Abstract
Myocardial Ca2+/calmodulin-dependent protein kinase II (CaMKII) inhibition improves cardiac function following myocardial infarction (MI), but the CaMKII-dependent pathways that participate in myocardial stress responses are incompletely understood. To address this issue, we sought to determine the transcriptional consequences of myocardial CaMKII inhibition after MI. We performed gene expression profiling in mouse hearts with cardiomyocyte-delimited transgenic expression of either a CaMKII inhibitory peptide (AC3-I) or a scrambled control peptide (AC3-C) following MI. Of the 8,600 mRNAs examined, 156 were substantially modulated by MI, and nearly half of these showed markedly altered responses to MI with CaMKII inhibition. CaMKII inhibition substantially reduced the MI-triggered upregulation of a constellation of proinflammatory genes. We studied 1 of these proinflammatory genes, complement factor B (Cfb), in detail, because complement proteins secreted by cells other than cardiomyocytes can induce sarcolemmal injury during MI. CFB protein expression in cardiomyocytes was triggered by CaMKII activation of the NF-κB pathway during both MI and exposure to bacterial endotoxin. CaMKII inhibition suppressed NF-κB activity in vitro and in vivo and reduced Cfb expression and sarcolemmal injury. The Cfb–/– mice were partially protected from the adverse consequences of MI. Our findings demonstrate what we believe is a novel target for CaMKII in myocardial injury and suggest that CaMKII is broadly important for the genetic effects of MI in cardiomyocytes.
Introduction
The multifunctional Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by increased intracellular Ca2+ (1) and enhanced oxidant stress (2), both prominent features of myocardial disease. CaMKII inhibition protects against heart failure (3) and cardiomyocyte death (4) in response to myocardial infarction (MI). These findings suggest that improved understanding of the CaMKII pathway may lead to new therapies for structural heart disease. However, mechanisms for the beneficial effects of CaMKII inhibition after MI are not well understood. CaMKII regulates diverse cellular functions that are likely to be important for myocardial adaptation to stress, including Ca2+ homeostasis (5), membrane excitability (6), cell survival (2), and gene transcription (7). We used a genetic mouse model of CaMKII inhibition, in which AC3-I, a CaMKII inhibitory peptide, is expressed only in cardiomyocytes (3), to test the transcriptional consequences of myocardial CaMKII inhibition after MI. A second transgenic mouse expressing a scrambled, inactive version of AC3-I (AC3-C) in cardiomyocytes was used as a genetic control.
Our microarrays contained 8,600 cDNA probes. Only 128 genes were substantially upregulated after MI, but approximately half of these (54 out of 128) showed marked reduction upon CaMKII inhibition. We noticed a group of proinflammatory genes that were upregulated after MI in AC3-C mice but not in AC3-I mice. Complement factor B (Cfb) was one of the prominent proinflammatory genes identified in these experiments. Although inflammation is a recognized consequence of MI and a marker of poor patient outcomes in structural heart disease (8–10), the myocardial cells are not known to express Cfb. Since our model only expressed AC3-I inhibitory peptide in myocardial cells, these results suggested that myocardial cells expressed Cfb after MI and that CFB protein could contribute to myocardial injury.
Cfb was identified in our screenings to have markedly augmented expression after MI in AC3-C hearts but reduced expression in AC3-I hearts after MI. The potential for differential Cfb expression by CaMKII after MI was intriguing, because complement activation is an inflammatory response and inflammation contributes to myocardial dysfunction after stress, injury, or infection (11–13). Following cardiac injury, increased deposition of the membrane attack complex (MAC) occurs in damaged tissue areas (14–18). The complement cascade can be activated by 3 independent pathways: the classical, alternative, and lectin pathways (19). Components of the classical complement pathway are activated in patients with heart disease (20). However, to our knowledge, the alternative complement pathway has not been studied in normal or injured hearts. Here, we show that (a) Cfb is expressed in cardiomyocytes; (b) Cfb expression by cardiomyocytes is required for LPS-triggered sarcolemmal injury response; (c) LPS-mediated injury is attenuated by CaMKII inhibition; (d) CaMKII inhibition reduces NF-κB activity and Cfb expression; and (e) mice lacking Cfb have a trend toward improved mortality after MI. These findings identify what we believe to be a previously unrecognized mechanism for CaMKII-mediated proinflammatory signaling in cardiomyocytes and highlight the immune response capability of cardiomyocytes during pathological stress.
Results
Reduced proinflammatory gene expression after MI in mice with CaMKII inhibition.
We used cDNA arrays representing 8,600 genes to measure the steady-state level of mRNAs, as an approach to identify genes with induced expression after MI by permanent occlusion of the left coronary artery. Three weeks after MI, a total of 156 genes were modulated in AC3-C control hearts (out of 8,600 represented on the microarrays), of which 128 genes were induced and 28 genes were repressed (Figure 1A and Supplemental Tables 1 and 2; supplemental material available online with this article; doi:10.1172/JCI35814DS1). A 1.8-fold threshold value, based on statistical analyses of the results from a total of 21 pairwise comparisons, was used to identify differentially expressed genes (21). Thus, a small number of the total genes represented on the microarray were modulated by MI. To specifically identify the CaMKII-regulated genes, we used a set of microarrays to determine gene expression in post-MI AC3-I hearts compared with post-MI AC3-C hearts (Figure 1B). In these experiments, we identified 71 genes whose expression was reduced in AC3-I hearts compared with AC3-C hearts, suggesting that these genes were positively regulated by CaMKII. Similarly, expression of 36 genes was greater in AC3-I hearts compared with AC3-C hearts, indicating negative regulation by CaMKII.
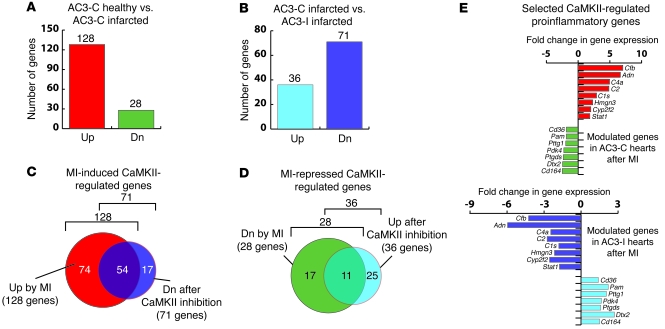
Figure 1. Microarray-based expression analyses of the genes induced by MI and regulated by CaMKII in mouse hearts.
(A) Cardiac genes modulated by MI. cDNA microarray comparison of healthy versus infarcted hearts expressing scrambled control peptide (AC3-C) identified 128 induced and 28 repressed genes out of 8,600 represented on the microarrays. Up, upregulated; Dn, downregulated. (B) Infarcted hearts expressing CaMKII inhibitory peptide (AC3-I) in microarray analyses showed repression of 71 genes and induction of 36 genes when compared with infarcted control (AC3-C) hearts, indicating CaMKII-regulated genes. (C) CaMKII-regulated genes induced by MI. Venn diagram displaying the genes that were induced by MI in AC3-C hearts but were repressed in post-MI AC3-I hearts. A total of 54 (42%) MI-induced genes in AC3-C hearts were regulated by CaMKII. (D) Venn diagram showing the genes that are repressed upon MI and are regulated by CaMKII. A total of 11 (39%) MI-repressed genes in AC3-C hearts were regulated by CaMKII. (E) A subset of proinflammatory genes that were either upregulated in AC3-C hearts (red bars) or downregulated in AC3-C hearts after MI (green bars). In CaMKII-inhibited AC3-I hearts, the same genes displayed reduced expression (blue bars) or increased expression (purple bars), respectively. Adn, complement factor D; C4a, complement component 4A; C2, complement component 2; C1s, complement component 1, s subcomponent; Hmgn3, high mobility group nucleosomal binding domain 3; Cyp2f2, cytochrome P450, family 2, subfamily f, polypeptide 2; Stat1, signal transducer and activator of transcription 1; Pam, peptidylglycine alpha-amidating monooxygenase; Pttg1, pituitary tumor-transforming gene 1; Pdk4, pyruvate dehydrogenase kinase, isoenzyme 4; Ptgds, prostaglandin D2 synthase; Dtx2, deltex 2 homolog.
To select the genes that are induced by CaMKII after MI, we compared the results from the 2 separate microarray experiments to identify genes that displayed regulation by CaMKII in both the experiments. We imposed the criteria of increased expression after MI in AC3-C hearts and reduced expression in infarcted AC3-I hearts compared with AC3-C hearts (Figure 1C). Similarly, to identify the genes whose expression is inhibited by CaMKII after MI, we selected those genes that were repressed in post-MI AC3-C hearts but induced in post-MI AC3-I hearts (Figure 1D). We identified 54 genes that are upregulated (42% of all MI-induced genes in AC3-C; Figure 1C) and 11 genes that are downregulated (39% of all MI-repressed genes in AC3-C; Figure 1D) by CaMKII after MI. Thus, a surprisingly large proportion of genes that were modulated in hearts after MI were also regulated by CaMKII, suggesting that CaMKII is of central importance for coordinating transcriptional responses to MI.
Upon inspection of these MI-induced CaMKII-regulated genes, we noticed a cluster of genes involved in inflammation (Figure 1E). We were surprised to find that expression of these genes was modulated in our microarray analyses by cardiomyocyte-specific CaMKII inhibition. This finding suggested that proinflammatory genes are expressed in ventricular myocytes and that Cfb expression was a myocardial response to MI.
We selected Cfb to study further as CFB is a crucial factor for initiating and sustaining activation of the alternative complement fixation pathway. Classical complement proteins are associated with sarcolemmal injury after MI (14–18), but the origin of these complements was attributed to extra-myocardial sources. To the best of our knowledge, Cfb expression or activation of the alternative complement pathway has not been described after MI. We hypothesized that Cfb suppression contributed to the benefits of CaMKII inhibition after MI, based on the finding that Cfb was an MI-induced, CaMKII-regulated gene and that complement proteins were known to participate in sarcolemmal injury after MI.
Cfb is expressed in cardiomyocytes.
We directly measured Cfb mRNA and protein expression in myocardium in order to validate our gene array results. We extracted RNA from WT hearts and livers (positive control) to confirm the expression of Cfb mRNA. RT-PCR products from both hearts and livers showed a band of the expected size for Cfb cDNA on the agarose gels (Figure 2A). We also tested the expression of CFB in mouse hearts by Western blot analysis. CFB was readily detected in mouse hearts (Figure 2B).
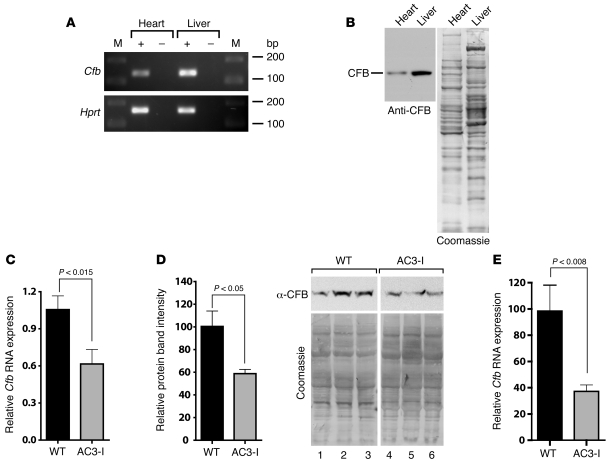
Figure 2. Cfb expression in heart.
(A) RT-PCR analyses using RNA show PCR products for Cfb (137 bp). PCR amplification of hypoxanthine-guanine phosphoryltransferase (Hprt) was used as a positive control (165 bp). M, lanes containing the DNA size markers (bp). The + and – indicate RT reactions with or without the RT, respectively. (B) Immunoblotting for CFB protein expression in heart and liver. Tissue homogenates were immunoblotted for CFB. After immunoblotting the blot was stained with Coomassie blue. (C) Reduced expression of Cfb mRNA in post-MI AC3-I hearts compared with WT controls (n = 3). The qRT-PCR results were normalized to Gapdh mRNA expression. (D) Reduced CFB protein in post-MI AC3-I hearts compared with WT hearts. Homogenates from WT and AC3-I–infarcted hearts were immunoblotted for CFB (n = 3). Following immunoblotting, total protein on the blots was visualized by Coomassie staining. The ratio of the CFB band to the total protein in each lane was determined and is presented as mean ± SEM. (E) Reduced Cfb mRNA expression in isolated cardiomyocytes from post-MI AC3-I hearts. RNA was prepared from cardiomyocytes isolated from WT and AC3-I hearts 7 days after MI (n = 3) and qRT-PCR was performed. Results were normalized to Gapdh mRNA expression.
Both mRNA and protein amounts in post-MI AC3-I hearts were reduced by approximately 40% when compared with the post-MI WT littermate hearts (Figure 2, C and D). However, in the absence of MI, we found that Cfb transcript levels were higher in AC3-I hearts compared with the WT. On the other hand, CFB protein levels were equivalent in AC3-I and WT hearts (Supplemental Figure 1). RNA derived from whole hearts represents both myocytes and non-myocytes. In AC3-I hearts, CaMKII is selectively inhibited in cardiomyocytes, because the transgenic expression of the inhibitory peptide is under the control of the myocyte-defining α myosin heavy chain promoter (22). Therefore, the proinflammatory genes displaying post-MI attenuation in AC3-I hearts were most likely expressed in cardiomyocytes. We tested this idea by performing RT-PCR analyses on the RNA from isolated adult cardiomyocytes and cultured neonatal cardiomyocytes. Cfb transcripts were detected in both adult and neonatal cardiomyocytes (Supplemental Figure 2A). Immunoblotting for CFB protein in cell homogenates further confirmed the expression of CFB protein in the isolated adult and cultured neonatal cardiomyocytes (Supplemental Figure 2B). Thus, a crucial component of the alternative complement pathway, CFB, is expressed in cardiomyocytes.
In order to test if reductions in Cfb mRNA observed in post-MI AC3-I hearts specifically occurred in cardiomyocytes, we performed quantitative RT-PCR (qRT-PCR) on the RNA from isolated cardiomyocytes from WT and AC3-I hearts 1 week after MI. In cardiomyocytes from post-MI AC3-I mice, expression of Cfb was reduced by approximately 60% compared with cardiomyocytes from post-MI WT mice (Figure 2E; P = 0.008). Taken together, these results demonstrated that Cfb mRNA and protein are expressed in the heart and that CaMKII inhibition results in reduced Cfb expression in cardiomyocytes following MI.
Complement factors are deposited on the sarcolemmal membranes in the ailing myocardium of human and rabbit models, in which they are believed to contribute to the MAC and induce sarcolemmal damage after MI (23). CFB is a proinflammatory protein that participates in the innate immune response and is upregulated under inflammatory conditions. To determine if inflammatory signals induced Cfb expression and caused membrane damage, we treated cultured cardiomyocytes with a potent inflammatory stimulus, bacterial LPS (E. coli). Cfb mRNA was strongly induced after LPS treatment (Figure 3A; P < 0.0001, compared with vehicle control). LPS treatment significantly (P < 0.01) increased CFB protein in the cardiomyocyte cultures compared with vehicle control (39 ± 10%; Figure 3B). Based on these findings, we hypothesized that LPS-triggered increases in CFB protein could induce cell membrane injury by participating in the MAC.
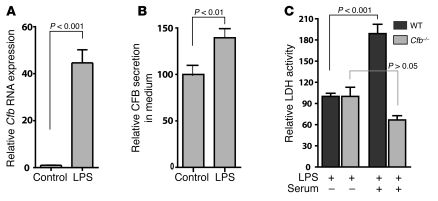
Figure 3. Cfb is induced by LPS in cardiomyocytes.
(A) LPS induces Cfb transcripts in neonatal cardiomyocytes. RNA from cultured mouse neonatal cardiomyocytes was isolated after 12 hours of treatment with 10 μg/ml LPS, and qRT-PCR was performed to detect Cfb RNA expression. Results were normalized to Hprt, and the relative abundance of Cfb RNA is displayed. (B) LPS-induced increase in CFB protein in cultured neonatal cardiomyocytes. Cells were grown in serum-free medium and treated with LPS (10 μg/ml) for 24 hours. (C) Membrane damage caused by complement fixation in neonatal cardiomyocyte cultures from WT and Cfb–/– mice was determined by LDH leakage in the culture medium after LPS treatment. LDH activity after LPS treatment (control) or LPS treatment in the presence of mouse serum (serum) was compared. Ratios of background subtracted LDH activity in the culture medium and total cellular content (Triton X-100 lysates) were determined after 24 hours of LPS treatment. All experiments were done with at least 3 samples.
Cardiomyocyte cell membrane damage is assayed clinically using intracellular proteins as markers of cardiomyocyte death or increased cell membrane permeability. One such marker protein is the cytosolic enzyme lactate dehydrogenase (LDH). We measured LDH enzyme activity released into the culture media from LPS-treated cardiomyocytes. In immune responsive cells, CFB is secreted and participates in forming the MAC by association with other complement factors in the serum (19). Neonatal cardiomyocytes grown in serum-free medium were challenged with LPS in the presence or absence of freshly prepared mouse serum. Addition of LPS in the presence of serum almost doubled the LDH activity in the medium, which is likely due to the increase in MAC formation, resulting from induced CFB production by the cardiomyocytes (Figure 3C; P < 0.001). To test that the increase in LDH activity specifically required increased CFB protein, we performed the same experiment on neonatal cardiomyocytes cultured from Cfb–/– mice (24). In these Cfb–/– cultures, treatment with LPS and mouse serum did not increase the LDH activity in the culture medium compared with treatment with serum and vehicle control (Figure 3C).
To determine whether the LDH release was a result of cell death or general cell injury, we measured cardiomyocyte uptake of a cell membrane impermeable vital fluorescent dye, propidium iodide (PI) (25). In this assay PI is taken up by mortally injured cells. LPS treatment did not significantly increase the number of PI-positive cells detected by flow cytometry compared with treatment with vehicle alone (P = 0.5; Supplemental Figure 3A). We also examined apoptosis in cultured cardiomyocytes using TUNEL staining. The number of TUNEL-positive cells in LPS-treated cells did not show a significant increase over the control cells (Supplemental Figure 3B). Taken together, our results show that increased Cfb expression by LPS treatment leads to cell membrane injury but did not significantly increase cell death under these conditions.
CaMKII regulates LPS-stimulated Cfb expression in cardiomyocytes.
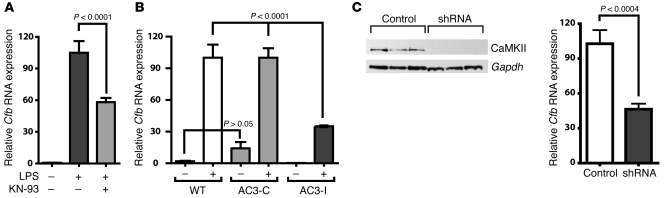
MI induces a complex signaling milieu that includes inflammatory and noninflammatory signaling pathways. Our microarray results showed that CaMKII regulates the expression of inflammatory genes. To delineate the role of CaMKII in cardiomyocyte inflammatory signaling, we inhibited CaMKII by small molecule and genetic methods and quantified Cfb mRNA expression upon LPS treatment. We treated neonatal WT cardiomyocytes with a pharmacological inhibitor of CaMKII (KN-93) prior to LPS treatment. Cfb transcript levels were strongly induced in LPS-treated cardiomyocytes (Figure 4A), whereas pretreatment with water-soluble KN-93 significantly (P < 0.001) blunted the induction of Cfb mRNA to 58% ± 4% of the control. We treated cultured neonatal cardiomyocytes from AC3-I mice with LPS. Similar to the KN-93–inhibited WT cells, AC3-I cardiomyocytes also showed a significant (P < 0.001) attenuation of Cfb induction (WT, 100.0% ± 12.0% versus AC3-I, 35.0% ± 0.8%) compared with the WT cells (Figure 4B). There were no differences, in either basal or LPS-induced Cfb RNA levels, between WT and AC3-C cells that expressed a noninhibitory control peptide (Figure 4B). Finally, we used lentivirus-mediated shRNA to knockdown CaMKII in cultured neonatal cardiomyocytes prior to LPS treatment. Induction of Cfb transcripts by LPS was significantly smaller in cells with CaMKII knockdown compared with controls (46% ± 5%; P < 0.001) (Figure 4C). Thus, CaMKII inhibition in cardiomyocytes, by a variety of independent approaches, consistently and significantly blunted Cfb induction in response to LPS. These results strongly support a concept that CaMKII is critical for Cfb induction in cardiomyocytes.
Figure 4. CaMKII regulates Cfb expression in cardiomyocytes by LPS stimulation.
(A) Neonatal cardiomyocytes were treated with LPS in the presence or absence of the CaMKII inhibitor KN-93 (2.5 μM). Twelve hours after LPS treatment, RNA was extracted and qRT-PCR was performed to determine Cfb RNA expression. KN-93 was added an hour prior to LPS induction. (B) Cfb RNA expression after LPS treatment of CaMKII-inhibited transgenic AC3-I cardiomyocytes. Neonatal cardiomyocytes from AC3-I mice and AC3-C and WT controls were induced with LPS, and Cfb transcripts were quantified using qRT-PCR. (C) Cultured cardiomyocytes were infected with empty vector control or shRNA lentivirus particles against CaMKII, followed by LPS treatment for 12 hours. RNA was isolated and qRT-PCR was performed to quantify the Cfb mRNA. Cell homogenates were analyzed to monitor knockdown of CaMKII by immunoblotting with CaMKII-specific antibodies (left panel).
CaMKII regulates Cfb expression through the NF-κB pathway.
LPS activates TLR-4 to induce the NF-κB signaling pathway (26–28). We used 3 different methods to test whether CaMKII regulates LPS-mediated Cfb expression in cardiomyocytes through the NF-κB signaling pathway. First, we transfected an NF-κB–luciferase reporter construct into neonatal cardiomyocytes and treated the cells with LPS. LPS treatment significantly (3.9- ± 0.1-fold; P < 0.0001) induced the luciferase reporter activity in cardiomyocytes (Figure 5A). Second, we performed EMSAs, using nuclear extracts from LPS-treated cultured cardiomyocytes and oligonucleotides with consensus NF-κB binding sites. In these experiments, treatment of cardiomyocytes with LPS induced specific binding to the NF-κB oligonucleotide probe (Figure 5B). Third, we infected cultured cardiomyocytes with lentivirus carrying a construct for expression of a dominant-negative form of IκB (IκB-DN) to inhibit NF-κB or the empty vector. Cfb mRNA induction in cells infected with IκB-DN was significantly inhibited compared with the cells infected with empty vector lentivirus (control, 103% ± 10% versus IκB-DN, 46% ± 8%; P = 0.0008; Figure 5C). Taken together, these results suggest that CaMKII regulates LPS-mediated Cfb induction in cardiomyocytes through the NF-κB pathway.
Figure 5. CaMKII regulates LPS-stimulated Cfb expression through the NF-κB pathway.
(A) Cultured neonatal cardiomyocytes were transfected with the NF-κB–luciferase reporter gene plasmid and treated with either PBS or LPS for 6 hours. Luciferase activity was measured and normalized to activity from a cotransfected Renilla luciferase plasmid. The data represent the mean ± SEM (n = 3). (B) EMSA showing NF-κB induction in cardiomyocytes upon LPS treatment. Cardiomyocytes were treated for 45 minutes with LPS and nuclear extracts were prepared. The specific NF-κB band is marked by the arrow. The lanes contain no nuclear extract (lane 1), untreated cells (lanes 2 and 3), LPS-treated cells (lanes 4 and 5), LPS-treated cells and 50- and 100-fold WT unlabeled competitor oligonucleotides (oligo) (lanes 6 and 7), and LPS-treated cells and 50- and 100-fold mutated unlabeled oligonucleotides (mut) (lanes 8 and 9). (C) A dominant-negative form of IκB (IκB-DN) attenuates LPS-induced Cfb mRNA expression. Control or dominant-negative IκB–expressing lentivirus particles were infected in cultured cardiomyocytes, followed by LPS induction for 12 hours. RNA was isolated and qRT-PCR performed. (D) In vivo CaMKII inhibition affects LPS-induced NF-κB activation. Luciferase activity of HLL or HLL crossed with CaMKIIN (HLL x CaMKIIN) hearts was measured 6 hours after intraperitoneal injection of LPS (2 μg/g body weight). Luciferase activity was normalized to the total protein concentration; at least 6 animals in each group were used in these experiments.
To test for an in vivo effect of CaMKII on LPS stimulation of the NF-κB pathway, we interbred a transgenic mouse strain that harbors an NF-κB–luciferase reporter gene (referred to as HIV–long terminal repeat/luciferase mice [HLL mice]) (29) with mice expressing the CaMKII inhibitor peptide (βCaMKIIN mice). These CaMKII inhibitor mice (CaMKIIN mice) had α-MHC promoter driven expression of a minigene encoding the CaMKIIN peptide βCaMKIIN (30) that is targeted to the cytoplasmic membranes by addition of a palmitoylation signal sequence. We engineered CaMKIIN with an HA epitope, and HA immunostaining confirmed that βCaMKIIN was expressed in cytoplasm but excluded from the nucleus in adult ventricular myocytes (Supplemental Figure 4). We measured the luciferase activity in heart extracts 6 hours after intraperitoneal injection of LPS. Compared with the HLL controls, HLL crossed with CaMKIIN coexpressing hearts showed significant reduction in luciferase activity upon LPS induction (141% ± 28% increase in HLL versus 40% ± 14% increase in HLL crossed with CaMKIIN hearts; P < 0.01; Figure 5D). These data show that the NF-κB pathway is critical for Cfb expression in cardiomyocytes and that CaMKII inhibition significantly attenuates NF-κB activity in response to LPS in myocardium in vivo.
CaMKII regulates TNF-α induced Cfb expression.
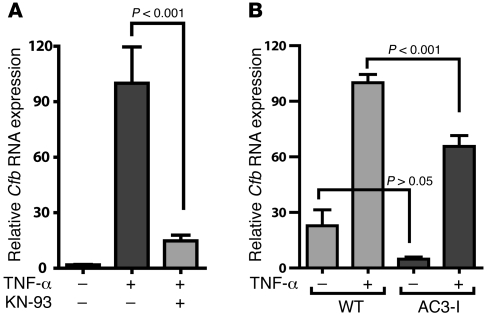
We reasoned that if NF-κB is a key control point for CaMKII effects on inflammatory signaling in cardiomyocytes, then CaMKII inhibition should also negatively regulate responses to other agonists that activate NF-κB. TNF-α is a proinflammatory cytokine that activates Cfb through NF-κB (28, 31). Furthermore, TNF-α expression is increased during MI (32), and increased systemic or local expression of TNF-α results in cardiomyopathy (33). We treated cultured neonatal cardiomyocytes with TNF-α in the presence of the CaMKII inhibitor KN-93 and measured Cfb induction using qRT-PCR (Figure 6A). TNF-α strongly induced Cfb expression in these experiments, and this induction was significantly blunted by CaMKII inhibition with KN-93 (TNF-α plus KN-93, 15% ± 3% versus TNF-α alone, 100% ± 20%; P < 0.001). We also tested the effect of TNF-α on Cfb expression in neonatal cardiomyocytes from AC3-I transgenic or WT mice. As expected, upon treatment with TNF-α, the Cfb expression level was significantly lower in the AC3-I cardiomyocytes (WT treated with TNF-α, 100% ± 4% versus AC3-I treated with TNF-α, 66% ± 6%; P < 0.001; Figure 6B). These results support the concept that the NF-κB pathway is regulated by CaMKII in cardiomyocytes in response to LPS and TNF-α stimulation.
Figure 6. TNF-α–mediated Cfb expression is regulated by CaMKII.
(A) Neonatal cardiomyocytes were treated with TNF-α (100 pg/ml) in the presence or absence of KN-93 (2.5 μM) and RNA was isolated after 12 hours. Cfb transcripts were quantified by qRT-PCR and normalized to Hprt. (B) Cultured AC3-I and WT neonatal cardiomyocytes were treated with TNF-α and qRT-PCR was performed on RNA isolated after 12 hours of treatment.
Cfb–/– mice are partially protected against MI.
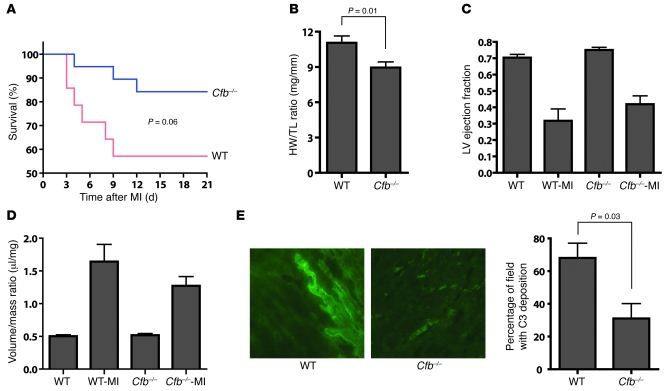
CaMKII has multiple downstream targets in cardiomyocytes. Therefore, the benefits of CaMKII inhibition after MI likely result from effects on a variety of protein substrates. In order to measure the functional importance of Cfb expression to MI outcomes, we compared the MI responses in Cfb–/– and WT mice. We observed a nonsignificant trend (P = 0.06) toward improved survival in Cfb–/– (84% survival) compared with the WT controls (57% survival) 21 days after MI (Figure 7A). Cfb–/– mice had significantly less cardiac hypertrophy (heart weight/tibia length [HW/TL] = 8.9 ± 0.5 mg/mm) compared with the WT controls (HW/TL = 11.1 ± 0.6 mg/mm; P = 0.01; Figure 7B). The baseline and post-MI ejection fractions (Figure 7C) and calculated LV volume/mass ratio (Figure 7D) did not differ between the WT and Cfb–/– mice. Our cellular studies identified increased sarcolemmal stress and LDH leak as a consequence of post-MI Cfb expression. In order to test if there was reduced complement deposition in Cfb–/– hearts after MI, we immunolabeled deposited complement C3 in frozen tissue sections. Compared with the WT myocardium, Cfb–/– hearts showed significantly reduced number of C3 deposits (Figure 7E; P = 0.03). These results show Cfb–/– mice have reduced mortality, hypertrophy, and myocardial complement deposition compared with WT controls after MI.
Figure 7. Improved cardiac function in Cfb–/– mice after MI.
(A) Kaplan-Meier graph of survival of Cfb–/– and WT mice after inducing MI. Cfb–/– mice showed a trend toward improved survival (84% survival, n = 14) compared with the WT mice (57% survival, n = 8) on 21 day after MI (P = 0.06). (B) Cardiac hypertrophy at 21 day after MI was measured as the ratio of heart weight to tibia length (HW/TL). (C) LV function was indexed as ejection fraction (EF). (D) The volume/mass ratio of LVs after MI was determined by echocardiography after MI (21 days). WT-MI, WT mice after MI; Cfb–/–-MI, Cfb–/– mice after MI. (E) Reduced complement (C3) deposition in myocardium of Cfb–/– mice 1 week after MI (original magnification, ×400).
Discussion
An unexpected role of CaMKII in myocardial inflammation.
MI, like other causes of tissue injury, is associated with inflammation (34–37). Activation of inflammatory pathways after MI is increasingly recognized as an important association with adverse patient outcomes (35, 38–40). CaMKII inhibition improves myocardial performance and protects against adverse structural effects after MI in mice (3), suggesting that CaMKII inhibition may be a successful approach for treating MI patients. However, a role of CaMKII in myocardial inflammation had not been previously suspected. Here, we provide new evidence that (a) cardiomyocytes express the inflammatory gene Cfb and that Cfb expression increases with pathological stress, while lack of Cfb improves the response to MI; (b) induction of Cfb results in sarcolemmal injury; (c) CaMKII inhibition reduces both Cfb expression and sarcolemmal injury; and (d) CaMKII is an activator of the NF-κB pathway in cardiomyocytes in vitro and in vivo. Because the CaMKII inhibitory peptide βCaMKIIN is targeted to cytoplasmic membranes in the CaMKIIN transgenic mice, our in vivo data suggests that CaMKII inhibition can suppress NF-κB independent of direct actions in the nucleus.
CaMKII regulates the activity of key proteins involved in cardiac excitation-contraction coupling (41). In addition, CaMKII regulates expression of hypertrophic genes by directly altering transcription activators or through histone-modifying enzymes (42). However, to our knowledge, a role of CaMKII in inflammatory gene regulation after cardiac injury was unprecedented. Using gene expression profiling, we confirmed that in response to MI, CaMKII increases the expression of inflammatory molecules in heart (Figure 1). Thus, our findings reveal what we believe to be a novel and unexpected role of CaMKII in favoring inflammatory responses in heart and highlight a specific pathway whereby CaMKII increases CFB and induces sarcolemmal injury in cardiomyocytes.
Calmodulin-activated protein kinases regulate NF-κB activity in neurons and T cells (43–46). In this report, we have demonstrated that CaMKII inhibition reduces Cfb responses to LPS, both in cultured cardiomyocytes and in vivo through regulation of NF-κB signaling pathway (Figures 4 and 5). These regulatory effects were also exerted on TNF-α–mediated gene expression (Figure 6, A and B), demonstrating a role of CaMKII in secondary effects on myocardial cytokine amplification upon injury (9). We observed a surprisingly large number of MI-regulated mRNAs (a total 65 modulated genes out of 156 genes modulated by MI) that were responsive to CaMKII inhibition. The possible preeminence of CaMKII as a regulator of post-MI gene changes, at least as detected in our microarray experiments, suggests that CaMKII participates in a wide range of transcriptional responses to myocardial injury.
Complement proteins in myocardial injury and inflammation.
Classical complement proteins, secreted from extra-myocardial tissue, are known to participate in myocardial injury responses to pathological stress, including MI. However, the involvement of the alternative pathway complement proteins and the possibility that complements were produced by myocardial cells under stress conditions was not previously considered. Our results do not discount the concept that inflammatory responses are amplified by blood-borne non-cardiac cells. However, our results do show that myocardium is capable of initiating and sustaining a defined local inflammatory response that leads to sarcolemmal injury in the absence of enabling non-cardiac cells. In the present study, we identified Cfb as a proinflammatory signal expressed in cardiomyocytes after MI and defined a new role for CaMKII in regulating the expression of a crucial component of the alternative complement pathway. Our experiments showed increased cell membrane damage upon LPS induction of CFB in WT cells but not in Cfb–/– cells (Figure 3C), consistent with the concept that the alternative complement pathway was required for sarcolemmal damage. However, cell death did not correspond to the increased membrane damage following complement activation (Supplemental Figure 3, A and B). Thus, cultured cardiomyocytes appear to resist cell death by complement fixation. CFB protein initiated MAC may cause cell lysis or form a sublytic complex; the latter stimulates cells to amplify the inflammatory response through cytokine induction (47–49).
Using Cfb–/– mice in our studies, we demonstrated that Cfb is a determinant of MI outcome. Cfb–/– mice showed significantly reduced myocardial complement deposition and hypertrophy after MI and had a trend toward improved survival compared with WT mice (Figure 7). These results directly demonstrate for what we believe is the first time a role of the alternate complement pathway in exacerbating myocardial injury. However, the beneficial effects of Cfb ablation on post-MI responses are smaller than the benefits globally attributed to CaMKII inhibition (3), consistent with the idea that reduced CFB expression is only one of many downstream CaMKII targets involved in pathological outcomes after MI. Although Cfb ablation did not result in gross phenotypic differences compared with WT littermates (24), it is possible that loss of Cfb causes uncharacterized effects.
In our experiments, LPS stimulation of CFB in cardiomyocytes increased cell membrane leakage, as determined by increased LDH activity in the culture supernatants (Figure 3C). Although the causes of initial injury and induction of inflammatory response in the myocardium are not well understood, TLR-4 has been implicated in several tissue injury models, including LPS infusion. TLR-4–deficient mice are resistant to myocardial ischemia/reperfusion injury and display reduced inflammatory response, including reduced myocardial complement fixation (50). In addition, blocking MyD88, the downstream adapter of TLR-4 signaling, displays similar protection against myocardial injury (51). Moreover, several studies have underlined the role of the NF-κB pathway in cardiac hypertrophy and inflammation (52–55). Since the NF-κB pathway is one of the major proinflammatory pathways to be induced by TLR activation, it is likely that a subgroup of inflammatory genes, including Cfb, is induced by TLR-4 activation upon myocardial injury. We have demonstrated that LPS-stimulated increase in Cfb expression is related to CaMKII activation of the NF-κB pathway, both in vitro and in vivo (Figure 5). Because the CaMKII inhibitory peptide βCaMKIIN is targeted to cytoplasmic membranes in the CaMKIIN transgenic mice, our in vivo data suggests that CaMKII inhibition can suppress NF-κB independent of direct actions in the nucleus. Our findings suggest an exciting new hypothesis that these inflammatory response pathways will be regulated by CaMKII. As the role of TLR-4 in sensing tissue injury is becoming evident, a broader role of CaMKII as a regulator of inflammatory responses to ischemia, infarction, and sepsis may emerge.
How many targets are needed to treat MI?
Patient outcomes after MI almost certainly depend upon the combined activation of several distinct but potentially interrelated signaling pathways. The plethora of deleterious signals activated during MI suggests that specific treatments exclusively targeted toward any single pathway will not be successful. Our results from post-MI cardiac functions suggest that although Cfb alone does not seem to be the determinant of the complex phenotype arising after MI, it certainly contributes adversely to the disease. On the other hand, CaMKII participates in several different downstream pathological outcomes in MI, by activating hypertrophic gene programs (56, 57), inducing proarrhythmic electrical remodeling (3, 58), stimulating cell death pathways (4, 59), and disturbing cellular Ca2+ homeostasis (60). CaMKII activity increases are initiated by a variety of upstream signals that lead to increased cellular Ca2+ and/or increased oxidation (2). Our finding that LPS activates a CaMKII pathway to induce Cfb contributes new evidence to illustrate the multifunctional capacity of CaMKII as a nodal signal for integrating and transducing upstream signals into diverse downstream pathological consequences in heart disease. The emerging concept that CaMKII participates in multiple critical disease processes raises hope that further exploration and understanding of the CaMKII pathway and CaMKII inhibition could provide an improved, “broad spectrum” therapy for MI patients.
Methods
Cell cultures.
All animal experiments were approved by the Institutional Animal Care and Use Committee of University of Iowa. Mouse neonatal cardiomyocytes were isolated from 1- to 3-day-old newborn mice, according to an established protocol in our laboratory, and detailed in Supplemental Methods.
Myocardial CaMKIIN mice.
Targeting of βCaMKIIN to cytoplasmic membranes in transgenic mice was achieved by fusion of tandem Cys residues from GAP-43 (neuromodulin). These Cys are palmitoylated, which causes the CaMKIIN to partition into detergent-resistant cellular membranes (61). The details of the generation of transgenic mice are provided in Supplemental Methods.
Pharmacological reagents.
Aqueous solutions of LPS from E. coli (Sigma-Aldrich), TNF-α (BD Biochemicals), and water-soluble KN-93 (Calbiochem) were used at 10 μg/ml, 300 pg/ml, and 2.5 μM final concentrations, respectively. In this study, we only used the water-soluble form of KN-93, because a water-soluble form of the KN-93 control drug, KN-92, is not available. We did not use the DMSO-soluble forms of KN-93 or KN-92, because DMSO at a concentration necessary to dissolve KN-92 markedly affected transcript levels in our experiments (data not shown).
cDNA microarray analysis.
Gene expression profiles were determined from cDNA microarrays, containing 8,600 elements derived from clones isolated from normalized cDNA libraries or purchased from ResGen (Invitrogen), as described in detail previously (62). Differential expression values were expressed as the ratio of the median of background-subtracted fluorescence intensity of the experimental RNA to the median of background-subtracted fluorescence intensity of the control RNA. In the present study, a 1.8-fold threshold value was used to identify differentially expressed genes. For ratios that were greater than or equal to 1.0, the ratio was expressed as a positive value. For ratios of less than 1.0, the ratio was expressed as the negative reciprocal (i.e., a ratio of 0.5 = –2.0). Median ratios were normalized to 1.0 using 2 pools of 3,000 randomly chosen cDNAs in each pool. Six replicates of each of the 2 pools were printed in 4 evenly distributed blocks of the array. A statistically significant differential expression threshold value was empirically determined according to the method of Yang et al. (21).
MI surgery and echocardiography.
Mouse hearts were infarcted by opening the thoracic cavity and placing a ligature on the left descending coronary artery, as described previously (3). Both male and female mice were infarcted and at 3 weeks after infarction, RNA was isolated from cardiac tissue using RNeasy kit (QIAGEN).
Transthoracic echocardiograms were recorded in conscious sedated mice, as described previously (63). Images were acquired and analyzed by an operator blinded to mouse genotype.
NF-κB reporter mice.
For evaluating NF-κB activation in mouse hearts in vivo, we used transgenic mice that were engineered to possess proximal 5′ HIV-1 long terminal repeat, driving the expression of Photinus luciferase cDNA (referred to as HLL mice) in each tissue (29). Both CaMKIIN and HLL mice have the same B6D2 background.
RNA isolation and qRT-PCR.
Total RNA was isolated from mouse tissues and cardiomyocytes using RNeasy RNA Isolation Kit (QIAGEN). A 500-ng aliquot of each RNA sample was used for cDNA synthesis in a 50 μl reaction mix, using oligo dT16 as primer and SuperScript III Reverse Transcriptase (Invitrogen). SYBR Green–based quantitative real-time PCR reactions were performed as described in Supplemental Methods.
CaMKII knockdown.
Cultured neonatal cardiomyocytes were infected with FIV, lentivirus containing shRNA against CaMKIIδ, as previously described (2). The shRNA were targeted to the conserved sequence of the mouse CaMKIIδ transcript. RNA isolation and cell lysates preparations were done 48 hours after viral infection.
Immunoblotting.
Tissue samples were homogenized in modified RIPA buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% v/v NP-40, and 0.5% w/v deoxycholate), containing a mixture of protease and phosphatase inhibitors. Equal amounts of protein were fractionated on NuPAGE gels (Invitrogen) and transferred onto PVDF membranes (Bio-Rad). After blocking nonspecific binding with 10% w/v non-fat milk powder in TBS-T (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 0.1% v/v Tween-20), blots were incubated in primary antibodies (rabbit anti-CFB, Atlas Antibodies; rabbit anti-actin, Sigma-Aldrich) overnight at 4°C. Antibodies to CaMKIIδ were a gift from Harold Singer (Albany Medical College, Albany, New York, USA). Blots were washed in TBS-T and incubated with appropriate HRP-conjugated secondary antibodies. Protein bands were detected using ECL reagent (Lumi-Light, Roche), and loading was routinely monitored by coomassie staining of the blots after antibody probing. For quantification, Quantity One software (Bio-Rad) was used.
Immunofluorescence labeling.
Cardiomyocytes from HLL, CaMKIIN, or HLL crossed with CaMKIIN interbred mice were fixed in 4% paraformaldehyde, and cells were permeabilized by PBS-0.1% Tween20. To visualize the HA-tagged peptide, a monoclonal anti-HA antibody (Covance) and Alexa Fluor 568–conjugated goat anti-mouse IgG (Molecular Probes) secondary antibody were used. The control labeling experiments were performed under identical conditions except the anti-HA Mab was omitted.
For C3 deposition in myocardium, frozen heart sections were labeled using specific antibody. Twenty-five randomly selected fields for each of the WT and Cfb–/– post-MI heart sections were scored for positive or negative signal in a blind experiment. Four WT and Cfb–/– mouse hearts were assessed, and the percentage of positive fields for C3 deposition are presented.
ELISA.
To determine the secreted CFB from neonatal cardiomyocytes, flat-bottom polystyrene plates (Costar Corning) were coated with 100 μl of culture medium for 18 hours at 4°C. Skim milk (2%) in PBS solution was used as a blocking reagent. After washing the wells with PBS containing 0.05% v/v Tween-20, a 1:1,000 dilution of affinity-purified antibody to CFB was added to each well (100 μl per each well). After washes, biotinylated anti-rabbit IgG (goat anti-rabbit IgG, Jackson ImmunoResearch Laboratories Inc.), at a 1:2,000 dilution, was incubated, and new washes were performed. An affinity-purified biotin-conjugated secondary antibody (1:2,000 dilution, goat anti-rabbit IgG, Sigma-Aldrich) was added to the wells. A second conjugate, Streptavidin-alkaline phosphatase (1:2,000 dilution, Jackson ImmunoResearch Laboratories Inc.), was incubated. p-Nitrophenyl phosphate tablets (SigmaFast, Sigma-Aldrich) in Tris-HCl buffer (200 mM, pH 8.0) were used as chromogen substrate. The chromogenic reaction was monitored by measuring the absorbance at 405 nm in a plate reader (Molecular Devices).
LDH assays.
A 100-μl aliquot of culture medium was used for LDH detection, using a commercial LDH assay kit (Clontech). The total LDH was determined after removing the culture medium and replenishing the wells with medium, containing 1% Triton X-100 to lyse the cells. The activity of released LDH in culture medium was normalized to the total cellular LDH activity to determine the effect of treatments.
TUNEL and PI assays.
TUNEL assays were performed using In Situ Cell Death Detection Kit, TMR red (Roche), according to the manufacturer’s recommended protocol, as described in Erickson et al. (2). To assay the cell death, we used the fluorescent vital dye PI (Molecular Probes), according to the manufacturer’s protocol. Cells were grown in serum-free medium, and control (serum) or treated (serum and LPS) cultured neonatal cardiomyocytes (24-hour treatment) were trypsinized and mixed with aqueous PI solution (1 μg/ml, final concentration). After 30 minutes of incubation on ice, cells were sorted by flow cytometry, using a 610-nm/20-nm band-pass filter (LSR II, BD).
EMSA.
EMSA experiments were performed using [32P]-labeled duplex oligodeoxynucleotides (probe), representing the binding sites for NF-κB (5′-AGTTGAGGGGACTTTCCCAGGC-3′) (Promega). Neonatal cardiomyocytes were treated with LPS for 45 minutes and nuclear extracts were prepared by a method described by Dignam and Roeder (64). Labeled probe was incubated with nuclear extracts in the presence of poly(dI-dC) to prevent nonspecific protein-DNA interactions. Bound DNA-protein complexes were separated by polyacrylamide gel electrophoresis and visualized by autoradiography. The specificity of protein-DNA interaction was tested by addition of 50- and 100-fold molar excess of unlabeled duplex WT or mutated oligos (5′-AGTTGAGTTGACTTTCCCAGGC-3′).
Statistics.
Statistical significance was determined by a Student’s t test (2-tailed) or ANOVA, as appropriate. Post-hoc analyses from multiple group comparisons were performed using the Bonferroni method. A P value of less than 0.05 was considered statistically significant. All results are presented as mean ± SEM.
Supplementary Material
Acknowledgments
We are grateful to Paul Rothman and John Colgan (University of Iowa) and Mollie Meffert (Johns Hopkins University) for helpful scientific discussions. We thank Kathy Walters (Central Microscopy Core, University of Iowa) for confocal imaging and George Rasmussen (Flow Cytometry Facility, University of Iowa) for FACS analyses. This work was funded by NIH grants R01 HL 079031, R01 HL 62494, and R01 HL 70250 (to M.E. Anderson); NIH grants R01 HL084583 and R01 HL083422 and Pew Scholars Trust (to P.J. Mohler); NIH grant R01 DK076690 (to J.M. Thurman); and the University of Iowa Research Foundation. This work was supported in part by the Fondation Leducq Award to the Alliance for Calmodulin Kinase Signaling in Heart Disease.
Footnotes
Conflict of interest: Madhu V. Singh and Mark E. Anderson are part of a pending patent application on use of CaMKII inhibitors to treat or prevent inflammation in heart muscle. Joshua M. Thurman is a consultant for Taligen Therapeutics on developing complement inhibitors.
Nonstandard abbreviations used: CaMKII, Ca2+/calmodulin-dependent protein kinase II; CaMKIIN mice, CaMKII inhibitor mice; Cfb, complement factor B; LDH, lactate dehydrogenase; MAC, membrane attack complex; MI, myocardial infarction; PI, propidium iodide; qRT-PCR, quantitative RT-PCR.
Citation for this article: J. Clin. Invest. 119:986–996 (2009). doi:10.1172/JCI35814
References
- 1.Schulman H., Greengard P. Ca2+-dependent protein phosphorylation system in membranes from various tissues, and its activation by “calcium-dependent regulator”. Proc. Natl. Acad. Sci. U. S. A. 1978;75:5432–5436. doi: 10.1073/pnas.75.11.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erickson J.R., et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang R., et al. Calmodulin kinase II inhibition protects against structural heart disease. Nat. Med. 2005;11:409–417. doi: 10.1038/nm1215. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., et al. Calmodulin kinase II inhibition protects against myocardial cell apoptosis in vivo. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H3065–H3075. doi: 10.1152/ajpheart.00353.2006. [DOI] [PubMed] [Google Scholar]
- 5.Grueter C.E., Abiria S.A., Wu Y., Anderson M.E., Colbran R.J. Differential regulated interactions of calcium/calmodulin-dependent protein kinase II with isoforms of voltage-gated calcium channel beta subunits. Biochemistry. 2008;47:1760–1767. doi: 10.1021/bi701755q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner S., et al. Ca2+/calmodulin-dependent protein kinase II regulates cardiac Na+ channels. J. Clin. Invest. 2006;116:3127–3138. doi: 10.1172/JCI26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X., et al. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J. Clin. Invest. 2006;116:675–682. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braunwald E. Biomarkers in heart failure. N. Engl. J. Med. 2008;358:2148–2159. doi: 10.1056/NEJMra0800239. [DOI] [PubMed] [Google Scholar]
- 9.Knuefermann P., Vallejo J., Mann D.L. The role of innate immune responses in the heart in health and disease. Trends Cardiovasc. Med. 2004;14:1–7. doi: 10.1016/j.tcm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Ridker P.M. Inflammatory biomarkers and risks of myocardial infarction, stroke, diabetes, and total mortality: implications for longevity. Nutr. Rev. 2007;65:S253–S259. doi: 10.1111/j.1753-4887.2007.tb00372.x. [DOI] [PubMed] [Google Scholar]
- 11.Mann D.L. Stress-activated cytokines and the heart: from adaptation to maladaptation. Annu. Rev. Physiol. 2003;65:81–101. doi: 10.1146/annurev.physiol.65.092101.142249. [DOI] [PubMed] [Google Scholar]
- 12.Malave H.A., Taylor A.A., Nattama J., Deswal A., Mann D.L. Circulating levels of tumor necrosis factor correlate with indexes of depressed heart rate variability: a study in patients with mild-to-moderate heart failure. Chest. 2003;123:716–724. doi: 10.1378/chest.123.3.716. [DOI] [PubMed] [Google Scholar]
- 13.Feldman A.M., et al. The role of tumor necrosis factor in the pathophysiology of heart failure. J. Am. Coll. Cardiol. 2000;35:537–544. doi: 10.1016/S0735-1097(99)00600-2. [DOI] [PubMed] [Google Scholar]
- 14.Yasojima K., Schwab C., McGeer E.G., McGeer P.L. Human heart generates complement proteins that are upregulated and activated after myocardial infarction. Circ. Res. 1998;83:860–869. doi: 10.1161/01.res.83.8.860. [DOI] [PubMed] [Google Scholar]
- 15.Yasojima K., Kilgore K.S., Washington R.A., Lucchesi B.R., McGeer P.L. Complement gene expression by rabbit heart: upregulation by ischemia and reperfusion. Circ. Res. 1998;82:1224–1230. doi: 10.1161/01.res.82.11.1224. [DOI] [PubMed] [Google Scholar]
- 16.Vakeva A.P., et al. Myocardial infarction and apoptosis after myocardial ischemia and reperfusion: role of the terminal complement components and inhibition by anti-C5 therapy. Circulation. 1998;97:2259–2267. doi: 10.1161/01.cir.97.22.2259. [DOI] [PubMed] [Google Scholar]
- 17.Rossen R.D., et al. Selective accumulation of the first component of complement and leukocytes in ischemic canine heart muscle. A possible initiator of an extra myocardial mechanism of ischemic injury. Circ. Res. 1985;57:119–130. doi: 10.1161/01.res.57.1.119. [DOI] [PubMed] [Google Scholar]
- 18.Pinckard R.N., et al. Consumption of classical complement components by heart subcellular membranes in vitro and in patients after acute myocardial infarction. J. Clin. Invest. 1975;56:740–750. doi: 10.1172/JCI108145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gros P., Milder F.J., Janssen B.J. Complement driven by conformational changes. Nat. Rev. Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 20.Afanasyeva M., Rose N.R. Cardiomyopathy is linked to complement activation. Am. J. Pathol. 2002;161:351–357. doi: 10.1016/S0002-9440(10)64189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang I., et al. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3:research0062.01–research0062.12. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulick J., Subramaniam A., Neumann J., Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J. Biol. Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 23.Homeister J.W., Satoh P., Lucchesi B.R. Effects of complement activation in the isolated heart. Role of the terminal complement components. Circ. Res. 1992;71:303–319. doi: 10.1161/01.res.71.2.303. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto M., et al. Abrogation of the alternative complement pathway by targeted deletion of murine factor B. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8720–8725. doi: 10.1073/pnas.94.16.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra V., et al. Changes in mitochondrial dynamics during ceramide-induced cardiomyocyte early apoptosis. Cardiovasc. Res. 2008;77:387–397. doi: 10.1093/cvr/cvm029. [DOI] [PubMed] [Google Scholar]
- 26.Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 27.Moynagh P.N. The NF-kappaB pathway. J. Cell Sci. 2005;118:4589–4592. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber J., et al. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackwell T.S., et al. Multiorgan nuclear factor kappa B activation in a transgenic mouse model of systemic inflammation. Am. J. Respir. Crit. Care Med. 2000;162:1095–1101. doi: 10.1164/ajrccm.162.3.9906129. [DOI] [PubMed] [Google Scholar]
- 30.Chang B.H., Mukherji S., Soderling T.R. Characterization of a calmodulin kinase II inhibitor protein in brain. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10890–10895. doi: 10.1073/pnas.95.18.10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Krein P.M., Muruve D.A., Winston B.W. Complement factor B gene regulation: synergistic effects of TNF-alpha and IFN-gamma in macrophages. J. Immunol. 2002;169:2627–2635. doi: 10.4049/jimmunol.169.5.2627. [DOI] [PubMed] [Google Scholar]
- 32.Irwin M.W., et al. Tissue expression and immunolocalization of tumor necrosis factor-alpha in postinfarction dysfunctional myocardium. Circulation. 1999;99:1492–1498. doi: 10.1161/01.cir.99.11.1492. [DOI] [PubMed] [Google Scholar]
- 33.Bryant D., et al. Cardiac failure in transgenic mice with myocardial expression of tumor necrosis factor-{alpha}. Circulation. 1998;97:1375–1381. doi: 10.1161/01.cir.97.14.1375. [DOI] [PubMed] [Google Scholar]
- 34.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 35.Frangogiannis N.G., Smith C.W., Entman M.L. The inflammatory response in myocardial infarction. Cardiovasc. Res. 2002;53:31–47. doi: 10.1016/S0008-6363(01)00434-5. [DOI] [PubMed] [Google Scholar]
- 36.Entman M.L., Smith C.W. Postreperfusion inflammation: a model for reaction to injury in cardiovascular disease. Cardiovasc. Res. 1994;28:1301–1311. doi: 10.1093/cvr/28.9.1301. [DOI] [PubMed] [Google Scholar]
- 37.Entman M.L., et al. Inflammation in the course of early myocardial ischemia. FASEB J. 1991;5:2529–2537. doi: 10.1096/fasebj.5.11.1868978. [DOI] [PubMed] [Google Scholar]
- 38.Vallance P., Collier J., Bhagat K. Infection, inflammation, and infarction: does acute endothelial dysfunction provide a link? Lancet. 1997;349:1391–1392. doi: 10.1016/S0140-6736(96)09424-X. [DOI] [PubMed] [Google Scholar]
- 39.Taqueti V.R., Mitchell R.N., Lichtman A.H. Protecting the pump: controlling myocardial inflammatory responses. Annu. Rev. Physiol. 2006;68:67–95. doi: 10.1146/annurev.physiol.68.040104.124611. [DOI] [PubMed] [Google Scholar]
- 40.Riedemann N.C., Ward P.A. Complement in ischemia reperfusion injury. Am. J. Pathol. 2003;162:363–367. doi: 10.1016/S0002-9440(10)63830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier L.S., Bers D.M. Role of Ca2+/calmodulin-dependent protein kinase (CaMK) in excitation-contraction coupling in the heart. Cardiovasc. Res. 2007;73:631–640. doi: 10.1016/j.cardiores.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Backs J., Song K., Bezprozvannaya S., Chang S., Olson E.N. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meffert M.K., Chang J.M., Wiltgen B.J., Fanselow M.S., Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat. Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 44.Jang M.K., et al. Ca2+/calmodulin-dependent protein kinase IV stimulates nuclear factor-kappa B transactivation via phosphorylation of the p65 subunit. J. Biol. Chem. 2001;276:20005–20010. doi: 10.1074/jbc.M010211200. [DOI] [PubMed] [Google Scholar]
- 45.Ishiguro K., et al. Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-kappaB activation. Mol. Cell. Biol. 2006;26:5497–5508. doi: 10.1128/MCB.02469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes K., Edin S., Antonsson A., Grundstrom T. Calmodulin-dependent kinase II mediates T cell receptor/CD3- and phorbol ester-induced activation of IkappaB kinase. J. Biol. Chem. 2001;276:36008–36013. doi: 10.1074/jbc.M106125200. [DOI] [PubMed] [Google Scholar]
- 47.Zwaka T.P., et al. Complement and dilated cardiomyopathy: a role of sublytic terminal complement complex-induced tumor necrosis factor-alpha synthesis in cardiac myocytes. Am. J. Pathol. 2002;161:449–457. doi: 10.1016/s0002-9440(10)64201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilgore K.S., et al. Sublytic concentrations of the membrane attack complex of complement induce endothelial interleukin-8 and monocyte chemoattractant protein-1 through nuclear factor-kappa B activation. Am. J. Pathol. 1997;150:2019–2031. [PMC free article] [PubMed] [Google Scholar]
- 49.Reiter Y., Ciobotariu A., Fishelson Z. Sublytic complement attack protects tumor cells from lytic doses of antibody and complement. Eur. J. Immunol. 1992;22:1207–1213. doi: 10.1002/eji.1830220515. [DOI] [PubMed] [Google Scholar]
- 50.Oyama J., et al. Reduced myocardial ischemia-reperfusion injury in toll-like receptor 4-deficient mice. Circulation. 2004;109:784–789. doi: 10.1161/01.CIR.0000112575.66565.84. [DOI] [PubMed] [Google Scholar]
- 51.Hua F., et al. Blocking the MyD88-dependent pathway protects the myocardium from ischemia/reperfusion injury in rat hearts. Biochem. Biophys. Res. Commun. 2005;338:1118–1125. doi: 10.1016/j.bbrc.2005.10.068. [DOI] [PubMed] [Google Scholar]
- 52.Gupta S., Purcell N.H., Lin A., Sen S. Activation of nuclear factor-kappaB is necessary for myotrophin-induced cardiac hypertrophy. J. Cell Biol. 2002;159:1019–1028. doi: 10.1083/jcb.200207149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H.-L., et al. Targeted cardiac overexpression of A20 improves left ventricular performance and reduces compensatory hypertrophy after myocardial infarction. Circulation. 2007;115:1885–1894. doi: 10.1161/CIRCULATIONAHA.106.656835. [DOI] [PubMed] [Google Scholar]
- 54.Misra A., et al. Nuclear factor-{kappa}B protects the adult cardiac myocyte against ischemia-induced apoptosis in a murine model of acute myocardial infarction. Circulation. 2003;108:3075–3078. doi: 10.1161/01.CIR.0000108929.93074.0B. [DOI] [PubMed] [Google Scholar]
- 55.Moss N.C., Stansfield W.E., Willis M.S., Tang R.-H., Selzman C.H. IKKbeta inhibition attenuates myocardial injury and dysfunction following acute ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H2248–H2253. doi: 10.1152/ajpheart.00776.2007. [DOI] [PubMed] [Google Scholar]
- 56.Zhang T., et al. The cardiac-specific nuclear delta B isoform of Ca2+/calmodulin-dependent protein kinase II induces hypertrophy and dilated cardiomyopathy associated with increased protein phosphatase 2A activity. J. Biol. Chem. 2002;277:1261–1267. doi: 10.1074/jbc.M108525200. [DOI] [PubMed] [Google Scholar]
- 57.Zhang T., Brown J.H. Role of Ca2+/calmodulin-dependent protein kinase II in cardiac hypertrophy and heart failure. Cardiovasc. Res. 2004;63:476–486. doi: 10.1016/j.cardiores.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 58.Li J., et al. Calmodulin kinase II inhibition shortens action potential duration by upregulation of K+ currents. Circ. Res. 2006;99:1092–1099. doi: 10.1161/01.RES.0000249369.71709.5c. [DOI] [PubMed] [Google Scholar]
- 59.Khoo M.S.C., et al. Death, cardiac dysfunction, and arrhythmias are increased by calmodulin kinase II in calcineurin cardiomyopathy. Circulation. 2006;114:1352–1359. doi: 10.1161/CIRCULATIONAHA.106.644583. [DOI] [PubMed] [Google Scholar]
- 60.Maier L.S., Bers D.M. Calcium, calmodulin, and calcium-calmodulin kinase II: heartbeat to heartbeat and beyond. J. Mol. Cell. Cardiol. 2002;34:919–939. doi: 10.1006/jmcc.2002.2038. [DOI] [PubMed] [Google Scholar]
- 61.Arni S., Keilbaugh S.A., Ostermeyer A.G., Brown D.A. Association of GAP-43 with detergent-resistant membranes requires two palmitoylated cysteine residues. J. Biol. Chem. 1998;273:28478–28485. doi: 10.1074/jbc.273.43.28478. [DOI] [PubMed] [Google Scholar]
- 62.Kapoun A.M., et al. B-type natriuretic peptide exerts broad functional opposition to transforming growth factor-beta in primary human cardiac fibroblasts: fibrosis, myofibroblast conversion, proliferation, and inflammation. Circ. Res. 2004;94:453–461. doi: 10.1161/01.RES.0000117070.86556.9F. [DOI] [PubMed] [Google Scholar]
- 63.Weiss R.M., Ohashi M., Miller J.D., Young S.G., Heistad D.D. Calcific aortic valve stenosis in old hypercholesterolemic mice. Circulation. 2006;114:2065–2069. doi: 10.1161/CIRCULATIONAHA.106.634139. [DOI] [PubMed] [Google Scholar]
- 64.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.