Abstract
Studies of the interaction between Bikunin proteins, tumor necrosis factor-stimulated gene-6 protein (TSG-6), and glycosaminoglycans have revealed a unique catalytic activity where TSG-6/heavy chain 2 transfer heavy chain subunits between glycosaminoglycan chains. The activity is mediated by TSG-6/heavy chain 2 and involves a transient SDS stable interaction between TSG-6 and the heavy chain to be transferred. The focus of this study was to characterize the molecular structure of this cross-link to gain further insight into the catalytic mechanism. The result showed that the C-terminal Asp residue of the heavy chains forms an ester bond to Ser28 β-carbon of TSG-6 suggesting that this residue plays a role during catalysis.
The Bikunin proteins, inter-α-inhibitor (IαI),2 pre-α-inhibitor (PαI), and heavy chain 2·bikunin (HC2·bikunin) are heterodimers or trimers composed of bikunin and one or two homologous heavy chains (HCs) (1, 2). The subunits are covalently linked by a chondroitin sulfate (CS) originating from Ser10 of bikunin forming an inter-chain protein-glycosaminoglycan-protein bond between the C-terminal Asp residues of the HCs C-6 of an internal N-acetylgalactosamine in the CS (2–5). The HCs have also been identified in complex with hyaluronan (HA) (6, 7). This cross-link is similarly mediated by an ester between the C-terminal Asp residue of the HCs and an internal C-6 of an N-acetylglucosamine in HA (8).
Tumor necrosis factor-stimulated gene-6 protein (TSG-6) is a 35-kDa extracellular matrix protein transiently expressed during inflammation and ovulation (9–11). TSG-6 is composed of a Link domain mediating binding to glycosaminoglycans (12, 13) and a CUB domain of uncertain function. In other proteins the CUB domain is often involved in protein-protein interactions, and the CUB domain in TSG-6 has been shown to interact with fibronectin (14).
TSG-6 and IαI interact spontaneously (15) in an SDS stable manner to form HC1·TSG-6 and HC2·TSG-6 complexes (16). During the formation of these complexes, TSG-6 cleaves the protein-glycosaminoglycan-protein cross-link of IαI, and HC2 is linked to TSG-6 through the C-terminal Asp residue of the HC (16). The HC1·TSG-6 attachment site has not been identified, but it has previously been suggested that it similarly is mediated by the C-terminal Asp residue (16). The HC1·TSG-6 or HC2·TSG-6 complexes are only transient as the HCs are further transferred to HA, if present (17). The transfer mechanism requires the presence of HC2 and involves two sequential divalent cation-dependent transesterifications, where the HC·TSG-6 complexes act as intermediates (16–18). In the absence of HA the HCs are transferred to the CS chain of the bikunin proteins, generating high molecular weight bikunin proteins (16, 18).
Free TSG-6, HC·TSG-6 intermediates, and the HC·HA complexes are present in synovial fluid from arthritis patients (8, 19, 20). The importance of TSG-6 during arthritis has been emphasized as knock-out mice develop the disease more rapidly, whereas mice over-expressing TSG-6 were protected (21–23). Mice models have also been used to demonstrate the importance of the HC·HA complexes during ovulation. Both TSG-6 knock-out mice and bikunin knock-out mice have impaired extracellular matrix formation around the oocyte resulting in female infertility (24, 25). Furthermore, cell studies have shown that the interaction between the HCs and HA produces cable-like structures mediating the binding of leukocytes (26, 27). Likewise the HC·HA structures increase cell binding and migration compared with unmodified HA (28, 29). In general, the interactions are important for formation and remodeling of many types of extracellular HA matrixes.
In the present study we have determined the molecular structure of the HC·TSG-6 cross-link. The results show that the β-carbon of Ser28 of TSG-6 (numbered according to the preprotein) forms an ester bond to the α-carbon of the C-terminal Asp residue of the HCs suggesting that Ser28 is part of a catalytic site. Site-directed mutagenesis similarly suggested that Ser28 was essential for the activity. The Asp-Ser28 bond is akin to the acyl-enzyme intermediate transiently formed during serine protease catalysis suggesting that TSG-6/HC2 similarly utilizes covalent catalysis.
EXPERIMENTAL PROCEDURES
Materials—Horseradish peroxidase-conjugated goat anti-rabbit IgG, Glu-fibrinopeptide B, and α-cyano-4-hydroxycinnamic acid were obtained from Sigma. ECL Western blotting reagents, Ettan™ CAF™ MALDI sequencing kit, HiTrap Q column (5 ml), HiTrap Blue column (5 ml), and Mono Q 5/50 GL column (1 ml) were obtained from GE Healthcare. MS grade trypsin was from Promega and Stagetips (C18) were from Proxeon Biosystems A/S. ZipTips (C18) and Amicon centriprep YM-10 were obtained from Millipore. The reverse phase (RP) column was from Vydac (C18, 2.1 × 250 mm, particle size 5 μm). The strong cation exchange PolySULFOETHYL A™ column (2.1 × 200 mm, particle size 5 μm) was obtained from PolyLC Inc. The RNeasy mini kit was from Qiagen and Herculase Enhanced Polymerase was from Stratagene. Superscript II reverse transcriptase, pcDNA5/FRT expression vector, and Zero Blunt TOPO PCR Cloning Kit were obtained from Invitrogen. All primers were from Eurofins MWG GmbH and restriction enzymes were from New England Biolabs. For the site-directed mutagenesis study recombinant human TSG-6 was expressed in mammalian cells, as described later. IαI was purified from human plasma basically as previously described (2). Recombinant human TSG-6 was expressed in insect cells and purified as described before (30). The resulting protein was used for the generation of rabbit anti-TSG-6 antibody (30). Human plasma was from Statens Serum Institut, Denmark, and human liver tissue was obtained from the Pathology Department, Aarhus University Hospital, Denmark. Protein concentrations were determined spectrophotometrically at 280 nm using theoretical extinction coefficients (GPMAW software, Lighthouse Data).
SDS-PAGE and Immunoblotting—Samples were boiled in SDS-sample buffer containing dithiothreitol and resolved in 5–15% gradient gels using the glycine/2-amino-2-methyl-1,3-propandiol-HCl system described previously (31). The resolved proteins were either visualized by Coomassie Blue staining or they were electrotransferred to polyvinylidene difluoride membranes (32) and processed for immunoblotting using chemiluminescence as described before (16).
Complex Formation between IαI and TSG-6 and Trypsinization—Human IαI and human recombinant TSG-6 (100 μg) expressed in insect cells were incubated in 2 mm MgCl2, 20 mm Tris-HCl, 150 mm NaCl, pH 8.0, at a 1:1 ratio (w/w) for 3 h at 37 °C. The complex was then denatured in 6 m guanidinium hydrochloride, pH 8, for 1 h at 25 °C and the disulfides reduced in the same buffer containing 10 mm dithiothreitol. Subsequently, the complex was carboxymethylated using 20 mm iodoacetamide and dialyzed into 10 mm NH4HCO3. The reduced and carboxymethylated sample was digested using 4 μg of trypsin for 20 h at 37 °C and lyophilized (SpeedVac, Savant).
Purification of Cross-linked Peptides—The lyophilized peptides were rehydrated in 10 mm KH2PO4, 20% acetonitrile, pH 2.8 (Buffer A), filtered, and applied to a PolySULFOETHYL A column connected to a Ettan LC system (GE Healthcare) equilibrated in buffer A. The bound material was eluted with buffer B (500 mm KCl in Buffer A) using a gradient from 0 to 15% B after 30 min, 50% B after 70 min, and 100% B after 95 min. The column was operated at 25 °C at a flow rate of 150 μl/min. Fractions of 300 μl were collected and the eluate monitored at 220 nm. The fractions of interest were either analyzed directly by RP-HPLC using a 2.1-mm Vydac C18 column equilibrated in 0.1% trifluoroacetic acid or pretreated with NaOH (final concentration 200 mm) for 15 min (3). The column was developed using a linear gradient from 0.1% trifluoroacetic acid to 90% acetonitrile containing 0.07% trifluoroacetic acid at 25 °C and a flow rate of 200 μl/min. The eluate was monitored at 220 nm and fractions were collected manually.
Sample Preparation for Matrix-assisted Laser Desorption/Ionization (MALDI)-Mass Spectrometry (MS)—Aliquots from the PolySULFOETHYL A column fractions were made 100 mm in NaOH, incubated for 15 min at 25 °C (3), and acidified. Peptides derived from these samples or from diluted aliquots of the column fractions were purified on C18 stage tips and eluted directly onto MALDI targets using 1 μl of matrix solution containing 70% (v/v) acetonitrile, 0.03% trifluoroacetic acid (v/v), and 0.4% recrystallized α-cyano-4-hydroxycinnamic acid. Aliquots from the reverse phase columns were concentrated to 5 μl (SpeedVac, Savant), diluted by adding 50 μl of 0.1% trifluoroacetic acid, purified by C18 stage tips, and eluted directly onto MALDI targets.
Chemically Assisted Fragmentation (CAF) Reagent Derivatization of Cross-linked Peptides—The derivatization of peptides was performed according to the manufacturer's recommendations (GE Healthcare). In short, tryptic peptides were purified on a C18 ZipTip and Lys side chains were converted to homoarginine to avoid sulfonation of Lys residues in the next step. Subsequently, the N-terminal of the peptides was sulfonated. Finally, the derivatized peptides were eluted from the ZipTip using the previously described matrix solution and spotted directly onto the MALDI target.
MALDI-MS Analyses—MS or MS/MS spectra were collected using a Q-TOF Ultima Global mass spectrometer (Micromass/Waters Corp.) calibrated over the m/z 50–3000 range using a polyethylene glycol mixture. Each MS spectrum was calibrated using Glu-fibrinopeptide B (m/z 1570.6774) for external calibration. The obtained MS and MS/MS data were processed using Masslynx 4.0 software (Micromass). The GPMAW software package (Lighthouse Data) was used to generate theoretical MS/MS fragment ions of peptides, to facilitate manual interpretation of the obtained MS/MS data.
N-terminal Sequence Analyses—Samples were applied directly to precycled Biobrene-coated micro-trifluoroacetic acid filter or adsorbed to a polyvinylidene difluoride membrane using a ProSorb device (Applied Biosystems). Automated Edman degradation was performed in a PROCISE™ 494 HT sequencer with on-line phenylthiohydantion-derivative HPLC analysis using a 140C Microgradient System (Applied Biosystems), operated according to the manufacturer's recommendations.
TSG-6 cDNA Cloning and Site-directed Mutagenesis of Ser28—Total RNA was isolated from 30 mg of human liver using the RNeasy mini kit from Qiagen. Subsequently, Superscript II reverse transcriptase was used to synthesize oligo(dT)-primed first strand cDNA from 3 μg of total RNA. Full-length TSG-6 cDNA was obtained from the cDNA library by PCR using Herculase Enhanced Polymerase and the following primer set spanning the open reading frame: 5′-AACTCCTAAAAAACCGCCACCATGATCATCTTAATTTACTTATTTCTCTT-3′ and 5′-TTATAAGTGGCTAAATCTTCCAGCTAAAAAG-3′ (start and stop (complementary) codons are bolded). The blunt end PCR product was initially cloned in the pCR-blunt II vector using the Zero Blunt TOPO PCR Cloning Kit. Later the insert was excised with XhoI and KpnI and ligated into the pcDNA5/FRT mammalian expression vector. The TSG-6 insert sequence was confirmed by sequencing both strands. Afterward the S28A mutation was introduced in the TSG-6 cDNA by in vitro mutagenesis using Herculase Enhanced Polymerase and the following complementary mutation primer set: 5′-CAAGGATGGAATTTTTCATAACGCCATATGGCTTGAACGAGCA-3′ and 5′-TGCTCGTTCAAGCCATATGGCGTTATGAAAAATTCCATCCTTG-3′ (the mutation is underlined). The mutation was confirmed by sequencing both strands.
HEK293 cells (a human embryonic kidney cell line) were grown in Dulbecco's modified Eagle's medium with glutamine supplemented with 10% fetal calf serum and 100 IU penicillin/streptomycin. Approximately 5.0 × 106 cells were seeded onto 10-cm dishes 1 day prior to transfection. Four hours before transfection the medium was replaced with fresh medium. Transfection was done using the calcium phosphate coprecipitation method with 20 μg of plasmid DNA per dish including a reporter plasmid containing the β-galactosidase gene for recording transfection efficiency. Three dishes with cells were transfected with mammalian expression vectors containing TSG-6 wild type cDNA, TSG-6 S28A mutant cDNA, or extracellular superoxide dismutase cDNA (control). Sixteen hours following transfection the medium was changed to the same as above but without calf serum. Finally conditioned serum-free medium was collected every 24 h over a period of 2 days and concentrated 50 times using Centriprep YM-10.
The presence and the concentration of wild type TSG-6 and TSG-6 S28A mutant in the media were evaluated by immunoblotting. In addition, the expression of TSG-6, both wild type and mutant, was confirmed by direct analysis of the proteins by tandem mass spectrometry (see below).
Complex Formation between IαI and TSG-6 S28A—Approximately 1 μg of purified human IαI was mixed with concentrated extracellular superoxide dismutase medium (control) or concentrated medium containing the same amount of either wild type or mutant TSG-6. MgCl2 was added to a final concentration of 2 mm and the samples incubated for 1 h at 37 °C. Subsequently, the proteins were resolved by SDS-PAGE and immunoblotting was performed using an anti-TSG-6 antibody.
Nanoelectrospray Mass Spectrometry—The concentrated medium derived from control, wild type TSG-6, and mutant TSG-6 expressions were subjected to reduced SDS-PAGE and stained for protein using Coomassie Blue. The proteins migrating in the area of interest were excised and digested with trypsin (16, 33). The resulting peptides were analyzed by nano-liquid chromatography (LC)-MS/MS using an EASY-nLC (Proxeon Biosystems) system connected in-line to a Q-TOF Ultima API (Micromass/Waters Corp.) mass spectrometer. The data were processed using Masslynx 4.0 (Micromass) and the generated peak list files were used to query the Swiss-Prot data base using the Mascot program (34). The searches were performed with propionamide as fixed modification, peptide mass, and fragment mass tolerances offset to 0.2 Da, and no missed tryptic cleavage sites were allowed.
RESULTS
HC·TSG-6 Cross-link Generation, Initial Purification, and Characterization—The cross-link was formed by incubating IαI and TSG-6, the reaction was followed by Coomassie Blue-stained SDS gels and immunoblotting using an anti-TSG-6 antibody (Fig. 1). The analyses confirmed that the covalent HC1·TSG-6 and HC2·TSG-6 complexes were formed. The cross-linked proteins were reduced and carboxymethylated, digested with trypsin, and subjected to strong cation exchange chromatography at pH 2.8 (supplemental data Fig. S1). Using these conditions each tryptic peptide was expected to contain two positive charges derived from the N-terminus and the C-terminal Lys or Arg residues. The only peptides containing more than two positive charges were either cross-linked and/or contain His residues. Consequently, the cross-linked HC·TSG-6 peptides were expected to elute later from the column than most peptides. In addition, the HC·TSG-6 cross-link readily dissociate during NaOH treatment (16), the mass of the cross-linked peptide is thus likely to be significantly reduced by NaOH treatment. These properties were exploited during the purification and detection of the cross-linked peptides.
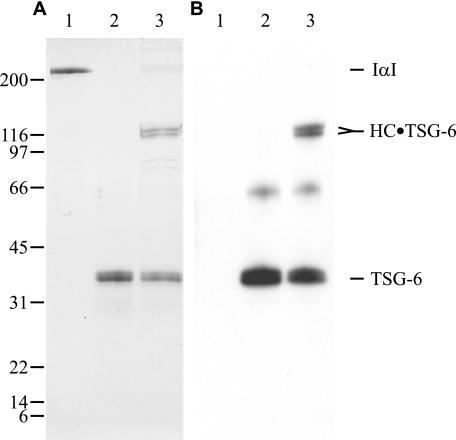
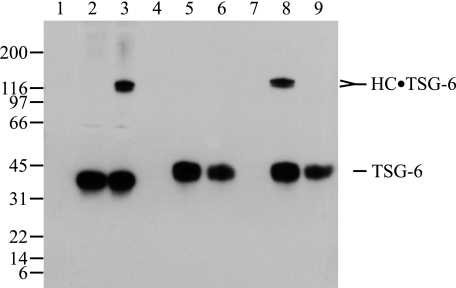
FIGURE 1.
Complex formation between IαI and TSG-6. Purified human IαI (lane 1), purified recombinant human TSG-6 (lane 2), and purified human IαI incubated with purified recombinant human TSG-6 (lane 3) were subjected to reduced SDS-PAGE. Subsequently the resolved proteins were visualized by Coomassie Blue staining (A) or subjected to immunoblotting using anti-TSG-6 antibody (B). The experiment shows the formation of the previously identified covalent HC1·TSG-6 and HC2·TSG-6 complexes.
The mass of the late eluting peptides (fractions 22–44) were determined before and after NaOH treatment. A significant difference between the MALDI-MS analyses was observed in fractions 31 and 34–35 (supplemental data Fig. S2). Without NaOH treatment protonated molecular ions (MH+) peaks of m/z 2174.05 and 1943.92 were observed in fractions 31 (pool 1) and 34–35 (pool 2), respectively. In the samples treated with NaOH these molecular ions disappeared and a new MH+ at m/z 1486.73 became apparent (supplemental data Fig. S2). This molecular ion corresponded to the tryptic TSG-6 peptide, 22DGIFHNSIWLER33 (numbered according to preprotein), with a theoretical monoisotopic MH+ of 1486.74 Da. It has been shown that the cross-link between TSG-6 and HC2 is mediated by an ester between the C-terminal Asp residue of the HCs (16) and thus suggested that the unknown TSG-6 residue has to contain a hydroxyl group (Ser, Thr, or Tyr) (16, 35). Recent studies have suggested that it also is the C-terminal Asp residue of HC1 that mediates the cross-link from HC1 to TSG-6 (16, 17). The theoretical monoisotopic MH+ of the C-terminal tryptic peptides derived from HC1 and HC2 are 706.33 Da (632V2TGVDTD638) and 476.20 Da (645VEND648), respectively. Because water is lost during the formation of an ester linkage the theoretical monoisotopic protonated mass of the cross-linked peptides are 2174.04 Da (HC1·TSG-6), and 1943.91 Da (HC2·TSG-6). These values correspond to the observed MH+ values in untreated pool 1 (m/z 2174.05) and untreated pool 2 (m/z 1943.92), respectively. In addition both molecular ions disappeared after NaOH treatment (supplemental data Fig. S2).
Taken together, these data suggests that 632VTGVDTD638 derived from HC1 and 645VEND648 derived from HC2 are cross-linked to the TSG-6 peptide 22DGIFHNS28IWLER33. The hydroxyl group of Ser28 appears to be the most likely candidate for the formation of the ester linkage as it is the only hydroxyl group containing residues in the peptide. All relevant fractions were analyzed and only the mentioned fractions contained NaOH-sensitive cross-links suggesting that Ser28 exclusively mediates the HC·TSG-6 cross-link.
RP-HPLC Purification of the HC1·TSG-6 and HC2·TSG-6 Cross-links—The HC·TSG-6 cross-links were further purified by RP-HPLC. Pool 1 containing HC1·TSG-6 and pool 2 containing the HC2·TSG-6 cross-link were subjected to RP-HPLC before and after NaOH treatment (Figs. 2, A and B, and 3, A and B). A visual comparison of the chromatograms revealed peptides sensitive to NaOH treatment (Figs. 2A, peak α, and 3A, peak γ). MS analysis confirmed that they contained the purified HC1·TSG-6 (Fig. 2C, m/z 2174.04) and purified HC2·TSG-6 cross-link (Fig. 3C, m/z 1943.92). Comparison of the absorbance at 220 nm of the two purified cross-links (Figs. 2A, peak α, and 3A, peak γ) indicates that the amounts of the purified complexes are similar. This was anticipated, because the amount of HC1·TSG-6 and HC2·TSG-6 generated during IαI/TSG-6 interaction is similar (Fig. 1).
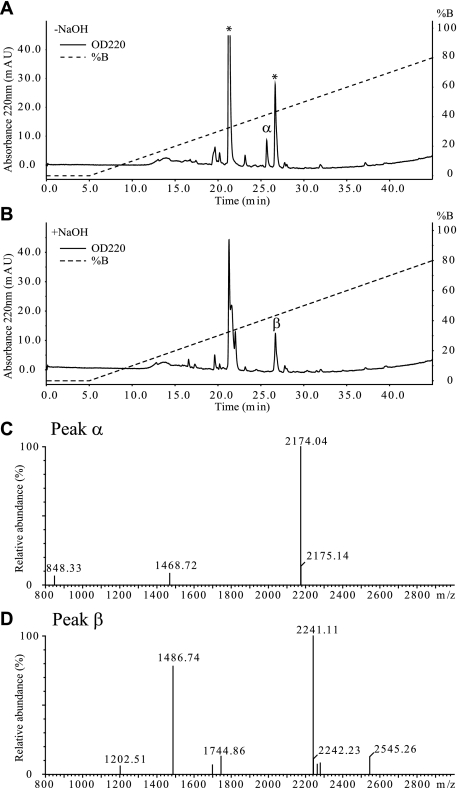
FIGURE 2.
Purification of HC1·TSG-6 complex. Pool 1, containing the HC1·TSG-6 complex, from the strong cation exchange-HPLC analysis was subjected to RP-HPLC analysis directly (A) or after treatment with NaOH (B). The asterisk-labeled peaks contained MH+ of m/z 1424.59 and m/z 2241.15, respectively. C, MALDI-TOF MS analysis of peak α from RP-HPLC of the untreated pool 1. The analysis shows that the peak contains the HC1·TSG-6 complex (theoretical monoisotopic MH+: m/z 2174.04). D, MALDI-TOF MS analysis of peak β from RP-HPLC of the NaOH-treated pool 1. The analysis shows that the peak contains the tryptic TSG-6 peptide (22DGIFHNSIWLER33, theoretical monoisotopic MH+: m/z 1486.73), that participates in the cross-link. The analyses demonstrate that HC1·TSG-6 (632VTGVDTD638 cross-linked to 22DGIFHNSIWLER33) has been purified and is present in peak α.
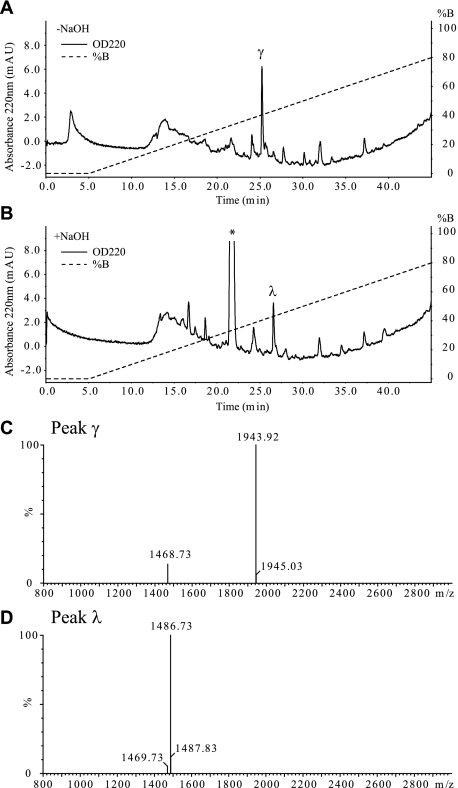
FIGURE 3.
Purification of HC2·TSG-6 complex. Pool 2, containing the HC2·TSG-6 complex, from the strong cation exchange-HPLC analysis was subjected to RP-HPLC analysis directly (A) or after treatment with NaOH (B). The asterisk-labeled peak contains a non-peptide derived contamination. C, MALDI-TOF MS analysis of peak γ from RP-HPLC analysis of the untreated pool 2 (panel A) shows that the peak contains the HC2·TSG-6 complex (theoretical monoisotopic MH+: m/z 1943.91). D, MALDI-TOF MS analysis of peak λ from RP-HPLC of the NaOH treated pool 2 (panel B) shows that the peak contains the tryptic TSG-6 peptide (22DGIFHNSIWLER33, theoretical monoisotopic MH+: m/z 1486.73), which participates in the cross-link. The analyses demonstrate that HC2·TSG-6 (645VEND648 cross-linked to 22DGIFHNSIWLER33) has been purified and is present in peak γ.
The chromatograms contained other peaks, the two dominating peptides in the separation of the untreated HC1·TSG-6 cross-link included peptides of m/z 1424.59 and 2241.15 (Fig. 2A, labeled with *). These were anticipated as they were present in the direct MS analysis of pool 1 (supplemental data Fig. S2A), but they were not affected by NaOH treatment (Fig. 2B). An analysis of all peaks generated after NaOH treatment of the HC1·TSG-6 cross-link revealed that the TSG-6 peptide 22DGIFHNS28IWLER33 co-elutes with one of the two major peaks in the chromatogram (Fig. 2, B and D, peak β). The C-terminal HC1 peptide 632VTGVDTD638 is very hydrophilic and was not detected. The reverse phase separation of the NaOH-treated HC2·TSG-6 cross-link contained a peak not related to the cross-link (Fig. 3B, labeled with *). MS analysis and N-terminal sequencing suggested that this peak did not contain peptides. The other dominating peak (Fig. 3B, peak λ) contained the TSG-6 peptide 22DGIFHNS28IWLER33 involved in cross-linking (m/z 1486.74) (Fig. 3D). Analogous to the HC1·TSG-6 cross-link, the liberated HC2 peptide (645VEND648) was not observed during RP-HPLC of the NaOH-treated sample.
Tandem MS Analyses and NH2-terminal Protein Sequencing of the HC·TSG-6 Cross-links—The purified cross-linked HC1·TSG-6 and HC2·TSG-6 peptides were analyzed by tandem MS using CAF (Fig. 4, A–D) (36). The CAF protocol simplifies spectra interpretation because y-ions are favored, although in this case the presence of two peptides complicated the spectra due to the increased number of possible fragment ions (supplemental data Tables S1 and S2). All the observed fragment ions were in agreement with an ester bond between the C-terminal Asp residue of HC1 or HC2 and the β-carbon of Ser28 of TSG-6 (supplemental data Tables S1 and S2). The results demonstrate that like in HC2·TSG-6 (16), the C-terminal Asp residue of HC1 mediates the cross-linking from HC1 to TSG-6. The fragmentation of the ester bond between the Asp and Ser residues apparently favored the loss of water from Ser28, generating a double bond between the Cα and Cβ of the Ser residue. The NaOH-released TSG-6 peptide (peak β and peak λ) was also subjected to CAF derivatization and MS/MS analysis (supplemental data Fig. S3). These analyses confirmed the TSG-6 peptide sequence, 22DGIFHNS28IWLER33. The fragmentation of the free TSG-6 peptide did not induce the loss of water from Ser28 (supplemental data Fig. S3) supporting, that Ser28 is modified and mediates the HC cross-linking.
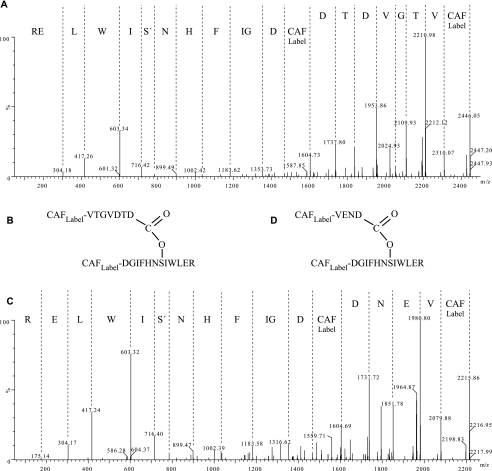
FIGURE 4.
Tandem MS analyses of the purified HC·TSG-6 complexes. The purified HC1·TSG-6 cross-link (Fig. 2A, peak α)(A and B) and the purified HC2·TSG-6 cross-link (Fig. 3A, peak γ)(C and D) were modified using the CAF derivatization protocol and subjected to tandem MS analyses. The MS/MS fragmentation of the ester bond favors a loss of water from Ser and the derived sequences shown above the spectra are based on dehydrated Ser. Analysis of the fragment ions (supplemental data Tables S1 and S2) correlates with an ester bond between the C-terminal Asp residue of the HCs and Ser28 in TSG-6.
Aliquots of the purified HC·TSG-6 complexes (peak α and peak γ) were subjected to N-terminal protein sequence analysis (supplemental data Table S3). In addition to the C-terminal sequence of HC1 and HC2, respectively, the following sequence was identified 22DGIFHN(S28)I(W)LE(R33) (parentheses indicates residues not identified during Edman degradation) in both samples (peak α and peak γ). All residues were positively identified except Ser28, Trp30, and the C-terminal Arg33. For comparison we sequenced the N-terminal of unreacted TSG-6 clearly detecting Ser28 (supplemental data Table S3). The failure to identify the Ser residue during the analyses of the cross-linked peptides further support that this residue is modified and involved in the cross-link (the C-terminal and Trp residues are usually not detected during Edman degradation).
In conclusion, tandem MS and NH2-terminal protein sequence analyses support a cross-link between Ser28 of the TSG-6 protein and the C-terminal Asp residue of the HC. These data also show that the nature of the cross-link between HC1 and TSG-6 and between HC2 and TSG-6 is the same.
Ser28 in TSG-6 Is Required for HC·TSG-6 Formation—Ser28 was mutated to an Ala residue and both wild type TSG-6 and the S28A mutant TSG-6 were expressed in a mammalian cell line. The expression of both forms was confirmed directly by LC-MS/MS analyses (supplemental data Table S4) and by immunoblotting using anti-TSG-6 antibody (Fig. 5). The possibility that the S28A mutation perturbs the wild type folding of TSG-6 could not be excluded. However, the fact that (i) only one amino acid residue is mutated, (ii) the substituted amino acid residue is similar in size, (iii) the TSG-6 mutant is soluble, and (iv) the mutation is positioned very close to the N-terminal of the protein, indicate that the S28A TSG-6 mutant has the native three-dimensional fold. Media containing the mutant or wild type TSG-6 were incubated with purified IαI and the HC·TSG-6 complex formation was evaluated by immunoblotting using an anti-TSG-6 antibody (Fig. 5). The wild type TSG-6 formed, as expected, covalent complexes with IαI (Fig. 5, lane 8). In contrast, no complex formation was detected between IαI and the S28A TSG-6 mutant (Fig. 5, lane 9). These results are consistent with Ser28 of TSG-6 being the site of covalent attachment and they indicate that no alternative cross-linking sites exist in TSG-6.
FIGURE 5.
Mutation of Ser28 in TSG-6 prevents covalent complex formation. The complex formation of wild-type and the TSG-6 S28A mutant were analyzed by reduced SDS-PAGE and immunoblotting using an anti-TSG-6 antibody. As controls purified IαI and purified wild type TSG-6 expressed in insect cells were analyzed alone (lanes 1 and 2), and following coincubation (lane 3). The medium from cells transfected with an unrelated protein (extracellular superoxide dismutase) was included as a negative control (lane 4). The cell culture medium from mammalian cells transfected with TSG-6 wild type cDNA and TSG-6 S28A mutant cDNA were analyzed alone (lanes 5 and 6). IαI was incubated with medium from mammalian cells transfected with cDNA encoding an unrelated protein (extracellular superoxide dismutase) (lane 7), medium from mammalian cells transfected with TSG-6 wild type cDNA (lane 8), and medium from mammalian cells transfected with the TSG-6 S28A mutant cDNA (lane 9). The immunoblot shows that both wild type and mutant TSG-6 were present in the cell culture medium, but only the wild type TSG-6 formed covalent complexes with IαI. The slightly shorter migration of TSG-6 expressed in mammalian cells compared with TSG-6 expressed in insect cells is most likely caused by differences in glycosylation. The results support that HC·TSG-6 complex formation requires Ser28 of TSG-6 and that no other residues are able to form the cross-link.
DISCUSSION
TSG-6/HC2 transfers HCs from the bikunin proteins to HA or unsulfated chondroitin in a reaction that involves covalent HC·TSG-6 intermediates (16–18, 37) (Fig. 6). The molecular structure of the transient cross-link is likely to lead to further insights into the mechanism of the transfer reaction. It has previously been shown that the C-terminal Asp residues of HC2 is covalently linked to TSG-6 (16). However, the involved TSG-6 residue was unknown leading to speculations regarding the nature of the cross-link. Because the Link domain does not interact covalently with the HCs (38), we have speculated that the cross-linked residue was positioned on the CUB domain (39), whereas others have suggested that the Link domain was involved (11, 40, 41). However, in the present study we show that the residue responsible for cross-linking is neither located in the Link domain nor in the CUB domain, but in the N-terminal part of TSG-6. An ester bond cross-links the C-terminal Asp residue of the HCs to the β-carbon of Ser28 in the TSG-6 protein (Fig. 6). Sequence similarity analyses using Basic Local Alignment Search Tool (BLAST) revealed that Ser28 was conserved throughout evolution and that the overall TSG-6 sequence was also highly conserved. In fact, the sequence HNSIWLE (residues 26–32 in human TSG-6 protein) is conserved from such distantly related species as Xenopus laevis and Danio rerio suggesting that the TSG-6/HC2-mediated HC transfer activity is maintained, underlining the importance of these interactions.
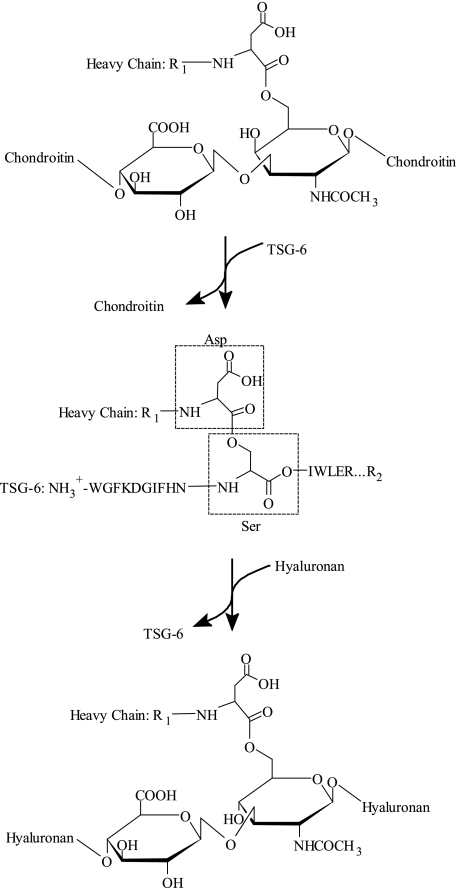
FIGURE 6.
An overview of the HC transfer mechanism. The schematic illustrates the reaction mechanism for the HC transfer and summarizes the relation between the different cross-links mediated by the ester bond from the C-terminal Asp residue of the HCs. In the bikunin proteins the HCs are cross-linked to a C-6 of an internal N-acetylgalactosamine in the CS chain originating from Ser10 of bikunin (top). In the presence of TSG-6 and HC2 this ester bond is cleaved and a covalent intermediate is formed, where the C-terminal of the HCs are esterified to Ser28 of TSG-6 (middle). The HC from this intermediate is subsequently transferred to HA and TSG-6 released. In the last step a new ester bond is formed between the C-terminal of the HC and C-6 of an internal N-acetylglucosamine in HA (bottom). In addition to HA, chondroitin also acts as HC acceptor. The reaction mechanism contains two transesterification steps; both of these steps are divalent cation-dependent and require the presence of HC2. R1 represents the N-terminal sequence of the HCs and R2 represents the C-terminal sequence of TSG-6.
The catalytic mechanism utilized by TSG-6/HC2 is unknown, but the transfer activity is likely to employ several catalytic strategies including, acid-base catalysis, metal ion catalysis, transition state stabilization, and covalent catalysis. An acid-base catalytic strategy could produce an activated Ser28 residue, which in turn performs a nucleophilic attack on the C-terminal carbon of the HC, thereby cleaving the ester bond between the HC and CS chain (16). A previous study (39) has shown that the HC·TSG-6 formation includes a divalent cation-independent binding of TSG-6 to the CS of bikunin followed by a divalent cation-dependent transesterification, supporting that the catalytic strategy involves metal ions. HC2 and TSG-6 are likely to, in concert, form the active site and/or HC2 might stabilize the transition state and/or be involved in orientation of the reactants. Another possibility is that HC2 triggers a conformational change that in turns activates the active site Ser residue. Thus, the role of HC2 in the reaction mechanism requires further investigations. The HC·TSG-6 intermediate has similarity with the acyl-enzyme intermediate of the serine proteases in which there is a transient covalent ester bond between the active site Ser195 residue (chymotrypsin numbering system) and the C-terminal carbon of the substrate (42–44). In the serine proteases, an activated water molecule quickly hydrolyzes the ester bond to regenerate the enzyme. In the HC transfer mechanism, instead of water, HA performs the nucleophilic attack on the intermediate and the HC·HA complex is released.
The three-dimensional structure of the Link domain in TSG-6 has been solved (12) and the three-dimensional structure of the CUB domain has been predicted (11), but unfortunately the structure of the N-terminal part of TSG-6 remains unknown. Secondary structure prediction of the N-terminal peptide suggests that it is mainly random coiled (Jpred data base). Future studies solving the three-dimensional structure of full-length TSG-6 and the HC·TSG-6 intermediates are required to understand the unique enzymatic activity of TSG-6. Furthermore, the role of the different GAGs remains to be clarified. We have previously shown that the CS chain of bikunin remains associated with HC·TSG-6 complexes (39), but these experiments were performed in the absence of HA. It is possible that CS is released from the HC·TSG-6 complex when HA is bound to TSG-6, but CS and HA might also bind simultaneously to TSG-6, it remains to be investigated. In this context, it might be important that the Link domain of TSG-6 in fact contains two GAG binding sites (13). Interestingly, HA is not the only HC acceptor because both unsulfated chondroitin and the CS chain originating from bikunin (mainly unsulfated chondroitin), are HC acceptors as well (16, 37, 45). It demonstrates that the same type of GAG could be the HC donor and HC acceptor, and that HA is not required to induce the second transesterification of the reaction.
Supplementary Material
Acknowledgments
We thank Anna Julie Rasmussen for assisting during the IαI purification and Anne Gylling for technical assistance during the expression of wild type and S28A mutant TSG-6.
This work was supported by grants from the Danish National Science Foundation (to J. J. E.) and the Carlsberg Foundation (to K. W. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S4.
Footnotes
The abbreviations used are: IαI, inter-α-inhibitor; CAF, chemically assisted fragmentation; CS, chondroitin sulfate; GAG, glycosaminoglycan; HA, hyaluronan; HC, heavy chain; LC, liquid chromatography; MALDI, matrix-assisted laser desorption/ionization; MH+, protonated molecular ion; MS, mass spectrometry; RP, reverse phase; TOF, time-of-flight; and TSG-6, tumor necrosis factor-stimulated gene-6 protein.
References
- 1.Bost, F., Diarra-Mehrpour, M., and Martin, J. P. (1998) Eur. J. Biochem. 252 339–346 [DOI] [PubMed] [Google Scholar]
- 2.Enghild, J. J., Thogersen, I. B., Pizzo, S. V., and Salvesen, G. (1989) J. Biol. Chem. 264 15975–15981 [PubMed] [Google Scholar]
- 3.Enghild, J. J., Salvesen, G., Hefta, S. A., Thogersen, I. B., Rutherfurd, S., and Pizzo, S. V. (1991) J. Biol. Chem. 266 747–751 [PubMed] [Google Scholar]
- 4.Enghild, J. J., Salvesen, G., Thogersen, I. B., Valnickova, Z., Pizzo, S. V., and Hefta, S. A. (1993) J. Biol. Chem. 268 8711–8716 [PubMed] [Google Scholar]
- 5.Morelle, W., Capon, C., Balduyck, M., Sautiere, P., Kouach, M., Michalski, C., Fournet, B., and Mizon, J. (1994) Eur. J. Biochem. 221 881–888 [DOI] [PubMed] [Google Scholar]
- 6.Huang, L., Yoneda, M., and Kimata, K. (1993) J. Biol. Chem. 268 26725–26730 [PubMed] [Google Scholar]
- 7.Yoneda, M., Suzuki, S., and Kimata, K. (1990) J. Biol. Chem. 265 5247–5257 [PubMed] [Google Scholar]
- 8.Zhao, M., Yoneda, M., Ohashi, Y., Kurono, S., Iwata, H., Ohnuki, Y., and Kimata, K. (1995) J. Biol. Chem. 270 26657–26663 [DOI] [PubMed] [Google Scholar]
- 9.Lee, T. H., Wisniewski, H. G., and Vilcek, J. (1992) J. Cell Biol. 116 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milner, C. M., and Day, A. J. (2003) J. Cell Sci. 116 1863–1873 [DOI] [PubMed] [Google Scholar]
- 11.Milner, C. M., Higman, V. A., and Day, A. J. (2006) Biochem. Soc. Trans. 34 446–450 [DOI] [PubMed] [Google Scholar]
- 12.Kohda, D., Morton, C. J., Parkar, A. A., Hatanaka, H., Inagaki, F. M., Campbell, I. D., and Day, A. J. (1996) Cell 86 767–775 [DOI] [PubMed] [Google Scholar]
- 13.Mahoney, D. J., Mulloy, B., Forster, M. J., Blundell, C. D., Fries, E., Milner, C. M., and Day, A. J. (2005) J. Biol. Chem. 280 27044–27055 [DOI] [PubMed] [Google Scholar]
- 14.Kuznetsova, S. A., Mahoney, D. J., Martin-Manso, G., Ali, T., Nentwich, H. A., Sipes, J. M., Zeng, B., Vogel, T., Day, A. J., and Roberts, D. D. (2007) Matrix Biol. 27 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wisniewski, H. G., Burgess, W. H., Oppenheim, J. D., and Vilcek, J. (1994) Biochemistry 33 7423–7429 [DOI] [PubMed] [Google Scholar]
- 16.Sanggaard, K. W., Karring, H., Valnickova, Z., Thogersen, I. B., and Enghild, J. J. (2005) J. Biol. Chem. 280 11936–11942 [DOI] [PubMed] [Google Scholar]
- 17.Rugg, M. S., Willis, A. C., Mukhopadhyay, D., Hascall, V. C., Fries, E., Fulop, C., Milner, C. M., and Day, A. J. (2005) J. Biol. Chem. 280 25674–25686 [DOI] [PubMed] [Google Scholar]
- 18.Sanggaard, K. W., Sonne-Schmidt, C. S., Krogager, T. P., Lorentzen, K. A., Wisniewski, H. G., Thogersen, I. B., and Enghild, J. J. (2008) J. Biol. Chem. 283 18530–18537 [DOI] [PubMed] [Google Scholar]
- 19.Sandson, J., and Hamerman, D. (1964) Science 146 70–71 [DOI] [PubMed] [Google Scholar]
- 20.Wisniewski, H. G., Maier, R., Lotz, M., Lee, S., Klampfer, L., Lee, T. H., and Vilcek, J. (1993) J. Immunol. 151 6593–6601 [PubMed] [Google Scholar]
- 21.Bardos, T., Kamath, R. V., Mikecz, K., and Glant, T. T. (2001) Am. J. Pathol. 159 1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mindrescu, C., Dias, A. A., Olszewski, R. J., Klein, M. J., Reis, L. F., and Wisniewski, H. G. (2002) Arthritis Rheum. 46 2453–2464 [DOI] [PubMed] [Google Scholar]
- 23.Szanto, S., Bardos, T., Gal, I., Glant, T. T., and Mikecz, K. (2004) Arthritis Rheum. 50 3012–3022 [DOI] [PubMed] [Google Scholar]
- 24.Fulop, C., Szanto, S., Mukhopadhyay, D., Bardos, T., Kamath, R. V., Rugg, M. S., Day, A. J., Salustri, A., Hascall, V. C., Glant, T. T., and Mikecz, K. (2003) Development 130 2253–2261 [DOI] [PubMed] [Google Scholar]
- 25.Zhuo, L., Yoneda, M., Zhao, M., Yingsung, W., Yoshida, N., Kitagawa, Y., Kawamura, K., Suzuki, T., and Kimata, K. (2001) J. Biol. Chem. 276 7693–7696 [DOI] [PubMed] [Google Scholar]
- 26.de la Motte, C. A., Hascall, V. C., Drazba, J., Bandyopadhyay, S. K., and Strong, S. A. (2003) Am. J. Pathol. 163 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selbi, W., de la Motte, C. A., Hascall, V. C., Day, A. J., Bowen, T., and Phillips, A. O. (2006) Kidney Int. 70 1287–1295 [DOI] [PubMed] [Google Scholar]
- 28.Selbi, W., Day, A. J., Rugg, M. S., Fulop, C., de la Motte, C. A., Bowen, T., Hascall, V. C., and Phillips, A. O. (2006) J. Am. Soc. Nephrol. 17 1553–1567 [DOI] [PubMed] [Google Scholar]
- 29.Zhuo, L., Kanamori, A., Kannagi, R., Itano, N., Wu, J., Hamaguchi, M., Ishiguro, N., and Kimata, K. (2006) J. Biol. Chem. 281 20303–20314 [DOI] [PubMed] [Google Scholar]
- 30.Mindrescu, C., Le, J., Wisniewski, H. G., and Vilcek, J. (2005) Biochem. Biophys. Res. Commun. 330 737–745 [DOI] [PubMed] [Google Scholar]
- 31.Bury, A. F. (1981) J. Chromatogr. 213 491–500 [Google Scholar]
- 32.Matsudaira, P. (1987) J. Biol. Chem. 262 10035–10038 [PubMed] [Google Scholar]
- 33.Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V., and Mann, M. (2006) Nat. Protoc. 1 2856–2860 [DOI] [PubMed] [Google Scholar]
- 34.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Electrophoresis 20 3551–3567 [DOI] [PubMed] [Google Scholar]
- 35.Milner, C. M., Tongsoongnoen, W., Rugg, M. S., and Day, A. J. (2007) Biochem. Soc. Trans. 35 672–676 [DOI] [PubMed] [Google Scholar]
- 36.Keough, T., Lacey, M. P., and Youngquist, R. S. (2002) Rapid Commun. Mass Spectrom. 16 1003–1015 [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, D., Asari, A., Rugg, M. S., Day, A. J., and Fulop, C. (2004) J. Biol. Chem. 279 11119–11128 [DOI] [PubMed] [Google Scholar]
- 38.Getting, S. J., Mahoney, D. J., Cao, T., Rugg, M. S., Fries, E., Milner, C. M., Perretti, M., and Day, A. J. (2002) J. Biol. Chem. 277 51068–51076 [DOI] [PubMed] [Google Scholar]
- 39.Sanggaard, K. W., Sonne-Schmidt, C. S., Jacobsen, C., Thogersen, I. B., Valnickova, Z., Wisniewski, H. G., and Enghild, J. J. (2006) Biochemistry 45 7661–7668 [DOI] [PubMed] [Google Scholar]
- 40.Lesley, J., English, N. M., Gal, I., Mikecz, K., Day, A. J., and Hyman, R. (2002) J. Biol. Chem. 277 26600–26608 [DOI] [PubMed] [Google Scholar]
- 41.Ochsner, S. A., Day, A. J., Rugg, M. S., Breyer, R. M., Gomer, R. H., and Richards, J. S. (2003) Endocrinology 144 4376–4384 [DOI] [PubMed] [Google Scholar]
- 42.Carter, P., and Wells, J. A. (1988) Nature 332 564–568 [DOI] [PubMed] [Google Scholar]
- 43.Katona, G., Wilmouth, R. C., Wright, P. A., Berglund, G. I., Hajdu, J., Neutze, R., and Schofield, C. J. (2002) J. Biol. Chem. 277 21962–21970 [DOI] [PubMed] [Google Scholar]
- 44.Wilmouth, R. C., Clifton, I. J., Robinson, C. V., Roach, P. L., Aplin, R. T., Westwood, N. J., Hajdu, J., and Schofield, C. J. (1997) Nat. Struct. Biol. 4 456–462 [DOI] [PubMed] [Google Scholar]
- 45.Enghild, J. J., Thogersen, I. B., Cheng, F., Fransson, L. A., Roepstorff, P., and Rahbek-Nielsen, H. (1999) Biochemistry 38 11804–11813 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.