resin, and the flow-through containing Mcm7 was flash-frozen.
Radiolabeling and Annealing DNA—DNA was end-labeled with T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's instructions. To anneal DNA, 500 nm radiolabeled DNA was incubated overnight at 37 °C with 1 μm complementary DNA in 20 mm Tris-HCl, 4% glycerol, 0.1 mm EDTA, 40 μg/ml bovine serum albumin, 5 mm DTT, and 5 mm magnesium acetate in a final volume of 12 μl. Following the overnight incubation, the reaction was diluted to a final concentration of 50 nm (concentration of radiolabeled DNA) with 20 mm Tris-HCl, 0.1 mm EDTA.
Helicase Assay—Each unwinding reaction contained 25.4 mm Tris-HCl, pH 7.5, 9.9 mm magnesium acetate, 23.3% glycerol, 152 μm EDTA, 42.4 μg/ml bovine serum albumin, 5.01 mm DTT, 4.98 mm ATP, 4.98 mm creatine phosphate, 19.9 μg/ml creatine kinase, 1.2 nm32P-labeled DNA, and proteins as detailed in each figure legend, in a final volume of 11 μl. Unless otherwise indicated, the final concentration of each Mcm subunit was 709 nm, expressed as a monomer (850 ng of Mcm4, 897 ng of Mcm6, and 750 ng of Mcm7). When UvrD was used as a control, the final concentration was 3.7 μg/ml, or 45 nm monomer. All reaction samples were prepared on ice, and shifted to 37 °C to initiate the reaction. At the end of the reaction, 1 μl of proteinase K (10 mg/ml) was added, and the sample was incubated an additional 1 min at 37 °C. 5 μl of stop solution (2% SDS, 80 mm EDTA) was added, followed by 5 μl of 6× loading dye (15% Ficoll + 0.1% xylene cyanol), and finally the reaction was flash-frozen. For DNA containing a biotin moiety, 100 nm streptavidin was preincubated with the DNA prior to adding Mcm proteins. For experiments containing T4 gp32, 26 ng (73 nm) of T4 gp32 (New England Biolabs) was preincubated with the DNA prior to adding the Mcm proteins.
For experiments with oligonucleotide substrates, samples were analyzed by an 8% native polyacrylamide gel containing 1× TBE (90 mm Tris-HCl borate, 2 mm EDTA) at 225 V. The gel was dried for 2 h at 60 °C and exposed to a phosphorimaging screen overnight. For experiments with M13 circular ssDNA substrates, samples were analyzed by a 1% native agarose gel containing 1× TBE at 100 V. The gel was dried for 2 h at 60 °C and exposed to a phosphorimaging screen overnight.
Analytical Size-exclusion Chromatography—Mcm4, Mcm6, and/or Mcm7 in a total volume of 400 μl were incubated at 15 °C for 30 min. For single subunit experiments, 400 μg of each subunit was analyzed (9.17 μm Mcm4 monomer, 8.69 μm Mcm6 monomer, or 10.5 μm Mcm7 monomer); for the binary complex, 350 μg of Mcm4 and 400 μg of Mcm7 were analyzed (8.02 μm Mcm4 monomer and 10.5 μm Mcm7 monomer); and for the ternary complex, 300 μg of Mcm4, 300 μg of Mcm6, and 400 μg of Mcm7 were analyzed (6.88 μm Mcm4 monomer, 6.52 μm Mcm6 monomer, and 5.26 μm Mcm7 monomer). The samples were applied to a 24-ml Superose 6 column (GE Healthcare) pre-equilibrated in Buffer A + 150 mm NaCl. 250-μl fractions from the column were analyzed by SDS-PAGE followed by staining with Coomassie Blue. The elution peaks of molecular weight standards were determined under the same conditions. 5 μl of each fraction of the elution was further analyzed for helicase activity.
Electron Microscopy—Samples for electron microscopy contained 20 mm Tris-HCl, pH 7.5, 200 μm DTT, 5 mm magnesium acetate, 5 mm ATP, and Mcm proteins, each at a concentration of 709 nm (monomer), which is 77.3 μg/ml Mcm4, 81.5 μg/ml Mcm6, and 67.4 μg/ml Mcm7. The sample was allowed to incubate at 37 °C for 16 min. For samples containing glutaraldehyde, 0.04% glutaraldehyde was added after the initial incubation, and the sample was incubated at 37 °C for an additional 30 s as described previously (41). The reaction was quenched by the addition of 50 mm EDTA. The reactions were kept on ice until they could be fixed by negative staining. After 100-fold dilution into helicase reaction buffer, 20 μl of sample was applied to a copper mesh 300 grid for 30 s and wicked away, and 20 μl of 1% uranyl acetate was applied to the grid for 30 s and then wicked away. Images were taken with a Philips CM-12 transmission electron microscope at a voltage of 80 kV with a magnification of ×140,000. Scion Image software was used to create an average of selected images. Scion Image was also used to measure the outer distance of the observed ring particles.
ATP Hydrolysis Assay—The ATP hydrolysis assay in a total volume of 11 μl contained 25.4 mm Tris-HCl, pH 7.5, 9.9 mm magnesium acetate, 23.3% glycerol, 152 μm EDTA, 42.4 μg/ml bovine serum albumin, 4.98 mm DTT, 4.98 mm ATP, 0.73 μCi of [γ-32P]ATP (PerkinElmer Life Sciences BLU002Z00), and 709 nm (monomer) of each Mcm subunit (850 ng of Mcm4, 897 ng of Mcm6, and/or 750 ng of Mcm7). The samples were incubated at 37 °C, and 2 μl were removed at 0-, 5-, 10-, and 30-min intervals. The reaction was quenched with 2 μl of stop solution (2% SDS, 80 mm EDTA), and then 1 μl of each sample was spotted on thin layer chromatography plates (PEI-Cellulose F, EMD Chemicals, Inc.) Each TLC plate was developed in 1 m formic acid and 0.5 m LiCl. The plates were allowed to dry and exposed to phosphorimaging cassettes for 30 min. For reactions containing DNA, 1 μm of a 60(dT) oligomer was added to the reaction.
RESULTS
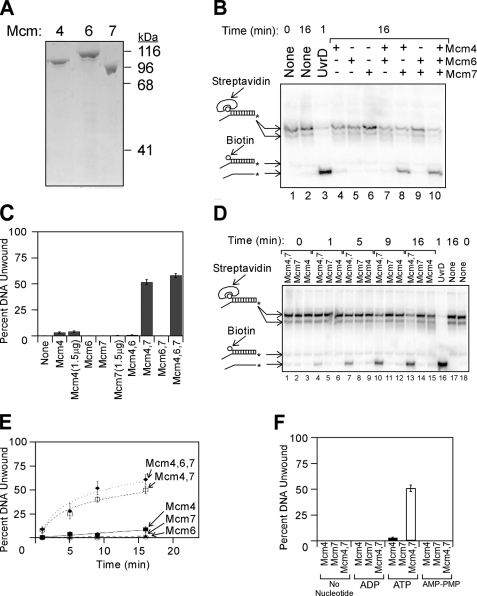
Mcm4 and Mcm7 Form an Active Helicase Assembly—Previously, the Mcm4-Mcm6-Mcm7 complex was studied by purification of the intact hexameric assembly (29, 35, 40, 42, 43). To study how the individual subunits assemble to form a functional helicase, we improved the production and purification of individual Mcm subunits, and SDS-PAGE analysis of the purified subunits is shown (Fig. 1A). The new preparations of Mcm subunits were incubated with one another and examined for helicase activity (Fig. 1, B and C). The DNA substrate used in this unwinding assay is duplex DNA bearing a single strand extension at a 3′ end and a biotin-streptavidin group attached to the 5′ end. The 3′-single strand extension serves as a loading strand for the helicase, whereas the biotin-streptavidin is a bulky group that blocks helicase translocation along dsDNA.
FIGURE 1.
Purified Mcm subunits can assemble into two different active unwinding complexes. A, purified Mcm4, Mcm6, and Mcm7 subunits were analyzed by SDS-PAGE and Coomassie Blue staining. The migration position of molecular weight standards is shown to the right of the gel. B, purified proteins were added to radiolabeled DNA substrate in the presence of ATP at 37 °C, and unwinding was determined by native gel analysis as described under “Experimental Procedures.” The DNA substrate is 32P-labeled duplex DNA bearing a 60(dT) 3′-single strand extension and a 5′-biotin bound to streptavidin. The radiolabeled DNA was incubated with no protein (lanes 1 and 2), UvrD (lane 3), or individually purified Mcm subunits (lanes 4-10) at 37 °C for the times indicated. DNA oligonucleotides used in these studies are described in supplemental Table 1, and the position of the radiolabel is indicated with an asterisk. DNA species in the gel were identified by comparison with standards analyzed in the same gel, and the mobility of each standard is indicated to the left of the gel. The streptavidin-bound oligonucleotides migrate in two different positions on the gel because streptavidin can form multimers. C, data from experiments similar to that shown in B were quantified, averaged, and plotted as mean ± S.E. (n ≥ 6). D, Mcm4, Mcm7, or an equimolar mixture of Mcm4/Mcm7 was incubated with radiolabeled DNA substrate for the time points indicated, and unwinding was monitored by native gel electrophoresis. E, data from experiments similar to that shown in D were averaged and plotted as mean ± S.E. as a function of time, and for each condition the data were then fit with a logarithmic or linear equation. The filled diamonds represent data with Mcm4, Mcm6, and Mcm7; the open squares are Mcm4 and Mcm7 data; the filled squares are Mcm4 data; the open circles are Mcm7 data; and the filled triangles are Mcm6 data. F, Mcm4, Mcm7, or an equimolar mixture of Mcm4/Mcm7 was mixed with the radiolabeled substrate in B for 16 min at 37 °C in the presence of different nucleotides. The fraction of substrate DNA unwound was determined and plotted as the mean ± S.E. for each condition (n ≥ 3).
Previously, only the Mcm4-Mcm6-Mcm7 complex has been shown to unwind DNA, and we fully expected helicase activity to require the presence all three Mcm subunits. When studied alone, Mcm6 or Mcm7 exhibited practically no unwinding activity (Fig. 1B, lanes 5 and 6, and Fig. 1C), whereas Mcm4 exhibited very weak activity (Fig. 1B, lane 4, and Fig. 1C). We then mixed the proteins in binary combinations to study if any of these complexes exhibit unwinding function. Surprisingly, Mcm4 in combination with Mcm7 exhibited helicase activity (Fig. 1B, lane 8, and Fig. 1C). In contrast, no unwinding was observed when Mcm4 was mixed with Mcm6 or Mcm6 was mixed with Mcm7 (Fig. 1B, lanes 7 and 9, and Fig. 1C). These results suggest that Mcm4 and Mcm7 can assemble to form an active, multimeric helicase complex. Furthermore, the concentration of Mcm4/Mcm7 used in our assays (77.3 μg/ml Mcm4 and 67.4 μg/ml Mcm7) is likely to be physiologic, because even higher concentrations of Mcm4 and Mcm7 (344 ng/μl) are present in yeast cells (9, 44). The addition of Mcm6 to Mcm4 and Mcm7 resulted in a slight increase in activity compared with Mcm4 with Mcm7 (Fig. 1B, lane 10, and Fig. 1C).
We next analyzed the time course of Mcm helicase action (Fig. 1, D and E). As expected from our previous observations, Mcm4 added to Mcm7 resulted in an active unwinding complex. We also added Mcm6 to Mcm4 and Mcm7 to determine the effect on helicase action. The addition of Mcm6 to Mcm4 and Mcm7 resulted in a slight increase in the unwinding rate compared with Mcm4 with Mcm7 (Fig. 1E, compare filled diamonds with open squares). Thus, we can recapitulate the observation that Mcm4/Mcm6/Mcm7 is an active helicase by adding the three individual components together and incubating them directly with DNA. Surprisingly, the Mcm4/Mcm7 mixture is nearly as active as Mcm4/Mcm6/Mcm7.
The observation that DNA is unwound by Mcm4 in combination with Mcm7 suggests that the two proteins are functioning as a helicase complex. An important characteristic of an active helicase is the hydrolysis of a nucleotide triphosphate to power DNA unwinding. To test if unwinding of DNA by the Mcm4 and Mcm7 proteins is dependent upon ATP hydrolysis, the unwinding reaction was performed in the presence of different nucleotide analogs (Fig. 1F). Mcm4 in combination with Mcm7 unwound DNA in the presence of ATP but not in the presence of the nonhydrolyzable analog AMP-PNP. Furthermore, no unwinding was observed in the presence of ADP or in the absence of nucleotide. These data demonstrate that the Mcm4 and Mcm7 proteins require a hydrolyzable nucleotide triphosphate to unwind DNA.
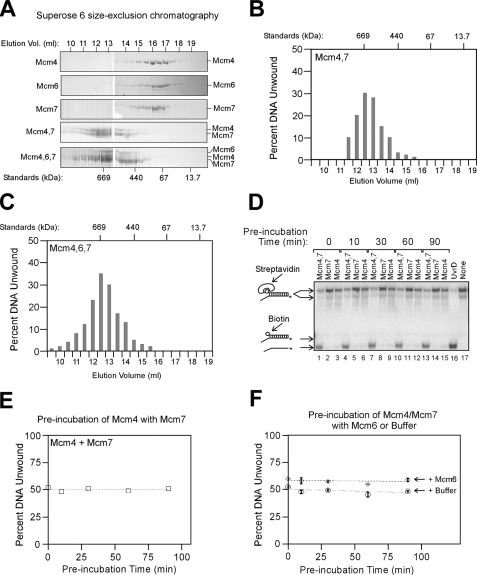
Mcm4 and Mcm7 Assemble to Form a Hexameric Helicase Complex—We next isolated an active Mcm4-Mcm7 complex by size-exclusion chromatography to approximate the size of the helicase assembly (Fig. 2, A and B). Mcm4 alone, Mcm6 alone, or Mcm7 alone had a peak elution consistent with a subunit monomer or dimer (Fig. 2A). When Mcm4 and Mcm7 were mixed together and then subjected to size-exclusion chromatography, there was a peak elution of Mcm4/Mcm7 at a Stokes radius that matched that of thyroglobulin, a 669-kDa protein (Fig. 2A). Fractions from the Mcm4/Mcm7 size-exclusion fractions were then analyzed for unwinding activity using a DNA substrate bearing a 3′-ssDNA extension and a 5′-biotin-streptavidin (Fig. 2B). Peak unwinding activity was observed at an elution volume that is consistent with a single hexamer of Mcm4/Mcm7 (Fig. 2B). These data suggest that a single hexamer of Mcm4/Mcm7 functions as a helicase.
FIGURE 2.
Mcm4 and Mcm7 assemble to form a hexameric helicase complex. A, Mcm4, Mcm6, and/or Mcm7 were mixed, incubated, and analyzed by Superose 6 size-exclusion chromatography as described under “Experimental Procedures.” 5 μl of each 250-μl fraction was then subjected to SDS-PAGE followed by staining with Coomassie Blue. The elution peaks of molecular weight standards are shown below the gels. B, 5 μl of Mcm4/Mcm7 fractions from the Superose 6 column shown in A were analyzed for helicase activity using the DNA substrate and methods described in Fig. 1B. Percent DNA unwound is plotted as a function of elution fraction number. The elution peaks of molecular weight standards are shown at the top of the graph. C, same as B, but for Mcm4/Mcm6/Mcm7. D, 850 ng of Mcm4 (709 nm monomer) and 750 ng of Mcm7 (709 nm monomer) were preincubated with one another at 15 °C for varying times up to 90 min. The Mcm4/Mcm7 mixture was then incubated with the DNA substrate at 37 °C for 16 min, and unwinding was monitored by native gel electrophoresis. The DNA substrate is composed of duplex DNA bearing a 3′-60(dT) ssDNA extension and a 5′-biotin. E, data from D were quantified and plotted as a function of preincubation time. F, 850 ng of Mcm4 and 750 ng of Mcm7 were first mixed with one another at 15 °C for 30 min. 897 ng of Mcm6 was then preincubated with Mcm4/Mcm7 for varying times up to 90 min. (The final concentration of each Mcm subunit is 709 nm.) The Mcm4/Mcm6/Mcm7 mixture was then incubated with the DNA substrate at 37 °C for 16 min, and unwinding was monitored by native gel electrophoresis. The DNA substrate is composed of duplex DNA bearing a 3′-60(dT) ssDNA extension and a 5′-biotin. The data presented are the mean ± S.D. (n = 2).
When mouse or Schizosaccharomyces pombe Mcm4/Mcm6/Mcm7 is copurified as a pre-assembled complex, an Mcm hexamer containing all three subunits is active as a helicase (29, 33). To determine whether mixing S. cerevisiae Mcm6 with Mcm4 and Mcm7 results in the formation of a hexameric helicase assembly containing all three Mcm subunits, we subjected the Mcm4/Mcm6/Mcm7 mixture to size-exclusion chromatography analysis (Fig. 2, A and C). The proteins eluted as a complex containing all three Mcm subunits (Fig. 2A). The elution volume of the assembly exhibited an activity peak that matched that of thyroglobulin (Fig. 2C), suggesting that mixing the three Mcm subunits results in a single hexameric helicase assembly that contains all three subunits.
To study how rapidly the Mcm4 and Mcm7 proteins interact with one another to form an active helicase complex, we next studied how preincubating the Mcm subunits with one another affects the unwinding rate (Fig. 2, D and E). In this experiment, Mcm4 was preincubated with Mcm7 for varying times to allow the formation of an active complex. The Mcm4-Mcm7 complex was then incubated with radiolabeled DNA substrate, and the fraction of unwound DNA was determined by native gel electrophoresis. As the preincubation time varied from 0 to 90 min, the fraction of unwound DNA product did not change (Fig. 2, D and E). Thus, Mcm4 assembles with Mcm7 rapidly relative to the unwinding time, and extended preincubation does not further promote complex formation.
We next studied how rapidly Mcm6 interacts with Mcm4/Mcm7 to form an Mcm4-Mcm6-Mcm7 complex (Fig. 2F). First, Mcm4 was mixed with Mcm7 to form Mcm4/Mcm7. Mcm6 was then preincubated with Mcm4/Mcm7 for varying times to allow for the formation of a ternary complex. The proteins were then incubated with DNA substrate for 16 min, and DNA unwinding was monitored. The data indicate that the fraction of unwound DNA did not vary as a function of preincubation time (Fig. 2F). Furthermore, at every time point, the unwinding percentage was significantly greater than in a control experiment, in which buffer was added instead of Mcm6. Therefore, the addition of Mcm6 to Mcm4/Mcm7 results in the rapid formation of an active Mcm4-Mcm6-Mcm7 complex.
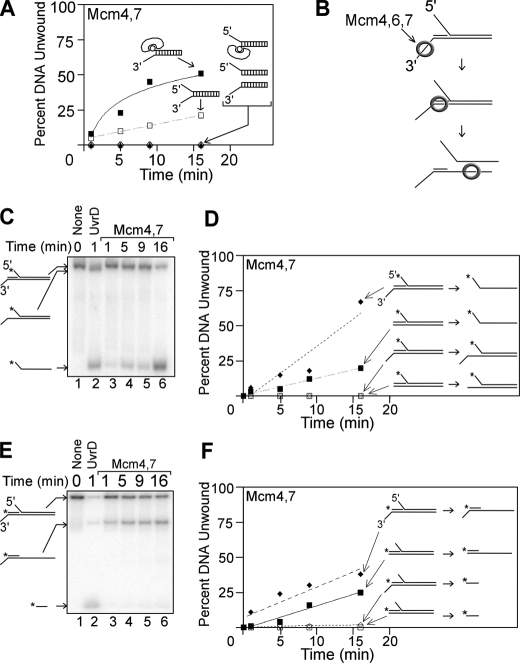
Mcm4/Mcm7 Unwinds DNA with 3′ to 5′ Polarity by a Steric Exclusion Mechanism—We next characterized how Mcm4/Mcm7 unwinds various DNA substrates to gain insight into its mechanism of action (Fig. 3A). It was previously observed that the Mcm4-Mcm6-Mcm7 complex requires duplex DNA bearing a 3′-ssDNA extension to observe unwinding, suggesting that Mcm4/Mcm6/Mcm7 unwinds DNA with 3′ to 5′ polarity (29, 33, 42). Similar to Mcm4/Mcm6/Mcm7, Mcm4/Mcm7 unwound a DNA substrate with a 3′-ssDNA extension and biotin-streptavidin positioned at the 5′ end (Fig. 3A, filled squares, and Fig. 1). However, Mcm4/Mcm7 did not unwind DNA with a reverse arrangement, because no activity was observed with DNA bearing a 5′-ssDNA extension and biotin-streptavidin positioned at the 3′ end (Fig. 3A, filled triangles). These data suggest that Mcm4/Mcm7, like Mcm4/Mcm6/Mcm7, unwinds DNA with 3′ to 5′ polarity.
FIGURE 3.
The Mcm4/Mcm7 assembly unwinds DNA substrates with 3′-to 5′-polarity in a manner consistent with a steric exclusion mechanism. A, equimolar mixture of Mcm4/Mcm7 was incubated at 37 °C with various DNA substrates for the time points indicated, and unwinding was monitored by native gel electrophoresis as described under “Experimental Procedures.” The DNA oligonucleotides used in these experiments are detailed in supplemental Table 1. The time course data for each DNA substrate were fit with a logarithmic or linear equation. Each substrate is composed of identical dsDNA sequence bearing the following extensions: a 3′-60(dT)-ssDNA extension and a 5′-biotin-streptavidin (filled squares), a 3′-60(dT) ssDNA extension and a 5′-60(dT) ssDNA extension (open squares), a 5′-60(dT) ssDNA extension and a 3′-biotin-streptavidin (open diamonds), a 5′-60(dT) extension (filled triangles), and a 3′-60(dT) ssDNA extension (open circles). B, model of how Mcm4/Mcm6/Mcm7 unwinds a tandem duplex DNA substrate by a steric exclusion mechanism (35). The bottom strand is continuous, and the top strand bears a nick, creating two DNA duplexes in tandem. The left duplex bears a 3′-60(dT) ssDNA extension, and the right duplex bears a 5′-60(dT) ssDNA extension. Mcm4/Mcm6/Mcm7 unwinds only the right strand of this tandem duplex substrate. C, equimolar mixture of Mcm4/Mcm7 was incubated with a tandem duplex substrate and ATP at 37 °C for the time points indicated (lanes 3-6). The left duplex bears a 3′-60(dT) ssDNA extension, and the right duplex bears a 5′-60(dT) ssDNA extension. UvrD was used as a SF1 helicase family control (lane 2), and a no protein control is also shown (lane 1). D, data from experiments similar to C were quantified and plotted as a function of time. Data are shown for the tandem duplex substrate bearing a 3′-ssDNA extension (diamonds) and for a similar substrate that lacks the 3′-ssDNA extension (squares). For each DNA product, the time point data were fit to a linear equation. E, this experiment is similar to C, except a different strand is radiolabeled. F, data from experiments similar to E were quantified and plotted as a function of time for each DNA product.
A key feature of many ring-shaped helicases is that they unwind DNA by a steric exclusion mechanism. For example, Mcm4/Mcm6/Mcm7 unwinds DNA only if it bears a 3′-single strand extension and a bulky group, such as ssDNA or biotin-streptavidin, attached to the 5′ end (35, 40). Mcm4/Mcm6/Mcm7 will not unwind DNA bearing only a 3′- or 5′-single strand extension. To test if Mcm4/Mcm7 unwinds DNA with a similar requirement for DNA substrate as Mcm4/Mcm6/Mcm7 and other ring-shaped helicases, the Mcm subunits were incubated with ATP and various DNA substrates (Fig. 3A). Mcm4 in combination with Mcm7 unwound DNA with a 5′- and 3′-single strand extension (Fig. 3A, open squares), but it did not unwind DNA bearing only a 5′- or 3′-single strand extension (open diamonds or open circles). Thus, the requirement of Mcm4/Mcm7 for a bulky group attached to the 5′ end of duplex DNA is very similar to that observed for Mcm4/Mcm6/Mcm7, suggesting a similar mode of action. The Mcm from the archaeal organism Sulfolobus solfataricus exhibits a similar preference for forked DNA substrates (45), suggesting a similar unwinding mechanism between the yeast and archaeal Mcms.
It was previously shown that Mcm4-Mcm6-Mcm7 complexes from S. pombe and S. cerevisiae can translocate along dsDNA with no unwinding (35, 40). In these previous studies, Mcm4/Mcm6/Mcm7 was incubated with the tandem duplex substrate shown in Fig. 3B. In this tandem substrate, the bottom strand is continuous, whereas the top strand bears a nick to create two duplexes in tandem. The duplex on the left bears a 3′-single strand extension for Mcm loading and no bulky group attached to the 5′ end. The duplex on the right bears a long 5′-single strand extension to promote steric exclusion of this strand. When Mcm4/Mcm6/Mcm7 is incubated with this tandem duplex DNA, the protein complexes load on the 3′-single strand extension and then move in the 3′ to 5′ direction toward the duplex (Fig. 3B). The Mcm4-Mcm6-Mcm7 complex does not unwind the duplex on the left, but it does unwind the duplex on the right. Based on these results, it was postulated that Mcm4/Mcm6/Mcm7 encircles two strands while translocating along the left duplex, and unwinding of the right duplex occurs by steric exclusion (35).
To determine whether Mcm4/Mcm7 unwinds DNA by a similar mechanism as Mcm4/Mcm6/Mcm7, Mcm4 and Mcm7 were incubated with the tandem duplex substrate (Fig. 3, C and E). The only difference between the substrates in Fig. 3, C and E, is the position of the radioactive label, and the rate of tandem duplex unwinding is plotted (Fig. 3, D and F). Mcm4/Mcm7 did not unwind the left duplex of the tandem duplex substrate (Fig. 3, C and E, lanes 3-6, and Fig. 3, D and F, open diamonds), but it did unwind the right duplex (Fig. 3, C and E, lanes 3-6, and Fig. 3, D and F, filled diamonds). This result contrasts with that of UvrD, an SF1 helicase that efficiently unwound both DNA strands of this tandem duplex (Fig. 3, C and E, lane 2). Thus, Mcm4/Mcm7 appears to have the ability to translocate along dsDNA with no unwinding, similar to Mcm4/Mcm6/Mcm7. Furthermore, the data suggest that Mcm4/Mcm7 may unwind DNA by a steric exclusion mechanism.
It was previously observed that Mcm4/Mcm6/Mcm7 does not unwind tandem duplex DNA when the 3′-loading strand is absent (35, 40), and we next examined if Mcm4/Mcm7 requires the 3′-single strand extension to load onto the tandem substrate. When the 3′-single strand extension of the tandem substrate was removed, the Mcm4/Mcm7 unwinding rate of the right duplex decreased by roughly one-half (Fig. 3, D and F, compare filled diamonds with filled squares), whereas the left duplex was still not unwound (Fig. 3, D and F, open squares). Thus, the 3′-single strand extension promotes the loading of Mcm4/Mcm7 onto duplex DNA, but it is not absolutely required. These data suggest that the Mcm4-Mcm7 complex, unlike the Mcm4-Mcm6-Mcm7 complex, has a weak ability to load directly onto dsDNA.
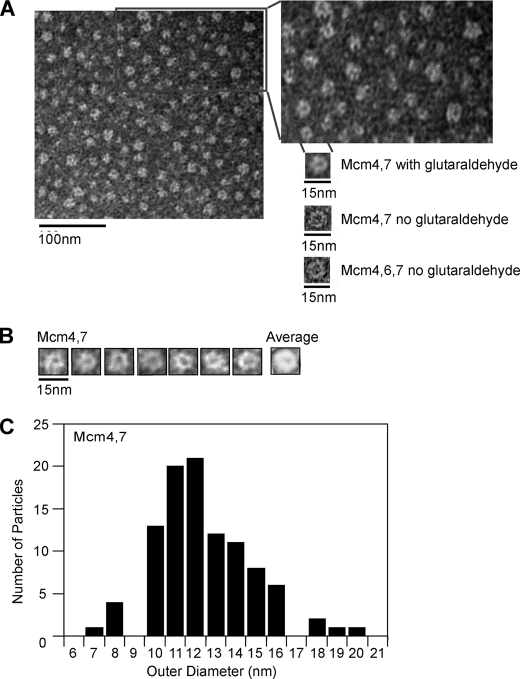
Mcm4/7 Forms Ring-shaped Particles—Mcm4/Mcm6/Mcm7 forms a ring-shaped hexamer that is a dimer of trimers, and it has been proposed that the ring-shaped nature of this protein may be important for unwinding (29, 33, 46). Thus, the observation that Mcm4/Mcm7 exhibits similar substrate dependence for DNA unwinding as Mcm4/Mcm6/Mcm7 led us to speculate that Mcm4/Mcm7 may assemble into a ring-shaped particle. The concentration of Mcm protein used in our unwinding assays (1.6 μg of Mcm4/Mcm7 in 11 μl total volume) was higher than that routinely used in electron microscopy studies. Therefore, we diluted the Mcm4/Mcm7 particles 100-fold prior to imaging by negative stain electron microscopy. Using this approach, we detected ring-shaped images of Mcm4/Mcm7 of a size and shape that is similar to that of Mcm4/Mcm6/Mcm7 (Fig. 4A). The Mcm4/Mcm7 rings were sparsely populated on the microscopy grid, and we postulated that upon dilution and deposition on the grid, many of the Mcm4/Mcm7 rings may disassemble. Thus, we cross-linked the Mcm4/Mcm7 sample with glutaraldehyde and then imaged the assembly with negative stain electron microscopy (Fig. 4A). This technique has been used in the past to identify ring-shaped particles of T7 gp4 helicase (41). Using this approach, a dense array of particles appeared that are very similar to those of Mcm4/Mcm6/Mcm7 (Fig. 4A) (47). We next used the identical technique that was successful in visualizing Mcm4/Mcm7 ring-shaped particles to examine particles of Mcm4 alone, Mcm6 alone, Mcm7 alone, Mcm4/Mcm6, and Mcm6/Mcm7. We were unable to detect hexameric or ring-shaped particles in any of these experiments, either in the presence or absence of glutaraldehyde (not shown).
FIGURE 4.
Mcm4-Mcm7 complex is ring-shaped. A, Mcm4 was incubated with equimolar Mcm7 in the absence and presence of glutaraldehyde as described under “Experimental Procedures.” The Mcm4-Mcm7 complex was then diluted and analyzed by negative stain electron microscopy. The boxed area is enlarged to show detail. For comparison, an equimolar mixture of Mcm4/Mcm6/Mcm7 was diluted and analyzed by negative stain electron microscopy using the same procedures. B, selected Mcm4/Mcm7 rings from experiments described in A were averaged using Scion Imaging software. C, outer diameters of 100 ring-shaped Mcm4/Mcm7 particles from experiments described in A were measured and plotted.
Mcm4/Mcm7 particles that appeared to be ring-shaped were selected for averaging, and the averaged image appears to be that of a single hexamer (Fig. 4B). We also measured the outer diameter of 100 of these ring-shaped particles, and we found that the mean diameter, 12.4 nm, is similar to that reported previously for Mcm4/Mcm6/Mcm7 (Fig. 4C) (46, 48). Double hexamers of Mcm4/Mcm7 or Mcm4/Mcm6/Mcm7 were not observed. Taken together, the data suggest that Mcm4/Mcm7 forms single hexameric rings at a concentration that is active in unwinding DNA.
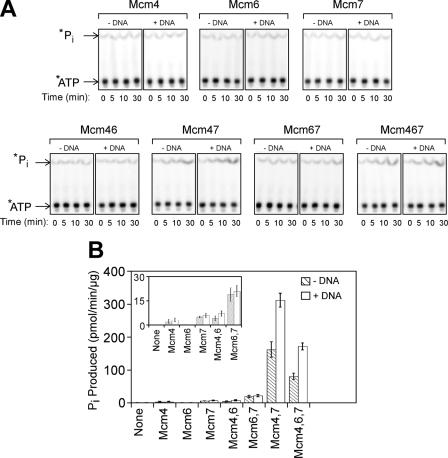
ssDNA Stimulates Mcm4/Mcm7 Hydrolysis of ATP—One hallmark of helicase activity is DNA-dependent stimulation of ATP hydrolysis, because helicases use the energy derived from ATP binding and hydrolysis to power unwinding. We first mixed individual Mcm subunits and measured the rate of ATP hydrolysis in the absence of DNA (Fig. 5, A and B). We found that combining Mcm4 with Mcm7 resulted in a dramatic increase in ATP hydrolysis (Fig. 5, A and B), similar to a previous report (27). However, we also found low levels of ATP hydrolysis for other binary combinations of Mcms (Fig. 5B, inset). Mcm6 in combination with Mcm7 resulted in an increase in ATP hydrolysis compared with Mcm6 or Mcm7 alone. A current model of Mcm2-7 architecture proposes that Mcm6 does not interact with Mcm7 (27, 49). Thus, the Mcm subunits may have an ability to productively interact with one another in vitro in a manner that is not currently modeled for the Mcm2-7 complex.
FIGURE 5.
Mixtures of Mcm4, Mcm6, and Mcm7 catalyze the hydrolysis of ATP. A, Mcm proteins were incubated with [γ-32P]ATP at 37 °C for the time points indicated, and the reaction products were then analyzed by thin layer chromatography. The experiments were conducted in the absence (-DNA) and presence (+DNA) of 1 μm 60-dT ssDNA. B, data from experiments in A were quantified, and the rates of ATP hydrolysis were determined by a linear fit of the data. Data for time 0 were subtracted as background, and the data shown are the calculated ATP hydrolysis rates + error. The inset allows the comparison of Mcm mixtures with slower ATP hydrolysis rates. Striped bars indicate the absence of DNA, and open bars indicate the presence of 60(dT) ssDNA.
We also found that upon mixing Mcm6 with Mcm4 and Mcm7, lower levels of ATP hydrolysis are observed compared with Mcm4/Mcm7 (Fig. 5B). These data suggest that Mcm4/Mcm6/Mcm7 hydrolyzes ATP at a slower rate compared with Mcm4/Mcm7. We then added linear ssDNA to each of the Mcm subunit mixtures. We found that ssDNA substantially stimulated the ATP hydrolysis of Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7, with a 1.9-fold and a 2.1-fold increase, respectively (Fig. 5, A and B). In contrast, the effect of DNA on ATP hydrolysis of other Mcm mixtures was modest (Fig. 5B, inset). Thus, the ability of DNA to stimulate ATP hydrolysis correlates with the ability of certain mixtures of Mcm components to unwind DNA.
The Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 Helicases Can Disassemble to Load onto Circular ssDNA—Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 can unwind DNA oligonucleotides that are annealed to one another provided the duplex bears a 3′-single strand extension and a bulky group positioned at the 5′ end (Fig. 1). Furthermore, these Mcm complexes are ring-shaped, and they may unwind DNA by a steric exclusion mechanism. There are two likely modes whereby ring-shaped particles can assemble onto the 3′-single strand extension of these DNA duplexes (50). In one model, the ring-shaped particle remains intact, and the single-stranded DNA is inserted into the helicase ring, much like the threading of a needle. In a second model, the helicase ring opens or disassembles, and the subunits then reassemble into a ring to surround the DNA strand.
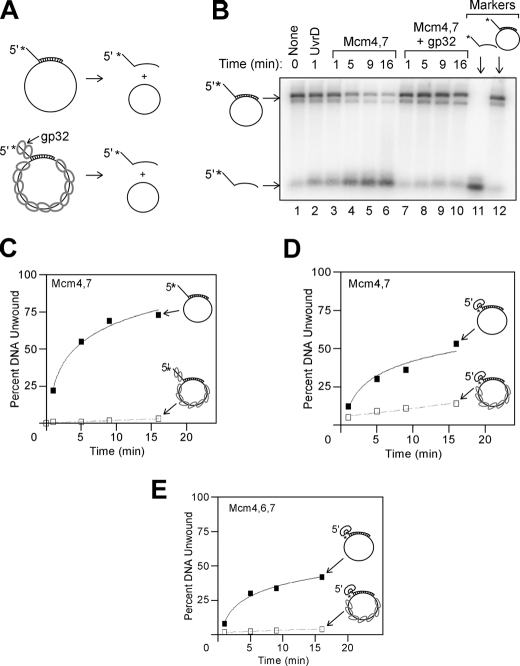
To determine which of these two models is more likely for Mcm4/Mcm7, the complex was incubated with circular ssDNA with an annealed radiolabeled oligonucleotide, and unwinding was monitored (Fig. 6, A, B, and C). The annealed oligonucleotide contains a 5′-single strand extension to prevent the helicase from encircling two DNA strands. Mcm4/Mcm7 rapidly unwound the radiolabeled oligonucleotide, suggesting that the complex efficiently loads onto circular ssDNA (Fig. 6, B and C). To determine whether ssDNA binding is required for the helicase activity observed, the DNA was preincubated with gp32 from T4 phage, a protein that binds tightly to ssDNA. Preincubation with T4-gp32 markedly inhibited the unwinding activity of the Mcm4-Mcm7 complex, suggesting that the Mcm4-Mcm7 complex requires loading onto ssDNA to unwind the annealed oligonucleotide.
FIGURE 6.
Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 helicase assemblies can load onto circular ssDNA. A, schematic for experiments to determine whether Mcm assemblies can load onto circular ssDNA. A radiolabeled DNA oligonucleotide is annealed to M13 circular ssDNA. The radiolabeled oligonucleotide bears a 5′-single strand extension to promote steric exclusion, and unwinding of the radiolabeled strand is monitored by native gel electrophoresis. The experiment is also performed in the presence of T4 gp32 to coat the free ssDNA with a nonspecific protein. B, equimolar mixture of Mcm4/Mcm7 was incubated with DNA substrate for the time points incubated at 37 °C in the absence (lanes 3-6) and presence (lanes 7-10) of T4 gp32. The position of free ssDNA and DNA annealed to M13 were determined by standards analyzed on the same gel (lanes 11 and 12). UvrD was also tested (lane 2), as well as a no protein control (lane 1). C, data from experiments similar to B were quantified and plotted as a function of time in the absence (filled squares) and presence (open squares) of T4 gp32. The data in the absence of T4 gp32 were fit to a logarithmic equation, and the data in the presence of T4 gp32 were fit to a linear equation. D, experiment is similar to that shown in C, except the radiolabeled oligonucleotide bears a 5′-biotin-streptavidin in the place of the ssDNA extension. Unwinding rates were determined in the absence (filled squares) and presence (open squares) of T4 gp32. E, experiment is similar to that shown in D, except Mcm4/Mcm6/Mcm7 replaced Mcm4/Mcm7.
To determine whether Mcm4/Mcm7 loading onto the 5′-single strand extension was required for unwinding of the annealed oligonucleotide, a biotin-streptavidin group was substituted for the 5′-single strand extension (Fig. 6D). Mcm4/Mcm7 unwound this oligonucleotide as well, and this unwinding was largely blocked by preincubation with T4 gp32. These data suggest that the Mcm4/Mcm7 ring likely can open or disassemble and then reform on circular ssDNA. Similar results were found with the Mcm4-Mcm6-Mcm7 complex (Fig. 6E). Thus, one feature of these Mcm helicase complexes is the ability of these ring-shaped particles to assemble around circular ssDNA.
DISCUSSION
In this study we find that the addition of Mcm4 with Mcm7 results in the formation of an active hexameric helicase complex at physiologically relevant concentrations. The addition of Mcm6 to Mcm4/Mcm7 results in the formation of an active Mcm4/Mcm6/Mcm7 helicase. The Mcm4-Mcm7 complex unwinds DNA with 3′ to 5′ polarity, and unwinding activity requires the presence of a bulky group attached to the 5′ end of the duplex. The Mcm4-Mcm7 complex can also actively translocate across dsDNA, much like the Mcm4-Mcm6-Mcm7 complex, and the Mcm4-Mcm7 complex appears by negative stain electron microscopy as a single hexameric ring that is similar in dimension and shape to the Mcm4/Mcm6/Mcm7 particle. The Mcm4-Mcm7 complex rapidly hydrolyzes ATP, and DNA further stimulates the rate of ATP hydrolysis. The mixture of Mcm4 with Mcm6 or Mcm6 with Mcm7 results in ATP hydrolysis as well, but helicase activity and ring formation are not observed for these other Mcm mixtures. The Mcm4-Mcm7 and the Mcm4-Mcm6-Mcm7 complexes can unwind DNA that is annealed to circular ssDNA, suggesting the helicase rings can partially disassemble and then load onto the circular DNA.
What Makes an Active Mcm Helicase?—One feature that Mcm helicases have in common is that they form ring-shaped structures. Other replication fork helicases are ring-shaped, including eukaryotic viral helicases such as SV40 T antigen, papillomavirus E1 helicase, the bacterial DnaB helicase, and T7 and T4 phage helicases (51-58). The ability of these protein complexes to form rings may be critical for the capacity of these complexes to function as helicases. In our study of Mcm4, Mcm6, and Mcm7, the ability of different mixtures to form rings correlated with their ability to unwind DNA. The ring-shaped nature of these complexes may be important for unwinding DNA by steric exclusion, as has been previously proposed (59). Additionally, the ring-shaped nature may be important for these helicases because they act through a cyclic mechanism to translocate along DNA. Different models for DNA translocation have been proposed for the ring-shaped helicases, including the “bucket brigade” mechanism for T7 gp4 (55) and the “concerted escort” model for papilloma E1 (56). In both of these models, the ring-shaped architecture enables the helicases to bind and hydrolyze ATP in a cyclic manner, and this activity is critical for DNA translocation and hence DNA unwinding.
Although formation of a ring-shaped particle correlates nicely with unwinding activity in this study, the hydrolysis of ATP is more complex. For example, we found that Mcm6 with Mcm7 hydrolyzes ATP at a greater rate than the sum of both components. Nevertheless, this complex is not active in unwinding DNA. Furthermore, Mcm4/Mcm6/Mcm7 hydrolyzes ATP more slowly than Mcm4/Mcm7, even though the Mcm4-Mcm6-Mcm7 complex exhibits a faster unwinding rate. Thus, whereas the hydrolysis of ATP is necessary for the formation of an active helicase complex, it is not sufficient. We found a tighter correlation between helicase activity and DNA-stimulated ATPase activity, as only the Mcm4-Mcm7 and Mcm4-Mcm6-Mcm7 complexes exhibited DNA-dependent stimulation of ATP hydrolysis. Nevertheless, although it may be necessary for a helicase to couple DNA binding with ATP hydrolysis, this property is unlikely to be sufficient for helicase activity.
The Mcm4-Mcm7 and the Mcm4-Mcm6-Mcm7 complexes unwind DNA that is annealed to circular ssDNA. These data suggest that the Mcm protein rings can open or disassemble and then reassemble to load onto circular ssDNA. This property reflects the dynamic nature of these ring-shaped helicases; they are not static rings. The dynamic nature of these Mcm rings is also reflected by the observation that the addition of Mcm6 to Mcm4/Mcm7 results in the formation of an active Mcm4/Mcm6/Mcm7 helicase.
Mcm Proteins Have a Surprising Ability to Interact with One Another to Form Different Architectures—The Mcm2-7 and Mcm4-Mcm6-Mcm7 complexes both form ring-shaped hexamers, and they therefore represent two different protein hexamers that arise from similar parts. We have now extended this observation to find that in the absence of Mcm6, Mcm4 and Mcm7 can form ring-shaped particles that are likely to be hexameric. This observation indicates that the Mcm proteins have considerable flexibility in their ability to interact with one another to form ring-shaped hexamers. Furthermore, Mcm4/Mcm6/Mcm7 and Mcm4/Mcm7 are both active as helicases, suggesting that the different architectures are biochemically functional. We also find in this study that Mcm6 and Mcm7 can interact with one another to hydrolyze ATP. According to the best available models, Mcm6 and Mcm7 do not contact each other within the Mcm2-7 ring (27). Thus, the ability of Mcm subunits to productively interact with one another is surprisingly flexible, and not strictly dictated by their arrangement in the Mcm2-7 ring.
Because only the Mcm2-7 complex is thought to act at a cellular replication fork, why do Mcm proteins exhibit a high degree of flexibility to form different complexes? One possibility is that Mcm4/Mcm6/Mcm7 and/or Mcm4/Mcm7 act in the cell in a process that is presently not known, as described below.
Possible in Vivo Roles of Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 Helicases—There is substantial evidence that the Mcm2-7 complex is the active replication fork helicase in eukaryotic cells (3, 4, 7, 8, 60). However, there is a 100-fold excess of Mcm proteins relative to replication origins in the cell, raising the possibility that Mcm proteins are involved in functions aside from DNA replication. Furthermore, it has been demonstrated that Mcm proteins function in activities other than DNA replication such as transcription (14, 15), chromatin remodeling (16, 17, 20, 21), and genome stability (23, 24). Intriguingly, a hypomorphic mutation in Mcm4 causes chromosome instability and mammary adenocarcinomas in mice (24), and deregulated minichromosomal maintenance protein MCM7 contributes to oncogene-driven tumorigenesis (23). These data suggest that Mcm4 and Mcm7 proteins may have roles in genome maintenance aside from their role as part of the replicative helicase. Thus, it is possible that the Mcm4-Mcm7 complex may function as a helicase in a process in the cell that maintains genome stability, such as DNA repair. Moreover, the Mcm4-Mcm6-Mcm7 complex is an active helicase in vitro using proteins from mouse, Drosophila, Xenopus, S. pombe, and S. cerevisiae (3, 29, 33, 35, 37). The observation that Mcm4/Mcm6/Mcm7 activity is conserved across species suggests that the complex may act as a DNA helicase in an important cellular process such as genome maintenance.
Potential Role of Eukaryotic Mcm Subunit Specialization in DNA Unwinding—Most replication fork helicases that have been studied are homohexameric. Because each subunit of the homohexamer is identical in its covalent structure, some models for activity attribute equivalent functional roles for each of the six subunits. For example, in both the bucket brigade model for T7 gp4 and the “coordinated escort” model for papilloma E1 virus, each subunit of the hexamer binds and hydrolyzes ATP in a sequential manner, and each subunit binds and later releases DNA in a sequential manner (55, 56). Thus, each subunit is functionally equivalent.
The Mcm helicase system is different from these homohexameric systems, in that each of the six subunits of the Mcm2-7 complex has a unique amino acid sequence. The high degree of sequence conservation across species for Mcm subunits suggests that each subunit has evolved for a particular function. There have been numerous reports ascribing specific biological functions to particular Mcm subunits. However, an important question that arises is whether different subunits within the Mcm2-7 complex function equivalently in unwinding DNA, consistent with the bucket brigade or coordinated escort models, or whether specific subunits within the Mcm complex are designated to perform particular functions as part of an unwinding machine.
In this study, we find that different Mcm subunits have distinct biochemical properties, in particular with regard to the ability of various mixtures to unwind DNA, form ring-shaped particles, and hydrolyze ATP. These observations extend the current literature that demonstrates specific functions for Mcm subunits. However, we also find that Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 function as active helicases. Although we do not know how the Mcm4, Mcm6, and Mcm7 subunits are arranged within these two complexes, the subunits are arranged differently, because one complex contains Mcm6 and the other does not. Because the position of the Mcms can be varied in different ways and still yield active complexes, we propose that the particular arrangement of the Mcm subunits is not essential for unwinding activity. In one sense, the eukaryotic Mcms may be functioning in a manner similar to the archaeal homohexamer, in which each subunit is identical. This idea is consistent with the evolution of the eukaryotic Mcms, because they share a common ancestor with the archaeal Mcms. It is also of interest that of the six eukaryotic Mcms, Mcm4 is most closely related to the archaeal Mcm, and Mcm4 is also present in each of the active unwinding complexes. However, one important difference between the archaeal and eukaryotic Mcms is that the archaeal Mcm readily forms double or single hexamers (61-65), whereas the eukaryotic Mcms form only single hexamers.
Supplementary Material
Acknowledgments
We thank Mike O'Donnell for providing Mcm expression constructs. Electron microscopy assistance was provided by Denny Kerr of the Vanderbilt University EM Core Facility. We thank Katherine Friedman and Ellen Fanning for critical comments of this manuscript.
This work was supported by Vanderbilt University start-up funds (to D. L. K.), a Vanderbilt University discovery grant (to D. L. K.), a pilot grant from the Vanderbilt Ingram Cancer Center (to D. L. K.), and American Cancer Society Research Scholar Grant SG-08-124-01-CCG (to D. L. K. and I. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
Footnotes
The abbreviations used are: ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; AMP-PNP, adenosine 5′-(β,γ-imido)triphosphate; DTT, dithiothreitol.
References
- 1.Singleton, M., Dillingham, M., and Wigley, D. (2007) Annu. Rev. Biochem. 76 23-50 [DOI] [PubMed] [Google Scholar]
- 2.Patel, S. S., and Picha, K. M. (2000) Annu. Rev. Biochem. 69 651-697 [DOI] [PubMed] [Google Scholar]
- 3.Moyer, S., Lewis, P., and Botchan, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10236-10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacek, M., Tutter, A., Kubota, Y., Takisawa, H., and Walter, J. (2006) Mol Cell. 21 581-587 [DOI] [PubMed] [Google Scholar]
- 5.Pacek, M., and Walter, J. C. (2004) EMBO J. 23 3667-3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambus, A., Jones, R. C., Sanchez-Diaz, A., Kanemaki, M., Deursen, F. V., Edmondson, R. D., and Labib, K. (2006) Nat. Cell Biol. 8 358-366 [DOI] [PubMed] [Google Scholar]
- 7.Labib, K., Tercero, J. A., and Diffley, J. F. X. (2000) Science 288 1643-1647 [DOI] [PubMed] [Google Scholar]
- 8.Bochman, M., and Schwacha, A. (2008) Mol. Cell 31 287-293 [DOI] [PubMed] [Google Scholar]
- 9.Donovan, S., Harwood, J., Drury, L., and Diffley, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5611-5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei, M., Kawasaki, Y., and Tye, B. (1996) Mol. Cell. Biol. 16 5081-5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrova, D., Todorov, I. T., Melendy, T., and Gilbert, D. M. (1999) J. Cell Biol. 146 709-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krude, T., Musahl, C., Laskey, R., and Knippers, R. (1996) J. Cell Sci. 109 309-318 [DOI] [PubMed] [Google Scholar]
- 13.Madine, M., Khoo, C., Mills, A., Musahl, C., and Laskey, R. (1995) Curr. Biol. 5 1270-1279 [DOI] [PubMed] [Google Scholar]
- 14.Fitch, M., Donato, J., and Tye, B. (2003) J. Biol. Chem. 278 25408-25416 [DOI] [PubMed] [Google Scholar]
- 15.Snyder, M., He, W., and Zhang, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14539-14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groth, A., Corpet, A., Cook, A., Roche, D., Bartek, J., Lukas, J., and Almouzni, G. (2007) Science 318 1928-1931 [DOI] [PubMed] [Google Scholar]
- 17.Tan, B., Chien, C., Hirose, S., and Lee, S. (2006) EMBO J. 25 3975-3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishimi, Y., Komamura, Y., You, Z., and Kimura, H. (1998) J. Biol. Chem. 273 8369-8375 [DOI] [PubMed] [Google Scholar]
- 19.Ishimi, Y., Komamura-Kohno, Y., Arai, K., and Masai, H. (2001) J. Biol. Chem. 276 42744-42752 [DOI] [PubMed] [Google Scholar]
- 20.Burke, T. W, Cook, J.-G., Asano, M., and Nevins, J. (2001) J. Biol. Chem. 276 15397-15408 [DOI] [PubMed] [Google Scholar]
- 21.Iizuka, M., and Stillman, B. (1999) J. Biol. Chem. 274 23027-23034 [DOI] [PubMed] [Google Scholar]
- 22.Ishimi, Y., Ichinose, S., Omori, A., Sato, K., and Kimura, H. (1996) J. Mol. Biol. 271 24115-24122 [DOI] [PubMed] [Google Scholar]
- 23.Honeycutt, K., Chen, Z., Koster, M., Miers, M., Nuchtern, J., Hicks, J., Roop, D., and Shohet, J. (2006) Oncogene 25 4027-4032 [DOI] [PubMed] [Google Scholar]
- 24.Shima, N., Alcaraz, A., Liachko, I., Buske, T., Andrews, C., Munroe, R., Hartford, S., Tye, B., and Schimenti, J. (2007) Nat. Genet. 39 93-98 [DOI] [PubMed] [Google Scholar]
- 25.Ge, X., Jackson, D., and Blow, J. (2007) Genes Dev. 21 3331-3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coue, M., Amariglio, F., Maiorano, D., Bocquet, S., and Mechali, M. (1998) Exp. Cell Res. 245 282-289 [DOI] [PubMed] [Google Scholar]
- 27.Davey, M. J., Indiani, C., and O'Donnell, M. (2003) J. Biol. Chem. 278 4491-4499 [DOI] [PubMed] [Google Scholar]
- 28.Holthoff, H., Baack, M., Richter, A., Ritzi, M., and Knippers, R. (1998) J. Biol. Chem. 273 7320-7325 [DOI] [PubMed] [Google Scholar]
- 29.Lee, J.-K., and Hurwitz, J. (2000) J. Biol. Chem. 275 18871-18878 [DOI] [PubMed] [Google Scholar]
- 30.Musahl, C., Schulte, D., Burkhart, R., and Knippers, R. (1995) Eur. J. Biochem. 230 1096-1101 [DOI] [PubMed] [Google Scholar]
- 31.Prokhorova, T., and Blow, J. (2000) J. Biol. Chem. 275 2491-2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman, D., and Forsburg, S. (1998) Nucleic Acids Res. 26 3955-3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishimi, Y. (1997) J. Biol. Chem. 272 24508-24513 [DOI] [PubMed] [Google Scholar]
- 34.You, Z., Komamura, Y., and Ishimi, Y. (1999) Mol. Cell. Biol. 19 8003-8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan, D. L., Davey, M. J., and O'Donnell, M. (2003) J. Biol. Chem. 278 49171-49182 [DOI] [PubMed] [Google Scholar]
- 36.Lee, J.-K., and Hurwitz, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 54-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying, C. Y., and Gautier, J. (2005) EMBO J. 24 4334-4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer, L., Leipe, D., Koonin, E., and Avardind, L. (2004) J. Struct. Biol. 146 11-31 [DOI] [PubMed] [Google Scholar]
- 39.Kaplan, D. L., and O'Donnell, M. (2004) Mol. Cell 15 453-465 [DOI] [PubMed] [Google Scholar]
- 40.Shin, J. H., Jiang, Y., Grabowski, B., Hurwitz, J., and Kelman, Z. (2003) J. Biol. Chem. 278 49053-49062 [DOI] [PubMed] [Google Scholar]
- 41.Crampton, D., Ohi, M., Qimron, U., Walz, T., and Richardson, C. (2006) J. Mol. Biol. 360 667-677 [DOI] [PubMed] [Google Scholar]
- 42.You, Z., and Masai, H. (2005) Nucleic Acids Res. 33 3033-3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You, Z., Ishimi, Y., Mizuno, T., Sugasawa, K., Hanaoka, F., and Masai, H. (2003) EMBO J. 22 6148-6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei, M., Kawasaki, Y., Young, M., Kihara, M., Sugino, A., and Tye, B. (1997) Genes Dev. 11 3365-3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothenberg, E., Trakselis, M., Bell, S., and Ha, T. (2007) J. Biol. Chem. 282 34229-34234 [DOI] [PubMed] [Google Scholar]
- 46.Sato, M., Gotow, T., You, Z., Komamura-Kohno, Y., Uchiyama, Y., Yabuta, N., Nojima, H., and Ishimi, Y. (2000) J. Mol. Biol. 300 421-431 [DOI] [PubMed] [Google Scholar]
- 47.Yabuta, N., Kajimura, N., Mayanagi, K., Sato, M., Gotow, T., Uchiyama, Y., Ishimi, Y., and Nojima, H. (2003) Genes Cells 8 413-421 [DOI] [PubMed] [Google Scholar]
- 48.Bochman, M., and Schwacha, A. (2007) J. Biol. Chem. 282 33795-33804 [DOI] [PubMed] [Google Scholar]
- 49.Yu, Z., Feng, D., and Liang, C. (2004) J. Mol. Biol. 340 1197-1206 [DOI] [PubMed] [Google Scholar]
- 50.Davey, M. J., and O'Donnell, M. (2003) Curr. Biol. [DOI] [PubMed]
- 51.Egelman, E. H., Yu, X., Wild, R., Hingorani, M. M., and Patel, S. S. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3869-3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norcum, M., Warrington, J., Spiering, M., Ishmael, F., Trakselis, M., and Benkovic, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3623-3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, S., Yu, X., VanLoock, M., Jezewska, M., Bujalowski, W., and Egelman, E. (2002) J. Mol. Biol. 321 839-849 [DOI] [PubMed] [Google Scholar]
- 54.Bailey, S., Eliason, W., and Steitz, T. (2007) Science 318 459-463 [DOI] [PubMed] [Google Scholar]
- 55.Singleton, M. R., Sawaya, M. R., Ellenberger, T., and Wigley, D. B. (2000) Cell 101 589-600 [DOI] [PubMed] [Google Scholar]
- 56.Enemark, E., and Joshua-Tor, L. (2006) Nature 442 270-275 [DOI] [PubMed] [Google Scholar]
- 57.Valle, M., Chen, X., Donate, L., Fanning, E., and Carazo, J. (2006) J. Mol. Biol. 357 1295-1305 [DOI] [PubMed] [Google Scholar]
- 58.Gai, D., Zhao, R., Li, D., Finkielstein, C., and Chen, X. (2004) Cell 119 47-60 [DOI] [PubMed] [Google Scholar]
- 59.Kaplan, D. L., and O'Donnell, M. (2002) Mol. Cell 10 647-657 [DOI] [PubMed] [Google Scholar]
- 60.Calzada, A., Hodgson, B., Kanemaki, M., Bueno, A., and Labib, K. (2005) Genes Dev. 19 1905-1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa, A., Pape, T., van Heel, M., Brick, P., Patwardhan, A., and Onesti, S. (2006) Nucleic Acids Res. 34 5829-5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa, A., Pape, T., van Heel, M., Brick, P., Patwardhan, A., and Onesti, S. (2006) J. Struct. Biol. 156 210-219 [DOI] [PubMed] [Google Scholar]
- 63.Gomez-Llorente, Y., Fletcher, R., Chex, X., Carazo, X., and San Martin, C. (2005) J. Biol. Chem. 280 40909-40915 [DOI] [PubMed] [Google Scholar]
- 64.Fletcher, R. J., Shen, J., Gomez-Llorente, Y., Martin, C. S, Carazo, J. M., and Chen, X. (2005) J. Biol. Chem. 280 42405-42410 [DOI] [PubMed] [Google Scholar]
- 65.Chong, J. P. J., Hayashi, M. K., Simon, M. N., Xu, R.-M., and Stillman, B. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1530-1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.