Abstract
Estrogen hormones play critical roles in the regulation of many tissue functions. The effects of estrogens are primarily mediated by the estrogen receptors (ER) α and β. ERs are ligand-activated transcription factors that regulate a complex array of genomic events that orchestrate cellular growth, differentiation and death. Although many factors contribute to their etiology, estrogens are thought to be the primary agents for the development and/or progression of target tissue malignancies. Many of the current modalities for the treatment of estrogen target tissue malignancies are based on agents with diverse pharmacology that alter or prevent ER functions by acting as estrogen competitors. Although these compounds have been successfully used in clinical settings, the efficacy of treatment shows variability. An increasing body of evidence implicates ERα polymorphisms as one of the contributory factors for differential responses to estrogen competitors. This review aims to highlight the recent findings on polymorphisms of the lately identified ERβ in order to provide a functional perspective with potential pharmacogenomic implications.
Keywords: Estrogen, estrogen receptor β, estrogen signaling, polymorphisms, pharmacogenetics
A. INTRODUCTION
Estrogen hormones, particularly the main circulating estrogen, 17β-estradiol (E2), are involved in many physiological processes concerning a wide range of tissues that include the reproductive organs, breast, skeletal, nervous, cardiovascular, digestive and immune systems [Deroo, B.J. and Korach, K.S. 2006; Heldring, N. et al. 2007]. Estrogens are also implicated in the ontogeny of target tissue malignancies, including cancers of reproductive organs, breast, and colorectum as well as osteoporosis, neurodegenerative diseases, cardiovascular disease, diabetes and obesity [Deroo, B.J. and Korach, K.S. 2006; Heldring, N. et al. 2007]. Although the etiology of estrogen target tissue malignancies is multifactorial in which a polygenic background is modulated by the integrated effects of hormonal, environmental and nutritional factors, aberrant estrogen signaling appears to be a basis for many of these diseases. The estrogen information is primarily conveyed by the members of a family of nuclear transcription factors, estrogen receptor (ER) α and β [Deroo, B.J. and Korach, K.S. 2006; Heldring, N. et al. 2007; Huang, J. et al. 2005]. Consequently, ERs are also targets for many therapeutic interventions [Deroo, B.J. and Korach, K.S. 2006; Heldring, N. et al. 2007; Huang, J. et al. 2005].
Despite major advances in counteractive measures, there is a significant heterogeneity in the efficacy and toxicity of treatment [McLeod, H.L. et al. 2003; Roden, D.M. et al. 2006]. While many clinical variables are potential modifiers of responses to interventions, genetic changes in ERs reflected as altered gene expression and/or protein structure could have a great impact on treatment outcome [Gennari, L. et al. 2005; Herrington, D.M. and Klein, K.P. 2001].
Pharmacogenomics is the study of how inheritance affects the individual response to therapeutic interventions [McLeod, H.L. et al. 2003; Watters, J.W. and McLeod, H.L. 2003]. Pharmacogenomics is proposed to have the potential to personalize medical therapies with greater efficacy and safety due to genetic testing for predisposition to disease or response to therapeutic intervention by combining pharmaceutical sciences with annotated knowledge of genes, proteins, and single nucleotide polymorphisms [McLeod, H.L. et al. 2003; Watters, J.W. and McLeod, H.L. 2003].
A better understanding of the mechanism of E2 action is critical for the development of effective therapeutic interventions with improved safety assessments. In this communication, we aimed to highlight recent findings on polymorphisms of the recently discovered ERβ in order to provide a functional perspective on ERβ-mediated E2 signaling with potential pharmacogenetic implications.
B. STRUCTURE AND FUNCTION OF ERβ
The mechanism of ERα action is comparatively better known than ERβ. To fully appreciate the potential contribution of ERβ variants in the pharmacogenetics of E2 signaling, we will initially review the structure-function of the wild-type ERβ in comparison with ERα.
B.1. The Expression of the ERβ Gene
ERα and ERβ are distinct gene products [Nilsson, S. and Gustafsson, J.A. 2002b]. The chromosomal location of the ERα gene is 6q25.1 while the ERβ gene is localized on chromosome 14q23.2 [Nilsson, S. and Gustafsson, J.A. 2002b]. ERs are expressed in the same tissue as well as in different tissues at varying levels [Nilsson, S. et al. 2001]. ERα is the dominant specie expressed in uterus, liver, adipose, skeletal muscle, pituitary and hypothalamus, whereas ERβ is the major form in ovary, testis and prostate, and other regions of the brain including the limbic system, cerebellum and cerebral cortex [Deroo, B.J. and Korach, K.S. 2006; Heldring, N. et al. 2007]. ERα and ERβ are co-expressed in breast tissue, the urogenital tract, bone and the cardiovascular system within the same cell-type as well as in different cell populations [Deroo, B.J. and Korach, K.S. 2006; Heldring, N. et al. 2007].
Genomic organization of the ERβ gene reveals that the gene spans 254 kb, while eight translated exons (1 through 8) extend more than 140 kb [Sand, P. et al. 2002] as depicted in Fig. (1). The 5’ flanking untranslated region of the ERβ gene is large and contains various regulatory elements [Li, L.C. et al. 2000; Zhang, X. et al. 2007; Zhu, X. et al. 2004]. This suggests a versatile utilization of regulatory signals and tissue-specific expression [Hirata, S. et al. 2001, 2003; Sand, P. et al. 2002]. It appears that differential splicing of 5’ untranslated regions of the ERβ gene generates at least seven ERβ transcripts with various sizes of untranslated 5’ exons (0K, 0X1, 0X2, 0X3, 0X4, 0X5 and 0N) [Hirata, S. et al. 2001, 2003; Sand, P. et al. 2002]. Additionally, an “intronic” exon, exon M, is reported between exons 4 and 5 [Shoda, T. et al. 2002]. One promoter region immediately upstream of the exon 0N contains putative binding sequences for transcription factors vERB/RARF/MYC-MAX, C-REl and SREBP-1/N-Myc/Amt/USF and a TATA box sequence. The sequences within the exon 0N also act as a promoter containing binding sites of GATA, SP1, AP-1, and AP-2, Oct-1, and AP-4/MyoD proteins [Li, L.C. et al. 2000; Zhang, X. et al. 2007].
Fig. (1).
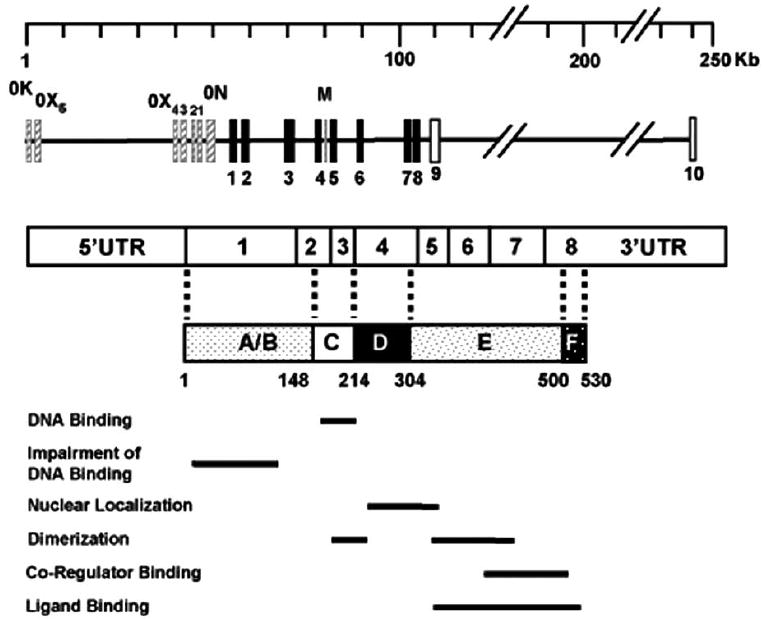
Genomic organization of the ERβ gene. Exons encoding the ERβ protein are indicated with black boxes. Exon M is indicated as a solid gray bar. The mRNA of the ERβ is schematized with the untranslated 5’ and 3’ regions together with encoding exons whose boundaries in the ERβ protein are shown with broken lines. Solid bars indicate functional features of the domains. In the ERβ protein, A/B indicates the amino-terminus. The A/B domain of ERβ impairs the ability of the receptor to interact with ERE. C is the DNA binding domain responsible for binding to ERE sequences. D region represents the hinge domain and contains a nuclear localization signal. The multifunctional carboxyl-terminus E and F domains are responsible for the ligand binding, dimerization and co-regulatory protein interactions, consequently for the ligand-dependent transactivation function of the receptor.
The promoter regions of ERβ are also GC-rich with a GC content of 65% [Sand, P. et al. 2002; Zhang, X. et al. 2007]. The abundance of GC-rich sites implies susceptibility to hypermethylation of the promoter regions, an event that is associated with altered gene expression and increased risk of disease. Studies suggest that the methylation status of an ERβ promoter region could be an important mechanism for ERβ gene silencing in breast cancer [Zhao, C. et al. 2003]. It was found that the expression of ERβ mRNA in breast tumors is inversely associated with the degree of methylation of promoter 0N, but not promoter 0K, which is unmethylated in normal breast tissue [Zhao, C. et al. 2003].
Moreover, epigenetic regulation of ERβ gene expression is shown to be a reversible and tumor stage-specific process in prostate cancer [Zhu, X. et al. 2004]. Similarly, studies suggest that a decrease in the ERβ gene expression may be associated with breast tumorigenesis, and that DNA methylation is an important mechanism for ERβ gene silencing in breast cancer [Zhao, C. et al. 2003]. Epigenetic dysregulation of the ERβ gene expression is also suggested to contribute to the development of atherosclerosis and vascular aging [Kim, J. et al. 2007]. This, in turn, raises the intriguing possibility that the lack of efficacy of estrogen as a protector of the cardiovascular system in particular [Manson, J.E. et al. 2003], and other target tissue in general, in women undergoing to hormone replacement therapy (HRT) may be related in part to age-related epigenetic silencing of estrogen receptors in critical cells, which would then be non-responsive to HRT [Kim, J. et al. 2007].
B.2. The Protein Structure of ERβ
ERs, as with other transcription factors, are modular in nature such that isolated structural domains display subsets of the functional activities of the parent receptor [Hall, J.M. et al. 2001; Nilsson, S. and Gustafsson, J.A. 2002a; Parker, M.G. 1998]. ERs share varying degrees of domain-specific structural/functional similarities. The amino-terminal A/B domains of ERs differ in both length and amino acid sequence, exhibiting only 30% amino acid identity. The central region of ER is the DNA binding domain (DBD) and is referred to as the C domain. ERs share a high amino-acid homology (97%) in their DBD domains. The hinge, or D, domain contains a nuclear localization function linking the DBD to the multi-functional carboxyl-terminal E/F region, or ligand binding domain (LBD). ERs share 56% amino acid homology at their E/F domains, which mediate ligand binding, dimerization, and the ligand-dependent transactivation function (AF-2).
Exon 1 encodes the A/B region of ERα and ERβ. Exons 2 and 3 encode part of the C region. Exon 4 encodes the remainder of the C region, all of D the domain, and the amino-terminus of the LBD, E. Exons 4–8 encode the E region. The remainder of exon 8 encodes region F [Green, S. et al. 1986a; Green, S. et al. 1986b; Green, S. et al. 1986c; Kuiper, G.G. and Gustafsson, J.A. 1997].
ERs bind to specific DNA sequences, estrogen responsive elements (EREs) [Klinge, C.M. 2001] mediated by the binding of the two-zinc finger motifs of each DBD of the dimer [Schwabe, J.W. et al. 1993; Schwabe, J.W. et al. 1990]. The nearly identical amino acid sequence of the DBD of ERβ to that of ERα allows the receptor to bind to the same spectrum of DNA sequences with similar affinities utilizing the same nucleotides [Li, X. et al. 2004a; Loven, M.A. et al. 2001; Yi, P. et al. 2002b].
Despite a 56% amino-acid identity between ERs, the LBDs display similar tertiary and quaternary architecture [Brzozowski, A.M. et al. 1997; Pike, A.C. et al. 1999]. These similar structural features are responsible for comparable binding affinities of both ER subtypes to E2 [Kuiper, G.G. et al. 1997]. The LBD displays a fold with 12 α-helices (numbered H1-H12). The binding of E2 to ERs is accompanied by a major structural reorganization of the LBD. E2 binding realigns the carboxyl terminal H12 across the LBD fold and buries the hormone within the cavity. This H12 repositioning on the hormone-binding cavity also creates a shallow hydrophobic groove that serves as the docking site for coregulator proteins [Pike, A.C. 2006].
In addition to estrogen, ERs also bind compounds that act as estrogen competitors [Jensen, E.V. and Jordan, V.C. 2003; Jordan, V.C. and Morrow, M. 1999; McDonnell, D.P. 1999]. Differences in ER conformations in the presence of different ligands constitute the structural basis for antagonism. Among compounds, tamoxifen, toremifene and raloxifene are referred to as selective estrogen receptor modulators, SERMs. SERMs can function as agonists or antagonists depending on ER subtypes, cells and tissues in which they operate [McDonnell, D.P. 1999; Wakeling, A.E. 2000]. Tamoxifen and raloxifene function as antagonists in breast tissue. While tamoxifen acts as an agonist in the uterus, bone and cardiovascular system, raloxifen functions as a pure antagonist in the uterus but as an agonist in bone. Binding of SERMs to ERs sterically interferes with H12 positioning in that H12 interacts with a hydrophobic groove distinctly, which partially buries residues in the groove necessary for AF-2 activity [Brzozowski, A.M. et al. 1997; Pike, A.C. et al. 1999]. This positioning alters co-factor recruitment and subsequent transcription activation [Brzozowski, A.M. et al. 1997; Pike, A.C. et al. 1999]. Interestingly, SERMs act as agonists only through ERα [McDonnell, D.P. 1999; Wakeling, A.E. 2000].
Compounds that act as pure antagonists for both ERα and ERβ include steroidal molecules ICI 164,384 and ICI 182,780, which are derivatives of estrogen and are devoid of any estrogenic activity in most experimental systems tested [McDonnell, D.P. 1999; Wakeling, A.E. et al. 2001]. The distinct pharmacological properties of these antiestrogens allow treatment regimens to be targeted to a specific tissue of interest to minimize unintended development of other tissue malignancies.
Although both receptors bind to estrogens, SERMs, and ICI similarly, certain estrogenic compounds exhibit ER subtype selectivity. For example, a plant-derived non-steroidal phytoestrogen, genistein, has a significantly higher binding affinity to ERβ than ERα [Kuiper, G.G. et al. 1997; Kuiper, G.G. et al. 1998]. Moreover, some tetrahydrochrysene compounds display agonistic properties when they bind to ERα but they are antagonists for ERβ [Sun, J. et al. 1999]. These observations also suggest differences in the function of LBDs depending upon the nature of the ER ligand. It appears that differences in the amino-acid sequences are critical for the formation of the ligand-binding pocket. Distinct residues outside the ligand-binding pocket make significant contributions to the subtype-specific positioning of the ligand [Nettles, K.W. et al. 2007]. Differences in secondary structural interactions, both in the hydrophobic core of each receptor and in the hydrogen bond networks on the surface, alter the shape of the ligand-binding pocket between the ER subtypes and the transcriptional output of both receptors [Nettles, K.W. et al. 2007].
C. MECHANISM OF ER ACTION
The integration of E2 effects mediated by ERs at various cellular locations, as depicted in Fig. (2), is critical for the regulation of responsive gene expression involved in cellular proliferation, motility and death [Chang, E.C. et al. 2006; Frasor, J. et al. 2003; Licznar, A. et al. 2003; Moggs, J.G. et al. 2005; Soulez, M. and Parker, M.G. 2001]. ERs are synthesized in the cytoplasm, then dimerize and translocate to the nucleus independent of E2 [Bai, Y. and Giguere, V. 2003]. Varying fractions of the ER population are also partitioned to the perimembrane and cytoplasm [Pedram, A. et al. 2002; Pedram, A. et al. 2006; Yang, S.H. et al. 2004]. A major structural reorganization of the LBD upon binding to E2 converts the inactive ERs to functionally active forms by generating surfaces that support protein-protein interactions [Brzozowski, A.M. et al. 1997; Pike, A.C. et al. 1999].
Fig. (2).
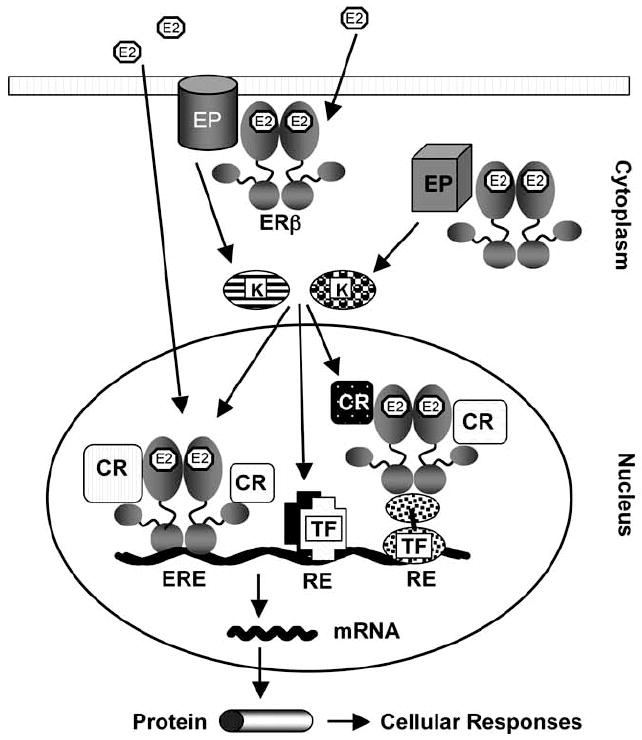
A model for E2-ERβ action. ERβ is localized at the perimemebrane/cytoplasm, but is primarily located in the nucleus. The binding of 17β-estradiol (E2), as the main circulating estrogen hormone, to ERβ induces conformational changes in the receptor that facilitate protein-protein interactions. The perimembrane/cytoplasmic E2-ERβ complex interacts with effector proteins (EP) to initiate signal transduction cascades that result in the activation of various kinases (K). Kinases then modulate the activities of transcription factors (TF) bound to cognate response elements (RE) to mediate transcription. Upon binding to E2 the nuclear ERβ interacts with estrogen responsive elements (EREs) on DNA, and recruits co-regulatory proteins to alter the expression of the estrogen responsive genes (the ERE-dependent E2-ERβ signaling pathway). The nuclear E2-ERβ also regulates the expression of the responsive gene transcription by functional interactions with transcription factors bound to cognate response elements (the ERE-independent E2-ERβ signaling pathway). Activated kinases could also influence the activity of the E2-ERβ complex through post-translational modifications of the complex or associated co-regulatory proteins. The expression of the responsive genes results in the cellular responses.
C.1. Membrane and Cytoplasmic ERs
The observations that E2 can induce rapid cellular responses, ranging in seconds to minutes, has led to the recognition of plasma membrane derived E2 signaling. Although these rapid effects of E2 are attributed to binding to perimembrane ERs, the structural nature, and therefore functional aspects of, perimembrane ERs is yet unclear. Studies suggest that perimembrane ERs are the products of the same gene encoding the nuclear ERs as well as different gene products that encode pharmacologically different proteins [Joe, I. and Ramirez, V.D. 2001; Nadal, A. et al. 2001; Razandi, M. et al. 1999]. Nevertheless, the E2 bound-membrane ERs associate with a variety of proximal signaling molecules that include G proteins [Razandi, M. et al. 1999]. This results in the activation of signal transduction cascades that lead to the intracellular mobilization of Ca2+, activation of protein kinase C, receptor kinase 2 and cAMP in a cell-type dependent manner. Perimembrane E2-ER complexes also initiate a cascade of protein–protein interactions involving Shc, GRB-2 and SOS to activate MAPK signaling [Ansonoff, M.A. and Etgen, A.M. 1998; Castoria, G. et al. 1999; Castoria, G. et al. 2001; Kousteni, S. et al. 2001; Morey, A.K. et al. 1997; Razandi, M. et al. 2000; Shevde, N.K. et al. 2000].
Moreover, recent studies also suggest a coupling between E2-ER and cytosolic signaling pathways. A fraction of E2-bound ERα in the cytoplasm can associate with the regulatory subunit of phosphatidylinositol-3-OH kinase. This interaction results in the activation of the AKT serine/threonine kinase [Simoncini, T. et al. 2000]. Interestingly, AKT can also directly phosphorylate ERα, resulting in enhanced potency of the receptor to induce transcription [Campbell, R.A. et al. 2001].
Perimembrane ERs convey E2 signaling to proximal molecules through the LBD with similar mechanisms and efficiencies [Levin, E.R. 2002; Razandi, M. et al. 2003].
C.2. Nuclear ER Signaling
The primary E2 signaling events take place in the nucleus. The interaction of E2-ERs with EREs [Klinge, C.M. 2001], constitutes one canonical signaling pathway, referred to as the ERE-dependent E2 signaling pathway [Hall, J.M. et al. 2001; Huang, J. et al. 2005; Nilsson, S. and Gustafsson, J.A. 2002b]. E2-ERs also regulate gene transcription through functional interaction with a transfactor(s) bound to cognate responsive elements on the regulatory regions of estrogen responsive genes. This E2 signaling is called the ERE-independent E2-ER signaling pathway [Hall, J.M. et al. 2001; Huang, J. et al. 2005; Nilsson, S. and Gustafsson, J.A. 2002b].
C.2.1. ERE-Dependent E2-ERα Signaling
EREs are permutations of a palindromic sequence separated by three non-specific nucleotides idealized with the so-called consensus ERE, 5’-GGTCAnnnTGACC-3’ [Klinge, C.M. 2001]. The binding of ERα to an ERE is the initial step in the regulation of estrogen-responsive gene transcription. The transcription of target genes is a dynamic event and is modulated cyclically [Chen, H.W. et al. 1999; Kraus, W.L. and Kadonaga, J.T. 1998; Kraus, W.L. and Wong, J. 2002; Shang, Y. et al. 2000].
Studies indicated that the binding of E2-ERα to the ERE initiates a series of interdependent events that result in an extended periodicity of cyclic promoter engagement in compared to the unliganded ERα [Metivier, R. et al. 2004; Metivier, R. et al. 2003; Reid, G. et al. 2003]. Following an initial transcriptionally silent cycle, E2 bound ERα through the amino and the carboxyl termini recruits various multi-subunit coactivator complexes, enzymes of the ubiquitin-proteasome pathway, and the basal transcription machinery together with Polymerase II (Pol II) to initiate transcription [Metivier, R. et al. 2004; Metivier, R. et al. 2003; Reid, G. et al. 2003]. An effective recruitment of coregulators by both the AF1 and AF2 domains of ERα appears to be necessary to form a stable platform for subsequent ordered and combinatorial recruitment of complexes for transcription.
At the end of a transcriptionally productive cycle, histone deacetylase enzyme complexes are recruited by the activated Pol II in association with chromatin remodelers to modify local chromatin structure [Metivier, R. et al. 2004; Metivier, R. et al. 2003; Reid, G. et al. 2003]. Activities of these complexes leads to the dissociation of associated factors from the promoter, transcription termination and chromatin modeling non-permissive to transcription [Metivier, R. et al. 2004; Metivier, R. et al. 2003; Reid, G. et al. 2003].
C.2.2. ERβ-Mediated Nuclear Signaling
In spite of the fact that ERs display similar ERE and E2 binding properties in vitro [Kuiper, G.G. et al. 1997; Loven, M.A. et al. 2001; Yi, P. et al. 2002b] arising from structural similarities between DBDs and LBDs, numerous studies have established that E2-ERβ is less effective than E2-ERα in inducing transcription from the ERE-dependent signaling pathway [Hall, J.M. et al. 2001; Nilsson, S. and Gustafsson, J.A. 2002b]. The mechanism of ER subtype-specific transactivation is unclear. However, a body of emerging evidence suggests that the receptor subtype-specific A/B domain defines the ER subtype transcriptional activity in the ERE-dependent signaling pathway [Huang, J. et al. 2006; Li, X. et al. 2008].
Studies assessing the ability of ERs to interact with ERE in situ showed that although unliganded ERs interact with ERE similarly, E2 enhances the interaction of only ERα with ERE without affecting the binding of ERβ to ERE [Hall, J.M. and McDonnell, D.P. 1999; Huang, J. et al. 2004]. Structural studies further revealed that the A/B domain of ERβ impairs the ability of the receptor to interact in situ with ERE independent of E2, in contrast to the A/B domain of ERα that does not affect the interaction of the receptor with ERE [Huang, J. et al. 2006; Li, X. et al. 2008].
Studies also indicated that the impairment of ERβ-ERE interactions by the A/B domain is a contributing factor, but is not sufficient to explain transcription inefficiency of the receptor. As discussed above, the amino terminal A/B domain of ERα contains an activation function that operates independently as well as in cooperation with the carboxyl-terminus in a cell and promoter context-dependent manner [Beekman, J.M. et al. 1993; Kraus, W.L. et al. 1995; McInerney, E.M. et al. 1996; Tora, L. et al. 1989; Tzukerman, M.T. et al. 1994]. The ability of the A/B domain to recruit co-regulatory proteins is critical for the AF1 function [Webb, P. et al. 1998; Yi, P. et al. 2002b]. The recruitment of co-regulatory proteins by AF1 is also critical for the functional integration of both AF1 and AF2 for ERα to mediate transcription at full capacity [Benecke, A. et al. 2000; Kraus, W.L. et al. 1995; Yi, P. et al. 2002a]. On the other hand, the A/B domain of ERβ lacks the ability to induce transcription [Cowley, S.M. and Parker, M.G. 1999]. Moreover, the A/B domain of ERβ is incapable of functionally integrating with AF2 to augment transcription in response to E2 [Yi, P. et al. 2002a].
Thus, it appears that the structurally distinct A/B domain is critical in defining the function of each receptor-subtype in the ERE-dependent E2 signaling.
Regardless of the underlying mechanism(s) for the E2-ERβ mediated transcription initiation, it appears that transcription termination from the ERE-dependent signaling pathway is dependent upon the ubiquitination and subsequent targeting of ERβ, as with ERα, to the ubiquitin-proteasome pathway [Tateishi, Y. et al. 2004; Tateishi, Y. et al. 2006].
C.2.3. ERE-Independent E2-ER Signaling
The E2-ER complex also mediates gene expression by tethering to a trans-acting factor bound to a cis-element, known as DNA-dependent and ERE-independent estrogen-ER signaling [Hall, J.M. et al. 2001; Kushner, P.J. et al. 2000; Safe, S. 2001]. However, agonist bound-ERα and -ERβ signal in opposite ways at a cis-acting activator protein-1 (AP-1) response element-bound AP-1 protein (i.e. the fos/jun complex). While E2-ERα activates gene transcription from an AP-1 site, E2-ERβ inhibits it. Moreover, ERα, but not ERβ, is suggested to interact with the Sp-1 transcription factor to augment transactivation of a variety of estrogen responsive genes [Safe, S. 2001]. This interaction is mediated by tethering of ERα to the GC-box response element bound Sp-1 protein [Safe, S. 2001]. Although the underlying mechanism remains unclear, the distinct features of the amino- and carboxyl-termini are thought to be responsible for the ER subtype-specific gene expression [Kushner, P.J. et al. 2000; Safe, S. 2001].
D. VARIATIONS IN THE ERβ GENE AND PROTEIN
ER functions can also be modulated by post-translational mechanisms as well as the heterogeneity in ER isoforms that affect ligand signaling and the initiation and/or the development of target tissue malignancies [Herynk, M.H. and Fuqua, S.A. 2004; Murphy, L.C. et al. 1998; Murphy, L.C. et al. 1997].
D.1. Post-Translational Modifications
Phosphorylation, acetylation, ubiquitylation, sumoylation and glycosylation of ERs are critical post-translational modifications that affect receptor stability and activity [Faus, H. and Haendler, B. 2006; Fu, M. et al. 2003; Fu, M. et al. 2004; Lannigan, D.A. 2003; Leader, J.E. et al. 2006]. Modifications also provide a potential mechanism for cell- or gene-specific regulation of ER functions. Aberrant post-translational modifications could adversely affect unliganded or liganded-ER signaling, consequently contributing to the initiation and/or development of estrogen target tissue malignancies. Post-translational modifications of ERs are certainly a critical component of ER functions. However, their importance in physiology and pathophysiology of E2 signaling is beyond the scope of this work and has been extensively addressed in recent review articles [Enmark, E. and Gustafsson, J.A. 1999; Faus, H. and Haendler, B. 2006; Hall, J.M. and Korach, K.S. 2002; Huang, J. et al. 2005].
D.2. Polymorphisms
The involvement of a genetic component in the etiology of a disease derives from population studies. However, only in rare cases the actual genes involved in the susceptibility and/or in responses to drugs have been identified. DNA variants are common, often defined as greater than 1% in a given population [Chakravarti, A. 1999]. The impact of nucleotide differences is variable and dependent upon the location of the polymorphism in the genome. The most common type of polymorphism is a single nucleotide (base pair) change in DNA sequence and is referred to as single nucleotide polymorphisms (SNPs). SNPs account for many well-characterized phenotypes, including disease susceptibility and resistance [Collins, F.S. et al. 1997; Lander, E.S. 1996] as well as drug response [Kleyn, P.W. and Vesell, E.S. 1998]. The use of SNPs in association with disease is primarily concerned with the propensity for such polymorphisms to affect the gene in which it is located, either through modifications in the expression or the protein product [Chakravarti, A. 1999; Collins, F.S. et al. 1997; Lander, E.S. 1996].
SNPs result from errors in DNA replication or repair. The frequency of SNPs varies between genomic regions, and between coding and non-coding sequences. The extent of nucleotide diversity ranges from 3 to 50 SNPs per 10 kb when two chromosomes are compared [Chakravarti, A. 1999].
SNPs can serve as genetic markers for identifying disease genes. This is primarily accomplished by linkage studies in families, linkage disequilibrium in isolated populations, association analysis of patients and controls, and loss-of-heterozygosity studies in tumors [Chakravarti, A. 1999; Collins, F.S. et al. 1997; Lander, E.S. 1996; Wang, D.G. et al. 1998].
The majority of SNPs occurs in the non-coding regions. One important role of 5’ untranslated regulatory regions of a gene is to control the amount of primary transcript in the basal state or in response to stimuli through the binding of transfactors to cognate responsive elements. Sequence variations in responsive elements could result in aberrant regulation of transcription. For example, the inhibitory receptor FcRIIb is a negative regulator of antibody production and inflammatory responses. The G→C polymorphism in the human FCGR2B gene promoter is associated with systemic lupus erythematosus (SLE). Functional studies revealed that the G→C transition could participate in the development of SLE by leading to a decrease in the binding of an AP-1 protein complex to the promoter by competitive interaction of the transcription factor, Yin-Yang 1 (YY1), with the mutant sequence [Olferiev, M. et al. 2007].
Similarly, polymorphisms at untranslated 3’ non-coding regions could affect the processing and/or stability of the transcript, altering the template effectiveness. For instance, it was shown that the G→A transition in the 3’untranslated region of the prothrombin mRNA generates a bifunctional polymorphism that alters the efficiency of mRNA processing and also the decay rate of the mRNA [Carter, A.M. et al. 2002].
Synonymous polymorphisms within the coding regions of a gene result in no amino acid change, whereas non-synonymous polymorphisms give rise to different residue (mis-sense) or to premature termination (non-sense) [Brinkman, B.M. 2004; Gingeras, T.R. 2007; Kalnina, Z. et al. 2005]. The impact of a mis-sense SNP also depends upon the properties of an amino acid and the functional and/or structural importance of that amino acid in the resulting protein. Polymorphisms at intron/exon boundaries could affect exonic or intronic splicing, enhancer/silencer positions, or particularly conserved GT donor or AG acceptor positions [Brinkman, B.M. 2004; Gingeras, T.R. 2007; Kalnina, Z. et al. 2005]. This gives rise to a remarkable heterogeneity in the resultant protein products. Polymorphisms at intron/exon boundaries could also play a significant role in malignancy development [Brinkman, B.M. 2004; Gingeras, T.R. 2007; Kalnina, Z. et al. 2005]. Loss of fidelity, variation of the splicing process, even controlled switching to specific splicing alternatives may occur during tumor progression [Brinkman, B.M. 2004; Gingeras, T.R. 2007; Kalnina, Z. et al. 2005]. The physiological activity of splice variant products may be completely different when compared with the wild-type counterpart. A functional domain may be added or deleted from the protein coding sequence, leading to dominant negative or positive phenotypes.
SNPs can also affect the folding of mRNA that influences mRNA splicing, processing and translational regulation [Shen, L.X. et al. 1999]. Moreover, an SNP may be in linkage disequilibrium (LD), which indicates the non-random associations of alleles at different loci. LD is generally caused by genetic linkage and the rate of mutation or recombination, random-drift or non-random mating and population size.
Other polymorphisms include insertion and deletion of multiple sequential nucleotides or large-scale duplications or deletions. Polymorphisms of short tandem repeats (STRs, or microsatellites) [Zhang, W, and Yu, Y.Y. 2007] could also contribute to disease processes. STRs are composed of multiple dinucleotide, trinucleotide or tetranucleotide repeats and are widely distributed in the genome. STRs may be present in the promoter regions, exons and/or introns [Li, Y.C. et al. 2004b; Zhang, W. and Yu, Y.Y. 2007]. STRs could affect DNA structure and genomic (in)stability. This in turn could lead to alterations in gene expression, RNA splicing, stability, or protein structure and function. STRs are commonly used as markers in initial genetic studies to identify genes or chromosomal regions that confer increased disease risk [Zhang, W. and Yu, Y.Y. 2007].
SNP genotyping strategies involve allele-discrimination and allele-detection [Gray, I.C. et al. 2000; Kim, S. and Misra, A. 2007; Nowotny, P. et al. 2001]. Allele discrimination is carried out with allele-specific biochemical reactions that include primer extension (nucleotide incorporation), hybridization, ligation, and enzymatic cleavage. In addition, single-strand conformation polymorphism (SSCP) which is based on the use of the secondary structure of single-stranded DNA and mis-match repair detection (MRD) that employs a bacterial mis-match repair system are currently being used for the discrimination of alleles. An allele discrimination approach is followed by an allele detection methodology. This detection methodology includes fluorescent signal detection or fluorescent polarization, chemiluminescence, fluorescence resonance energy transfer (FRET), denaturing high-performance liquid chromatograph linkage disequilibrium (DHPLC) and mass-spectrometry.
The human genome has a haplotype block structure with limited haplotype diversity [Zhang, K. et al. 2002; Lin, Z. and Altman, R.B. 2004]. In each block, a small fraction of SNPs, referred as haplotype tagging SNPs or htSNPs can be used to distinguish a large fraction of the haplotypes [Zhang, K. et al. 2002; Lin, Z. and Altman, R.B. 2004]. htSNPs have the potential to be extremely useful for association studies without genotype all SNPs and have gained a wide use in cost-effective genotyping and, subsequently, genotype-phenotype association studies.
D.3. ERβ Polymorphisms and Disease
Since SNPs could account for disease susceptibility and responses to drugs, discovery efforts related to SNPs of the ERβ gene have exponentially increased in recent years. More than 650 SNPs of the ERβ gene have been complied in the National Center for Biotechnology Information Database (http://www.ncbi.nlm.nih.gov/SNP). It appears that while the majority of the SNPs are located in introns, there are more than 30 SNPs located at the 5’untranslated region including promoters and more than 40 SNPs present at the 3’ untranslated region of the ERβ gene.
Estrogen target tissue malignancies are multifactorial involving genetic heterogeneity, population admixture, gene-environment and gene-gene interactions [Feigelson, H.S. and Henderson, B.E. 2000; Gennari, L. et al. 2005; Herrington, D.M. 2003]. The number of reports on the contribution of genetic variability in the ERβ gene based on the carriage of low-penetrance high-frequency polymorphisms to estrogen target tissue malignancies is increasing. These polymorphisms could contribute to malignancies by affecting the expression and/or processing of the ERβ transcript, consequently altering E2 signaling where ERβ is synthesized. We will attempt here to evaluate some of the findings on polymorphisms of the ERβ gene, depicted in Fig. (3), and their potential impact on the initiation and/or development of estrogen target tissue disorders.
Fig. (3).
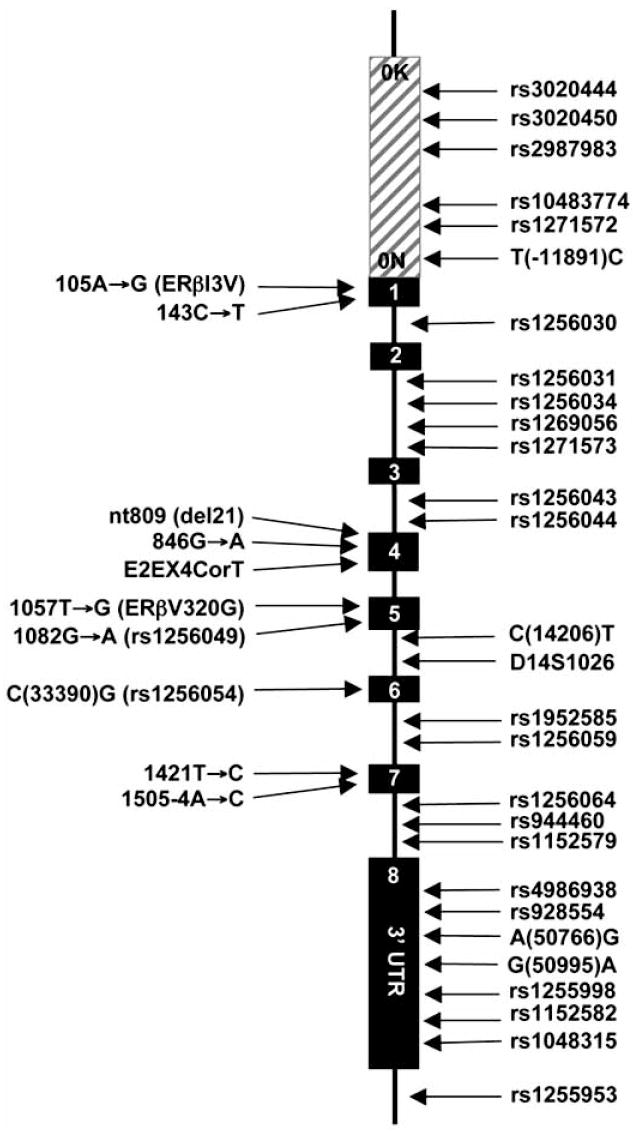
Locations of polymorphisms in the ERβ gene reviewed in the text.
D.3.1. ERβ Polymorphisms in Reproductive System Disorders of Women
Ovulatory Disorders and Ovarian Cancer
E2 orchestrates menstrual cycles that culminate in ovulation. The expression of ERβ gene in the human ovarian granulosa [Enmark, E. et al. 1997] and surface epithelial cells [Brandenberger, A.W. et al. 1998; Lau, K.M. et al. 1999] implicates ERβ as one of the important participants in the ovulation process, which is also supported by observations with the ERβ knock-out mice model [Krege, J.H. et al. 1998]. In addition, it appears that a dysregulated expression of the ERβ gene could be involved in the initiation and/or progression of ovarian cancer, particularly those are granulose cell-derived [Bardin, A. et al. 2004; Brandenberger, A.W. et al. 1998; Pujol, P. et al. 1998].
Systematic scanning of the entire ERβ exons for the first time identified several polymorphisms [Rosenkranz, K. et al. 1998]. These include the nt809(del21) polymorphism, which is a 21-nucleotide deletion encompassing codon 238 and 244 in exon 4. This results in the deletion of seven amino acids from the D domain of the ERβ protein. The SNP 846G→A in exon 4 is a rare non-synonymous change that generates a CfrI restriction enzyme site and encodes a serine residue replacing glycine at codon 250. A rare synonymous T→C transition was also found to be located at codon 1421 in exon 7. One of the common synonymous changes was at position 1082 in exon 5 within the LBD of ERβ, or rs1256049, which replaces G with A resulting in the generation of an RsaI restriction enzyme site. In addition, SNP rs4986938, which is also a common A→G transition at position 1730 in the 3’-untranslated region (UTR) of exon 8, generates an AluI restriction enzyme site [Rosenkranz, K. et al. 1998].
A study exploring the role of the ERβ rs1256049 and rs4986938 polymorphisms in a population of Chinese women with menstrual disorders suggests that patients homozygous for the polymorphisms display ovulatory dysfunctions without etiologic pathology [Sundarrajan, C. et al. 2001]. While rs1256049 and rs4986938 polymorphisms in the ERβ gene may associate with ovulatory dysfunctions, a comprehensive evaluation of variations at the ERβ gene locus using a haplotype tagging SNP approach (htSNPs), which includes intronic rs3020450, rs1256031 and rs994046 polymorphisms together with rs1256049 and rs4986938, suggests that the individual htSNPs or the related haplotypes are not associated with ovarian cancer risk in a large population of multiethnic group of women in the United States [Pearce CL, 2008].
Endometrial Dysfunctions and Cancer
While conclusions are variable, there are also reports examining the association of SNPs in the ERβ gene with endometriosis [Luisi, S. et al. 2006]. Displaying heritable tendencies with polygenic/multifactorial etiology [Simpson, J.L. and Bischoff, F.Z. 2002], endometriosis is a condition in which endometrial tissue grows outside the uterus and attaches to other organs in the abdominal cavity [Kitawaki, J. et al. 2002; Practice Committee 2004]. As a progressive disease, endometriosis is one of the most common causes of pelvic pain and infertility in women. Expression of ERs in endometriotic lesions [Fujishita, A. et al. 1997] reinforces a role for E2 signaling in the pathogenesis of the disease [Kitawaki, J. et al. 2002].
Studies on genotype distribution and allele frequency of rs1256049 and rs4986938 polymorphisms in a Japanese population report that only rs4986938 is associated with a late stage of endometriosis [Wang, Z. et al. 2004]. In contrast, the rs4986938 polymorphism showed no association with endometriosis in cohorts of Greek [Georgiou, I. et al. 1999], Italian [Luisi, S. et al. 2006] or Korean [Lee, G.H. et al. 2007] women. Studies also suggest no association of the rs1256049 polymorphism or intronic cytosine-adenine dinucleotide repeat in intron 5, known as (CA)n polymorphism or D14S1026, of the ERβ gene with endometrial cancer risk in a population of postmenopausal women in the United States [Setiawan, V.W. et al. 2004].
Preeclampsia
Preeclampsia (PE) is a pregnancy disorder that affects both the mother and fetus. PE is a rapidly progressing condition characterized by high blood pressure and the presence of proteins in the urine after 20 weeks of pregnancy. The pathophysiologic process for PE involves the reduced placental perfusion that affects trophoblast invasion and placental function. This subsequently leads to maternal inflammatory, metabolic, and thrombotic responses, converging to alter vascular and endothelial cell injury. Injury in turn results in a dysfunctional cascade of coagulation, vasoconstriction, and intravascular fluid redistribution that constitute the clinical syndromes of PE [Gammill, H.S. and Roberts, J.M. 2007; Roberts, J.M. and Catov, J.M. 2008]. The expression of ERβ in differentiating trophoblast cells and stimulation of trophoblast differentiation by estrogens, suggests a role of ERβ in the regulation of placental function [Bukovsky, A. et al. 2003]. This together with the observations that the ERβ gene expression is reduced in trophoblast cells in placentas of growth-restricted pregnancies and augmented in preeclamptic placentas implies the involvement of ERβ in the pathophysiology of pregnancy as well [Schiessl, B. et al. 2005]. A study investigating the relationships between SNPs in the ERβ gene and PE in a small group of Japanese woman suggests that SNPs rs125630, rs928554 and rs1048315 are not associated with PE. However, the allelic distribution of rs928554 displays a significant correlation with family history of hypertension. This suggests that a mutation(s) in the ERβ gene could prescribe a genetic predisposition for PE in patients with a family history of hypertension [Maruyama, A. et al. 2004].
ERβ Polymorphisms and Breast Cancer
The involvement of E2 in the development and the maintenance of the breast tissue functions is well established [Ali, S. and Coombes, R.C. 2002; Huang, J. et al. 2005]. E2 is also involved in the initiation and progression of breast cancer, which is the consequence of uncontrolled growth and division of breast epithelial cells [Ali, S. and Coombes, R.C. 2002; Huang, J. et al. 2005]. While many factors contribute to its etiology, aberrations in the metabolism of E2 and/or in E2-ER signaling appear to be two critical mechanisms for breast cancers. ERs are synthesized in normal breast luminal epithelial cells and in some breast tumors [Ali, S. and Coombes, R.C. 2002; Huang, J. et al. 2005]. Importantly, ERs are the primary target for both chemoprevention and endocrine therapy of breast cancer [Ali, S. and Coombes, R.C. 2002; Huang, J. et al. 2005; Murphy, L.C. and Watson, P.H. 2006]. The ER status of breast tumors provides prognostic information and is an important predictor of response to endocrine therapy [Ali, S. and Coombes, R.C. 2002; Huang, J. et al. 2005; Murphy, L.C. and Watson, P.H. 2006].
Studies compared the variant allele frequencies at the polymorphic sites in the ERβ gene in a small group of woman with sporadic breast cancer to the allele frequencies in the general population by using ethnically and regionally matched male controls in Finland [Forsti, A. et al. 2003]. Results suggest that polymorphic rs1256049, rs4986938, 846G→A, 1505-4A→G (located at a splice acceptor site at the exon/intron junction of exon 7), D14S1026 or nt805(del21) are not associated with the breast cancer risk. Another study examined the potential association of SNPs rs1256049, rs4986938 and rs928554 (or Cx+56 polymorphism located 56 bases 3’ of the coding region of the alternatively spliced exon 9 [Nilsson, M. et al. 2004]), with breast cancer in a large cohort of sporadic and familial breast cancer cases from Sweden [Maguire, P. et al. 2005]. While no association was found for any of the single polymorphisms, one haplotype of the SNP rs1256049, rs4986938 and rs928554 suggested an association with increased risk of sporadic breast cancer.
In another large study conducted in Shanghai consisting of 1459 incident cases of breast cancer, allele frequencies of SNP T(-11891)C (which is located at the promoter region), C(14206)T (which is located in the intron 5) and rs1256049 together with SNPs A(50766)G and G(50995)A, both of which are located at the 3’ UTR, showed no differences compared to those observed with the control groups [Zheng, S.L. et al. 2003]. However, allele frequency of the intronic SNP C(14206)T as well as the synonymous SNP C(33390)G, or rs1256054, that latter of which is located in exon 6 containing an exonic splicing enhancer motif that signifies the binding site for splice factor SC35, was found to be associated with breast cancer in post-menopausal women who had prolonged estrogen exposure due to a greater number of menstrual years [Zheng, S.L. et al. 2003].
The association of the ERα, ERβ and progesterone receptor gene SNPs and haplotypes with breast cancer risk was also examined in a large multiethnic group of women with breast cancer in the United States [Gold, B. et al. 2004]. SNPs rs1271572, rs1256030, E2EX4CorT (located in exon 4), rs1256049, rs986938, rs1152579, rs928554 and rs1255998 were not associated with breast cancer risk. However, haplotype analysis of SNPs rs1271572, rs1256030, E2EX4CorT, rs1256049 and rs986938 revealed a significant association with an increased risk for breast cancer only in a group of Ashkenazi Jewish patients [Gold, B. et al. 2004].
In a large study, The National Cancer Institute Breast and Prostate Cancer Cohort Consortium (BPC3) has systematically selected haplotype tagging SNPs in genes along the steroid hormone synthesis, metabolism and binding pathways, including the ERβ gene. Four htSNPs [rs3020450, rs1256031, rs1256049 and rs4986938] tag the 6 major (>5% frequency) haplotypes of the ERβ gene. These polymorphisms have been genotyped in 5,789 breast cancer cases and 7,761 controls nested within the American Cancer Society Cancer Prevention Study II, European Prospective Investigation into Cancer and Nutrition, Multiethnic Cohort, Nurses’ Health Study and Women’s Health Study cohorts. Results revealed that SNPs independently or as haplotypes were not associated with breast cancer risk [Cox, D.G. et al. 2008].
D.3.2. ERβ Polymorphisms in Reproductive System Disorders of Men
Testicular Dysgenesis Syndrome and Prostate Cancer
E2-ER signaling plays a significant role in the development and maintenance of male fertility as well [Jones, M.E. and Simpson, E.R. 2000; O’Donnell, L. et al. 2001]. E2-ER signaling is further suggested to participate in the development of the testicular dysgenesis syndrome (TDS) that includes hypospadias, cryptorchidism, low semen quality and testicular cancer [Skakkebaek, N.E. et al. 2001]. A study aimed at investigating the rs1256049 and rs4986938 ERβ polymorphisms in the etiology of TDS in groups of men with various disorders suggests that only the rs1256049 polymorphism may have a modulatory effect on spermatogenesis, while neither polymorphism shows an association with hypospadias, cryptorchidism or testicular cancer [Aschim, E.L. et al. 2005]. On the other hand, intronic (CA)n ERβ polymorphism with a longer CA stretch (larger than 24 nucleotides) as well as rs10483774 and rs1271572 polymorphisms in the promoter region of the ERβ gene in a Swedish cohort of boys is reported to be associated with hypospadias [Beleza-Meireles, A. et al. 2007].
Interestingly, longer (CA)n repeat polymorphism also appears to be associated with reduced testosterone levels and increased sex hormone binding globulin [Westberg, L. et al. 2001], prolactin hormone release [Westberg, L. et al. 2004b] and lower bone mineral density [Ogawa, S. et al. 2000; Scariano, J.K. et al. 2004] in women of various ethnic background.
On the other hand, (CA)n repeat polymorphism in a cohort of men in the United States shows no effect on the risk of prostate cancer [McIntyre, M.H. et al. 2007] which is a multigenic/multifactorial disease. The ERβ gene is expressed at high levels in prostate epithelium [Taylor, A.H. and Al-Azzawi, F. 2000]. This finding together with the observations that the ERβ gene expression diminishes in prostate cancer [Horvath, L.G. et al. 2001; Ji, Q. et al. 2005; Leav, I. et al. 2001] and re-appears in metastatic lesions [Leav, I. et al. 2001] reinforces the suggestion that ERβ-mediated E2 signaling is involved in the prostate pathophysiology as well [Prins, G.S. and Korach, K.S. 2008]. Alterations in the ERβ gene expression in prostate cancer could be associated with polymorphisms in the promoter regions of the ERβ gene [Hedelin, M. et al. 2006; Thellenberg-Karlsson, C. et al. 2006]. Large population-based case-control studies using htSNP spanning the entire ERβ gene suggest that the htSNP rs2987983 at the ERβ gene promoter is associated with prostate cancer [Hedelin, M. et al. 2006; Thellenberg-Karlsson, C. et al. 2006]. On the other hand, nested case-control studies conducted BPC3 utilizing a very large of pool of prostate cancer cases from various ethnicities suggest that htSNPs in the promoter and coding regions of the ERβ gene does not affect prostate cancer risk [Chen, Y.C. et al. 2007; Cox, D.G. et al. 2008].
D.3.3. ERβ Polymorphisms and Skeletal System
Osteoporosis is characterized by low bone mass and micro-architectural deterioration of bone tissue leading to bone fragility and susceptibility to fracture in both women and men [Carbonell Sala, S. et al. 2005; Deroo, B.J. and Korach, K.S. 2006; Gennari, L. et al. 2005]. Bone mineral density (BMD) is one of the most important determinants of bone strength and is the best clinical predictor of osteoporotic fractures. BMD is a complex quantitative trait regulated by both genetic and environmental factors.
Involvement of gonadal steroid hormones, particularly E2 and testosterone, in the development and the maintenance of the skeletal system is well established [Carbonell Sala, S. et al. 2005; Deroo, B.J. and Korach, K.S. 2006; Gennari, L. et al. 2005]. ERα and ERβ are synthesized in osteoblasts, osteoclasts, and several types of cells from the bone marrow [Braidman, I.P. et al. 2001] and are co-localized in adult bone [Batra, G.S. et al. 2003]. The ERβ synthesis also shows variability in a gender, age and cell-type dependent manner [Batra, G.S. et al. 2003]. These observations as well as experimental studies utilizing knock-out mice models [Deroo, B.J. and Korach, K.S. 2006; Imamov, O. et al. 2005] suggest that in addition to ERα, ERβ is involved in the mediation of E2 signaling in the skeletal system.
Studies aimed at examining the effects of ERβ rs1256049 polymorphism on BMD suggest that this variation is not associated with lumbar BMD in a group of post-menopausal Italian [Gennari, L. et al. 2002] or Slovenian [Arko, B. et al. 2002] women. Similarly intronic rs1256031 and rs1256059 SNPs of the ERβ gene showed no association with femoral BMD in post-menopausal woman of he Framingham Heart Study’s offspring cohort in the United States [Shearman, A.M. et al. 2004]. Interestingly, SNPs rs1256031 and rs 1256059 exhibit an association with femoral BMD in men [Shearman, A.M. et al. 2004].
The presence of D14S1026 in the ERβ gene in a cohort of post-menopausal Japanese women suggests that women possessing at least one allele with 26CA repeats have significantly higher BMD at the lumbar spine compared with those without the 26CA repeat allele [Ogawa, S. et al. 2000]. A large-scale analysis of D14S1026 within the Framingham Study offspring cohort also shows a significant association between the number of CA repeats and femoral BMD, but not spinal BMD, in both post-menopausal women and older men [Shearman, A.M. et al. 2004]. In contrast, no association between different CA repeat allelic variants and lumbar or femoral BMD was found in a group of post-menopausal Italian [Gennari, L. et al. 2002] or Chinese [Lau, H.H. et al. 2002] women. However, in the same cohort of Chinese women, the 20CA repeat allele was found to be associated with high BMD in pre-menopausal women [Lau, H.H. et al. 2002].
Although rs1256031 and rs1256059 polymorphisms showed a gender-specific association with femoral BMD in the Framingham Heart Study’s offspring cohort as discussed above, the analysis of rs1256031 and rs1256059 polymorphisms together with D14S1026 indicated that the haplotype exhibits a significant association with femoral BMD in both sexes [Shearman, A.M. et al. 2004]. Similarly, a haplotype based on rs1256031 and rs498638 SNPs was shown to be associated with increased risk of vertebral and incident fragility fracture in post-menopausal participants of the Rotterdam Study, which is a large prospective population-based cohort study of 55-years-old men and women from the Netherlands [Rivadeneira, F. et al. 2006].
D.3.4. ERβ Polymorphisms and Cardiovascular System
Disorders of the cardiovascular system are polygenic and multifactorial as a result of the interaction of genetic and environmental influences. However, clinical and experimental studies indicate that E2 acting primarily through ERs plays an important role in the biology of the cardiovascular system [Deroo, B.J. and Korach, K.S. 2006]. An examination of the potential association of various ERβ polymorphisms with hypertension in the community-based Framingham Heart Study offspring cohort suggests that only rs944460 SNP of the ERβ gene is associated with pulse pressure in men but not in women [Peter, I. et al. 2005b]. In an age-adjusted, case-control study involving small number of women and men with multiethnic background in Brazil, the analysis of ERβ polymorphisms revealed that the rs4986938 SNP shows higher frequency in patients with premature coronary artery disease (CAD) compared to healthy subjects [Mansur Ade, P. et al. 2005]. Interestingly, homozygosis for this mutation also shows an increased body mass index, reduced HDL-cholesterol, elevation in serum triglycerides and apolipoprotein B, all of which are phenotypic characteristics associated with CAD [Assmann, G. and Schulte, H. 1992; Miller, M. et al. 1998]. In contrast, there was no association of the rs4986938 SNP with atherothrombotic cardiovascular disease [myocardial infarction (MI) or ischemic stroke (IS)] in a cohort of mixed gender Caucasian subjects from the Women’s Health Study and from the Physician’s Health Study [Rexrode, K.M. et al. 2007]. However, in the same cohort, rs1271572 and rs1256049 polymorphisms were found to be linked to MI, but not IS, in women but not in men. A potential gender-specific variation in the ERβ gene was also observed with rs1256031 and rs1256059 SNPs, which showed an association with left ventricular mass and wall thickness in women but not in men according to the Framingham Heart Study [Peter, I. et al. 2005a].
D.3.5. ERβ Polymorphisms and Nervous System Disorders
E2 has an important role in maintaining neural functions and in protecting against damage in the normal adult brain in both sexes [Craig, M.C. and Murphy, D.G. 2007; McEwen, B. 2002]. E2 is also involved in the regulation of mood, memory and cognition [Craig, M.C. and Murphy, D.G. 2007; McEwen, B. 2002]. Alzheimer’s disease (AD) is the most common type of dementia within the aged population [Tang, M.X. et al. 1996]. E2 is suggested to provide a protective effect against neurodegenerative diseases, including AD [Craig, M.C. and Murphy, D.G. 2007; Tang, M.X. et al. 1996; Waring, S.C. et al. 1999].
A study investigating whether variations in the ERβ gene are genetically associated with the risk of AD used five intronic SNPs among 387 AD patients from eastern Finland [Pirskanen, M. et al. 2005]. It was found that intronic SNPs rs1271573 and rs1256043 individually and as a diplotype are associated with AD in women but not in men. Examination of the association of SNPs rs4986938 and rs1255953 with AD in a small population of mixed gender AD patients of German or Austrian descent revealed that although individual SNPs showed no effect, diplotypes were highly associated with the disease [Luckhaus, C. et al. 2006]. Another study examining the association of rs4986938 with the risk of developing AD in a small mixed-gender Caucasian population in the United Kingdom found no association in the total sample or within either gender. However, there was a significant correlation of allelic distribution in the patient cohort compared to the control group when rs4986938 was analyzed with polymorphisms of the ERα gene [Lambert, J.C. et al. 2001].
Similar to AD, E2 is also suggested to have protective effect on Parkinson’s disease (PD), which is a progressive neurodegenerative disorder primarily affecting nigrostriatal dopaminergic neurons [Craig, M.C. and Murphy, D.G. 2007; McEwen, B. 2002]. ERβ rs1256049 and rs4986938 polymorphisms were not associated with an increased risk for PD, but rs4986938 was found to influence the age of onset of PD [Westberg, L. et al. 2004a]. Similarly, the rs4986938 SNP was found to be strongly associated with the age of onset of PD in conjunction with polymorphisms of the IL-6 gene in a small cohort from Sweden [Hakansson, A. et al. 2005].
ERβ polymorphisms were also studied in anorexia nervosa, (AN), bulimia nervosa (BN) and chronic fatigue syndrome (CFS), disorders that are largely observed in women. SNP rs1256049 of the ERβ gene showed an association with CFS in a small group of Caucasian young women in the United Kingdom [Eastwood, H. et al. 2002]. Similarly, studies suggest that SNPs rs4986938 and rs928554 are associated with bulimia in a small cohort of Swedish patients [Nilsson, M. et al. 2004]. In contrast, there was no association of SNP rs4986938 or rs928554 with chronic fatigue syndrome in a group of Swedish women and men [Grans, H. et al. 2007]. Neither did rs1256049, rs4986938, nt809(del21), 846G→A and 1421T→C polymorphisms show an association with extreme obesity, AN or BN in a small cohort of German children and adolescence [Rosenkranz, K. et al. 1998].
D.3.6. Remarks
The allelic variants of the ERβ gene certainly have the potential to contribute to the etiology of, or disposition towards, estrogen target tissue malignancies. While conclusions regarding the association of these polymorphisms with malignancies are often variable and conflicting, it is also evident that the majority of polymorphisms are located in the untranslated regions or introns of the ERβ gene that do not contribute to the synthesis of ERβ variants themselves. However, these polymorphisms could differentially influence the expression and/or processing of the ERβ transcript, consequently affecting E2 signaling where ERβ is synthesized. Importantly, these polymorphisms could constitute important marker alleles that are in linkage disequilibrium with a mutation(s) elsewhere within or in the vicinity of ERβ gene or other genes involved in the estrogen target tissue disorders. More evidence based on meta-analyses of large case-control studies together with functional dissection of the underlying mechanisms would be critical in defining the importance of the ERβ gene polymorphisms in disease process.
D.4. Variant ERβ Proteins
In addition to SNPs, the existence of variant ERβ proteins, depicted in Fig. (4), suggest that ERβ isoforms could contribute to the pathophysiology of estrogen target tissues as well as responses to therapies. We will therefore review studies on the expression and potential functions of the some of ERβ variants.
Fig. (4).
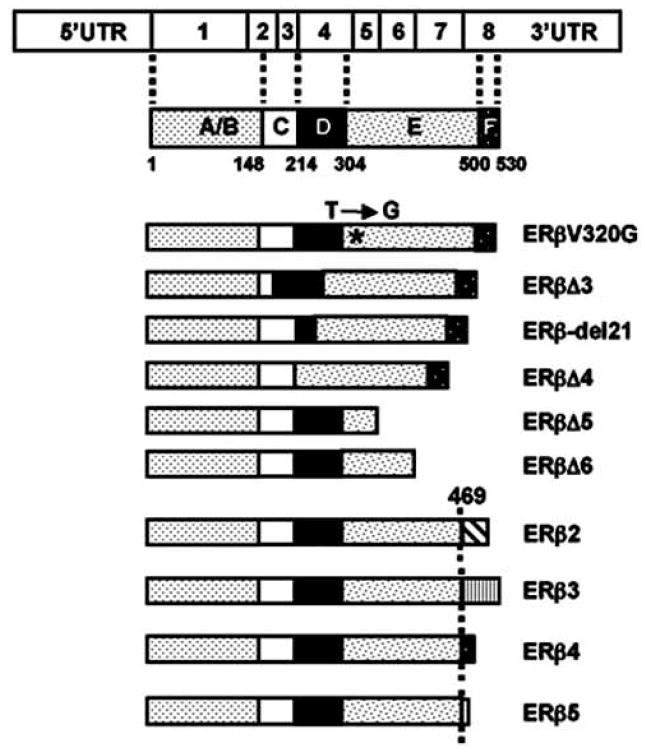
Schematics of ERβ isoforms with potential importance in E2 signaling. The T→G transition (*) leads to the replacement of valine with a glycine residue at position 320 resulting in the synthesis of ERβV320G. The exon 3 deleted (Δ) ERβ isoform (ERβΔ3) lacks the critical residues in the DBD for interaction with ERE. Due to exon 4 skipping, ERβΔ4 lacks the D domain critical for the nuclear localization of the receptor. Deletion of exon 5 or exon 6 results in a frame shift mutation causing premature termination of translation. This generates carboxyl-terminally truncated ERβΔ5 or ERβΔ6 lacking the LBD functions of the parent receptor. Designation of ERβ2-5 indicates various ERβ isoforms generated by the differential splicing of exon 8. A common divergence at amino acid 469, together with the novel amino acid stretches at various lengths, is indicated. In ERβ2, a novel 26 amino acid stretch replaces the last 61 amino acids of the parent ERβ of 530 amino acid in length. ERβ3 is generated by the replacement of the last 61 amino acid region with a novel 44 amino acid peptide, leading to a 510 amino acid long protein. The last 61 amino acid stretch of ERβ is replaced by novel 12 and 3 amino acid lengths in ERβ4 and ERβ5 isoforms, respectively.
D.4.1. Distinct Translation Initiation Sites
Cloning of a novel isoform of human ERβ (ERβ548) from human testis cDNA and genomic DNA was reported [Wilkinson, H.A. et al. 2002]. ERβ548 results from the presence of an additional A-T base pair in the 5’ untranslated region of the ERβ gene that generates an ATG initiation codon that extends the amino-terminus of ERβ by 18 additional amino acids [Wilkinson, H.A. et al. 2002]. Interestingly, ERβ548 appears to have more robust activity than the wild-type ERβ in inducing transcription in an ERE-dependent reporter system [Wilkinson, H.A. et al. 2002]. Moreover, tamoxifen and raloxifen act as agonists for ERβ548, in contrast with their action as antagonists for the wild-type ERβ [Wilkinson, H.A. et al. 2002]. However, subsequent studies have revealed that this amino-terminally extended ERβ isoform is rare and is not a common variant among various ethnic groups [Xu, L. et al. 2003].
Additionally, an ERβ isoform is encoded by the alternative utilization of exon M located in the intron positioned between exon 4 and 5 [Shoda, T. et al. 2002]. This isoform is expressed in testis, ovary and endometrium. Although the biological functions are unknown, the transcript is expected to encode the entire LBD, the E and F domains, of the ERβ. This truncated ERβ could bind to an ER ligand without transactivation function due to the absence of the DNA binding domain. This could interfere with E2 signaling as the variant may squelch away ligands or block the function of the wild-type receptor as a heterodimer.
D.4.2. ERβ Variants with Point Mutations
Studies on polymorphisms of the ERβ gene (see also Fig. 3) in an African population have identified SNPs that change the primary amino acid sequence in the ERβ protein [Zhao, C. et al. 2004]. One of the SNPs involves the 105A→G transition that changes the amino acid isoleucine at position 3 to a valine residue in exon 1, referred to as ERβI3V. However, this mutation does not alter the functional properties of the resultant variant compared to the wild-type ERβ [Zhao, C. et al. 2004]. Although effects on protein function are yet to be explored, a rare SNP 846G→A in exon 4 replaces a glycine residue with serine at codon 250 [Rosenkranz, K. et al. 1998]. Additionally, the SNP with the 1057 T→G transition substitutes valine at position 320 to glycine in exon 5, which corresponds to the Helix 4 of the ligand-binding domain of the receptor, represents the first functional polymorphism in the ERβ gene [Zhao, C. et al. 2004]. Referred to as ERβV320G, this variant ERβ shows reduced transcription activity due to decreased interaction with co-regulatory proteins [Zhao, C. et al. 2004].
As discussed above, the SNP nt809 (del21) is a rare variant in which the 21-nucleotides encompassing codon 238 and 244 in exon 4 are deleted [Rosenkranz, K. et al. 1998]. This results in the removal of seven amino acids (QLHCAGK) from the D domain of the ERβ protein functional consequences that are yet unknown. Since the hinge region is responsible for nuclear localization, the deletion could adversely affect transregulatory activity of ERβ by preventing the localization of the protein to the nucleus.
D.4.3. Splice Variants of ERβ
The best-characterized isoforms of ERβ are the splice variants. Splice variants are expressed as single or multiple exon-deleted or truncated transcripts expressed alone or together with the wild-type ER transcripts in both normal and neoplastic estrogen target tissues. Whether polymorphisms of the ERβ gene are involved in the generation of these splice variants is yet to be determined, these ERβs appear largely to be the products of alternative splicing [Hirata, S. et al. 2003].
ERβ Variants with Single Exon Deletion
An ERβ transcript containing an exon 2 deletion (ERβΔ2) was initially identified by a splice targeted primer approach expressed in the ovary, some breast cancers and breast cancer cell lines [Poola, I. et al. 2002a]. Deletion of exon 2 causes a frame shift mutation in the ERβ transcript that results in premature termination of translation. If synthesized, this transcript would give rise to a carboxy-terminally truncated receptor specie that lacks the known functions of ERβ.
The ERβ transcript with exon 3 deletion (ERβΔ3) was initially identified in the ovary. The deletion of exon 3 does not alter the open reading frame of the ERβ transcript [Poola, I. et al. 2002a]. The protein product is expected to result in an in-frame loss of 117 nucleotides that encode 39 amino acid in the carboxyl-terminal half of the DBD including the second zinc finger. Experimental studies utilizing heterologous expression systems indicate that ERβΔ3 localizes to discrete spots within the nucleus in the presence of ER agonists [Price, R.H. et al. 2001]. In the presence of ER antagonists, the variant receptor shows a diffuse distribution in the nucleus. Importantly, ERβΔ3 can activate transcription from reporter constructs emulating the ERE-independent signaling pathway but not from the ERE-dependent signaling pathway [Price, R.H. et al. 2001]. Thus, it appears that the variant translocates to the nucleus and has the ability to selectively regulate gene transcription from the ERE-independent signaling pathway.
The ERβΔ4 transcript with deleted exon 4 is primarily detected in the ovary [Poola, I. et al. 2002a]. Although this deletion does not alter the open reading frame, this variant, when synthesized, would lack the nuclear localization signal.
Deletion of exon 5 (ERβΔ5) or exon 6 (ERβΔ5) in ERβ mRNA causes a frame shift mutation resulting in premature termination of translation, thereby generating a diverse class of carboxyl-terminally truncated receptor species [Poola, I. et al. 2002a]. While, both ERβΔ5 and ERβΔ6 are expressed in ovary, uterus and breast tissue, only ERβΔ6 is expressed in breast cancer and in some cell lines derived from breast carcinoma as well [Poola, I. et al. 2002a]. The absence of the exon 5 or exon 6 predicts that the synthesized protein would lack a part of the LBD and consequently would not bind to a ligand. Indeed, heterologous expression studies indicate that while the ERβΔ5 is capable of translocating to the nucleus, it lack the activity to induce transcription [Inoue, S. et al. 2000]. However, the variant receptor effectively represses transcription induced by the wild type ERα or ERβ. These findings indicate that ERβΔ5 has the potential to act as a dominant negative regulator for E2 signaling mediated by both ER subtypes. This is also consistent with an observation that the higher expression of the ERβΔ5 transcript is associated with low aggressiveness of breast tumors [Mandusic, V. et al. 2007]. This in turn suggests that the uncontrolled local tumor growth may occur as the expression of ERβΔ5 mRNA decreases in estrogen-dependent breast cancer [Mandusic, V. et al. 2007].
ERβ Variants with Multiple Exon Deletions
In addition to ERβ transcripts with single exon deletions, studies have also identified ERβ transcripts with multiple exon deletions in ovary, uterus, breast and bone as well as in various breast cancer cell lines [Poola, I. et al. 2002a; Poola, I. et al. 2005b; Treeck, O. et al. 2007a; Treeck, O. et al. 2007b]. These include exon 1 and 2 (ERβΔ1,2); exon 2 and 5 (ERβΔ2,5); exon 2 and 6 (ERβΔ2,6); exon 2, 3 and 4 (ERβΔ2,3,4); exon 2, 3 and 6 (ERβΔ2,3,6); exon 2, 5 and 6 (ERβΔ2,5, 6); exon 5 and 6 (ERβΔ5,6); exon 1, 2 and 5 (ERβΔ1,2,5); exon 1, 2, 5 and 6 (ERβΔ1,2,5,6). Although functional studies remain to be carried out, the protein products of these multiple exon deleted ERβ transcripts are expected to play very little, if any, role in modulating E2 signaling due to lack of large functional domains [Treeck, O. et al. 2007a; Treeck, O. et al. 2007b].
Exon 8 Splice Variants of ERβ
In addition to single and multiple exon deleted ERβ variants, studies also identified various ERβ isoforms (ERβ2-5) resulting from the differential splicing of exon 8 [Moore, J.T. et al. 1998; Ogawa, S. et al. 1998]. Predicted amino acid sequences of these isoforms show a common divergence at amino acid 469, which correspond to helix 10 within the LBD, of the 530 amino acid in length wild-type ERβ. The nucleotide boundaries of these isoforms are consistent with a 5’ splice junction sequence. This indicates that these ERβ isoforms are generated by differential exon splicing. In ERβcx, or ERβ2, a 26 novel amino acid stretch replaces the last 61 amino acid of the wild type ERβ, hence 490 amino acid in length; in ERβ3 the last 61 amino acid region is replaced by a 44 novel amino acid peptide, leading to a 510 amino acid long protein. On the other hand, in ERβ4 and ERβ5, the last 61 amino acids of ERβ are replaced by 12 and 3 novel amino acids respectively.
Among the exon 8 splice variants of ERβ, expression and function of ERβ2 is well documented. The ERβ2 transcript is expressed in the testis, ovary, thymus, uterus, prostate and colon as well as in several E2 target tissue cancers and cell lines derived from different tissue [Campbell-Thompson, M. et al. 2001; Chi, A. et al. 2003; Critchley, H.O. et al. 2002; Fujimura, T. et al. 2001; Girault, I. et al. 2004; Moore, J.T. et al. 1998; Ogawa, S. et al. 1998]. Molecular modeling and experimental studies suggest that the helix 12 in ERβ2 displays a disorderly position with a marked shrinkage in the coregulator binding surface [Leung, Y.K. et al. 2006]. It appears that ERβ2 does not bind to ERβ ligands [Ogawa, S. et al. 1998], but translocates to the nucleus [Price, R.H. et al. 2001] and interacts, albeit at a lesser efficiency compared to the wild-type ERβ, with ERE [Moore, J.T. et al. 1998; Peng, B. et al. 2003]. Since the dimerization surface within the LBD of ERβ2 remains functional, this variant binds to ERE as a homodimer and as a heterodimer with either with the wild type ERα or ERβ [Critchley, H.O. et al. 2002]. This ERβ isoform lacks transactivation function as examined in experimental systems [Ogawa, S. et al. 1998; Peng, B. et al. 2003] and consequently does not alter endogenous gene expression [Secreto, F.J. et al. 2007]. Although the underlying mechanism is not clear, ERβ2 shows a dominant negative activity on transactivation mediated only by ERα [Ogawa, S. et al. 1998; Omoto, Y. et al. 2003; Peng, B. et al. 2003].
Analysis of the ERβ2 expression in the endometrium has shown that the protein levels, as determined by an isoform-specific monoclonal antibody, fluctuate during the menstrual cycle [Critchley, H.O. et al. 2002]. This suggests a physiological role for ERβ2 in normal tissues, at least in the endometrium. Moreover, it appears that the expression of ERβ2 increases in breast tumors, which is suggested to correlate with favorable response to endocrine therapy [Omoto, Y. et al. 2002; Palmieri, C. et al. 2004].
The isoforms of ERβ3, 4, and 5 are expressed in the testis, breast, uterus and ovary [Moore, J.T. et al. 1998; Poola, I. et al. 2005a]. ERβ4 and ERβ5 along with wild-type ERβ are also expressed in adipose tissues [Pedersen, S.B. et al. 2001]. Studies also showed that ERβ5 is expressed in breast tumors and in ERα-negative breast cancer cell lines [Poola, I. et al. 2002b]. Moreover, elevated levels of the ERβ5 appear to be associated with high tumor grade and postmenopausal status [Poola, I. et al. 2002b].
While very little is known for the potential function of ERβ3 in E2 signaling, recent studies suggest that ERβ4 and ERβ5 mimic the intracellular behaviors of ERβ2 in experimental systems [Leung, Y.K. et al. 2006; Peng, B. et al. 2003; Poola, I. et al. 2005a]. This is not surprising given the fact that all differentially spliced exon 8 isoforms of ERβ result from the same divergence point [Moore, J.T. et al. 1998; Ogawa, S. et al. 1998]. It appears that both ERβ4 and ERβ5 do not bind to E2, yet they interact with ERE and induce transcription from heterologous promoters emulating the ERE-dependent signaling pathway [Poola, I. et al. 2005a]. Importantly, both receptor isoforms are capable of heterodimerizing with ERα and negatively regulating its transcriptional activity [Peng, B. et al. 2003; Poola, I. et al. 2005a].
D.4.4. Remarks
The importance of ERβ in the homeodynamic regulation of many tissue functions is withstanding, the expression and synthesis of ERβ isoforms alone or together with the wild-type receptor could alter estrogen signaling thereby contributing to the initiation and/or the development of tissue malignancies. Importantly, these isoforms have the potential to adversely affect responses to therapies leading to resistance. Since the expression of, at least some, the ERβ isoforms are also altered during tumorigenesis, these isoforms could also be used as markers for disease or as prognostic indicators for counteractive measures.
E. PHARMACOGENOMIC IMPLICATIONS
Hormone replacement therapy (HRT) is an approach that utilizes one or more sex steroid hormones, usually estrogen alone (ERT) or together with progesterone, to alleviate discomfort and health problems by replacing the otherwise diminished endogenous steroid hormones in a variety of conditions [Grady, D. et al. 1998]. Early clinical observations suggested that ERT in post-menopausal women decreases total and low-density lipoprotein cholesterol levels as well as increases high-density lipoprotein cholesterol and triglyceride levels [Godsland, I.F. 2001]. These alterations were predicted to result in a significant decline in coronary heart disease [Kuller, L.H. 2003]. The Women’s Health Initiative (WHI) is a large clinical investigation of strategies for the prevention and control of some of the most common causes of morbidity and mortality among postmenopausal women, including cancer, cardiovascular disease and skeletal disease. WHI studies revealed that responses to hormone therapies are variable and the treatment does not appear to provide a significant cardiovascular benefit or may even lead to cardiovascular disease, as it may also increase the risk of invasive breast cancer [Rossouw, J.E. et al. 2002]. Heart and Estrogen/Progestin Replacement Study Group also reached similar conclusions [Hulley, S. et al. 2002].
Although the underlying mechanisms are unclear, variable responses to hormone therapies could stem from genetic variations in ER genes that alter the efficacy or toxicity of therapies [Herrington, D.M. 2003; Herrington, D.M. and Klein, K.P. 2001]. While studies on the association of ERβ gene polymorphisms with estrogen target tissue malignancies are extensive, the involvement of these genetic variants in altering responses to therapeutic approaches is very limited. One study, assessed the potential involvement of the common rs1256049 and rs4986938 SNPs of the ERβ gene in lipoprotein levels in a cross-sectional study consisted of a group of pre- menopausal women and groups of post-menopausal women without or with exposure to HRT for at least six months in Brazil [Almeida, S. et al. 2005]. Results revealed that low-density lipoprotein cholesterol levels were associated with only the rs1256049 polymorphism in pre-menopausal women as well as post-menopausal women who were exposed to HRT. These finding suggest that the rs1256049 genotype may influence the concentration of low-density lipoprotein cholesterol in women in response to HRT. On the other hand, a cross-sectional segregation analysis of the effects of the ERβ rs4986938 SNP in 280 post-menopausal Caucasian Danish women on the response to one year of HRT (estrogen + progesterone) therapy found that allelic frequency of the rs4986938 SNP is associated with total cholesterol levels mediated by HRT [Silvestri, S. et al. 2006].
As with E2, tamoxifen also alters serum lipid levels with an effect that exhibits variability. Another study tested the prediction that tamoxifen-induced changes in serum lipid profiles is associated with genetic variants in the ERβ gene in a small group of largely Caucasian pre- and post-menopausal breast cancer patients who were exposed to tamoxifen as an adjuvant therapy for at least four months in a multi-center prospective observational trial [Ntukidem, N.I. et al. 2008]. Results showed that tamoxifen-mediated alterations in triglyceride levels were associated with the rs498693 SNP in the ERβ gene in both pre- and post-menopausal patients. However, these tamoxifen-mediated changes were not reflected by total, low- or high-density cholesterol concentrations.
Despite the harmful effects of HRT on breast tissue and the cardiovascular system, ERT has beneficial effects on the skeletal system in post-menopausal women [Rossouw, J.E. et al. 2002]. In the same Caucasian Danish study group of women who showed an association of rs4986938 with total cholesterol levels in response to one year HRT, [Silvestri, S. et al. 2006], the rs4986938 ERβ gene polymorphism was also found to be associated with the prevention of bone loss in the forearm without affecting the rate of bone loss in the spine during hormone therapy.
Like E2, tamoxifen displays estrogenic activity in the skeletal system in that tamoxifen preserves or increases BMD by lowering bone turnover in healthy post-menopausal women as well as in post-menopausal with breast cancer [Gennari, L. et al. 2007]. A study examined the relationship of the effect of tamoxifen on BMD and burn turnover markers in 21 post-menopausal Japanese breast cancer patients who received an adjuvant tamoxifen treatment for 6 to 12 months following conserving breast surgery or mastectomy [Yoneda, K. et al. 2002]. It was found that tamoxifen increases BMD of the lumbar spine by reducing the bone turnover in patients. This bone restoring effect of tamoxifen is more pronounced at 12 months in the dinucleotide (CA)n polymorphism (D14S1026) of the ERβ gene allele carriers compared to non-carriers.
The effects of the ERβ rs1256049 polymorphism on lumbar BMD and responses to bisphosphonate alendronate, which is a potent inhibitor of bone resorption, was studied in 79 post-menopausal Slovenian women [Arko, B. et al. 2002]. It was found that the rs1256049 polymorphism is not associated with lumbar BMD nor does it affect the treatment outcome after one year of alendronate therapy.
Phytoestrogens, as discussed, are plant compounds that are structurally related to estrogens. Although they bind and regulate the activities of both ER subtypes, phytoestrogens display higher binding affinities to ERβ than ERα [Kuiper, G.G. et al. 1997; Kuiper, G.G. et al. 1998]. Phytoestrogens may provide some protection against prostate cancer, the incidence of which shows geographical and ethnic variations [Ganry, O. 2005; Von Low, L.C. et al. 2007]. In a population based case-control study of prostate cancer etiology in Sweden, high intake of phytoestrogens was found to substantially reduce prostate cancer risk among men with the rs2987983 SNP located at the promoter region of the ERβ gene [Hedelin, M. et al. 2006].
E.1. Remarks
While these findings underscore the potential of the ERβ genetic variants to modulate responses to therapeutic approaches, the roles of ERβ protein variants in altering efficacy and/or toxicity of therapies are unknown. As we have reviewed here, the presence of various ERβ transcripts with corresponding proteins in some cases, i.e. in normal breast tissue and breast neoplasms, provides a heterogeneity that could alter E2 signaling and could affect treatment outcomes as well. This could also be the situation for diseases of other estrogen target tissues wherein ERβ, in addition to ERα, is involved in mediating the effects of E2. An altered response or resistance to treatment regimens could therefore potentially result from ERβ polymorphisms, as suggested for ERα [Murphy, L.C. et al. 1997].
F. CONCLUSION
In this review we aimed to highlight the recent findings on polymorphisms of ERβ in order to provide a functional perspective with potential pharmacogenomic implications. It is clear that ERs regulate various cellular processes that are central to the mediation of estrogen target tissue functions. Accordingly, ERs represent an attractive target for clinical intervention in disease states. In estrogen target tissue malignancies, the presence of ERα has been used as a predictor for responsiveness to therapies that target the receptor’s function. The subsequent identification of ERβ has led to the recognition that E2 signaling in physiology and pathophysiology utilizes mechanisms conveyed by both ERα and ERβ. There are a large number of studies that address the importance of the polymorphisms of the ERα gene in the pharmacogenetics of estrogens (reviewed in [Carbonell Sala, S. et al. 2005; Gennari, L. et al. 2005; Herrington, D.M. 2003]), which is a rapidly evolving area of investigation with the potential to influence disease prevention and treatment choices. ERβ polymorphisms could also be a source of differential responses to various interventions. Insufficient information however, renders this possibility only speculative at this juncture, making recommendations premature.
A better understanding of the clinical impact of genetic and protein variants of ERs in general and ERβ in particular, in addition to the development of target tissue- and subtype-selective therapeutic agents, could help to elucidate the importance of altered estrogen signaling in the initiation and/or development of target tissue malignancies. Moreover, along with ERs as a target for therapeutic interventions, polymorphisms in genes encoding for drug transporters and drug metabolizing enzymes have the potential to contribute to individual variations in responses to treatments. This necessitates the integrations of various aspects of therapeutic interventions. This, in turn, could provide multigenic models with genetic topographies of malignancies, as recently described [Wood, L.D. et al. 2007], providing foundations of novel avenues in tailoring the selection of appropriate treatment for individual patients.
Acknowledgments
We thank Dr. Russell Hilf for critical reading of the manuscript. The work in the laboratory is supported by an NIH grant, CA113682 (MM), grants from the Susan G. Komen Foundation (MM) and the University of Rochester Clinical and Translational Science Institute (MM).
Footnotes
DUALITY/CONFLICTS OF INTERESTS None Declared
References
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- Almeida S, Franken N, Zandona MR, Osorio-Wender MC, Hutz MH. Estrogen receptor 2 and progesterone receptor gene polymorphisms and lipid levels in women with different hormonal status. Pharmacogenomics J. 2005;5:30–34. doi: 10.1038/sj.tpj.6500272. [DOI] [PubMed] [Google Scholar]
- Ansonoff MA, Etgen AM. Estradiol elevates protein kinase C catalytic activity in the preoptic area of female rats. Endocrinology. 1998;139:3050–3056. doi: 10.1210/endo.139.7.6088. [DOI] [PubMed] [Google Scholar]
- Arko B, Prezelj J, Komel R, Kocijancic A, Marc J. No major effect of estrogen receptor β gene RsaI polymorphism on bone mineral density and response to alendronate therapy in post-menopausal osteoporosis. J Steroid Biochem Mol Biol. 2002;81:147–152. doi: 10.1016/s0960-0760(02)00061-4. [DOI] [PubMed] [Google Scholar]
- Aschim EL, Giwercman A, Stahl O, Eberhard J, Cwikiel M, Nordenskjold A, Haugen TB, Grotmol T, Giwercman YL. The RsaI polymorphism in the estrogen receptor-β gene is associated with male infertility. J Clin Endocrinol Metab. 2005;90:5343–5348. doi: 10.1210/jc.2005-0263. [DOI] [PubMed] [Google Scholar]
- Assmann G, Schulte H. Role of triglycerides in coronary artery disease: lessons from the Prospective Cardiovascular Munster Study. Am J Cardiol. 1992;70:10H–13H. doi: 10.1016/0002-9149(92)91084-h. [DOI] [PubMed] [Google Scholar]
- Bai Y, Giguere V. Isoform-selective interactions between estrogen receptors and steroid receptor coactivators promoted by estradiol and ErbB-2 signaling in living cells. Mol Endocrinol. 2003;17:589–599. doi: 10.1210/me.2002-0351. [DOI] [PubMed] [Google Scholar]
- Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P. Loss of ERβ expression as a common step in estrogen-dependent tumor progression. Endocr Relat Cancer. 2004;11:537–551. doi: 10.1677/erc.1.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra GS, Hainey L, Freemont AJ, Andrew G, Saunders PT, Hoyland JA, Braidman IP. Evidence for cell-specific changes with age in expression of oestrogen receptor (ER) α and β in bone fractures from men and women. J Pathol. 2003;200:65–73. doi: 10.1002/path.1332. [DOI] [PubMed] [Google Scholar]
- Beekman JM, Allan GF, Tsai SY, Tsai MJ, O’Malley BW. Transcriptional activation by the estrogen receptor requires a conformational change in the ligand binding domain. Mol Endocrinol. 1993;7:1266–1274. doi: 10.1210/mend.7.10.8264659. [DOI] [PubMed] [Google Scholar]
- Beleza-Meireles A, Kockum I, Lundberg F, Soderhall C, Nordenskjold A. Risk factors for hypospadias in the estrogen receptor 2 gene. J Clin Endocrinol Metab. 2007;92:3712–3718. doi: 10.1210/jc.2007-0543. [DOI] [PubMed] [Google Scholar]
- Benecke A, Chambon P, Gronemeyer H. Synergy between estrogen receptor α activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 2000;1:151–157. doi: 10.1093/embo-reports/kvd028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidman IP, Hainey L, Batra G, Selby PL, Saunders PT, Hoyland JA. Localization of estrogen receptor β protein expression in adult human bone. J Bone Miner Res. 2001;16:214–220. doi: 10.1359/jbmr.2001.16.2.214. [DOI] [PubMed] [Google Scholar]
- Brandenberger AW, Tee MK, Jaffe RB. Estrogen receptor α (ERα) and β (ERβ) mRNAs in normal ovary, ovarian serous cystadenocarcinoma and ovarian cancer cell lines: down-regulation of ERβ in neoplastic tissues. J Clin Endocrinol Metab. 1998;83:1025–1028. doi: 10.1210/jcem.83.3.4788. [DOI] [PubMed] [Google Scholar]
- Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Bukovsky A, Caudle MR, Cekanova M, Fernando RI, Wimalasena J, Foster JS, Henley DC, Elder RF. Placental expression of estrogen receptor β and its hormone binding variant--comparison with estrogen receptor α and a role for estrogen receptors in asymmetric division and differentiation of estrogen-dependent cells. Reprod Biol Endocrinol. 2003;1:36. doi: 10.1186/1477-7827-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor α: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- Campbell-Thompson M, Lynch IJ, Bhardwaj B. Expression of estrogen receptor (ER) subtypes and ERβ isoforms in colon cancer. Cancer Res. 2001;61:632–640. [PubMed] [Google Scholar]
- Carbonell Sala S, Masi L, Marini F, Del Monte F, Falchetti A, Franceschelli F, Brandi ML. Genetics and pharmacogenetics of osteoporosis. J Endocrinol Invest. 2005;28:2–7. [PubMed] [Google Scholar]
- Carter AM, Sachchithananthan M, Stasinopoulos S, Maurer F, Medcalf RL. Prothrombin G20210A is a bifunctional gene polymorphism. Thromb Haemost. 2002;87:846–853. [PubMed] [Google Scholar]
- Castoria G, Barone MV, Di Domenico M, Bilancio A, Ametrano D, Migliaccio A, Auricchio F. Non-transcriptional action of oestradiol and progestin triggers DNA synthesis. EMBO J. 1999;18:2500–2510. doi: 10.1093/emboj/18.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoria G, Migliaccio A, Bilancio A, Di Domenico M, de Falco A, Lombardi M, Fiorentino R, Varricchio L, Barone MV, Auricchio F. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20:6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A. Population genetics--making sense out of sequence. Nat Genet. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of estrogen receptor β on gene networks regulated by estrogen receptor α in breast cancer cells. Endocrinology. 2006;147:4831–4842. doi: 10.1210/en.2006-0563. [DOI] [PubMed] [Google Scholar]
- Chen HW, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen YC, Kraft P, Bretsky P, Ketkar S, Hunter DJ, Albanes D, Altshuler D, Andriole G, Berg CD, Boeing H, Burtt N, Bueno-de-Mesquita B, Cann H, Canzian F, Chanock S, Dunning A, Feigelson HS, Freedman M, Gaziano JM, Giovannucci E, Sanchez MJ, Haiman CA, Hallmans G, Hayes RB, Henderson BE, Hirschhorn J, Kaaks R, Key TJ, Kolonel LN, LeMarchand L, Ma J, Overvad K, Palli D, Pharaoh P, Pike M, Riboli E, Rodriguez C, Setiawan VW, Stampfer M, Stram DO, Thomas G, Thun MJ, Travis RC, Virtamo J, Trichopoulou A, Wacholder S, Weinstein SJ. Sequence variants of estrogen receptor β and risk of prostate cancer in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Cancer Epidemiol Biomarkers Prev. 2007;16:1973–1981. doi: 10.1158/1055-9965.EPI-07-0431. [DOI] [PubMed] [Google Scholar]
- Chi A, Chen X, Chirala M, Younes M. Differential expression of estrogen receptor β isoforms in human breast cancer tissue. Anticancer Res. 2003;23:211–216. [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Cowley SM, Parker MG. A comparison of transcriptional activation by ERα and ERβ. J Steroid Biochem Mol Biol. 1999;69:165–175. doi: 10.1016/s0960-0760(99)00055-2. [DOI] [PubMed] [Google Scholar]
- Cox DG, Bretsky P, Kraft P, Pharoah P, Albanes D, Altshuler D, Amiano P, Berglund G, Boeing H, Buring J, Burtt N, Calle EE, Canzian F, Chanock S, Clavel-Chapelon F, Colditz GA, Feigelson HS, Haiman CA, Hankinson SE, Hirschhorn J, Henderson BE, Hoover R, Hunter DJ, Kaaks R, Kolonel L, LeMarchand L, Lund E, Palli D, Peeters PH, Pike MC, Riboli E, Stram DO, Thun M, Tjonneland A, Travis RC, Trichopoulos D, Yeager M. Haplotypes of the estrogen receptor β gene and breast cancer risk. Int J Cancer. 2008;122:387–392. doi: 10.1002/ijc.23127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig MC, Murphy DG. Estrogen: effects on normal brain function and neuropsychiatric disorders. Climacteric. 2007;10(Suppl 2):97–104. doi: 10.1080/13697130701598746. [DOI] [PubMed] [Google Scholar]
- Critchley HO, Henderson TA, Kelly RW, Scobie GS, Evans LR, Groome NP, Saunders PT. Wild-type estrogen receptor (ERβ1) and the splice variant (ERβcx/β2) are both expressed within the human endometrium throughout the normal menstrual cycle. J Clin Endocrinol Metab. 2002;87:5265–5273. doi: 10.1210/jc.2002-020502. [DOI] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood H, Brown KM, Markovic D, Pieri LF. Variation in the ESR1 and ESR2 genes and genetic susceptibility to anorexia nervosa. Mol Psychiatry. 2002;7:86–89. doi: 10.1038/sj.mp.4000929. [DOI] [PubMed] [Google Scholar]
- Enmark E, Gustafsson JA. Oestrogen receptors - an overview. J Intern Med. 1999;246:133–138. doi: 10.1046/j.1365-2796.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- Enmark E, Pelto-Huikko M, Grandien K, Lagercrantz S, Lagercrantz J, Fried G, Nordenskjold M, Gustafsson JA. Human estrogen receptor β-gene structure, chromosomal localization, and expression pattern. J Clin Endocrinol Metab. 1997;82:4258–4265. doi: 10.1210/jcem.82.12.4470. [DOI] [PubMed] [Google Scholar]
- Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60:520–528. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- Feigelson HS, Henderson BE. Future possibilities in the prevention of breast cancer: role of genetic variation in breast cancer prevention. Breast Cancer Res. 2000;2:277–282. doi: 10.1186/bcr69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsti A, Zhao C, Israelsson E, Dahlman-Wright K, Gustafsson JA, Hemminki K. Polymorphisms in the estrogen receptor β gene and risk of breast cancer: no association. Breast Cancer Res Treat. 2003;79:409–413. doi: 10.1023/a:1024020609833. [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Zhang X, Pestell R. Nuclear receptor modifications and endocrine cell proliferation. J Steroid Biochem Mol Biol. 2003;85:133–138. doi: 10.1016/s0960-0760(03)00223-1. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Zhang X, Pestell RG. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem Pharmacol. 2004;68:1199–1208. doi: 10.1016/j.bcp.2004.05.037. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Takahashi S, Urano T, Ogawa S, Ouchi Y, Kitamura T, Muramatsu M, Inoue S. Differential expression of estrogen receptor β (ERβ) and its C-terminal truncated splice variant ERβcx as prognostic predictors in human prostatic cancer. Biochem Biophys Res Commun. 2001;289:692–699. doi: 10.1006/bbrc.2001.6038. [DOI] [PubMed] [Google Scholar]
- Fujishita A, Nakane PK, Koji T, Masuzaki H, Chavez RO, Yamabe T, Ishimaru T. Expression of estrogen and progesterone receptors in endometrium and peritoneal endometriosis: an immunohistochemical and in situ hybridization study. Fertil Steril. 1997;67:856–864. doi: 10.1016/s0015-0282(97)81397-0. [DOI] [PubMed] [Google Scholar]
- Gammill HS, Roberts JM. Emerging concepts in preeclampsia investigation. Front Biosci. 2007;12:2403–2411. doi: 10.2741/2242. [DOI] [PubMed] [Google Scholar]
- Ganry O. Phytoestrogens and prostate cancer risk. Prev Med. 2005;41:1–6. doi: 10.1016/j.ypmed.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Gennari L, Becherini L, Falchetti A, Masi L, Massart F, Brandi ML. Genetics of osteoporosis: role of steroid hormone receptor gene polymorphisms. J Steroid Biochem Mol Biol. 2002;81:1–24. doi: 10.1016/s0960-0760(02)00043-2. [DOI] [PubMed] [Google Scholar]
- Gennari L, Merlotti D, De Paola V, Calabro A, Becherini L, Martini G, Nuti R. Estrogen receptor gene polymorphisms and the genetics of osteoporosis: a HuGE review. Am J Epidemiol. 2005;161:307–320. doi: 10.1093/aje/kwi055. [DOI] [PubMed] [Google Scholar]
- Gennari L, Merlotti D, Valleggi F, Martini G, Nuti R. Selective estrogen receptor modulators for postmenopausal osteoporosis: current state of development. Drugs Aging. 2007;24:361–379. doi: 10.2165/00002512-200724050-00002. [DOI] [PubMed] [Google Scholar]
- Georgiou I, Syrrou M, Bouba I, Dalkalitsis N, Paschopoulos M, Navrozoglou I, Lolis D. Association of estrogen receptor gene polymorphisms with endometriosis. Fertil Steril. 1999;72:164–166. doi: 10.1016/s0015-0282(99)00198-3. [DOI] [PubMed] [Google Scholar]
- Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- Girault I, Andrieu C, Tozlu S, Spyratos F, Bieche I, Lidereau R. Altered expression pattern of alternatively spliced estrogen receptor β transcripts in breast carcinoma. Cancer Lett. 2004;215:101–112. doi: 10.1016/j.canlet.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Godsland IF. Effects of postmenopausal hormone replacement therapy on lipid, lipoprotein, and apolipoprotein (a) concentrations: analysis of studies published from 1974-2000. Fertil Steril. 2001;75:898–915. doi: 10.1016/s0015-0282(01)01699-5. [DOI] [PubMed] [Google Scholar]
- Gold B, Kalush F, Bergeron J, Scott K, Mitra N, Wilson K, Ellis N, Huang H, Chen M, Lippert R, Halldorsson BV, Woodworth B, White T, Clark AG, Parl FF, Broder S, Dean M, Offit K. Estrogen receptor genotypes and haplotypes associated with breast cancer risk. Cancer Res. 2004;64:8891–8900. doi: 10.1158/0008-5472.CAN-04-1256. [DOI] [PubMed] [Google Scholar]
- Grady D, Applegate W, Bush T, Furberg C, Riggs B, Hulley SB. Heart and Estrogen/progestin Replacement Study (HERS): design, methods, and baseline characteristics. Control Clin Trials. 1998;19:314–335. doi: 10.1016/s0197-2456(98)00010-5. [DOI] [PubMed] [Google Scholar]
- Grans H, Nilsson M, Dahlman-Wright K, Evengard B. Reduced levels of oestrogen receptor β mRNA in Swedish patients with chronic fatigue syndrome. J Clin Pathol. 2007;60:195–198. doi: 10.1136/jcp.2005.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray IC, Campbell DA, Spurr NK. Single nucleotide polymorphisms as tools in human genetics. Hum Mol Genet. 2000;9:2403–2408. doi: 10.1093/hmg/9.16.2403. [DOI] [PubMed] [Google Scholar]
- Green S, Kumar V, Krust A, Walter P, Chambon P. Structural and functional domains of the estrogen receptor. Cold Spring Harbor Symp Quan Biol. 1986a;51:751–758. doi: 10.1101/sqb.1986.051.01.088. [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Greene G, Krust A, Goffin C, Jensen E, Scrace G, Waterfield M, Chambon P. Cloning of the human oestrogen receptor cDNA. J Steroid Biochem. 1986b;24:77–83. doi: 10.1016/0022-4731(86)90035-x. [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986c;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- Hakansson A, Westberg L, Nilsson S, Buervenich S, Carmine A, Holmberg B, Sydow O, Olson L, Johnels B, Eriksson E, Nissbrandt H. Interaction of polymorphisms in the genes encoding interleukin-6 and estrogen receptor β on the susceptibility to Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005;133:88–92. doi: 10.1002/ajmg.b.30136. [DOI] [PubMed] [Google Scholar]
- Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- Hall JM, Korach KS. Analysis of the molecular mechanisms of human estrogen receptors αand β reveals differential specificity in target promoter regulation by xenoestrogens. J Biol Chem. 2002;277:44455–44461. doi: 10.1074/jbc.M200849200. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- Hedelin M, Balter KA, Chang ET, Bellocco R, Klint A, Johansson JE, Wiklund F, Thellenberg-Karlsson C, Adami HO, Gronberg H. Dietary intake of phytoestrogens, estrogen receptor-β polymorphisms and the risk of prostate cancer. Prostate. 2006;66:1512–1520. doi: 10.1002/pros.20487. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Herrington DM. Role of estrogen receptor-α in pharmacogenetics of estrogen action. Curr Opin Lipidol. 2003;14:145–150. doi: 10.1097/00041433-200304000-00005. [DOI] [PubMed] [Google Scholar]
- Herrington DM, Klein KP. Pharmacogenetics of estrogen replacement therapy. J Appl Physiol. 2001;91:2776–2784. doi: 10.1152/jappl.2001.91.6.2776. [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- Hirata S, Shoda T, Kato J, Hoshi K. The multiple untranslated first exons system of the human estrogen receptor β (ERβ) gene. J Steroid Biochem Mol Biol. 2001;78:33–40. doi: 10.1016/s0960-0760(01)00071-1. [DOI] [PubMed] [Google Scholar]
- Hirata S, Shoda T, Kato J, Hoshi K. Isoform/variant mRNAs for sex steroid hormone receptors in humans. Trends Endocrinol Metab. 2003;14:124–129. doi: 10.1016/s1043-2760(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Horvath LG, Henshall SM, Lee CS, Head DR, Quinn DI, Makela S, Delprado W, Golovsky D, Brenner PC, O’Neill G, Kooner R, Stricker PD, Grygiel JJ, Gustafsson JA, Sutherland RL. Frequent loss of estrogen receptor-β expression in prostate cancer. Cancer Res. 2001;61:5331–5335. [PubMed] [Google Scholar]
- Huang J, Li X, Hilf R, Bambara RA, Muyan M. Molecular basis of therapeutic strategies for breast cancer. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:379–396. doi: 10.2174/156800805774912944. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Qiao T, Bambara RA, Hilf R, Muyan M. A tale of two estrogen receptors (ERs): how differential ER-estrogen responsive element interactions contribute to subtype-specific transcriptional responses. Nucl Recept Signal. 2006;4:e015. doi: 10.1621/nrs.04015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li X, Yi P, Hilf R, Bambara RA, Muyan M. Targeting estrogen responsive elements (EREs): design of potent transactivators for ERE-containing genes. Mol Cell Endocrinol. 2004;218:65–78. doi: 10.1016/j.mce.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Hulley S, Furberg C, Barrett-Connor E, Cauley J, Grady D, Haskell W, Knopp R, Lowery M, Satterfield S, Schrott H, Vittinghoff E, Hunninghake D. Noncardiovascular disease outcomes during 6.8 years of hormone therapy: Heart and Estrogen/progestin Replacement Study follow-up (HERS II) JAMA. 2002;288:58–66. doi: 10.1001/jama.288.1.58. [DOI] [PubMed] [Google Scholar]
- Imamov O, Shim GJ, Warner M, Gustafsson JA. Estrogen receptor β in health and disease. Biol Reprod. 2005;73:866–871. doi: 10.1095/biolreprod.105.043497. [DOI] [PubMed] [Google Scholar]
- Inoue S, Ogawa S, Horie K, Hoshino S, Goto W, Hosoi T, Tsutsumi O, Muramatsu M, Ouchi Y. An estrogen receptor β isoform that lacks exon 5 has dominant negative activity on both ERα and ERβ. Biochem Biophys Res Commun. 2000;279:814–819. doi: 10.1006/bbrc.2000.4010. [DOI] [PubMed] [Google Scholar]
- Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–1989. [PubMed] [Google Scholar]
- Ji Q, Liu PI, Elshimali Y, Stolz A. Frequent loss of estrogen and progesterone receptors in human prostatic tumors determined by quantitative real-time PCR. Mol Cell Endocrinol. 2005;229:103–110. doi: 10.1016/j.mce.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Joe I, Ramirez VD. Binding of estrogen and progesterone-BSA conjugates to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the effects of the free steroids on GAPDH enzyme activity: physiological implications. Steroids. 2001;66:529–538. doi: 10.1016/s0039-128x(00)00220-8. [DOI] [PubMed] [Google Scholar]
- Jones ME, Simpson ER. Oestrogens in male reproduction. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14:505–516. doi: 10.1053/beem.2000.0094. [DOI] [PubMed] [Google Scholar]
- Jordan VC, Morrow M. Tamoxifen, raloxifene, and the prevention of breast cancer. Endocr Rev. 1999;20:253–278. doi: 10.1210/edrv.20.3.0368. [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JY, Song KS, Lee YH, Seo JS, Jelinek J, Gold-schmidt-Clermont PJ, Issa JP. Epigenetic changes in estrogen receptor β gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim Biophys Acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- Kitawaki J, Kado N, Ishihara H, Koshiba H, Kitaoka Y, Honjo H. Endometriosis: the pathophysiology as an estrogen-dependent disease. J Steroid Biochem Mol Biol. 2002;83:149–155. doi: 10.1016/s0960-0760(02)00260-1. [DOI] [PubMed] [Google Scholar]
- Kleyn PW, Vesell ES. Genetic variation as a guide to drug development. Science. 1998;281:1820–1821. doi: 10.1126/science.281.5384.1820. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousteni S, Bellido T, Plotkin LI, O’Brien CA, Bodenner DL, Han L, Han K, DiGregorio GB, Katzenellenbogen JA, Katzenellenbogen BS, Roberson PK, Weinstein RS, Jilka RL, Manolagas SC. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, McInerney EM, Katzenellenbogen BS. Ligand-dependent, transcriptionally productive association of the amino- and carboxyl-terminal regions of a steroid hormone nuclear receptor. Proc Natl Acad Sci USA. 1995;92:12314–12318. doi: 10.1073/pnas.92.26.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Wong J. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur J Biochem. 2002;269:2275–2283. doi: 10.1046/j.1432-1033.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA, Smithies O. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci USA. 1998;95:15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Gustafsson JA. The novel estrogen receptor-β subtype: potential role in the cell- and promoter-specific actions of estrogens and anti-estrogens. FEBS Lett. 1997;410:87–90. doi: 10.1016/s0014-5793(97)00413-4. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, Gustafsson JA. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- Kuller LH. Hormone replacement therapy and risk of cardiovascular disease: implications of the results of the Women’s Health Initiative. Arterioscler Thromb Vasc Biol. 2003;23:11–16. doi: 10.1161/01.atv.0000046033.32478.6d. [DOI] [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Harris JM, Mann D, Lemmon H, Coates J, Cumming A, St-Clair D, Lendon C. Are the estrogen receptors involved in Alzheimer’s disease? Neurosci Lett. 2001;306:193–197. doi: 10.1016/s0304-3940(01)01806-7. [DOI] [PubMed] [Google Scholar]
- Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Lau HH, Ho AY, Luk KD, Kung AW. Estrogen receptor β gene polymorphisms are associated with higher bone mineral density in premenopausal, but not postmenopausal southern Chinese women. Bone. 2002;31:276–281. doi: 10.1016/s8756-3282(02)00827-x. [DOI] [PubMed] [Google Scholar]
- Lau KM, Mok SC, Ho SM. Expression of human estrogen receptor-α and -β, progesterone receptor, and androgen receptor mRNA in normal and malignant ovarian epithelial cells. Proc Natl Acad Sci USA. 1999;96:5722–5727. doi: 10.1073/pnas.96.10.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader JE, Wang C, Popov VM, Fu M, Pestell RG. Epigenetics and the estrogen receptor. Ann N Y Acad Sci. 2006;1089:73–87. doi: 10.1196/annals.1386.047. [DOI] [PubMed] [Google Scholar]
- Leav I, Lau KM, Adams JY, McNeal JE, Taplin ME, Wang J, Singh H, Ho SM. Comparative studies of the estrogen receptors β and α and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am J Pathol. 2001;159:79–92. doi: 10.1016/s0002-9440(10)61676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GH, Kim SH, Choi YM, Suh CS, Kim JG, Moon SY. Estrogen receptor β gene +1730 G/A polymorphism in women with endometriosis. Fertil Steril. 2007;88:785–788. doi: 10.1016/j.fertnstert.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Leung YK, Mak P, Hassan S, Ho SM. Estrogen receptor (ER)-β isoforms: a key to understanding ER-β signaling. Proc Natl Acad Sci USA. 2006;103:13162–13167. doi: 10.1073/pnas.0605676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ER. Cellular functions of plasma membrane estrogen receptors. Steroids. 2002;67:471–475. doi: 10.1016/s0039-128x(01)00179-9. [DOI] [PubMed] [Google Scholar]
- Li LC, Yeh CC, Nojima D, Dahiya R. Cloning and characterization of human estrogen receptor β promoter. Biochem Biophys Res Commun. 2000;275:682–689. doi: 10.1006/bbrc.2000.3363. [DOI] [PubMed] [Google Scholar]
- Li X, Huang J, Fluharty BR, Huang Y, Nott SL, Muyan M. What are comparative studies telling us about the mechanism of ERβ action in the ERE-dependent E2 signaling pathway? J Steroid Biochem Mol Biol. 2008;109:266–272. doi: 10.1016/j.jsbmb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Huang J, Yi P, Bambara RA, Hilf R, Muyan M. Single-chain estrogen receptors (ERs) reveal that the ERα/β heterodimer emulates functions of the ERα dimer in genomic estrogen signaling pathways. Mol Cell Biol. 2004a;24:7681–7694. doi: 10.1128/MCB.24.17.7681-7694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004b;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Licznar A, Caporali S, Lucas A, Weisz A, Vignon F, Lazennec G. Identification of genes involved in growth inhibition of breast cancer cells transduced with estrogen receptor. FEBS Lett. 2003;553:445–450. doi: 10.1016/s0014-5793(03)01090-1. [DOI] [PubMed] [Google Scholar]
- Lin Z, Altman RB. Finding haplotype tagging SNPs by use of principal components analysis. Am J Hum Genet. 2004;75:850–861. doi: 10.1086/425587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven MA, Wood JR, Nardulli AM. Interaction of estrogen receptors α and β with estrogen response elements. Mol Cell Endocrinol. 2001;181:151–163. doi: 10.1016/s0303-7207(01)00491-9. [DOI] [PubMed] [Google Scholar]
- Luckhaus C, Spiegler C, Ibach B, Fischer P, Wichart I, Sterba N, Gatterer G, Rainer M, Jungwirth S, Huber K, Tragl KH, Grünblatt E, Riederer P, Sand PG. Estrogen receptor β gene (ESRβ) 3’-UTR variants in Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20:322–323. doi: 10.1097/01.wad.0000213861.12484.33. [DOI] [PubMed] [Google Scholar]
- Luisi S, Galleri L, Marini F, Ambrosini G, Brandi ML, Petraglia F. Estrogen receptor gene polymorphisms are associated with recurrence of endometriosis. Fertil Steril. 2006;85:764–766. doi: 10.1016/j.fertnstert.2005.08.038. [DOI] [PubMed] [Google Scholar]
- Maguire P, Margolin S, Skoglund J, Sun XF, Gustafsson JA, Borresen-Dale AL, Lindblom A. Estrogen receptor β (ESR2) polymorphisms in familial and sporadic breast cancer. Breast Cancer Res Treat. 2005;94:145–152. doi: 10.1007/s10549-005-7697-7. [DOI] [PubMed] [Google Scholar]
- Mandusic V, Nikolic-Vukosavljevic D, Tanic N, Kanjer K, Neskovic-Konstantinovic Z, Celeketic D, Dimitrijevic B. Expression of estrogen receptor β wt isoform (ERβ1) and ERβΔ5 splice variant mRNAs in sporadic breast cancer. J Cancer Res Clin Oncol. 2007;133:571–579. doi: 10.1007/s00432-007-0209-x. [DOI] [PubMed] [Google Scholar]
- Manson JE, Hsia J, Johnson KC, Rossouw JE, Assaf AR, Lasser NL, Trevisan M, Black HR, Heckbert SR, Detrano R, Strickland OL, Wong ND, Crouse JR, Stein E, Cushman M. Estrogen plus progestin and the risk of coronary heart disease. N Engl J Med. 2003;349:523–534. doi: 10.1056/NEJMoa030808. [DOI] [PubMed] [Google Scholar]
- Mansur Ade P, Nogueira CC, Strunz CM, Aldrighi JM, Ramires JA. Genetic polymorphisms of estrogen receptors in patients with premature coronary artery disease. Arch Med Res. 2005;36:511–517. doi: 10.1016/j.arcmed.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Maruyama A, Nakayama T, Sato N, Mizutani Y, Furuya K, Yamamoto T. Association study using single nucleotide polymorphisms in the estrogen receptor β (ESR2) gene for preeclampsia. Hypertens Res. 2004;27:903–909. doi: 10.1291/hypres.27.903. [DOI] [PubMed] [Google Scholar]
- McDonnell DP. The Molecular Pharmacology of SERMs. Trends Endocrinol Metab. 1999;10:301–311. doi: 10.1016/s1043-2760(99)00177-0. [DOI] [PubMed] [Google Scholar]
- McEwen B. Estrogen actions throughout the brain. Recent Prog Horm Res. 2002;57:357–384. doi: 10.1210/rp.57.1.357. [DOI] [PubMed] [Google Scholar]
- McInerney EM, Tsai MJ, O’Malley BW, Katzenellenbogen BS. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre MH, Kantoff PW, Stampfer MJ, Mucci LA, Parslow D, Li H, Gaziano JM, Abe M, Ma J. Prostate cancer risk and ESR1 TA, ESR2 CA repeat polymorphisms. Cancer Epidemiol Biomarkers Prev. 2007;16:2233–2236. doi: 10.1158/1055-9965.EPI-07-0481. [DOI] [PubMed] [Google Scholar]
- McLeod HL, Papageorgio C, Watters JW. Using genetic variation to optimize cancer chemotherapy. Clin Adv Hematol Oncol. 2003;1:107–111. [PubMed] [Google Scholar]
- Metivier R, Penot G, Carmouche RP, Hubner MR, Reid G, Denger S, Manu D, Brand H, Kos M, Benes V, Gannon F. Transcriptional complexes engaged by apoestrogen receptor-α isoforms have divergent outcomes. EMBO J. 2004;23:3653–3666. doi: 10.1038/sj.emboj.7600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Miller M, Seidler A, Moalemi A, Pearson TA. Normal triglyceride levels and coronary artery disease events: the Baltimore Coronary Observational Long-Term Study. J Am Coll Cardiol. 1998;31:1252–1257. doi: 10.1016/s0735-1097(98)00083-7. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Murphy TC, Lim FL, Moore DJ, Stuckey R, Antrobus K, Kimber I, Orphanides G. Anti-proliferative effect of estrogen in breast cancer cells that re-express ERα is mediated by aberrant regulation of cell cycle genes. J Mol Endocrinol. 2005;34:535–551. doi: 10.1677/jme.1.01677. [DOI] [PubMed] [Google Scholar]
- Moore JT, McKee DD, Slentz-Kesler K, Moore LB, Jones SA, Horne EL, Su JL, Kliewer SA, Lehmann JM, Willson TM. Cloning and characterization of human estrogen receptor β isoforms. Biochem Biophys Res Commun. 1998;247:75–78. doi: 10.1006/bbrc.1998.8738. [DOI] [PubMed] [Google Scholar]
- Morey AK, Pedram A, Razandi M, Prins BA, Hu RM, Biesiada E, Levin ER. Estrogen and progesterone inhibit vascular smooth muscle proliferation. Endocrinology. 1997;138:3330–3339. doi: 10.1210/endo.138.8.5354. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Dotzlaw H, Leygue E, Coutts A, Watson P. The pathophysiological role of estrogen receptor variants in human breast cancer. J Steroid Biochem Mol Biol. 1998;65:175–180. doi: 10.1016/s0960-0760(98)00012-0. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Dotzlaw H, Leygue E, Douglas D, Coutts A, Watson PH. Estrogen receptor variants and mutations. J Steroid Biochem Mol Biol. 1997;62:363–372. doi: 10.1016/s0960-0760(97)00084-8. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Watson PH. Is oestrogen receptor-β a predictor of endocrine therapy responsiveness in human breast cancer? Endocr Relat Cancer. 2006;13:327–334. doi: 10.1677/erc.1.01141. [DOI] [PubMed] [Google Scholar]
- Nadal A, Ropero AB, Fuentes E, Soria B. The plasma membrane estrogen receptor: nuclear or unclear? Trends Pharmacol Sci. 2001;22:597–599. doi: 10.1016/s0165-6147(00)01846-0. [DOI] [PubMed] [Google Scholar]
- Nettles KW, Bruning JB, Gil G, O’Neill EE, Nowak J, Guo Y, Kim Y, DeSombre ER, Dilis R, Hanson RN, Joachimiak A, Greene GL. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007;8:563–568. doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Naessen S, Dahlman I, Linden Hirschberg A, Gustafsson JA, Dahlman-Wright K. Association of estrogen receptor β gene polymorphisms with bulimic disease in women. Mol Psychiatry. 2004;9:28–34. doi: 10.1038/sj.mp.4001402. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Gustafsson JA. Biological role of estrogen and estrogen receptors. Crit Rev Biochem Mol Biol. 2002a;37:1–28. doi: 10.1080/10409230290771438. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Gustafsson JA. Estrogen receptor action. Crit Rev Eukaryot Gene Expr. 2002b;12:237–257. doi: 10.1615/critreveukaryotgeneexpr.v12.i4.10. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- Nowotny P, Kwon JM, Goate AM. SNP analysis to dissect human traits. Curr Opin Neurobiol. 2001;11:637–641. doi: 10.1016/s0959-4388(00)00261-0. [DOI] [PubMed] [Google Scholar]
- Ntukidem NI, Nguyen AT, Stearns V, Rehman M, Schott A, Skaar T, Jin Y, Blanche P, Li L, Lemler S, Hayden J, Krauss RM, Desta Z, Flockhart DA, Hayes DF Consortium on Breast Cancer Pharmacogenomics. Estrogen receptor genotypes, menopausal status, and the lipid effects of tamoxifen. Clin Pharmacol Ther. 2008;83:702–710. doi: 10.1038/sj.clpt.6100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L, Robertson KM, Jones ME, Simpson ER. Estrogen and spermatogenesis. Endocr Rev. 2001;22:289–318. doi: 10.1210/edrv.22.3.0431. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Hosoi T, Shiraki M, Orimo H, Emi M, Muramatsu M, Ouchi Y, Inoue S. Association of estrogen receptor β gene polymorphism with bone mineral density. Biochem Biophys Res Commun. 2000;269:537–541. doi: 10.1006/bbrc.2000.2285. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, Muramatsu M. Molecular cloning and characterization of human estrogen receptor βcx: a potential inhibitor ofestrogen action in human. Nucleic Acids Res. 1998;26:3505–3512. doi: 10.1093/nar/26.15.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olferiev M, Masuda E, Tanaka S, Blank MC, Pricop L. The role of activating protein 1 in the transcriptional regulation of the human FCGR2B promoter mediated by the -343 G -> C polymorphism associated with systemic lupus erythematosus. J Biol Chem. 2007;282:1738–1746. doi: 10.1074/jbc.M605808200. [DOI] [PubMed] [Google Scholar]
- Omoto Y, Eguchi H, Yamamoto-Yamaguchi Y, Hayashi S. Estrogen receptor (ER) β1 and ERβcx/β2 inhibit ERα function differently in breast cancer cell line MCF7. Oncogene. 2003;22:5011–5020. doi: 10.1038/sj.onc.1206787. [DOI] [PubMed] [Google Scholar]
- Omoto Y, Kobayashi S, Inoue S, Ogawa S, Toyama T, Yamashita H, Muramatsu M, Gustafsson JA, Iwase H. Evaluation of oestrogen receptor β wild-type and variant protein expression, and relationship with clinicopathological factors in breast cancers. Eur J Cancer. 2002;38:380–386. doi: 10.1016/s0959-8049(01)00383-5. [DOI] [PubMed] [Google Scholar]
- Palmieri C, Lam EW, Mansi J, MacDonald C, Shousha S, Madden P, Omoto Y, Sunters A, Warner M, Gustafsson JA, Coombes RC. The expression of ERβcx in human breast cancer and the relationship to endocrine therapy and survival. Clin Cancer Res. 2004;10:2421–2428. doi: 10.1158/1078-0432.ccr-03-0215. [DOI] [PubMed] [Google Scholar]
- Parker MG. Transcriptional activation by oestrogen receptors. Biochem Soc Symp. 1998;63:45–50. [PubMed] [Google Scholar]
- Pedersen SB, Bruun JM, Hube F, Kristensen K, Hauner H, Richelsen B. Demonstration of estrogen receptor subtypes α and β in human adipose tissue: influences of adipose cell differentiation and fat depot localization. Mol Cell Endocrinol. 2001;182:27–37. doi: 10.1016/s0303-7207(01)00557-3. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Aitkenhead M, Hughes CC, Levin ER. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J Biol Chem. 2002;277:50768–50775. doi: 10.1074/jbc.M210106200. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Lu B, Leygue E, Murphy LC. Putative functional characteristics of human estrogen receptor-β isoforms. J Mol Endocrinol. 2003;30:13–29. doi: 10.1677/jme.0.0300013. [DOI] [PubMed] [Google Scholar]
- Peter I, Shearman AM, Vasan RS, Zucker DR, Schmid CH, Demissie S, Cupples LA, Kuvin JT, Karas RH, Mendelsohn ME, Housman DE, Benjamin EJ. Association of estrogen receptor β gene polymorphisms with left ventricular mass and wall thickness in women. Am J Hypertens. 2005a;18:1388–1395. doi: 10.1016/j.amjhyper.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Peter I, Shearman AM, Zucker DR, Schmid CH, Demissie S, Cupples LA, Larson MG, Vasan RS, D’Agostino RB, Karas RH, Mendelsohn ME, Housman DE, Levy D. Variation in estrogen-related genes and cross-sectional and longitudinal blood pressure in the Framingham Heart Study. J Hypertens. 2005b;23:2193–2200. doi: 10.1097/01.hjh.0000188728.66183.92. [DOI] [PubMed] [Google Scholar]
- Pike AC. Lessons learnt from structural studies of the oestrogen receptor. Best Pract Res Clin Endocrinol Metab. 2006;20:1–14. doi: 10.1016/j.beem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor β in the presence of a partial agonist and a full antagonist. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirskanen M, Hiltunen M, Mannermaa A, Helisalmi S, Lehtovirta M, Hanninen T, Soininen H. Estrogen receptor β gene variants are associated with increased risk of Alzheimer’s disease in women. Eur J Hum Genet. 2005;13:1000–1006. doi: 10.1038/sj.ejhg.5201447. [DOI] [PubMed] [Google Scholar]
- Poola I, Abraham J, Baldwin K. Identification of ten exon deleted ERβ mRNAs in human ovary, breast, uterus and bone tissues: alternate splicing pattern of estrogen receptor beta mRNA is distinct from that of estrogen receptor alpha. FEBS Lett. 2002a;516:133–138. doi: 10.1016/s0014-5793(02)02521-8. [DOI] [PubMed] [Google Scholar]
- Poola I, Abraham J, Baldwin K, Saunders A, Bhatnagar R. Estrogen receptors β4 and β5 are full length functionally distinct ERβ isoforms: cloning from human ovary and functional characterization. Endocrine. 2005a;27:227–238. doi: 10.1385/ENDO:27:3:227. [DOI] [PubMed] [Google Scholar]
- Poola I, Clarke R, DeWitty R, Leffall LD. Functionally active estrogen receptor isoform profiles in the breast tumors of African American women are different from the profiles in breast tumors of Caucasian women. Cancer. 2002b;94:615–623. doi: 10.1002/cncr.10274. [DOI] [PubMed] [Google Scholar]
- Poola I, Fuqua SA, De Witty RL, Abraham J, Marshallack JJ, Liu A. Estrogen receptor α-negative breast cancer tissues express significant levels of estrogen-independent transcription factors, ERβ1 and ERβ5: potential molecular targets for chemoprevention. Clin Cancer Res. 2005b;11:7579–7585. doi: 10.1158/1078-0432.CCR-05-0728. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. Endometriosis and infertility. Fertil Steril. 2004;82(Suppl 1):S40–45. doi: 10.1016/j.fertnstert.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Price RH, Jr, Butler CA, Webb P, Uht R, Kushner P, Handa RJ. A splice variant of estrogen receptor β missing exon 3 displays altered subnuclear localization and capacity for transcriptional activation. Endocrinology. 2001;142:2039–2049. doi: 10.1210/endo.142.5.8130. [DOI] [PubMed] [Google Scholar]
- Prins GS, Korach KS. The role of estrogens and estrogen receptors in normal prostate growth and disease. Steroids. 2008;73:233–244. doi: 10.1016/j.steroids.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol P, Rey JM, Nirde P, Roger P, Gastaldi M, Laffargue F, Rochefort H, Maudelonde T. Differential expression of estrogen receptor-α and -β messenger RNAs as a potential marker of ovarian carcinogenesis. Cancer Res. 1998;58:5367–5373. [PubMed] [Google Scholar]
- Razandi M, Pedram A, Greene GL, Levin ER. Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERα and ERβ expressed in Chinese hamster ovary cells. Mol Endocrinol. 1999;13:307–319. doi: 10.1210/mend.13.2.0239. [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Levin ER. Plasma membrane estrogen receptors signal to antiapoptosis in breast cancer. Mol Endocrinol. 2000;14:1434–1447. doi: 10.1210/mend.14.9.0526. [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Park ST, Levin ER. Proximal events in signaling by plasma membrane estrogen receptors. J Biol Chem. 2003;278:2701–2712. doi: 10.1074/jbc.M205692200. [DOI] [PubMed] [Google Scholar]
- Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, protea-some-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Rexrode KM, Ridker PM, Hegener HH, Buring JE, Manson JE, Zee RY. Polymorphisms and haplotypes of the estrogen receptor-β gene (ESR2) and cardiovascular disease in men and women. Clin Chem. 2007;53:1749–1756. doi: 10.1373/clinchem.2007.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira F, van Meurs JB, Kant J, Zillikens MC, Stolk L, Beck TJ, Arp P, Schuit SC, Hofman A, Houwing-Duistermaat JJ, van Duijn CM, van Leeuwen JP, Pols HA, Uitterlinden AG. Estrogen receptor β (ESR2) polymorphisms in interaction with estrogen receptor α (ESR1) and insulin-like growth factor I (IGF1) variants influence the risk of fracture in postmenopausal women. J Bone Miner Res. 2006;21:1443–1456. doi: 10.1359/jbmr.060605. [DOI] [PubMed] [Google Scholar]
- Roberts JM, Catov JM. Preeclampsia more than 1 disease: or is it? Hypertension. 2008;51:989–990. doi: 10.1161/HYPERTENSIONAHA.107.100248. [DOI] [PubMed] [Google Scholar]
- Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, Krauss RM, McLeod HL, Ratain MJ, Relling MV, Ring HZ, Shuldiner AR, Weinshilboum RM, Weiss ST. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz K, Hinney A, Ziegler A, Hermann H, Fichter M, Mayer H, Siegfried W, Young JK, Remschmidt H, Hebebrand J. Systematic mutation screening of the estrogen receptor β gene in probands of different weight extremes: identification of several genetic variants. J Clin Endocrinol Metab. 1998;83:4524–4527. doi: 10.1210/jcem.83.12.5471. [DOI] [PubMed] [Google Scholar]
- Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA, Howard BV, Johnson KC, Kotchen JM, Ockene J Writing Group for the Women’s Health Initiative Investigators. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women’s Health Initiative randomized controlled trial. JAMA. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- Safe S. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- Sand P, Luckhaus C, Schlurmann K, Gotz M, Deckert J. Untangling the human estrogen receptor gene structure. J Neural Transm. 2002;109:567–583. doi: 10.1007/s007020200047. [DOI] [PubMed] [Google Scholar]
- Scariano JK, Simplicio SG, Montoya GD, Garry PJ, Baumgartner RN. Estrogen receptor β dinucleotide (CA) repeat polymorphism is significantly associated with bone mineral density in postmenopausal women. Calcif Tissue Int. 2004;74:501–508. doi: 10.1007/s00223-003-0170-x. [DOI] [PubMed] [Google Scholar]
- Schiessl B, Mylonas I, Hantschmann P, Kuhn C, Schulze S, Kunze S, Friese K, Jeschke U. Expression of endothelial NO synthase, inducible NO synthase, and estrogen receptors α and β in placental tissue of normal, preeclamptic, and intrauterine growth-restricted pregnancies. J Histochem Cytochem. 2005;53:1441–1449. doi: 10.1369/jhc.4A6480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Neuhaus D, Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- Secreto FJ, Monroe DG, Dutta S, Ingle JN, Spelsberg TC. Estrogen receptor α/β isoforms, but not βcx, modulate unique patterns of gene expression and cell proliferation in Hs578T cells. J Cell Biochem. 2007;101:1125–1147. doi: 10.1002/jcb.21205. [DOI] [PubMed] [Google Scholar]
- Setiawan VW, Hankinson SE, Colditz GA, Hunter DJ, De Vivo I. Estrogen receptor β (ESR2) polymorphisms and endometrial cancer (United States) Cancer Causes Control. 2004;15:627–633. doi: 10.1023/B:CACO.0000036170.28502.5f. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shearman AM, Karasik D, Gruenthal KM, Demissie S, Cupples LA, Housman DE, Kiel DP. Estrogen receptor β polymorphisms are associated with bone mass in women and men: the Framingham Study. J Bone Miner Res. 2004;19:773–781. doi: 10.1359/JBMR.0301258. [DOI] [PubMed] [Google Scholar]
- Shen LX, Basilion JP, Stanton VP., Jr Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc Natl Acad Sci USA. 1999;96:7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevde NK, Bendixen AC, Dienger KM, Pike JW. Estrogens suppress RANK ligand-induced osteoclast differentiation via a stromal cell independent mechanism involving c-Jun repression. Proc Natl Acad Sci USA. 2000;97:7829–7834. doi: 10.1073/pnas.130200197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoda T, Hirata S, Kato J, Hoshi K. Cloning of the novel isoform of the estrogen receptor β cDNA (ERβ isoform M cDNA) from the human testicular cDNA library. J Steroid Biochem Mol Biol. 2002;82:201–208. doi: 10.1016/s0960-0760(02)00186-3. [DOI] [PubMed] [Google Scholar]
- Silvestri S, Thomsen AB, Gozzini A, Bagger Y, Christiansen C, Brandi ML. Estrogen receptor α and β polymorphisms: is there an association with bone mineral density, plasma lipids, and response to postmenopausal hormone therapy? Menopause. 2006;13:451–461. doi: 10.1097/01.gme.0000182804.14385.a2. [DOI] [PubMed] [Google Scholar]
- Simoncini T, Hafezi-Moghadam A, Brazil DP, Ley K, Chin WW, Liao JK. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Bischoff FZ. Heritability and molecular genetic studies of endometriosis. Ann N Y Acad Sci. 2002;955:239–251. doi: 10.1111/j.1749-6632.2002.tb02785.x. [DOI] [PubMed] [Google Scholar]
- Skakkebaek NE, Rajpert-De Meyts E, Main KM. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum Reprod. 2001;16:972–978. doi: 10.1093/humrep/16.5.972. [DOI] [PubMed] [Google Scholar]
- Soulez M, Parker MG. Identification of novel oestrogen receptor target genes in human ZR75-1 breast cancer cells by expression profiling. J Mol Endocrinol. 2001;27:259–274. doi: 10.1677/jme.0.0270259. [DOI] [PubMed] [Google Scholar]
- Sun J, Meyers MJ, Fink BE, Rajendran R, Katzenellenbogen JA, Katzenellenbogen BS. Novel ligands that function as selective estrogens or antiestrogens for estrogen receptor-α or estrogen receptor-β. Endocrinology. 1999;140:800–804. doi: 10.1210/endo.140.2.6480. [DOI] [PubMed] [Google Scholar]
- Sundarrajan C, Liao WX, Roy AC, Ng SC. Association between estrogen receptor-β gene polymorphisms and ovulatory dysfunctions in patients with menstrual disorders. J Clin Endocrinol Metab. 2001;86:135–139. doi: 10.1210/jcem.86.1.7098. [DOI] [PubMed] [Google Scholar]
- Tang MX, Jacobs D, Stern Y, Marder K, Schofield P, Gurland B, Andrews H, Mayeux R. Effect of oestrogen during menopause on risk and age at onset of Alzheimer’s disease. Lancet. 1996;348:429–432. doi: 10.1016/S0140-6736(96)03356-9. [DOI] [PubMed] [Google Scholar]
- Tateishi Y, Kawabe Y, Chiba T, Murata S, Ichikawa K, Murayama A, Tanaka K, Baba T, Kato S, Yanagisawa J. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004;23:4813–4823. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi Y, Sonoo R, Sekiya Y, Sunahara N, Kawano M, Wayama M, Hirota R, Kawabe Y, Murayama A, Kato S, Kimura K, Yanagisawa J. Turning off estrogen receptor β-mediated transcription requires estrogen-dependent receptor proteolysis. Mol Cell Biol. 2006;26:7966–7976. doi: 10.1128/MCB.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AH, Al-Azzawi F. Immunolocalisation of oestrogen receptor β in human tissues. J Mol Endocrinol. 2000;24:145–155. doi: 10.1677/jme.0.0240145. [DOI] [PubMed] [Google Scholar]
- Thellenberg-Karlsson C, Lindstrom S, Malmer B, Wiklund F, Augustsson-Balter K, Adami HO, Stattin P, Nilsson M, Dahlman-Wright K, Gustafsson JA, Gronberg H. Estrogen receptor β polymorphism is associated with prostate cancer risk. Clin Cancer Res. 2006;12:1936–1941. doi: 10.1158/1078-0432.CCR-05-0269. [DOI] [PubMed] [Google Scholar]
- Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- Treeck O, Pfeiler G, Horn F, Federhofer B, Houlihan H, Vollmer A, Ortmann O. Novel estrogen receptor β transcript variants identified in human breast cancer cells affect cell growth and apoptosis of COS-1 cells. Mol Cell Endocrinol. 2007a;264:50–60. doi: 10.1016/j.mce.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Treeck O, Pfeiler G, Mitter D, Lattrich C, Piendl G, Ortmann O. Estrogen receptor β1 exerts antitumoral effects on SK-OV-3 ovarian cancer cells. J Endocrinol. 2007b;193:421–433. doi: 10.1677/JOE-07-0087. [DOI] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- Von Low EC, Perabo FG, Siener R, Muller SC. Facts and fiction of phytotherapy for prostate cancer: a critical assessment of preclinical and clinical data. In Vivo. 2007;21:189–204. [PubMed] [Google Scholar]
- Wakeling AE. Similarities and distinctions in the mode of action of different classes of antioestrogens. Endocr Relat Cancer. 2000;7:17–28. doi: 10.1677/erc.0.0070017. [DOI] [PubMed] [Google Scholar]
- Wakeling AE, Nicholson RI, Gee JM. Prospects for combining hormonal and nonhormonal growth factor inhibition. Clin Cancer Res. 2001;7:4350s–4355s. discussion 4411s-4412s. [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yoshida S, Negoro K, Kennedy S, Barlow D, Maruo T. Polymorphisms in the estrogen receptor β gene but not estrogen receptor α gene affect the risk of developing endometriosis in a Japanese population. Fertil Steril. 2004;81:1650–1656. doi: 10.1016/j.fertnstert.2004.02.094. [DOI] [PubMed] [Google Scholar]
- Waring SC, Rocca WA, Petersen RC, O’Brien PC, Tangalos EG, Kokmen E. Postmenopausal estrogen replacement therapy and risk of AD: a population-based study. Neurology. 1999;52:965–970. doi: 10.1212/wnl.52.5.965. [DOI] [PubMed] [Google Scholar]
- Watters JW, McLeod HL. Cancer pharmacogenomics: current and future applications. Biochim Biophys Acta. 2003;1603:99–111. doi: 10.1016/s0304-419x(03)00003-9. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen MP, Chen D, Huang SM, Subramanian S, McKinerney E, Katzenellenbogen BS, Stallcup MR, Kushner PJ. Estrogen receptor activation function 1 works by binding p160 co-activator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- Westberg L, Baghaei F, Rosmond R, Hellstrand M, Landen M, Jansson M, Holm G, Bjorntorp P, Eriksson E. Polymorphisms of the androgen receptor gene and the estrogen receptor β gene are associated with androgen levels in women. J Clin Endocrinol Metab. 2001;86:2562–2568. doi: 10.1210/jcem.86.6.7614. [DOI] [PubMed] [Google Scholar]
- Westberg L, Hakansson A, Melke J, Shahabi HN, Nilsson S, Buervenich S, Carmine A, Ahlberg J, Grundell MB, Schulhof B, Klingborg K, Holmberg B, Sydow O, Olson L, Johnels EB, Eriksson E, Nissbrandt H. Association between the estrogen receptor β gene and age of onset of Parkinson’s disease. Psychoneuroendocrinology. 2004a;29:993–998. doi: 10.1016/j.psyneuen.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Westberg L, Ho HP, Baghaei F, Nilsson S, Melke J, Rosmond R, Holm G, Bjorntorp P, Eriksson E. Polymorphisms in oestrogen and progesterone receptor genes: possible influence on prolactin levels in women. Clin Endocrinol (Oxf) 2004b;61:216–223. doi: 10.1111/j.1365-2265.2004.02083.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson HA, Dahllund J, Liu H, Yudkovitz J, Cai SJ, Nilsson S, Schaeffer JM, Mitra SW. Identification and characterization of a functionally distinct form of human estrogen receptor β. Endocrinology. 2002;143:1558–1561. doi: 10.1210/endo.143.4.8829. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Xu L, Pan-Hammarstrom Q, Forsti A, Hemminki K, Hammarstrom L, Labuda D, Gustafsson JA, Dahlman-Wright K. Human estrogen receptor β548 is not a common variant in three distinct populations. Endocrinology. 2003;144:3541–3546. doi: 10.1210/en.2002-0118. [DOI] [PubMed] [Google Scholar]
- Yang SH, Liu R, Perez EJ, Wen Y, Stevens SM, Jr, Valencia T, Brun-Zinkernagel AM, Prokai L, Will Y, Dykens J, Koulen P, Simpkins JW. Mitochondrial localization of estrogen receptor β. Proc Natl Acad Sci USA. 2004;101:4130–4135. doi: 10.1073/pnas.0306948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Bhagat S, Hilf R, Bambara RA, Muyan M. Differences in the abilities of estrogen receptors to integrate activation functions are critical for subtype-specific transcriptional responses. Mol Endocrinol. 2002a;16:1810–1827. doi: 10.1210/me.2001-0323. [DOI] [PubMed] [Google Scholar]
- Yi P, Driscoll MD, Huang J, Bhagat S, Hilf R, Bambara RA, Muyan M. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ERα and ERβ. Mol Endocrinol. 2002b;16:674–693. doi: 10.1210/mend.16.4.0810. [DOI] [PubMed] [Google Scholar]
- Yoneda K, Tanji Y, Ikeda N, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S. Influence of adjuvant tamoxifen treatment on bone mineral density and bone turnover markers in postmenopausal breast cancer patients in Japan. Cancer Lett. 2002;186:223–230. doi: 10.1016/s0304-3835(02)00345-2. [DOI] [PubMed] [Google Scholar]
- Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype block structure and its applications to association studies: power and study designs. Am J Hum Genet. 2002;71:1386–1394. doi: 10.1086/344780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yu YY. Polymorphisms of short tandem repeat of genes and breast cancer susceptibility. Eur J Surg Oncol. 2007;33:529–534. doi: 10.1016/j.ejso.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Zhang X, Leung YK, Ho SM. AP-2 regulates the transcription of estrogen receptor (ER)-β by acting through a methylation hotspot of the 0N promoter in prostate cancer cells. Oncogene. 2007;26:7346–7354. doi: 10.1038/sj.onc.1210537. [DOI] [PubMed] [Google Scholar]
- Zhao C, Lam EW, Sunters A, Enmark E, De Bella MT, Coombes RC, Gustafsson JA, Dahlman-Wright K. Expression of estrogen receptor β isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene. 2003;22:7600–7606. doi: 10.1038/sj.onc.1207100. [DOI] [PubMed] [Google Scholar]
- Zhao C, Xu L, Otsuki M, Toresson G, Koehler K, Pan-Hammarstrom Q, Hammarstrom L, Nilsson S, Gustafsson JA, Dahlman-Wright K. Identification of a functional variant of estrogen receptor β in an African population. Carcinogenesis. 2004;25:2067–2073. doi: 10.1093/carcin/bgh215. [DOI] [PubMed] [Google Scholar]
- Zheng SL, Zheng W, Chang BL, Shu XO, Cai Q, Yu H, Dai Q, Xu J, Gao YT. Joint effect of estrogen receptor β sequence variants and endogenous estrogen exposure on breast cancer risk in Chinese women. Cancer Res. 2003;63:7624–7629. [PubMed] [Google Scholar]
- Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, McNeal JE, Ho SM. Dynamic regulation of estrogen receptor-β expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


