Abstract
GRIM-19 (Gene associated with Retinoid-Interferon-induced Mortality 19) is a novel tumor suppressor regulated by Interferon/retinoid combination. We have recently shown that GRIM-19 inhibits v-Src-induced oncogenic transformation and metastatic behavior of cells. Oncogenic v-Src induces cell motility by cytoskeletal remodeling especially the formation of podosomes and. Here we show that GRIM-19 inhibited the v-Src-induced cell motility by inhibiting cytoskeletal remodeling i.e., podosome formation. We also show that the N-terminus of GRIM-19 played a major role in this process and identified critical residues in this region. More importantly, we show that tumor-associated GRIM-19 mutations disrupted its ability to inhibit v-Src-induced cell motility. These actions appear to occur independently of STAT3, a known target of GRIM-19-mediated inhibition. Lastly, tumor-associated GRIM-19 mutants significantly lost their ability to control v-Src-induced metastases in vivo, indicating the biological and pathological significance of these observations.
Introduction
The Interferon (IFN) group of cytokines regulates antitumor, antiviral and immune responses. Clinical studies demonstrated the efficacy of IFN in treatment of different cancers such as chronic myelogenous leukemia (CML), Kaposi's sarcoma, lymphomas, hairy cell leukemia, melanoma and renal cell carcinoma (Jonasch and Haluska, 2001). More strikingly the anti-proliferative effects of IFN are augmented when combined with retinoids, a class of vitamin A metabolites and synthetic homologues. For example, co-treatment of MCF-7 breast cancer cells with retinoic acid (RA) and IFN-β leads to synergistic upregulation of a set of apoptotic genes (TRAIL, DR3, FAS etc) (Ma et al., 2001) and suppression of tumor growth via apoptosis (Kolla et al., 1996; Lindner et al., 1997a). Based on these observation, we hypothesized that cell death by co-treatment of RA and IFN-β is induced through stimulation of a group of genes at the molecular level. To identify these apoptotic genes, we applied a genetic method, and isolated a novel group of genes and were collectively named Genes associated with Retinoid-Interferon-induced Mortality (GRIMs) (Angell et al., 2000). Knock-down of any GRIM ablated IFN-β/RA-induced cell death response. One such novel gene product, GRIM-19, upon ectopic expression in cells, manifested as slower growth to apoptosis of cells depending on cell lines and expression levels. Subsequently, GRIM-19 was shown to been a component of mitochondrial ETS complex I (Fearnley et al., 2001). The first direct evidence to implicate GRIM-19 as a potential tumor suppressor was illustrated by direct suppression of the transcriptional activity of STAT3 (Lufei et al., 2003; Zhang et al., 2003). Additionally, blockade of GRIM-19 by viral encoded products (Seo et al., 2002), mutations in thyroid tumors (Maximo et al., 2005) and loss of expression in renal cell carcinoma (Alchanati et al., 2006) and in some prostate cancers (Zhanget al., 2008) indicate a typical tumor suppressor-like character for GRIM-19. Recently, we have demonstrated that the anti-tumor effect of GRIM-19 was exerted at multiple levels on oncogenic v-Src through inhibition of cellular transformation, injury-induced and mitogen-driven migration of cells and in vivo tumor formation (Kalakonda et al., 2007a).
Expression of v-Src, controls several cellular activities including cell morphology, adhesion, motility, invasion etc (Martin, 2001) by interacting with plasma membrane and inducing tyrosine phosphorylation of membrane-associated and other cellular proteins (Frame et al., 2002). By associating with cytoskeleton, v-Src causes changes in cell morphology and structure. For example, cells transformed by v-Src convert from a well-spread shape to an extremely rounded shape, which is a characteristic of high motility and invasiveness of transformed cells. Most of these changes were associated with a reorganization of the cytoskeleton, including loss of stress fibers, adhesion plaques and formation of podosomes (dot-like structures rich with bundled F-actin) (Hakak et al., 2000). One of the most important substrates of v-Src is cortactin, a filamentous actin-bundling protein (Wu and Parsons, 1993) which serves as a scaffold protein linking the v-Src signaling pathway to the organization of cytoskeleton. For example, tyrosine phosphorylation of cortactin by v-Src is essential for the formation of podosomes. The proteolytic activity of podosomes causes cellular matrix degradation, leading to invasive capability of transformed cells. Tyrosyl phosphorylation of cortactin by v-Src occurs in a progressive manner (Head et al., 2003). Studies have shown that phosphorylation of cortactin by v-Src reduced the affinity of cortactin for actin and its ability to cross-link actin filaments (Huang et al., 1997). Depletion of cortactin lead to a specific loss of podosomes and re-expression of cortactin mutants lacking phospho-accepting residues (Y421, Y466 and Y482) does not restore podosome formation (Tehrani et al., 2006).
This attenuating effect of GRIM-19 on v-Src-induced cell migration lead us to explore the mechanism by which GRIM-19 suppresses cancer cell metastasis and whether tumor-associated GRIM-19 mutations affect the function in this aspect. In the present study, we demonstrate that wild-type GRIM-19 can reverse v-Src-induced cytoskeleton remodeling, especially formation of podosome. In addition, we demonstrate that the N-terminus of GRIM-19 is required for suppressing v-Src-induced cell motility. Experimental and tumor-associated mutations in the N-terminal region of GRIM-19 significantly lost their ability to suppress v-Src-induced cell cytoskeleton restructuring and cortactin phosphorylation, compared to wild-type GRIM-19.
Results
GRIM-19 suppresses v-Src-induced cellular morphology change and podosome formation
Our previous study demonstrated that GRIM-19 inhibits v-Src-induced transformation at multiple levels, including cell motility. Cells transformed by v-Src go through a series of morphologic and cytoskeletal changes, which permits loss of cell adhesion and a potential for motility. Therefore, we first studied whether GRIM-19 affected v-Src-induced morphologic change and cytoskeletal restructuring. Since most cancer cells possess multiple oncogenic changes, we used a non-oncogenic rat fibroblast line 3Y1, where introduction of a single oncogene like v-Src is sufficient to cause cellular transformation. Cells transfected with expression vector coding v-Src and control vector were used for this study. After infecting these cells with lentivirus coding for GRIM-19, cells were selected for 5 days with puromycin to eliminate uninfected cells (usually less than 5% under these conditions). In presence of v-Src around 70% of cells appeared rounded and ready to detach from the substratum compared to the controls that were well-spread in shape indicative of strong adherence (Fig. 1A). This rounded appearance of v-Src-transformed cells returned to the morphology of naïve 3Y1 cells upon expression of GRIM-19. However, expression of GRIM-19 alone did not cause any morphologic change (Fig. 1A). The expression of exogenous v-Src and GRIM-19 were confirmed by Western blot analysis (Fig. 2).
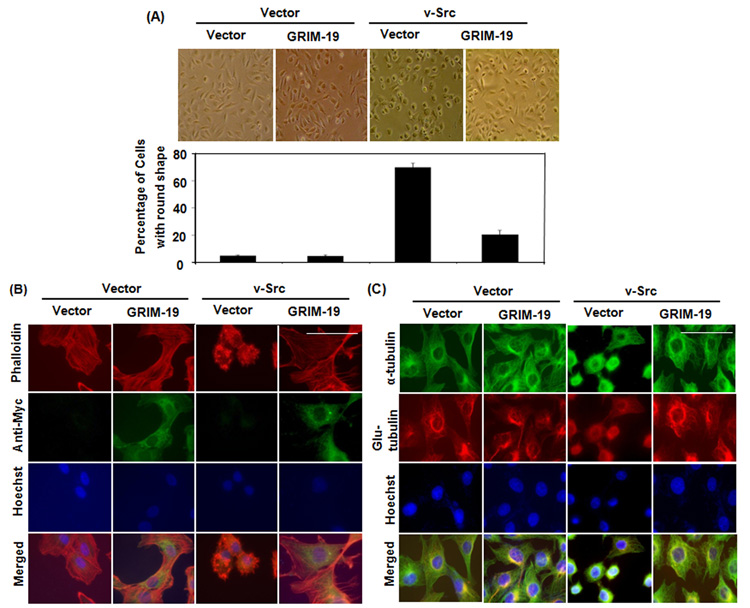
Fig. 1. GRIM-19 suppressed v-Src-induced morphologic changes and cytoskeleton remodeling.
(A) Phase contrast photomicrographs of 3Y1 cells expressing GRIM-19, v-Src, v-Src/GRIM-19 and control vector. The graph below show the percentage of cells with rounded morphology as quantified from various fields. Magnification: 10×. Mean percentages and SE were plotted. (B) Immunofluorescent images of cells expressing, control vector GRIM-19, v-Src, and v-Src/GRIM-19 showing actin network (Red), exogenous GRIM-19 (green) and nucleus (Blue). Magnification: 100×. Scale bar = 50µm (C) Immunofluorescent images of cells expressing control vector, GRIM-19, v-Src, and v-Src/GRIM-19 showing dynamic microtubules (green), stable microtubules (red) and nucleus (blue). Magnification: 100×. Scale bar = 50µm.
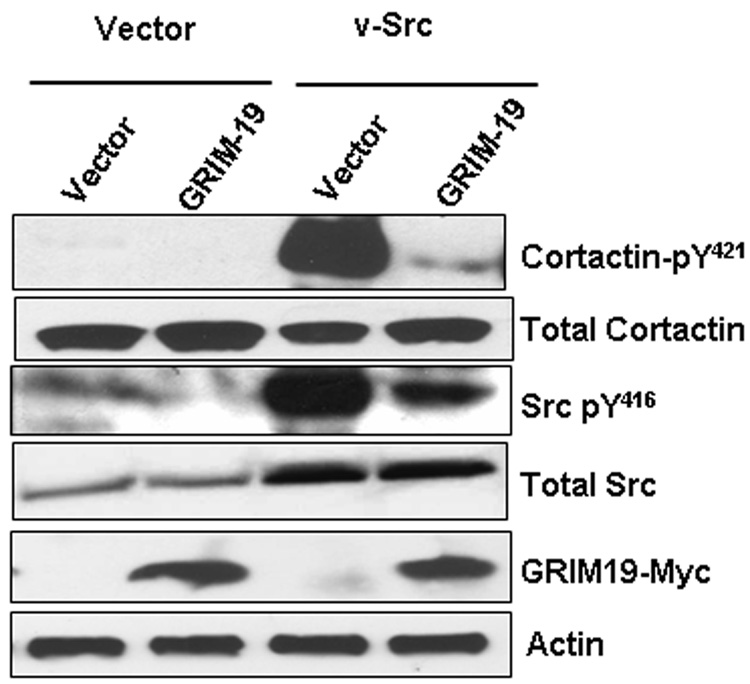
Fig. 2. GRIM-19 suppressed phosphorylation of cortactin by v-Src.
Representative Western blot profiles of total cellular lysates probed with the indicated antibodies and signals generated using ECL. Typically, duplicate gels were run in parallel and after transfer, the membrane was cut horizontally aided by protein molecular weight marker front (pre-stained) and incubated with the following antibodies separately; a) cortactin (1:2000) or phosphorylated cortactin (1:1000), b) Myc-tag (1:1000), c) Actin (1:2000) and d) Src (1:1000) or phosphorylated Src (1:1000). In some cases, a membrane was first probed with an antibody that detects the phosphorylated form of the protein (pY421 Cortactin or pY416 v-Src), then striped and reprobed with an antibody to detect the total protein (Cortactin or Src).
The cytoskeleton, including actin and tubulin, maintains cell morphology and modulates motility. Therefore, we further examined the effect of GRIM-19 on v-Src-induced cytoskeletal restructuring. As shown in Fig 1B, expression of v-Src led to loss of stress fibers and formation of podosomes, compared to the control cells upon phalloidin staining. The podosomes are structures formed by bundled actin, whose proteolytic activity enables the transformed cells to lose adhesion and enhance motility. Expression of GRIM-19 restored the formation of stress fibers and inhibited the formation of podosomes in v-Src transformed cells (Fig 1B). This probably explains the assumption of flat and adherent morphology of v-Src-transformed cells in the presence of GRIM-19. There was no significant change in the tubulin network in the v-Src-transformed cells (Fig 1C). However, the tubulin staining pattern was more condensed in the v-Src-transformed cells because of the round shape of these cells. Expression of GRIM-19 recovered the tubulin staining pattern similar to that found in control cells. Surprisingly, in the v-Src transformed cells, there was a slight decrease in the density of detyrosinated tubulin. Although all cells were photographed at the same magnification, the v-src expressing cells appear to have a smaller size nucleus. This could be due an impact of cytoskeletal reorganization on the nucleus.
GRIM-19 reduced the phosphorylation status of cortactin catalyzed by v-Src
The effect of v-Src on actin matrix is mediated by cortactin, an F-actin-bundling protein that localizes to podosomes and lamellipodia. Tyrosyl phosphorylation of cortactin is required for the formation of podosomes, which impart high motility to v-Src-transformed cells. In the next experiment, we examined whether GRIM-19 interfered with v-Src-induced tyrosyl phosphorylation of cortactin. There are three critical tyrosine residues (Y421, Y466 and Y482) located in the prolyl-rich region of cortactin. Sequential phosphorylation of these three residues, in which phosphorylation of Y421 is required for phosphorylation of the remaining two, occurs in v-Src-transformed cells. Therefore, we performed Western blot analyses with Y421-specific antibodies. Notably, v-Src induced a robust phosphorylation of cortactin, which was strongly suppressed in presence of GRIM-19 (Fig 2). The difference in the tyrosyl phosphorylation was not attributable to a differential loading of proteins in these lanes, as revealed by equivalent levels of total cortactin in these lanes (Fig. 2B, bottom blots). Surprisingly, GRIM-19 also suppressed the tyrosyl phosphorylation of v-Src. This difference was not due to different level of exogenous v-Src protein in the cells, since a Src monoclonal antibody detected comparable level of total Src proteins, in the presence and absence of GRIM-19. As expected, the v-Src transfectants had a higher (~2 fold) level of total Src protein compared to the empty vector-expressing cells. Thus, GRIM-19 inhibits the activity of v-Src by reducing the tyrosyl phosphorylation status of cortactin. However, we were unable to detect an interaction between either v-Src and GRIM-19 or cortactin and GRIM-19 upon immunoprecipitation analysis (data not shown).
STAT3 did not influence the suppression of v-Src-induced podosome formation
We and others have shown earlier that STAT3 is inhibited by GRIM-19. Therefore, we next examined if v-Src-induced podosome formation required STAT3 because of the following observations: 1) STAT3 is a prominent substrate of v-Src and phosphorylation of STAT3 plays a very important role in v-Src-induced transformation (Bromberg et al., 1998); 2) GRIM-19 inhibits STAT3-induced gene expression as well as cell growth (Zhang et al., 2003); and 3) STAT3 has been suggested to regulate the assembly of cytoskeleton (Ng et al., 2006). Therefore, we knocked down the endogenous STAT3 using lentiviral vectors coding for STAT3 shRNA. The specific loss of STAT3 protein expression was verified by Western blot analysis of cellular lysates (Fig S1A). STAT3 shRNA significantly promoted the loss of endogenous STAT3 compared to the controls. However, knockdown of STAT3 neither affected the podosome formation in the v-Src-transformed 3Y1 cells, nor reversed the inhibition of podosome formation by GRIM-19 (Fig S1B). These results suggest that the inhibitory effect of GRIM-19 on v-Src-induced podosome formation occurs through a STAT3-independent pathway. Further, we checked the effect of STAT3 knockdown on the microtubule (MT) network. As shown in Fig S1C, knockdown of STAT3 severely impaired the MT network, as demonstrated by α-tubulin and detyrosinated tubulin staining in both v-Src and v-Src/GRIM-19 expressing cells. Thus, the loss of STAT3 expression severely impaired only the MT network and does not seem to affect podosome formation induced by v-Src. However, STAT3 down-regulation also perturbed the MT network in naïve 3Y1 cells (data not shown).
The N-terminus of GRIM-19 is necessary for suppressing v-Src-induced podosome formation
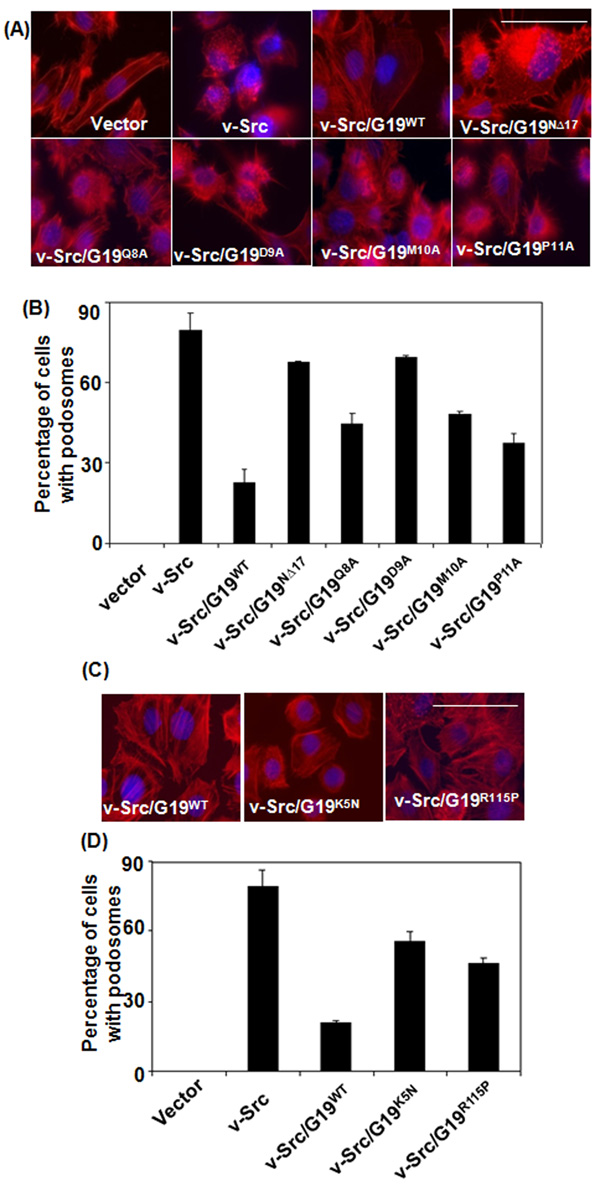
To determine the functional domains required for the GRIM-19 protein to suppress the v-Src induced effects, we used a deletion mutant lacking the N-terminal 17 amino acids and compared its effects on v-Src-induced podosome formation to wildtype GRIM-19. Eighty percent of the v-Src expressing cells possessed podosomes, which was robustly suppressed by wildtype GRIM-19. Unlike wildtype GRIM-19, the NΔ17 mutant lost its ability to suppress “dot-like” podosome formation (Figs 3A & B). To further narrowly define the residues critical for GRIM-19-mediated suppression of podosome formation we searched for conserved motifs in the N-terminus of GRIM-19 protein. These studies identified a small predicted motif within the N-terminus of GRIM-19 similar to the one found in some ECHO- and Human influenza viral proteins. Notably, 4 amino acids located between 8–11 were conserved among these proteins. Therefore, we individually mutated amino acids 8 to 11 and constructed lentiviral expression vectors. The lentiviral particles coding for mutant GRIM-19 (Q8A, D9A, M10A and P11A) were used to infected v-Src-transformed cells and stable lines were generated. Compared to wildtype GRIM-19, the mutants were less effective in inhibiting the formation of podosomes although to a varying extent (Figs 3A & B). Especially, mutation of the ninth residue of GRIM-19 (D9A) almost totally lost the inhibitory effect on v-Src-induced podosome formation. The differential activities of mutants were not due a differential expression of the mutants. Expression of all these GRIM-19 mutants alone in 3Y1 cells, in the absence of v-Src, did not affect actin filaments (Figs S2A & B).
Fig. 3. Podosome formation in v-Src-transformed cells expressing GRIM-19 proteins.
(A&C) Immunofluorescent images of actin network (Red) and nucleus (blue) in 3Y1 cells expressing v-Src and v-Src/GRIM-19 combinations. Magnification: 100×. Scale bar = 50µm. (B&D) A semi-quantitative representation of cells showing podosome-like feature in the indicated gene combinations. The percentage of cells with a rounded appearance, based on actin network, was counted manually from atleast 5 independent fields, each field containing ~70 cells. Rounded appearance of cells was counted as positive for podosome.
Tumor-associated GRIM-19 mutations impair the inhibitory effect on v-Src-induced cytoskeletal changes
Recently, somatic and germ-line mutations of GRIM-19 were identified in Hürthle cell thyroid tumors. However, the effects of these mutations on GRIM-19 function have not been examined. Therefore, we engineered these mutations into GRIM-19 to generate K5N and R115P constructs (see Table S1). Lentiviral particles coding for wildtype or mutants infected into v-Src-transformed 3Y1 cells to generate stable cell lines. We first investigated podosome formation in these cells using AlexaFluo555-conjugated phalloidin. As shown in Figs 3C and 3D, v-Src induced the formation of podosomes and altered cellular morphology to a round shape in ~80% of cells. As expected, wildtype GRIM-19 inhibited the formation of podosomes in almost all the v-Src-transformed cells and clearly restored stress fibers. In presence of GRIM-19 most of the v-Src-expressing cells changed from a round shape to a typical adherent cell shape, like naïve 3Y1 cells. The mutants were significantly impaired in their ability to the restore the morphologic changes and inhibit podosome formation, compared to wildtype GRIM-19 (Figs 3C & D). Quantification of cells demonstrated only 20% of v-Src-transformed cells still contained podosomes in presence of wild-type GRIM-19, while both GRIM-19 mutants were significantly defective in inhibiting podosome formation in v-Src-transformed cells. The K5N and R115P mutants allowed the formation of podosomes by 55% and 46%, respectively (Fig. 3D). Thus, tumor-associated mutations impaired the anti-v-Src effect of GRIM-19.
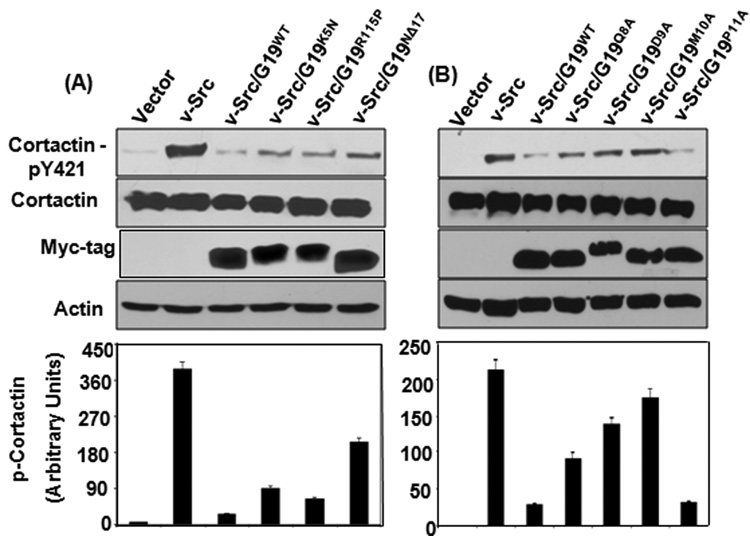
As mentioned earlier (see Fig. 2) tyrosyl phosphorylation of cortactin by v-Src is implicated in the formation of podosomes. Therefore, we examined if GRIM-19 mutants have lost their ability to suppress cortactin phosphorylation. Lysates prepared from cells expressing GRIM-19 mutants were probed for total cortactin and phosphorylated cortactin. GRIM-19 mutants (K5N, R115P, Q8A, D9A, M10A and NΔ17) were significantly less competent at suppressing v-Src-induced phosphorylation of cortactin (Figs. 4A & B) while P11A retained its ability to suppress phosphorylation of cortactin, which was in accordance with the ability of P11A in inhibiting podosome formation (see Fig. 3A). Expression of GRIM-19 mutants alone in 3Y1 cells did not effect phosphorylation of cortactin. (Figs S3A & B). Thus, the ability of GRIM-19 to suppress cortactin phosphorylation directly correlated with inhibition of v-Src-induced actin filament restructuring and podosome formation. All mutants expressed to an equivalent to extent. Unusually, the D9A mutant ran slower than other mutants. The reason for this anomalous migration is unclear, although there are no other differences in the sequence except for the mutant residue in this construct. In summary, clinical and experimental mutations in the N-terminus of GRIM-19 suppressed its ability to inhibit v-Src-induced podosome formation and cortactin phosphorylation.
Fig. 4. Effect of GRIM-19 on v-Src-induced phosphorylation of cortactin.
(A&B) Representative Western blots of total and phosphorylated cortactin in v-Src-transformed 3Y1 cells co-expressing wildtype or mutant GRIM-19 proteins. A semi-quantitative representation of phosphorylated cortactin levels in the respective cell lines. The band intensity of phosphorylated cortactin was normalized using band intensity of total cortactin. Bar represents mean band intensity ±SE from 4 independent samples is shown in each case.
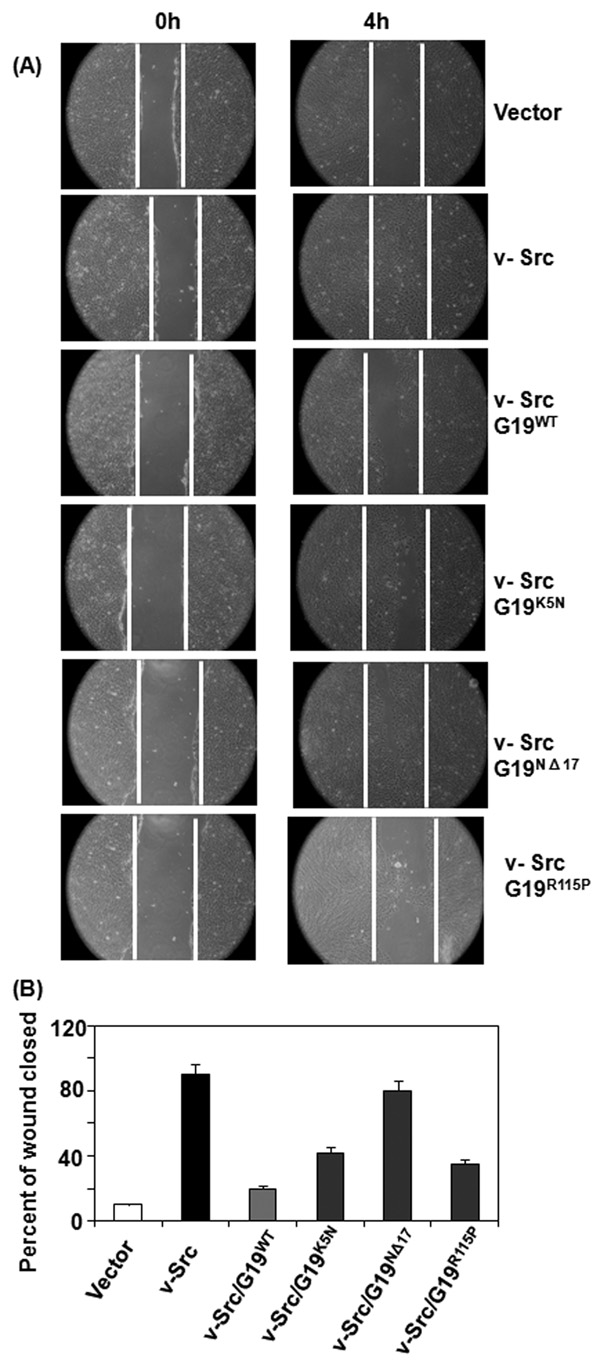
GRIM-19 mutants fail to suppress v-Src-induced motility
To determine the biological relevance of the above observations, we performed cell motility assays. Confluent monolayers expressing v-Src, GRIM-19 mutants and their combinations were used for this study. A scratch was induced into these monolayers and the sloughed cells were removed. Cell monolayers were fed with growth medium and the migration of cells into the denuded area was monitored at 4h. Representative data are shown in Fig. 5A. Since the wildtype GRIM-19 or mutants alone were no different from vector control (data not shown), we have only shown the effect of GRIM-19 cell motility in this figure. It is clear from this study that v-Src robustly activates cell motility following the injury, as evidenced by reformation of monolayer. Wildtype GRIM-19 strongly suppressed the v-Src-induced motility. In contrast, the K5N and NΔ17 mutants significantly lost their ability to suppress v-Src-induced cell motility. The R115P mutant was slightly stronger at suppressing v-Src-induced motility than K5N and NΔ17 mutants. Fig.5B shows the quantified view of these data.
Fig. 5. Effect of GRIM-19 on v-Src-dependent cell motility.
(A) Injury-induced migration of cells into the wounded area was monitored. Magnification: 100×. White line indicates the edge of injured site. Note the rapid migration of v-Src-expressing cells, but not the controls, into the wounded area. The wildtype and mutant GRIM-19 proteins were expressed using lentiviral vectors and their impact on cell motility was measured. Note suppression of v-Src-induced cell motility in the presence of wildtype GRIM-19. The mutant proteins have lost such suppressive activity to varying degrees. (B) A quantified view of the inhibitory effects of GRIM-19 on cell motility. Distance migrated from the edge of the monolayer to the center of the denuded area 4h after the induction of injury was calculated from a number of independent samples (n=8) and plotted.
Mutants defective in N-terminus lost their ability to suppress v-Src-induced metastasis in vivo
To study the pathological relevance and biological significance of the above observations in vivo, we performed metastatic assays. Cell lines expressing wildtype GRIM-19, K5N (clinical mutant) and NΔ17 (experimental mutant) in the presence of v-Src were used for these studies. Cell lines expressing v-Src alone and GRIM-19 alone were used as controls for this study. Athymic nude mice (n=10) were injected with 50,000 cells per mouse via the tail vein. Mice were then evaluated for the presence of tumor masses in lungs after 7 weeks. As shown in Table 1, no metastases were found in mice injected with control cells and GRIM-19 or its mutants. In contrast, v-Src-transformed cells formed a significant number of metastases. However, in presence of wildtype GRIM-19 there were fewer metastases. Although the K5N mutant was effective at suppressing tumor metastases, it was significantly weaker than wildtype GRIM-19. The experimental mutant NΔ17 significantly lost the anti-v-Src effect and behaved more like v-Src-transformed cells. These data for the first time show the in vivo importance of GRIM-19 mutants to tumor metastasis.
Table 1.
Effect of GRIM-19 mutants on metastases induced by v-Src.
| Cells expressing | No. of metastases | Average per mouse | Percent |
|---|---|---|---|
| Vector | 0 | 0 | 0.00 |
| GRIM-19 WT | 0 | 0 | 0.00 |
| v-Src | 32* | 3.2* | 100.00 |
| v-Src/GRIM-19 WT | 4 | 0.4 | 12.5 |
| GRIM-19 NΔ17 | 0 | 0 | 0.00 |
| v-Src/GRIM-19 NΔ17 | 26* | 2.6* | 81.25 |
| GRIM-19 K5N | 0 | 0 | 0.00 |
| v-Src/GRIM-19 K5N | 15** | 1.5** | 46.87 |
Athymic nude mice were injected 0.5×105 in a volume of 0.1 ml via the tail vein. n=10 mice/group. Seven weeks later mice were euthanized and necropsy was performed. Metastases in the lungs of each group were counted.
p>0.001
p>0.01 with respect to vector controls.
Discussion
In a recent study, we identified GRIM-19 inhibits v-Src-induced oncogenic transformation at multiple levels (Kalakonda et al., 2007a). GRIM-19 not only exerted its inhibitory effect on v-Src-induced gene expression via STAT3, but also significantly suppressed the v-Src-induced invasion and cell motility. In this study, we explored a mechanism by which GRIM-19 suppresses v-Src-induced cell motility through its effects on cytoskeletal remodeling. We mapped the N-terminus of GRIM-19 as an important regulator of this process. More importantly, we demonstrated for the first time that tumor-associated mutations cause dysfunction of its anti-oncogenic activity. Underlining the various cellular mechanical process, it is the spatial distribution and dynamic rearrangement of cytoskeleton, which determines cellular fate, including mitosis, invasion, morphologic changes, and anoikis (Rodriguez et al., 2003). GRIM-19 rescued the v-Src-induced aberrant cellular morphology, which endows transformed cells with a potential for anchorage-independent expansion. GRIM-19 not only slightly affected the v-Src-induced microtubulin structuring, but also, in a big part, suppressed the formation of podosomes. The dot-like podosomes in the transformed cells result from the bundled actin filaments, and its proteolytic activity, involving a matrix metalloproteinase (MMP)-mediated pathway (Monsky et al., 1993), that degrades extracellular matrix (ECM) and basement membrane (Mizutani et al., 2002). These enable tumor cells to detach from primary sites, intravasate into the blood vessel and disseminate to distant organs (Coussens et al., 2002). Nonetheless, this is not the only mechanism by which GRIM-19 can suppress proteolytic activity of MT1-MMP1. We and other groups had demonstrated that the direct association of GRIM-19 with STAT3 leads to the inhibition of STAT3-driven transcription of genes implicated in tumorigenesis, including, Bcl-XL, Cyclin B1 etc (Kalakonda et al., 2007b; Zhang et al., 2003) and MMP1 (Itoh et al., 2006). In this sense, GRIM-19 can regulate upstream events in the MT1-MMP-mediated invasion and metastasis through multiple signaling pathways. Apart from the formation of podosomes, expression of v-Src also disrupted stress fibers and perturbed the polymerization of microtubules (Figs 1B & C). This may involve the Rho family of small GTPase, RhoA, which regulates both actin and tubulin polymerization (Wittmann and Waterman-Storer, 2001) and this activity is antagonized by RhoGAP, a prominent downstream substrate of v-Src (Ellis et al., 1990). Tyrosyl phosphorylation of RhoGAP by v-Src negatively regulates the function of RhoA and downstream factors of RhoA-mediated pathway, the latter leading to the disruption of stress fibers and reduction of tubulin polymerization in v-Src-transformed cells. Expression of GRIM-19 antagonizes the v-Src-induced aberrant cytoskeletal change, which are implicated in the enhanced motility of transformed cells.
Considering links between STAT3 to v-Src, GRIM-19 and cytoskeletal network, we speculated that STAT3 might be involved in GRIM-19-mediated suppression of v-Src-induced cytoskeletal restructuring. In contrast to this hypothesis, our results demonstrated that STAT3 was not required for v-Src-induced podosome formation and its loss did not block the inhibitory effect of GRIM-19. Cortactin, an actin-associated protein, is an emerging scaffold protein, which couples v-Src signaling to cytoskeletal remodeling (Wu et al., 1991). Phosphorylation of cortactin by v-Src and other Src family kinases initiates a sequential cellular dynamic process: 1) neutralizes the cross-linking activity of filamentous actin (Huang et al., 1997); 2) enhances the actin assembly in vivo (Tehrani et al., 2007); and 3) plays an essential role in formation of podosome (Tehrani et al., 2006). Our result demonstrated that GRIM-19 dramatically suppressed v-Src-induced tyrosyl phosphorylation of Cortactin. This could be one of the mechanisms that explains the suppressive role of GRIM-19 on v-Src-induced podosome formation.
The pathological relevance of our studies is highlighted by the observations that experimental and tumor-associated mutations resulted in a loss of GRIM-19 effects on v-Src-induced podosome formation and cortactin phosphorylation. One of the clinical mutants falls into amino terminus domain mapped in this study. Our result showed that other residues in GRIM-19 (D9, Q8 and M10) also play a major role in the suppression of podosome formation and cortactin phosphorylation. Whether mutations also occur at these positions of GRIM-19 in primary tumors is unknown at this stage. We will be looking at this issue in our future studies.
Lastly, the biological and pathological significance of our observations were highlighted with the data shown in Fig.5 and Table1. Both in vitro and in vivo GRIM-19 appears to ablate cell motility and metastatic behavior of cells. The clinical mutant (K5N) and the experimental mutant (NΔ17) were weaker compared to wildtype GRIM-19. These data for the first time show a clinical importance of GRIM-19 mutations to tumor metastases. Whether similar mutations occur in other primary tumors need to be studied in the future. This is one of our top priorities for the future.
Materials and Methods
Plasmids and Reagents
Wild-type and mutants of GRIM-19 were generated using PCR and subsequently cloned into pIRES-Puro2 vector and expressed as C-terminally Myc-tagged proteins. Point mutations, K5N and R115P, in GRIM-19 were originally reported in thyroid tumors (Maximo et al., 2005) was reconstructed using PCR-directed mutagenesis. The primers used for generation of mutants are presented in Table S1. Sequence-verified constructs were used to generate lentiviral particles which were used in all experiments. The mammalian expression vectors for v-Src and lentiviral STAT3 shRNA vector and their controls have been described (Kalakonda et al., 2007a). Antibodies specific for Src, phospho-Y416 Src, Myc-tag, cortactin, phospho-Y421 cortactin, STAT3 (Cell signaling Technology, Beverly, MA); FITC-conjugated α-tubulin monoclonal antibody (Sigma), AlexaFluor555-conjugated Phalloidin (Invitrogen), rabbit detyrosinated tubulin antibody (Millipore), FITC-conjugated anti-mouse IgG (KPL) and Texas red-conjugated anti-rabbit IgG (Vector Laboratory ) were used in these studies.
Lentiviral particles
For generating viral particles, GRIM-19 inserts were cloned into the XhoI and XbaI sites of pLVX-Puro (Clontech, Inc). To produce lentiviral particles coding for expression constructs (GRIM-19) or knock-down constructs (STAT3), HEK-293T cells (3×106) were transfected with 5 µg of transfer vector (e.g. pLVX-Puro-GRIM-19-Myc or pLKO1-sh-STAT3), 3.75 µg of packaging plasmid (pCMV-dR8.2 dvpr) and 1.5 µg of envelope plasmid (pVSV-G) (Addgene, Inc) using Lipofectamine-Plus reagent (Invitrogen, Inc) for 6h in serum-free medium. Cells were then fed with 10 ml of regular growth medium (DMEM with 10% FBS). The viral particles from the growth medium were collected from 48–96 h post-transfection. The medium was centrifuged to remove debris, filtered through 0.45 µm filter and centrifuged at 50,000× g for 90 min at 4 °C. The viral particle pellet was suspended in 200µl TNE buffer (50 mM Tris.Cl pH 7.8, 130 mM NaCl, 1mM EDTA). Expression or knockdown of the target genes was assessed by performing Western blot analysis with specific antibodies.
Establishment of Stable Cell lines
A non-oncogenic immortalized rodent fibroblast cell line (3Y1) was employed for clearly defining the anti-v-Src effect of GRIM-19. The 3Y1 and 3Y1/v-Src cells were described in the previous study (Kalakonda et al., 2007a). 3Y1/GRIM-19 and 3Y1/GRIM-19/v-Src dual expression cells were established by infecting 3Y1 and 3Y1/v-Src cells with GRIM-19 coding lentiviral particles, respectively and selecting with puromycin (2µg/ml) for 10–12 days. In the STAT3 knockdown experiment, v-Src expressing cells were infected with lentiviral particles coding STAT3 shRNA and treated with puromycin (2µg/ml) for 7 days. All studies were conducted using pools of colonies (n=75–100) to avoid a clonal bias.
Immunofluorescence microscopy
Cells were cultured on sterile cover glass (Fisher Scientific, Inc) in 24-well tissue culture plate for 24 h before staining and fixed for 15 min using 3.7% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS and blocked in 5% BSA. Primary and secondary antibodies were diluted in PBS containing 5% BSA, incubated for 1h and washed. Hoechst was used to stain nuclei. Direct immunofluorescence was employed to visualize α-tubulin and actin network using FITC-conjugated anti-α-tubulin (1:400) and AlexaFluor555-conjugated phalloidin (1:400), respectively. Indirect immunofluorescence was used to visualize detyrosinated tubulin and GRIM-19 using rabbit antibodies (1:200) and anti-Myc-tag IgG (1:500), respectively with corresponding fluorescent-labeled secondary antibody listed above. Images were captured using a fluorescence microscope (Olympus BX-FLA, Osaka) fitted with digital camera (QICAM) and processed by Q-capture pro 5.1 (Q-Imaging corporation). Cell-counting was performed on captured immunofluorescent images from 5 randomly selected fields with each field containing ~70 cells and subjected to statistical analysis with Student’s t-test.
In vitro wound healing assay
Cells lines expressing various genes were grown in DMEM with 5% FBS and Penicillin-Streptomycin in a 6-well plate. Confluent monolayers were scratched with a 200 µl pipette tip to make a wound. The monolayer was washed twice with PBS to remove the detached cells, supplemented with full medium and incubated for 4h at 37°C. We first marked the injured front and measured space between the injured edges. Images were captured using a Nikon Eclipse TS-100 microscope fitted with a digital camera to monitor the cell movement into the wounded area. All micrographs were taken at the same time at the same magnification. For quantification of wound closure, the width of each wound was measured after 4h and expressed as a percentage of original wound size as described elsewhere (Scott et al., 2004). Mean values from 6 different plates (i.e photomicrographs.) per cell line were used for calculating these data.
Metastatic assays
Three to four-week-old athymic nude (nu/nu) NCr mice (Taconic) were used for these studies as described earlier (Lindner et al., 1997b). Procedures involving animals and their care were conducted in conformity with an institutionally approved protocol compliant with United States national and international laws and policies. For metastasis experiments, ~50,000 cells/mouse were injected via the tail vein. After 7 weeks, animals (n=10/group) were euthanized and evaluated for lung metastases by performing necropsy. Tumors were found only in the lungs. Student's t-test was used to assess the statistical significance of difference between pairs of mean numbers of tumors formed.
Supplementary Material
Acknowledgments
DVK thanks National Cancer Institute (CA105005) for grant support.
References
- Alchanati I, Nallar SC, Sun P, Gao L, Hu J, Stein A, et al. A proteomic analysis reveals the loss of expression of the cell death regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene. 2006 doi: 10.1038/sj.onc.1209708. [DOI] [PubMed] [Google Scholar]
- Angell JE, Lindner DJ, Shapiro PS, Hofmann ER, Kalvakolanu DV. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J Biol Chem. 2000;275:33416–33426. doi: 10.1074/jbc.M003929200. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Ellis C, Moran M, McCormick F, Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990;343:377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- Fearnley IM, Carroll J, Shannon RJ, Runswick MJ, Walker JE, Hirst J. GRIM-19, a cell death regulatory gene product, is a subunit of bovine mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2001;276:38345–38348. doi: 10.1074/jbc.C100444200. [DOI] [PubMed] [Google Scholar]
- Frame MC, Fincham VJ, Carragher NO, Wyke JA. v-Src's hold over actin and cell adhesions. Nat Rev Mol Cell Biol. 2002;3:233–245. doi: 10.1038/nrm779. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Hsu YS, Martin GS. Shp-2 mediates v-Src-induced morphological changes and activation of the anti-apoptotic protein kinase Akt. Oncogene. 2000;19:3164–3171. doi: 10.1038/sj.onc.1203655. [DOI] [PubMed] [Google Scholar]
- Head JA, Jiang D, Li M, Zorn LJ, Schaefer EM, Parsons JT, et al. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol Biol Cell. 2003;14:3216–3229. doi: 10.1091/mbc.E02-11-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J Biol Chem. 1997;272:13911–13915. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- Itoh M, Murata T, Suzuki T, Shindoh M, Nakajima K, Imai K, et al. Requirement of STAT3 activation for maximal collagenase-1 (MMP-1) induction by epidermal growth factor and malignant characteristics in T24 bladder cancer cells. Oncogene. 2006;25:1195–1204. doi: 10.1038/sj.onc.1209149. [DOI] [PubMed] [Google Scholar]
- Jonasch E, Haluska FG. Interferon in oncological practice: review of interferon biology, clinical applications, and toxicities. Oncologist. 2001;6:34–55. doi: 10.1634/theoncologist.6-1-34. [DOI] [PubMed] [Google Scholar]
- Kalakonda S, Nallar SC, Gong P, Lindner DJ, Goldblum SE, Reddy SP, et al. Tumor suppressive protein gene associated with retinoid-interferon-induced mortality (GRIM)-19 inhibits src-induced oncogenic transformation at multiple levels. Am J Pathol. 2007a;171:1352–1368. doi: 10.2353/ajpath.2007.070241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda S, Nallar SC, Lindner DJ, Hu J, Reddy SP, Kalvakolanu DV. Tumor-suppressive activity of the cell death activator GRIM-19 on a constitutively active signal transducer and activator of transcription 3. Cancer Res. 2007b;67:6212–6220. doi: 10.1158/0008-5472.CAN-07-0031. [DOI] [PubMed] [Google Scholar]
- Kolla V, Lindner DJ, Xiao W, Borden EC, Kalvakolanu DV. Modulation of interferon (IFN)-inducible gene expression by retinoic acid. Up-regulation of STAT1 protein in IFN-unresponsive cells. J Biol Chem. 1996;271:10508–10514. doi: 10.1074/jbc.271.18.10508. [DOI] [PubMed] [Google Scholar]
- Lindner DJ, Borden EC, Kalvakolanu DV. Synergistic antitumor effects of a combination of interferons and retinoic acid on human tumor cells in vitro and in vivo. Clin Cancer Res. 1997a;3:931–937. [PubMed] [Google Scholar]
- Lindner DJ, Borden EC, Kalvakolanu DV. Synergistic antitumor effects of a combination of interferons and retinoic acid on human tumor cells in vitro and in vivo. Clin Cancer Res. 1997b;3:931–937. [PubMed] [Google Scholar]
- Lufei C, Ma J, Huang G, Zhang T, Novotny-Diermayr V, Ong CT, et al. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 2003;22:1325–1335. doi: 10.1093/emboj/cdg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Karra S, Guo W, Lindner DJ, Hu J, Angell JE, et al. Regulation of interferon and retinoic acid-induced cell death activation through thioredoxin reductase. J Biol Chem. 2001;276:24843–24854. doi: 10.1074/jbc.M100380200. [DOI] [PubMed] [Google Scholar]
- Martin GS. The hunting of the Src. Nat Rev Mol Cell Biol. 2001;2:467–475. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- Maximo V, Botelho T, Capela J, Soares P, Lima J, Taveira A, et al. Somatic and germline mutation in GRIM-19, a dual function gene involved in mitochondrial metabolism and cell death, is linked to mitochondrion-rich (Hurthle cell) tumours of the thyroid. Br J Cancer. 2005;92:1892–1898. doi: 10.1038/sj.bjc.6602547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Miki H, He H, Maruta H, Takenawa T. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 2002;62:669–674. [PubMed] [Google Scholar]
- Monsky WL, Kelly T, Lin CY, Yeh Y, Stetler-Stevenson WG, Mueller SC, et al. Binding and localization of M(r) 72,000 matrix metalloproteinase at cell surface invadopodia. Cancer Res. 1993;53:3159–3164. [PubMed] [Google Scholar]
- Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, et al. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol. 2006;172:245–257. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Scott LA, Vass JK, Parkinson EK, Gillespie DA, Winnie JN, Ozanne BW. Invasion of normal human fibroblasts induced by v-Fos is independent of proliferation, immortalization, and the tumor suppressors p16INK4a and p53. Mol Cell Biol. 2004;24:1540–1559. doi: 10.1128/MCB.24.4.1540-1559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo T, Lee D, Shim YS, Angell JE, Chidambaram NV, Kalvakolanu DV, et al. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus interacts with a cell death regulator, GRIM19, and inhibits interferon/retinoic acid-induced cell death. J Virol. 2002;76:8797–8807. doi: 10.1128/JVI.76.17.8797-8807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. Cortactin has an essential and specific role in osteoclast actin assembly. Mol Biol Cell. 2006;17:2882–2895. doi: 10.1091/mbc.E06-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. Src phosphorylation of cortactin enhances actin assembly. Proc Natl Acad Sci U S A. 2007;104:11933–11938. doi: 10.1073/pnas.0701077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way? J Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Reynolds AB, Kanner SB, Vines RR, Parsons JT. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991;11:5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang J, Roy SK, Tininini S, Hu J, Bromberg JF, et al. The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3. Proc Natl Acad Sci U S A. 2003;100:9342–9347. doi: 10.1073/pnas.1633516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gao L, Li Y, Lin G, Shao Y, Ji K, et al. Effects of Plasmid-Based Stat3-Specific Short Hairpin RNA and GRIM-19 on PC-3M Tumor Cell Growth. Clin Cancer Res. 2008;14:559–568. doi: 10.1158/1078-0432.CCR-07-1176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.