Abstract
Mutations in CCAAT/enhancer binding protein α (CEBPA) are seen in 5% to 14% of acute myeloid leukemia (AML) and have been associated with a favorable clinical outcome. Most AMLs with CEBPA mutations simultaneously carry 2 mutations (CEBPAdouble-mut), usually biallelic, whereas single heterozygous mutations (CEBPAsingle-mut) are less frequently seen. Using denaturing high-performance liquid chromatography and nucleotide sequencing, we identified among a cohort of 598 newly diagnosed AMLs a subset of 41 CEBPA mutant cases (28 CEBPAdouble-mut and 13 CEBPAsingle-mut cases). CEBPAdouble-mut associated with a unique gene expression profile as well as favorable overall and event-free survival, retained in multivariable analysis that included cytogenetic risk, FLT3-ITD and NPM1 mutation, white blood cell count, and age. In contrast, CEBPAsingle-mut AMLs did not express a discriminating signature and could not be distinguished from wild-type cases as regards clinical outcome. These results demonstrate significant underlying heterogeneity within CEBPA mutation-positive AML with prognostic relevance.
Introduction
Mutations in the transcription factor CCAAT/enhancer binding protein α (CEBPA) are found in 5% to 14% of acute myeloid leukemia (AML).1–9 CEBPA mutations have been associated with a relatively favorable outcome and have therefore gained interest as a prognostic marker.4–6,10 Although variable sequence variations have been described, 2 prototypical classes of mutations are most frequent. N-terminal mutations are located between the major translational start codon and a second ATG in the same open reading frame. These mutations introduce a premature stop of translation of the p42 CEBPA protein while preserving translation of a p30 isoform that has been reported to inhibit the function of full-length protein.2 Mutations in the C-terminal basic leucine zipper (bZIP) region, in contrast, are in-frame and may impair DNA binding and/or homodimerization and heterodimerization.8 The remaining mutations are mostly found between the N-terminus and the bZIP region.11
Most CEBPA mutant AMLs exhibit 2 mutations, which most frequently involves a combination of an N-terminal and a bZIP gene mutation.7,8,11,12 In AMLs with 2 CEBPA mutations, the mutations are typically on different alleles.11 Hence, in these cases, no wild-type CEBPA protein is expressed. A similar condition is found in AMLs carrying a homozygous CEBPA mutation.13 However, there are also AMLs that only show one single heterozygous mutation and thus retain expression of a wild-type allele.7,11,12
To obtain better insight into the distribution of the various types of CEBPA mutations in de novo adult AML and their impact on clinical outcome, we examined a cohort of 598 cases. After denaturing high-performance liquid chromatography (dHPLC) and nucleotide sequencing, we distinguished cases with 2 different mutations or one homozygous mutation (further referred to as double mutations; CEBPAdouble-mut) as well as cases with only one single heterozygous mutation (CEBPAsingle-mut). Genome-wide gene expression profiling revealed that CEBPAdouble-mut AMLs expressed a highly characteristic signature, whereas CEBPAsingle-mut cases did not. In addition, favorable prognosis appeared uniquely associated with CEBPAdouble-mut AML.
Methods
AML samples, mRNA isolation, dHPLC analysis, and nucleotide sequencing
Bone marrow aspirates or peripheral blood samples of 598 cases of de novo AML were collected, blast cells were purified, and mRNA was isolated as reported.14 The entire CEBPA coding region was investigated by dHPLC and selected regions also by agarose gel analysis and/or nucleotide sequencing. For details on patient characteristics and experimental procedures, see Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). All studies were approved by the Erasmus University Medical Center Institutional Review Board (Rotterdam, The Netherlands), and patients' informed consent was obtained in accordance with the Declaration of Helsinki.
Statistical analysis
Survival was estimated according to the method of Kaplan and Meier. The log rank test was used to assess statistical significance. Multivariable analysis was performed using Cox proportional hazards models. Definitions of outcome parameters and cytogenetic risk groups have been described.15 Further details are given in Document S1. P values less than .05 were considered statistically significant.
Gene expression profiling analysis
Gene expression profiles were obtained using Affymetrix (Santa Clara, CA) HGU133Plus2.0 GeneChips. Details on data processing and analysis are given in Document S1.
Results and discussion
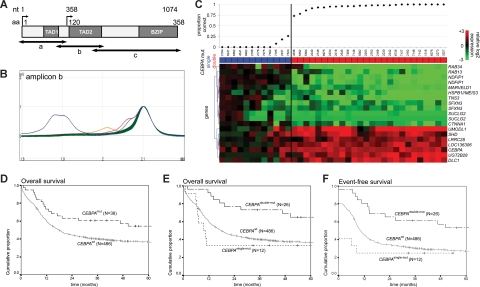
In a cohort of 598 cases of adult de novo AML, we identified 65 cases with an aberrant profile in at least 1 of the 3 investigated amplicons of the CEBPA coding sequence (Figure 1A,B). The presence of a CEBPA sequence variation was confirmed by nucleotide sequencing. Cases that only carried an insertion polymorphism16,17 or variation(s) that did not lead to amino acid changes were considered wild-type. Two additional specimens were not considered in further analysis because they carried in-frame variations of unknown significance in the N-terminus (Table S1). As a result, 41 of 598 unambiguous CEBPAmut AML cases (6.9%) were considered. These included 13 CEBPAsingle-mut cases and 28 CEBPAdouble-mut cases. Four of the CEBPAdouble-mut cases carried homozygous mutations, whereas the remaining 24 cases showed 2 heterozygous mutations (Table S1). Additional screening of the remaining AML cases, using a combination of agarose gel analysis and nucleotide sequencing as described,6 did not reveal mutations that had been missed by dHPLC.
Figure 1.
Schematic overview of dHPLC analysis, gene expression profiling analysis, and survival estimates. (A) Schematic representation of the CEBPA gene and location of amplicons a, b, and c for polymerase chain reaction, used for dHPLC analysis. Functional regions are depicted: 2 transactivation domains (TAD1 and TAD2) in the N-terminal part, and the bZIP region in the C-terminal part. Nucleotide (nt) position is indicated relative to the main translation start site. Amino acid (aa) numbering and the alternative translation start site at position nt 358 (aa 120) are also depicted. (B) Representative profiles of dHPLC analysis of 1 of the 3 investigated fragments (ie, amplicons b) in a random selection of approximately 90 samples. Heteroduplexes (various colors) are released earlier than homoduplexes (green) and can therefore be recognized as distinct peaks. Time is depicted on the x-axis, and absorbance on the y-axis. (C) A gene expression prediction signature for CEBPAmut AML (irrespective of single- or double-mutant status) was derived in a dataset of 524 AMLs, including 38 CEBPAmut cases. Prediction accuracy for each of the 38 CEBPAmut cases was estimated using repeated 10-fold cross-validation, as detailed in supplemental data. The proportion of correct predictions for the selected 38 CEBPAmut specimens is indicated (top panel). Mutation status is color coded (CEBPAsingle-mut, blue; CEBPAdouble-mut, red). The heatmap in the bottom panel depicts the 19 probe sets in the resulting CEBPAmut gene expression classifier (Table S2, probe set information). Intensity values (log2) were mean centered over the cohort of 524 AML cases; and for visualization purposes, the genes were hierarchically clustered (Euclidian distance, average linkage). Cells represent relative log 2 expression values and have been color coded on a scale ranging from bright green (−3) to bright red (+3), with black indicating no change relative to the mean. (D) Kaplan-Meier estimates of overall survival among CEBPAmut and CEBPAwt AML (log rank test, P = .027). (E) Overall survival among CEBPAdouble-mut versus CEBPAwt AML (P = .004) and versus CEBPAsingle-mut AML (P = .005; pooled P = .012). (F) Event-free survival (EFS) among CEBPAdouble-mut and CEBPAwt AML (P = .005) and versus CEBPAsingle-mut AML (P = .004; pooled P = .008). The cumulative proportion of survival at the intercept (the point where a line crosses the y-axis) reflects the proportion of patients reaching complete remission. Analyses similar to those depicted in panels D-F were performed after splitting the group of CEBPAwt AMLs into those with favorable cytogenetics and those with other cytogenetics. These additional analyses can be found in Figure S4.
To investigate whether CEBPA mutations related to gene expression, we examined genome-wide gene expression data of 524 AML cases, which included 26 CEBPAdouble-mut and 12 CEBPAsingle-mut cases. Clinical and molecular characteristics of the AML cases are reported in Tables S4 and S5. Using prediction analysis for microarrays,18 according to a supervised approach, we derived a 19-probe set signature predictive of CEBPA mutations (Figure 1C). This classifier showed a high specificity (99%) but a limited sensitivity (67%) in cross-validation, indicating a limited ability to recognize all CEBPAmut specimens. Strikingly, misclassification was almost entirely the result of CEBPAsingle-mut cases, whereas CEBPAdouble-mut AMLs were predicted with an accuracy that was near perfect (Figures 1C, S1). In line with this, we were able to derive a specific 21-probe set classifier for CEBPAdouble-mut AMLs within the entire AML cohort with a cross-validated sensitivity of 100% (specificity, 98%; Table S3). In further support, unsupervised analysis of the expression data derived from the CEBPAmut subset indicated an underlying variability in gene expression that correlated with either double or single mutation status (Figure S2).
We next assessed how these differences between CEBPAdouble-mut and CEBPAsingle-mut related to clinical outcome. In line with previous data, overall survival and event-free survival were significantly better for CEBPAmut cases compared with cases with wild-type CEBPA (CEBPAwt) (Figure 1D; and data not shown). Separate analyses for the CEBPAdouble-mut and CEBPAsingle-mut subgroups, however, revealed a favorable outcome that was specific for CEBPAdouble-mut cases. We failed to find a favorable prognostic effect in relation to the CEBPAsingle-mut cases. Indeed, CEBPAsingle-mut AMLs showed a significantly worse outcome than CEBPAdouble-mut cases, including a poor rate of complete remission (Figure 1E,F). These findings were also apparent in multivariable analysis (Table 1). When only patients younger than 60 years or only patients with normal cytogenetics were considered, similar results were found, although in the latter subgroup with smaller numbers only the pairwise comparison for overall survival between CEBPAdouble-mut and CEBPAsingle-mut reached statistical significance (Figure S3; Table S6).
Table 1.
Multivariable analysis of CEBPAdouble-mut and CEBPAsingle-mut as prognostic markers for overall and event-free survival
| Variable | Overall survival |
Event-free survival |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| CEBPAsingle-mut* | 1.18 (0.58-2.40) | .65 | 1.61 (0.82-3.17) | .16 |
| CEBPAdouble-mut* | 0.32 (0.17-0.61) | < .001 | 0.35 (0.20-0.62) | < .001 |
| Intermediate† | 2.21 (1.52-3.22) | < .001 | 2.05 (1.46-2.87) | < .001 |
| Poor† | 3.35 (2.27-4.94) | < .001 | 2.85 (2.00-4.06) | < .001 |
| Age, decades | 1.17 (1.08-1.28) | < .001 | 1.10 (1.02-1.19) | .014 |
| WBC‡ | 1.33 (1.05-1.68) | .019 | 1.29 (1.03-1.62) | .025 |
| FLT3 ITD§ | 1.56 (1.20-2.03) | < .001 | 1.46 (1.14-1.89) | .003 |
| NPM1 mutation∥ | 0.55 (0.41-0.74) | < .001 | 0.51 (0.39-0.67) | < .001 |
Complete data for multivariable analysis were available for 511 cases.
HR indicates hazard ratio; CI, confidence interval; WBC, white blood cell count; FLT3, fms-related tyrosine kinase 3; ITD, internal tandem duplication; and NPM1, nucleophosmin.
CEBPA status versus CEBPAwt.
Cytogenetic risk versus cytogenetic good risk.
WBC greater than 20 × 109/L versus less than 20 × 109/L.
FLT3 ITD versus no FLT3 ITD.
NPMI mutation versus no NPMI mutation.
Based on our previous analyses6 and on the literature,11 it is probable that, in the majority of the CEBPAdouble-mut AML studied, both CEBPA alleles were affected. A plausible hypothesis is therefore that absence of wild-type CEBPA mRNA is directly involved in the CEBPAdouble-mut gene expression profile. This may be further supported by our previous and current observations that indicate a high degree of similarity between the profiles of CEBPAdouble-mut AML and a specific subgroup of leukemias characterized by epigenetic CEBPA silencing (Figure S1).19 It is possible that analysis of larger patient series will lead to further refinement of this subclassification, for instance, based on the location of the mutations. For example, our data indicated a tendency of CEBPAsingle-mut cases with mutations in the bZIP region to be potentially less distinct from the CEBPAdouble-mut AMLs (case nos. 7185, 7324, and 2237; Figures 1C, S2). Of note, a subset of the CEBPAmut AMLs studied here was included in the cohort of 285 cases of AML that we previously investigated by gene expression profiling.14 In that study, all CEBPAdouble-mut AMLs were found in 2 particular clusters, whereas CEBPAsingle-mut AMLs did not specifically aggregate.14,19
Studies to date have associated CEBPA mutations with outcome4–6,9 but have not applied subdivisions into single and double mutants. It is unclear why CEBPAdouble-mut AMLs would have a better outcome than those with a single heterozygous mutation. One explanation could be that a single-mutant CEBPA allele is not sufficient for leukemogenesis and requires cooperating mutations, which may be in CEBPA itself or in other genes. Of note, recent data indicate that germline CEBPA mutations predispose to AML, and the acquisition of a second, somatic CEBPA mutation may then contribute to AML development.20 Indeed, we found a tendency toward more FLT3-ITD, FLT3-TKD, and NPM1 mutations in CEBPAsingle-mut compared with CEBPAdouble-mut cases (Table S5). Yet unknown abnormalities may associate with CEBPAsingle-mut AML as well and predispose to inferior outcome. It is however evident that these findings and their clinical significance warrant confirmation in independent cohorts of AML.
In conclusion, the data presented here indicate that CEBPAmut AML should at least be distinguished according to the presence of CEBPAdouble-mut and CEBPAsingle-mut. Screening using dHPLC, followed by nucleotide sequencing, appears useful for rapidly identifying mutant cases. In addition, gene expression-based classification, for instance, using the classifiers described here, enables the accurate identification of CEBPAdouble-mut AML cases.
Supplementary Material
Acknowledgments
The authors thank Gert J. Ossenkoppele (Free University Medical Center, Amsterdam, The Netherlands), Jaap Jan Zwaginga (Sanquin, The Netherlands), Edo Vellenga (University Hospital, Groningen, The Netherlands), Leo F. Verdonck (University Hospital, Utrecht, The Netherlands), Gregor Verhoef (Hospital Gasthuisberg, Leuven, Belgium), and Matthias Theobald (Johannes Gutenberg-University Hospital, Mainz, Germany) who provided AML samples; Sonja van der Poel for help with dHPLC analysis; and our colleagues from the Bone Marrow Transplantation Group and Molecular Diagnostics Group in the Department of Hematology of Erasmus University Medical Center for storage of samples and molecular analysis, respectively.
This work was supported by grants from the Dutch Cancer Society (Amsterdam, The Netherlands) Koningin Wilhelmina Fonds and the National Institutes of Health (Bethesda, MD).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.J.W. and C.A.J.E.-V. performed research; B.J.W., C.A.J.E.-V., W.L.J.v.P., P.J.M.V., and R.D. analyzed data; and B.J.W., B.L., P.J.M.V., and R.D. wrote the paper.
Conflict-of-interest disclosure: B.L., P.J.M.V., and R.D. have declared ownership interests in Skyline, a spin-off company of Erasmus University Medical Center (Erasmus MC), held in a Special Purpose Foundation of Erasmus MC. The remaining authors declare no competing financial interests.
Correspondence: Ruud Delwel, Erasmus University Medical Center, Department of Hematology, Room Ee1342, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: h.delwel@erasmusmc.nl.
References
- 1.Snaddon J, Smith ML, Neat M, et al. Mutations of CEBPA in acute myeloid leukemia FAB types M1 and M2. Genes Chromosomes Cancer. 2003;37:72–78. doi: 10.1002/gcc.10185. [DOI] [PubMed] [Google Scholar]
- 2.Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 3.Gombart AF, Hofmann WK, Kawano S, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 4.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 5.Frohling S, Schlenk RF, Stolze I, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 6.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Meijer J, et al. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4:31–40. doi: 10.1038/sj.thj.6200216. [DOI] [PubMed] [Google Scholar]
- 7.Mueller BU, Pabst T. C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr Opin Hematol. 2006;13:7–14. doi: 10.1097/01.moh.0000190110.08156.96. [DOI] [PubMed] [Google Scholar]
- 8.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 9.Bienz M, Ludwig M, Leibundgut EO, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 10.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 11.Pabst T, Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007;26:6829–6837. doi: 10.1038/sj.onc.1210765. [DOI] [PubMed] [Google Scholar]
- 12.Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 13.Wouters BJ, Sanders MA, Lugthart S, et al. Segmental uniparental disomy as a recurrent mechanism for homozygous CEBPA mutations in acute myeloid leukemia. Leukemia. 2007;21:2382–2384. doi: 10.1038/sj.leu.2404795. [DOI] [PubMed] [Google Scholar]
- 14.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 15.Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 16.Wouters BJ, Louwers I, Valk PJ, Lowenberg B, Delwel R. A recurrent in-frame insertion in a CEBPA transactivation domain is a polymorphism rather than a mutation that does not affect gene expression profiling-based clustering of AML. Blood. 2007;109:389–390. doi: 10.1182/blood-2006-08-042325. [DOI] [PubMed] [Google Scholar]
- 17.Lin LI, Chen CY, Lin DT, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–1379. doi: 10.1158/1078-0432.CCR-04-1816. [DOI] [PubMed] [Google Scholar]
- 18.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wouters BJ, Jorda MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pabst T, Eyholzer M, Haefliger S, Schardt J, Mueller BU. Somatic CEBPA mutations are a frequent second event in families with germline CEBPA mutations and familial acute myeloid leukemia. J Clin Oncol. 2008;26:5088–5093. doi: 10.1200/JCO.2008.16.5563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.