Abstract
Invasion of human erythrocytes by the malaria parasite Plasmodium falciparum utilizes multiple ligand-receptor interactions involving erythrocyte receptors and parasite erythrocyte binding proteins of the Duffy binding-like family. Erythrocyte binding antigen 175 (EBA-175) binds to glycophorin A, the most abundant protein on the human erythrocyte surface and EBA-140 (also known as BAEBL) binds to glycophorin C, while the receptor for EBA-181 (also known as JESEBL) remains unknown. EBA binding is mediated via region II, a highly structured extracellular domain that shows a degree of sequence variability between different laboratory strains/isolates. Here, we determined the influence of region II polymorphisms on host cell receptor binding and overall function during invasion of EBA-140, EBA-175, and EBA-181. Polymorphisms in the binding domains of EBA-140 and EBA-181 have been suggested previously to alter their respective receptor specificities. In our hands, these polymorphisms affected the levels of EBA-140 and EBA-181 binding to receptors but, critically, not the receptor specificities of these proteins. The degree of EBA-140 binding to glycophorin C correlates with the level of function for this ligand-receptor interaction in merozoite invasion. In contrast, EBA-175, which is highly polymorphic in region II, shows no variability in its ability to bind to its receptor, glycophorin A. Combined, these data highlight the importance of sequence variability in EBAs as driven by immune selection but not by receptor specificity.
Erythrocyte invasion by the blood-stage malaria parasites is a multistep process involving specific interactions between ligands of the merozoites and receptors on the red blood cell (3). Two protein families play central roles in this process: the Duffy binding-like (DBL) and reticulocyte binding-like protein families, binding to specific red blood cell receptors and initiating a cascade of molecular events resulting in merozoite entry (1, 2). Plasmodium falciparum, the most virulent human malaria agent, has a higher degree of flexibility in utilization of specific receptor-ligand interactions than other plasmodia, such as P. vivax, which relies on interaction of Duffy binding protein to reticulocytes (20, 32). In contrast, P. falciparum has multiple members of the DBL (1, 2) and reticulocyte binding-like (1, 40) proteins, enabling interaction with independent receptors, defined as alternative invasion pathways (10, 31, 33, 38, 39).
Three functional P. falciparum DBL proteins have been characterized: erythrocyte binding antigen 175 (EBA-175; MAL7P1.176) (43), EBA-140 (also known as BAEBL; MAL13P1.60) (28, 45), and EBA-181 (19) (also known as JESEBL; PFA0125c) (1). These proteins consist of structured domains, including the F region (region II) subdivided into two related domains, F1 and F2, involved in receptor binding (1). EBA-175 binds to glycophorin A (GYP A) in a sialic acid-dependent manner (43) and plays an important role in invasion, as evidenced by the ability of antibodies to partially inhibit invasion (37). The three-dimensional structure of the F1/F2 region has been solved, and the presence of three putative glycan binding sites that form between the interfaces of a protein dimer has been demonstrated (46). EBA-175 is not absolutely essential for merozoite invasion, as the gene can be disrupted, which in some strains selects for a switch in receptor utilization to a process not requiring sialic acid (14, 41).
The EBA-140 ligand binds to glycophorin C (GYP C) (26, 27, 29), mediating a distinct invasion pathway into human erythrocytes (27). Functional analysis of EBA-140 knockout parasites suggests that this ligand is promiscuous, binding to GYP A and glycophorin B (GYP B), although at greatly reduced levels compared to its binding to GYP C (27). However, despite this promiscuity, EBA-140's interactions with GYP A and B do not contribute measurably to invasion (27). Expression of the F1 and F2 region of EBA-140 on COS cells suggested that polymorphisms within this ligand, characterized for different strains of P. falciparum, alter receptor binding specificity (29). Certain expressed variant forms were reported to bind to neuraminidase-treated erythrocytes, suggesting that the receptor was not GYP C and that different P. falciparum strains might have multiple invasion pathways dependent on polymorphisms in EBA-140. The same experimental approach has been used to demonstrate that polymorphisms of EBA-181 alter its receptor specificity (30). While the exact identity of the EBA-181 receptor on the erythrocyte is unknown, its receptor specificity can be defined based on enzyme sensitivity of its binding to red blood cells (neuraminidase sensitive, trypsin resistant, and chymotrypsin sensitive but distinct from GYP B) (18). EBA-181 binds to erythrocyte membrane protein 4.1, although the significance of this observation in vivo is not known, since protein 4.1 is located inside the erythrocyte (23, 24).
To address the role of EBA-140 and EBA-181 polymorphisms in merozoite invasion, we analyzed parasite strains expressing distinct allelic forms of these ligands. We show that polymorphisms in EBA-140 and EBA-181 alter the level of binding to receptors but, critically, do not alter specificity. Furthermore, and consistent with this observation, we show a direct correlation between decreased EBA-140 binding and the importance of its function during merozoite invasion.
MATERIALS AND METHODS
Parasites, parasite culture supernatant, and binding assays.
P. falciparum 3D7 and HB3 (8) were obtained from David Walliker (University of Edinburgh), MCAMP was obtained from Queensland Institute of Medical Research, PF120 and BT3 (Papua New Guinea) were obtained from Karen Day (New York University), CSL2 was obtained from CSL Pty. Ltd., and 7G8, FCR3, and W2mef have been described previously (11, 22, 36). The genotypes of these parasite strains were confirmed (15). 3D7ΔEBA-140, W2mefΔEBA-140 (27), 3D7ΔEBA-175, and W2mefΔEBA-175 have been described previously (14). EBA-140 and EBA-175 were equalized in serial dilutions of supernatants by immunoblotting. For binding assays, 500 μl of culture supernatant was mixed with 100 μl of erythrocytes for 30 min at room temperature and analyzed as described previously (17).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblot analysis, erythrocyte ghosts, and overlay assay.
Proteins from parasite culture supernatants were separated on 3 to 8% Tris-acetate gels (Invitrogen). Western blotting onto nitrocellulose (0.45 μm; Schleicher and Schuell) was performed according to standard protocols. Antibodies used for antigen detection in immunoblots were rabbit anti-EBA-175 (raised against the region between the F2 and the C-terminal cysteine-rich domain, amino acids 761 to 1271; 1:250) (41) and rabbit anti-EBA-140 (corresponding region, amino acids 746 to 1048; 1:250) (45). The anti-EBA-181 antibodies were raised to a region between the F2 domain and the C-terminal cysteine-rich region encompassing amino acids 755 to 1339 of EBA-181 (19). Human GYP A/B and GYP C were detected with the mouse monoclonal antibodies E3 (1:1,500) and E5 (1:1,000) (Sigma), respectively. Horseradish peroxidase-coupled sheep anti-rabbit immunoglobulin and sheep anti-mouse immunoglobulin (Silenus) were used as the secondary antibody to visualize the antigens with a chemiluminescence system (ECL; Amersham). In order to detect binding of ligands to erythrocyte membrane proteins, a binding assay was used as described previously (27, 35). To quantify binding of EBAs, band intensities on immunoblots were determined with Quantity One software (Bio-Rad). Background was subtracted and values were adjusted for differences in EBA contents in supernatants. All bands representing each specific EBA ligand that bound to red blood cells were quantitated in each experiment. Bars represent the means of three independent experiments.
Preparation of supernatant.
Synchronized infected red blood cells at 5 to 10% parasitemia were neuraminidase/trypsin treated at the trophozoite stage to prevent reinvasion. The parasite culture was continued until schizont rupture and culture supernatant was harvested by centrifugation at 1,500 × g for 5 min. This supernatant was further cleared from cellular debris by centrifugation at 3,000 × g for 10 min at 4°C and stored at −20°C.
Antibody inhibition assay.
Parasite erythrocyte invasion assays were performed to characterize the invasions of five different parasite lines (3D7, HB3, PF120, MCAMP, and W2mef) into normal and chymotrypsin-treated cells in the presence or absence of protein G-purified rabbit serum raised against EBA-140. Synchronized parasites were treated with or without chymotrypsin as follows. Ring-stage cultures at a 4% hematocrit were adjusted to 0.5% parasitemia. Two hundred milliliters of packed parasitized erythrocytes was resuspended in 4 ml incomplete medium (RPMI-1640, HEPES, 0.2% NaHCO3) and was subsequently split, and one half was treated with 1.5 mg/ml chymotrypsin. Both treated and untreated cells were incubated for 1 h at 37°C on a rotating shaker. After being washed twice with incomplete medium, cells were incubated with trypsin-chymotrypsin inhibitor (1 mg/ml in incomplete medium) for 15 min at 37°C on a rotating shaker. These were subsequently washed three times and resuspended in 5 ml complete medium (RPMI-1640, HEPES, 0.2% NaHCO3 with 9% Albumax and 1% heat-inactivated human serum). Invasion assays were prepared with treated and untreated parasitized erythrocytes at a final hematocrit of 4% and a parasitemia level of 0.5%. One hundred-milliliter aliquots of each culture were placed into 96-well flat-bottom microtiter plates in three sets of triplicate wells. Protein G-purified rabbit serum (raised against the EBA-140 F2 domain) was added to the first triplicate wells of both treated and untreated cultures at a final concentration of 10 mg/ml in human tonicity phosphate-buffered saline (HTPBS). Protein G-purified normal rabbit serum was added to the second set of triplicate wells of both treated and untreated cultures at a final concentration of 10 mg/ml in HTPBS. The final set of triplicates did not contain antibody and were normal invasion controls. Control wells were smeared 72 h after assay preparation to determine the life cycle stage (mid-stage to late-stage trophozoites postreinvasion). Ten microliters of each resuspended assay well was added to 190 μl of 10 mg/ml ethidium bromide in HTPBS. This was mixed and incubated for 5 min at room temperature. The level of parasitemia for each well was determined by use of FACScan. Assays were performed on at least three separate occasions to determine statistical significance.
Modeling of EBA-140 and EBA-181 and gene sequences.
Models of EBA-140 and EBA-181 were created with the MODELLER (6v2) program (16) by using the X-ray crystal structure of EBA-175 (PDB accession code 1ZRL) as a template (46) and sequence alignments obtained from the FUGUE sequence-structure alignment program (42). From 25 models of each molecule, the structure with lowest modeler objective function was used for analysis. The EBA-175 (MAL7P1.176), EBA-181 (PFA0125c), and EBA140 (MAL13P1.60) gene sequences are available at http://plasmodb.org/.
Nucleotide sequence accession numbers.
The sequences for the F1/F2 region of EBA-140 have been submitted to GenBank with the following accession numbers: 3D7, FJ655420; W2mef, FJ655421; HB3, FJ655422; 7G8, FJ655423; Pf120, FJ65542; CSL2, FJ655425; BT3, FJ655426; FCR3, FJ655427; MCAMP, FJ655428. The sequences for the F1/F2 region of EBA-175 have been submitted to GenBank with the following accession numbers: 3D7, FJ655429; MCAMP, FJ655430; Pf120, FJ655431; CSL2, FJ655432, BT3, FJ655433; HB3, FJ655434; 7G8, FJ655435; FCR3, FJ655436; W2mef, FJ655437. The sequences for the F1/F2 region of EBA-181 have been submitted to GenBank with the following accession numbers: 3D7, FJ655438; MCAMP, FJ655439; Pf120, FJ655440; CSL2, FJ655441; BT3, FJ655442; HB3, FJ655443; 7G8, FJ655444; FCR3; FJ655445; W2mef, FJ655446.
RESULTS
Polymorphisms in EBA ligands and erythrocyte binding.
Previously, polymorphisms in EBA-140 and EBA-181 that have been suggested to alter host receptor specificity have been identified (29). We sequenced the EBA-140, EBA-181, and EBA-175 genes, across the F1 and F2 regions (Fig. 1A), of nine P. falciparum strains to identify those that expressed variant forms of EBA-140, EBA-181, and EBA-175 (Fig. 1A and Table 1). No additional polymorphisms were identified in comparison to previous studies (27, 29). Furthermore, sequencing across the F1/F2 region of the EBA-175 gene identified a high level of polymorphism, also in agreement with previous studies (Fig. 1A and Table 1) (6).
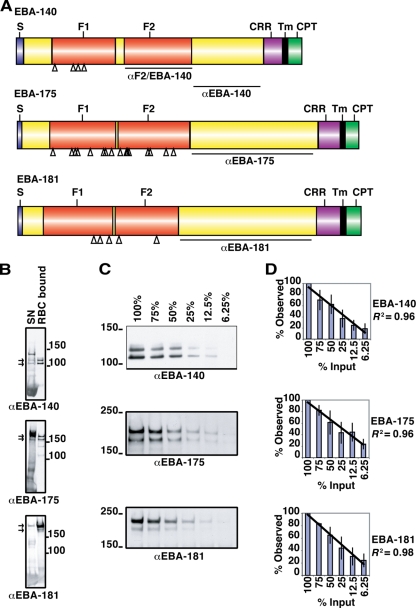
FIG. 1.
Schematic representation of the domain structures and quantitative erythrocyte binding assays for EBA-140, EBA-175, and EBA-181. (A) The signal sequence (S), F1 and F2 domains, C-terminal cysteine-rich region (CRR), transmembrane domain (Tm), and cytoplasmic tail (CPT) are shown. The regions against which antibodies were raised are indicated as αF2/EBA-140 (27), αEBA-140 (27), αEBA-175, and αEBA-181. Open triangles indicate the positions of strain-specific mutations within the F1/F2 domain. (B) Supernatant obtained from a culture of ruptured schizonts (SN) was incubated with erythrocytes, unbound supernatant was removed, the cells were washed, and EBAs bound to the erythrocytes were eluted with high-salt medium (RBC bound). Supernatant and eluted EBAs were subjected to Western blotting and probed with antibodies against EBA-140, EBA-175, and EBA-181 to compare binding patterns. Molecular mass markers in kDa are indicated. The arrows for each gel show the two specific bands for each ligand used for quantitation of erythrocyte binding. (C) Titration of different amounts of culture supernatant used in erythrocyte binding assays. Culture supernatant was diluted with culture media before an erythrocyte binding assay was performed on erythrocytes. Obtained eluants were subjected to Western blotting and probed with antibodies against different EBAs. Percentages indicate the amounts of culture supernatant used. The two bands shown in panel C (arrows in panel B) were used throughout this study for quantitation in densitometry, as these were the major bands observed. The ratio of these two bands can vary in different supernatant preparations; however, the sum totals of binding remain the same. This occurs due to different amounts of processing of the larger band that yields the smaller protein band. Molecular mass markers (in kDa) are indicated. α, anti. (D) Densitometric analysis of three titration experiments. Error bars indicate standard deviations. The diagonal lines indicate linear regressions with an R2 coefficient of determination of >0.95.
TABLE 1.
P. falciparum strain-specific polymorphisms in EBA-140, EBA-181, and EBA-175
| Strain | Origina | Polymorphism in F1/F2 of indicated ligand (by amino acid position)
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EBA-140
|
EBA-181
|
EBA-175
|
|||||||||||||||||||||||
| 185 | 239 | 261 | 285 | 359 | 363 | 414 | 443 | 637 | 891b | 157 | 274 | 279 | 286 | 336 | 388 | 390 | 403 | 478 | 481 | 577 | 584 | 644 | 664 | ||
| 3D7 | Africa? | I | N | K | K | R | V | N | K | N | − | N | E | K | K | D | K | P | E | K | K | N | Q | M | S |
| MCAMP | Malaysia | I | N | R | E | R | V | N | K | N | − | N | E | K | E | D | K | P | E | N | I | N | K | M | S |
| Pf120 | SE Asia | I | S | K | K | K | V | N | Q | N | + | N | E | K | E | D | K | P | E | K | K | N | Q | M | S |
| CSL2 | SE Asia | I | S | K | K | R | V | I | Q | K | + | N | E | K | E | D | K | P | E | K | K | N | Q | K | R |
| BT3 | SE Asia | I | S | K | K | R | V | I | Q | N | − | N | E | K | E | D | K | P | E | K | K | N | Q | M | R |
| HB3 | Honduras | V | S | K | K | R | V | N | Q | N | − | N | E | K | K | D | K | P | E | K | K | N | Q | M | R |
| 7G8 | Brazil | V | S | T | K | K | D | I | Q | N | − | S | K | E | K | D | K | S | E | N | I | K | K | M | S |
| FCR3 | Uganda | V | S | T | K | K | V | I | Q | N | + | N | K | K | E | D | K | P | E | K | K | N | Q | M | R |
| W2mef | Vietnam | V | S | T | K | R | V | I | Q | N | − | N | K | E | K | Y | N | S | K | K | I | N | K | M | S |
SE Asia, southeastern Asia.
Contains a nine-amino-acid insert.
To measure the strain-specific binding of EBAs, we employed an erythrocyte binding assay using supernatants containing released EBA-140, EBA-181, and EBA-175 (17). Anti-EBA-140 antibodies detected three specific bands in supernatant, while two specific bands for EBA-175 and EBA-181 were observed (Fig. 1B) (45). Only the lower bands of EBA-140 bound to red blood cells. Dilution of the supernatants established a linear range of erythrocyte binding for EBA-140, EBA-175, and EBA-181 (Fig. 1C). Antibodies used to detect binding were raised against a conserved region between the F2 domain and a distal cysteine-rich domain (Fig. 1), sequencing of which showed that this region was conserved except for a nine-amino-acid insert in EBA-181 (Fig. 1A and Table 1). Antibody reactivity to each EBA in supernatants provided a means to equalize the concentrations of ligands. To confirm that the binding detected was linear, we quantified the signal by using densitometry (Fig. 1D). Coefficients of determination (R2 > 0.95) indicated that there was a direct linear relationship between the amount of EBA in the supernatant, the amount bound to erythrocytes, and their detection in eluants.
Polymorphisms in EBA-140 and EBA-181 affect binding.
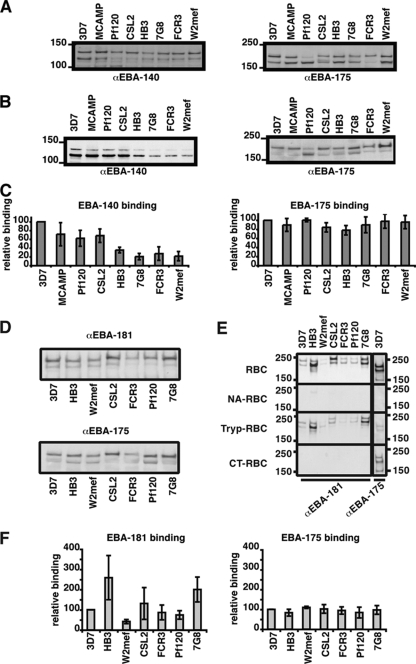
To determine the effect of EBA-140 F1/F2 polymorphisms on erythrocyte binding, we used supernatants from 3D7, MCAMP, Pf120, CSL2, HB3, 7G8, FCR3, and W2mef that represented five major allelic types (27, 29). The amount of EBA-140 was equalized by Western blotting (Fig. 2A), and the antibodies used have been shown previously to react specifically to this parasite ligand by using EBA-140 knockout parasites that lack its expression (28). The same supernatants were also probed with an antibody to EBA-175, with the levels of reactivity for the strains found to be similar (Fig. 2A). Supernatants containing approximately equal concentrations of EBA-140 were used to test binding to erythrocytes (Fig. 2B). EBA-140 from 3D7 (INKK) bound to erythrocytes at higher levels than other EBA-140 variants (Fig. 2B and C). Overall, the strains fell into two groups with respect to binding of EBA-140: the first group (3D7, MCAMP, Pf120, and CSL2) bound at >60% of 3D7 binding, whereas the second group (HB3, 7G8, FCR3, and W2mef) bound at <40% of 3D7 binding (Fig. 2C). Within the high-level-binding group, the EBA-140 protein from Pf120 and CSL2 (ISKK) differs by one amino acid from that of 3D7 (INKK) and showed a small but observable reduction in binding. In comparison, EBA-175 binding for the same supernatants showed no statistically significant difference between strains, despite the fact EBA-175 F1/F2 domains contain numerous variant residues (Fig. 2B and C and Table 1). This demonstrates that polymorphisms in the F1/F2 domain of EBA-140, but not EBA-175, affect binding affinity to their corresponding receptor.
FIG. 2.
EBA-140 and EBA-181 from different strains bind with different affinities to whole erythrocytes. Molecular mass markers (in kDa) are indicated to the left of the gels. (A) For loading controls, supernatants used for the erythrocyte binding assays were probed for the presence of EBA-140 and EBA-175. Differences in EBA-140 contents were adjusted by adding culture medium. The same supernatants were used for EBA-140 and EBA-175 erythrocyte binding assays. α, anti. (B) The amount of EBA-140 present in eluants from erythrocyte binding assays performed with strain-specific culture supernatant was determined via Western blotting, and this was probed with anti-EBA-140 antibodies (left panel). In this panel, the supernatants are ordered by descending ability of EBA-140 of each strain to bind to red blood cells. This order was then used for the panels presented for EBA-181 binding. The same filer was used for the blot shown for anti-EBA-175 (αEBA-175), but the tracks have been reordered digitally so that they can be easily compared with the corresponding blots for αEBA-140 and αEBA-181. For comparison, the same samples were probed for the presence of EBA-175 (right panel). (C) Densitometric analysis of erythrocyte binding assays. The levels of binding of EBA-140 (left) and EBA-175 (right) were quantified by densitometry and expressed as percentages relative to the levels of binding of 3D7 EBA-140 and EBA-175, respectively (which were set to 100%). Error bars indicate standard deviations for three experiments. (D) For loading controls, supernatants used for the erythrocyte binding assays were probed for the presence of EBA-181 and EBA-175. Differences in EBA-181 contents were adjusted by adding culture medium. The same supernatants were used for EBA-181 and EBA-175 erythrocyte binding assays. The apparent differences in mobility of EBA-181 were probably due to different amounts of EBA-181 eluted from the red blood cells in some. These experiments were done at least three times using different supernatant preparations, and the EBA-181 bands have the same molecular mass in independent experiments. (E) Binding of EBA-181 from culture supernatant of different strains to normal red blood cells (RBC) and red blood cells which were treated with neuraminidase (NA-RBC), trypsin (Tryp-RBC), and chymotrypsin (CT-RBC). For a comparison, the binding of 3D7 EBA-175 to these red blood cells is shown. (F) Quantification of EBA-181 and EBA-175 binding from different strains. Western blots detecting the binding of the EBAs were densitometrically analyzed, and binding was expressed as a percentage relative to binding of 3D7 EBA-181 (left) or 3D7 EBA-175 (right). Error bars show standard deviations for three experiments.
Sequence variants for EBA-181 have also been reported to affect receptor specificity in different parasite strains (29). To determine the effect of EBA-181 F1/F2 polymorphisms, we used supernatants from 3D7, HB3, W2mef, CSL2, FCR3, Pf120, and 7G8 strains (representing seven different polymorphisms) (Table 1) equalized by Western blotting (Fig. 2D). As described above, by using knockout parasites that lack EBA-181 expression, these antibodies have been shown to specifically react with EBA-181. As with EBA-140, binding affinity of EBA-181 from different strains varied greatly (Fig. 2E). A fivefold difference in the level of EBA-181 binding for the different strains could be observed compared to the level of 3D7 EBA-181 binding (Fig. 2E and F). This was in contrast to EBA-175, for which binding was consistent regardless of the variant. This demonstrates that polymorphisms in the F1/F2 domain of EBA-181 affect binding affinity.
Binding of EBA-181 variants to erythrocytes.
The identity of the EBA-181 receptor, though unknown, was originally defined by its sensitivity to neuraminidase and chymotrypsin, its resistance to trypsin treatment, and the fact that it was distinct from GYP B, which shares the same characteristics (19). Mayer et al. (30) looked at the binding of different variant forms of EBA-181 (F1/F2 region), using transient expression on CHO cells. Six polymorphic sites in the F1/F2 region gave rise to four different erythrocyte binding patterns as defined by enzyme treatment, suggesting that EBA-181 may bind four distinct receptors. We tested the ability of native EBA-181 proteins from different strains to bind to neuraminidase-, trypsin-, and chymotrypsin-treated erythrocytes (Fig. 2E). Contrary to the case for Mayer et al., native EBA-181 from culture supernatant for all strains bound to untreated and trypsin-treated cells, while binding was abolished when red blood cells were pretreated with neuraminidase or chymotrypsin (Fig. 2E). Binding of 3D7 EBA-175 was determined and, as expected, identified a neuraminidase- and trypsin-sensitive but chymotrypsin-resistant receptor consistent with GYP A, the established receptor for EBA-175 (12). This demonstrates that EBA-181 variants do not display differences in the enzyme profile of their binding to the erythrocyte surface.
Binding of EBA-140 variants to enzyme-treated erythrocytes.
Polymorphisms in the F1/F2 region of EBA-140 have been reported to affect receptor specificity (30). EBA-140 binds to GYP C, with its interaction defined by sensitivity to neuraminidase and trypsin and resistance to chymotrypsin treatment (45). To investigate if F1/F2 sequence variations in EBA-140 mediate binding to erythrocyte receptors other than GYP C, we looked at binding of the different variants to enzyme-treated erythrocytes (Fig. 3, left column). Although the intensities of the EBA-140 bands differed for the different strains, reflecting the affinity to the erythrocyte (Fig. 2B), the susceptibility patterns remained the same irrespective of the variant (Fig. 3). Significantly, none of the EBA-140 ligands from the different P. falciparum strains bound to neuraminidase-treated erythrocytes, confirming that detectable EBA-140 binding is dependent on sialic acid. Some binding to trypsin-treated erythrocytes was observed, although this was reduced compared to the levels of binding to neuraminidase- and chymotrypsin-treated erythrocytes. Binding of EBA-140 from W2mef to trypsin-treated erythrocytes was at the lowest level in comparison to binding by other variant forms of the protein. However, this was observed for binding to normal and chymotrypsin-treated erythrocytes and reflects the lower apparent affinity of binding of this protein and not its specificity for an alternative receptor. Some variation in the ability of EBA-140 to bind to trypsin-treated erythrocytes has been reported (26, 28, 34, 45), ranging between no binding and decreased binding; in our hands, this binding was dependent on the batch of trypsin and length of protease treatment. It is likely that some binding occurs through the interaction of EBA-140 with the trypsin-resistant receptor GYP B; however, this is clearly much less than the observed binding to GYP C. Despite the quantitative differences in binding seen between native EBA-140 proteins from different strains, all variants bound to GYP C as defined by receptor sensitivity. Therefore, the amino acid polymorphisms in the F1 domain are important for interaction between EBA-140 and GYP C, altering the apparent affinity of the ligand for the receptor. These results suggest that GYP C is the major receptor for the five polymorphic variants of EBA-140.
FIG. 3.
Binding of EBA-140 to enzyme-treated and mutant erythrocyte (erythroc.) membranes. (Left panel) Binding of EBA-140 to intact erythrocytes determined by using erythrocyte binding assays. The erythrocytes were left untreated (nRBC) or treated with neuraminidase (NA-RBC), trypsin (Try-RBC), or chymotrypsin (CT-RBC) and incubated with supernatants from different parasite strains (3D7, MCAMP, Pf120, HB3, and W2mef). Parasite proteins that bound to the intact erythrocytes were eluted off with a salt solution, separated on a SDS-PAGE gel, and analyzed via Western blotting using anti-EBA-140 (αEBA-140) antibodies. The strain-specific variations in the F1 domain of EBA-140 are indicated underneath the strain name (Table 1). (Middle panel) Binding of EBA-140 from these culture supernatants to ghost erythrocyte membranes derived from NA-, Try-, or CT-treated or normal red blood cells by using overlay assays. (Right panel) Binding of EBA-140 from 3D7, MCAMP, Pf120, HB3, and W2mef culture supernatant to ghost erythrocyte membranes from normal, Yus (Ge −2, 3, 4), Gerbich (Ge −2, −3, 4), and Leach (Ge −2, −3, −4) homozygote cells and Yus/Gerbich heterozygote erythrocytes by using overlay assays. Binding of EBA-175 to the same solubilized membranes and detection of GYP A/B with anti-GYP A/B (αGYP A/B) as a positive control are shown at the bottom of the figure.
In order to assess the role of polymorphisms in interaction between EBA-140 and GYP C or other receptors, we investigated EBA binding with culture supernatants and solubilized erythrocyte membranes (Fig. 3, middle panel) (27). For normal cells, a specific band at 35 kDa, the expected size for GYP C, could be detected. The intensity of that band varied depending on the supernatant used, consistent with binding observed for intact red blood cells (Fig. 2). EBA-140 from 3D7 (INKK), MCAMP (INRE), Pf120 (ISKK), HB3 (VSKK), and W2mef (VSTK) were not able to bind to GYP C from neuraminidase-treated erythrocytes, consistent with the requirement for sialic acid on this receptor. Similarly, the ligands from all parasite strains were unable to bind GYP C from trypsin-treated erythrocytes, in line with the receptor's known sensitivity to this protease (Fig. 3, middle panel). In contrast, EBA-140 bound to GYP C on chymotrypsin-treated erythrocytes, in line with this receptor's resistance to this protease. No differences were observed in the overall properties of the different polymorphic forms of EBA-140 binding to GYP C from solubilized erythrocyte membranes, although there were apparent differences in binding consistent with that seen for intact erythrocytes.
Binding of EBA-140 variants to GYP C mutant erythrocytes.
It has been reported that EBA-140 can bind to mutant GYP C of Gerbich (Ge −2, −3, 4) and Yus (Ge −2, 3, 4) erythrocytes but not to Leach erythrocytes (Ge −2, −3, −4), the latter lacking expression of GYP C (26). To analyze this and determine the properties of EBA-140 binding to normal GYP C and altered forms of this receptor, we used supernatants expressing different EBA-140 polymorphic variants in an overlay assay (Fig. 3, right panel). The EBA-140 ligands from 3D7 (INKK), MCAMP (INRE), Pf120 (ISKK), HB3 (VSKK), and W2mef (VSTK) all bind GYP C from normal erythrocytes; however, in contrast to previous reports, none of these ligands showed erythrocyte binding in Yus, Ge-negative, Leach, or Yus/Gerbich heterozygote red blood cells, expressing altered forms or lacking GYP C entirely. In contrast, EBA-175 bound efficiently to GYP A from all of the different erythrocytes. These results show that EBA-140 cannot bind to the altered GYP C in Ge-negative erythrocytes and suggest that protein components of exon 2 and 3 of GYP C are important for EBA-140 binding.
Pathway usage in EBA knockout cell lines.
To extend our observations across strains, we also explored the specificity of the red blood cell binding using supernatant from parasites deficient in EBA-140 or EBA-175. These transgenic cell lines were generated using both 3D7 and W2mef parental background, representing the spectrum of high and low levels of EBA-140 binding (Fig. 2B). In overlay experiments, levels of binding of EBA-140 (Fig. 4A) and EBA-175 (Fig. 4B) to red blood cell membranes, purified glycophorins, and membranes of the GYP C mutant Gerbich (Ge −2, −3, 4) erythrocytes were determined. EBA-140 from 3D7 (INKK) binds to GYP C as shown in Fig. 4A in the lanes showing the results of overlay experiments for the parental line and 3D7Δ175 (a parasite lacking EBA-175 but not EBA-140). In contrast, for 3D7Δ140, which does not express EBA-140, there was no binding to GYP C. Similar assays were performed with a parallel series of wild-type and knockout W2mef parasite lines (W2mef has the VSTK haplotype of EBA-140), which, while showing decreased binding to the GYP C receptor, showed the same pattern of binding seen for 3D7. This suggests that W2mef EBA-140 (VSTK) binds less effectively to GYP C than 3D7 EBA-140 (INKK) does. Again, neither of these EBA-140 allelic forms was able to bind to mutant GYP C from the blood of Gerbich-negative individuals (Fig. 4A), in agreement with results obtained previously (27).
FIG. 4.

Invasion pathways in EBA-deficient cell lines. (A) Binding of EBA-140 to GYP C from erythrocyte ghost membranes (RBC), from purified glycophorins (GYP), or from Gerbich erythrocyte ghost membranes (Ge) by using overlay assays. The erythrocyte membranes or purified glycophorins after SDS-PAGE and transfer to nitrocellulose membrane were incubated with culture supernatant, and bound EBA-140 was detected with anti-EBA-140 (αEBA-140) as indicated. The GYP C was detected in nitrocellulose membranes with anti-GYP C (αGYP C) antibodies as a control. Supernatants were derived from parental or EBA-140- and EBA-175-deficient cell lines from both the 3D7 and W2mef backgrounds. The parasite strains from which the culture supernatant was taken for incubation with the filter are indicated on top of each panel. PBST lanes are negative controls in which PBS-Tween 20 rather than culture supernatant was incubated with the nitrocellulose membrane. EBA-140 does not bind to Ge-negative red blood cells as has been shown previously (28). 3D7Δ140 and W2mefΔ140 lack expression of EBA-140 and therefore show no binding to GYP C. The far-right column corresponds to antibodies against GYP C after incubation with PBS-Tween 20 (PBST) as a control. trunc. C, truncated GYP C in Ge-negative red blood cells. (B) Binding of EBA-140 to GYP A from erythrocyte ghost membranes (RBC) or from purified glycophorins (GYP). The lanes are identical to those in panel A except that binding to GYP A is shown. Binding of EBA-140 to GYP A can be detected only when the supernatant used contained EBA-140, i.e., not with supernatant from 3D7Δ140 or W2mefΔ140. The binding of EBA-140 in W2mef and W2mefΔEBA-175 is barely visible, consistent with its decreased binding affinity. (C) Binding of EBA-175 to GYP A. The lanes are identical to those in panel A except that the nitrocellulose membranes were probed with anti-EBA-175 (αEBA-175) or anti-GYP A/B (αGYP A/B) antibodies and detect binding of EBA-175 to GYP A. A/A, GYP A homodimer; A/B, GYP A/B heterodimer.
In contrast to the results of EBA-140 variant binding to GYP C, the divergent allelic forms of EBA-175 from 3D7 and W2mef showed no differences in binding to GYP A (Fig. 4C). The EBA-175 ligands from both strains bound GYP A equally for solubilized erythrocyte membranes and glycophorins (Fig. 4C). Similar binding was observed for Gerbich-negative erythrocytes, which express normal GYP A. This is consistent with the selection of EBA-175 polymorphisms for immune evasion rather than changes in affinity to its erythrocyte receptor (6). The few polymorphic sites that separate EBA-140 in 3D7 and this protein in W2mef do not contain any signatures of positive selection as observed for EBA-175 and therefore do not appear to have been influenced by immune pressures (6).
Promiscuous EBA-140 binding.
Previously, it has been shown that EBA-140 can bind to GYP A and GYP B in addition to GYP C (27). Supporting this concept of promiscuity, EBA-140 from 3D7 (INKK) showed clear binding to homodimers of GYP A (Fig. 4B, upper band of doublet) and heterodimers of GYP A/B (Fig. 4B, lower band of doublet) compared to 3D7Δ140, which lacked expression of the ligand. In contrast, EBA-140 from W2mef (VSKK) showed barely detectable levels of binding (Fig. 4B). Therefore, the amino acid polymorphisms in EBA-140, associated with decreased binding to GYP C, also decrease the binding to the GYP A homodimer and the GYP A/B heterodimer.
Function of variant EBA-140 ligands in merozoite invasion.
Antibodies to the F2 region of EBA-140 have been shown to inhibit the function of this ligand in merozoite invasion (28). Chymotrypsin treatment of erythrocytes enhances this effect by limiting receptors available on the erythrocyte surface, thereby reducing alternative invasion options (4, 27) and testing the importance of the chymotrypsin-resistant GYP C. Previously, using EBA-140-deficient parasites and Gerbich-negative erythrocytes, it was shown that these antibodies specifically inhibit EBA-140 function and block utilization of the EBA-140-GYP C invasion pathway (4, 27). Inhibition of merozoite invasion by 3D7 parasites with anti-EBA-140 antibodies reduced invasion of untreated erythrocytes by approximately 20%. When these antibodies were used with chymotrypsin-treated erythrocytes, the inhibitory effect increased to 60%, similar to previous results (Fig. 5) (27). In contrast, the level of inhibition by these antibodies was decreased in MCAMP, Pf120, HB3, and W2mef, strains that express allelic forms of EBA-140. Although MCAMP and Pf120 showed significant inhibition with respect to invasion of chymotrypsin-treated erythrocytes, it was less than that observed for 3D7. Antibody inhibition was completely abolished in HB3 and W2mef. There was a direct correlation with respect to EBA-140 binding to GYP C (Fig. 2B) and the ability of anti-EBA-140 antibodies to inhibit invasion. The region of the EBA-140 gene that encodes the recombinant protein to which the antibodies were raised was sequenced and shown to be identical in the six different parasite strains tested. Therefore, the differential inhibition cannot be explained by EBA-140 polymorphisms in the antibody binding region. As such, higher-level affinity binding to GYP C correlates with increased functionality of the EBA-140-GYP C invasion pathway. It also demonstrates functionally that EBA-140 allelic forms showing decreased binding to GYP C do not alter the receptor specificity of this invasion ligand. Analysis of 38 field isolates from The Gambia showed that there was a statistically significant association, suggesting that parasites containing valine (rather than isoleucine) at amino acid position 185 were more dependent on sialic acid (see Fig. S1 in the supplemental material) (5).
FIG. 5.
Invasion of P. falciparum into human erythrocytes. Inhibition of merozoite invasion by an anti-EBA-140 F2 antibody (Fig. 1). Levels of invasion inhibition in the presence of nonspecific antibodies (immunoglobulin G [IgG]) from normal rabbit serum (Pre IgG) and in the presence of EBA-140 F2 antibodies were compared. Error bars indicate confidence levels (α = 0.1) of three independent experiments performed in triplicate. The anti-EBA-140 F2 antibodies have been described previously, and they have been proven to specifically inhibit the function of EBA-140 in these experiments (27).
Location of the polymorphisms in EBA-140, EBA-181, and EBA-175 three-dimensional structure.
X-ray crystal structure of the F1 and F2 region from EBA-175 has been solved with three putative glycan binding sites identified on each of these domains, formed at the interfaces of a protein dimer (46). The molecular surface of the residues putatively involved in contact with the glycan are shown in Fig. 6. To rationalize the ability of polymorphic variants to alter receptor affinity or specificity, polymorphisms listed in Table 1 were mapped to this surface (Fig. 6). The EBA-175 polymorphisms in F1 are located far from the putative glycan binding site, while in contrast, several of the polymorphisms in F2 occur in residues in close proximity to the glycan binding residues, although they do not interrupt the continuous surface of the putative glycan binding site. This is consistent with these polymorphisms not having any measurable effect on binding to GYP A. In comparison, the polymorphisms identified in EBA-140 all occur in F1 in depressions on the protein surface (Fig. 6). The central exposed surface forms part of a deep pocket in the model of EBA-140 F1, and this residue would be juxtaposed with some of the putative glycan binding residues in an EBA-140 dimer. Polymorphisms in the model of EBA-181 are located away from the putative glycan binding residues (Fig. 6). The polymorphisms of both EBA-140 and EBA-181 are therefore in regions unlikely to have a direct effect on receptor binding and more likely exert minor changes on the structure of the protein that may be responsible for altering the affinity of binding.
FIG. 6.
Comparison of the structures of EBA-175, EBA-140, and EBA-181. The X-ray crystal structure of EBA-175 (yellow) and homology models of EBA-140 (light blue) and EBA-181 (light green) are presented in ribbon form. Arrows point to the beta-hairpin loops on the F1 and F2 domains of EBA-175. The molecular surfaces of polymorphic residues are shown in red, while the molecular surface of the putative glycan contact residues on EBA-175 are shown in orange. Circled on the structures of EBA-140 and EBA-181 are the molecular surfaces (red) resulting from the polymorphisms listed in Table 1.
DISCUSSION
EBA-140 and EBA-181 are important ligands involved in the mediation of invasion pathways used by P. falciparum when invading erythrocytes, interacting with specific sialic acid-containing receptors on erythrocytes (26, 27, 29). Polymorphisms within the F1/F2 domain of these ligands have been identified previously (27, 29), and it has been suggested that they mediate changes in receptor specificity (29). We addressed this by analyzing variants of EBA-140 and EBA-181 for their abilities to bind to red blood cells and receptors and determining the effect of these residues, for EBA-140, on ligand function in invasion. We have shown that polymorphisms in EBA-140 alter overall binding to GYP C and there is a direct correlation between binding and function of the EBA-140-GYP C invasion pathway. The polymorphisms in EBA-140 and EBA-181 play a role in determining overall binding to erythrocytes and consequently their function in invasion.
The ability of EBA-140 variants to bind to GYP C was in contrast to previous results suggesting that polymorphic forms may mediate binding to different receptors (29, 30). The five variant EBA-140 ligands all bound GYP C and to some extent GYP A/B, with the amino acid differences determining the binding affinity to the receptor. The variants showing the lowest binding affinity to GYP C also demonstrated little or no detectable function in merozoite invasion assays, consistent with the requirement of a minimal affinity of binding for measurable participation in invasion. The function of EBA-140 appeared to be strongest in 3D7 (INKK), and the levels of function were decreased for MCAMP (INRE) and Pf120 (ISKK) and were essentially not measurable for the other variants. It has been suggested that the EBA-140 INRE, ISKK, and VSKK variants bind to receptors other than GYP C, as the F1/F2 domain expressed on COS cells had binding properties different from those of the VSTK form (29). The results presented here for native EBA-140 show that GYP C is the major receptor for the variant EBA-140 forms. Different EBA-140 polymorphisms clearly affect the extent of binding to GYP C, and as a consequence, the overall function in merozoite invasion through the GYP C-EBA-140 invasion pathway.
The different results obtained in our study compared to those of previous studies (29, 30) may be partially attributed to the experimental systems. Previously, red blood cell binding had been assessed by expression of EBA-140 and EBA-181 F1/F2 domains on COS cells. It is difficult to verify correct folding of the F1/F2 domains, and this appears to have been an issue in expression of the paralogous domain from EBA-181 and its ability to bind to red blood cells (30). Incorrect folding of the EBA-140 and EBA-181 domains on COS cells is likely to give inconsistent results in binding assays. Additionally, it is possible that other regions of EBA-140 and EBA-181 may contribute to the overall affinity of binding in a cooperative interaction. The binding of erythrocytes to COS cells expressing the F1/F2 domain of EBA-140 was scored as positive when five or more red blood cells were bound (29). Subtle differences in binding due to altered affinity might be ablated and an artificial threshold might be created, leading to an equivocal result not allowing the measure of the true degree of binding. For comparison, we used native EBA-140 and EBA-181 from P. falciparum supernatants that include most of the ectodomain in direct binding assays with receptors as well as red blood cells. Importantly, we measured the function of the variant forms of EBA-140 in overall merozoite invasion by specific inhibition with anti-EBA-140 antibodies. The polymorphisms in EBA-140 clearly affect the ability to bind to GYP C and thus the ability to invade using this receptor. These results provide conclusive evidence that the different variant forms of EBA-140 bind and function in merozoite invasion through GYP C as the receptor but also that no other receptors contribute. The lack of measurable participation of the EBA-140 VSKK and INRE variants in merozoite invasion implies that even if there was binding to another receptor, it is not functionally relevant.
In this study, we showed that the EBA-140 variants do not bind to mutant GYP C in Gerbich-negative erythrocytes. Previous results, however, suggested that the INKK form of EBA-140 bound to these cells (26). These experiments were performed by solubilization of EBA-140 bound to receptor on erythrocytes followed by immunoprecipitation of the complex by using anti-EBA-140 antibodies. This type of experiment would not take into account the ability of EBA-140 to bind to GYP A/B. We demonstrated here that the INKK form in particular binds to alternative glycophorins significantly much more than other variants, because of its generally increased receptor affinity. It is therefore likely that the observed binding to Gerbich-negative erythrocytes in the former study was due to binding to GYP A/B. This would be consistent with the reduced binding of EBA-140 to Gerbich-negative erythrocytes described previously (29).
The effect of EBA-140 and EBA-181 amino acid polymorphisms on binding to red blood cells was in contrast to that observed for EBA-175. EBA-175 shows extensive polymorphisms in the F1 and F2 regions, and these amino acid changes have no effect on its binding to its receptor, GYP A. This was consistent with EBA-175, where this ligand is under strong diversifying selection from host immune responses (6, 48). In contrast, there was no departure of neutrality for EBA-140 or EBA-181, indicating no immunoselection of these gene products. Intriguingly, an association of expression levels between EBA-140 and EBA-175 was observed, and this may reflect a shared mechanism (7). Any EBA-175 polymorphisms selected appear to be neutral with respect to the affinity of ligand-receptor binding. This suggests that EBA-175 is an important parasite ligand in merozoite invasion in many strains; however, it is not absolutely essential (14), suggesting that other parasite ligands, such as EBA-140 and EBA-181, play a supporting role.
The lack of diversifying selection for EBA-140 and EBA-181 (6) suggests that these ligands may be less dominant in merozoite invasion than in other ligand-receptor interactions. We have found no evidence for alternate receptors for the variant forms of EBA-181 and EBA-140, although they do bind to erythrocytes and receptors to different extents. It is possible that there is a selective advantage in allowing variability in affinity of EBA-140 to GYP C and EBA-181 to its receptor and therefore their participation in invasion. This is supported by the observation that parasites with V-185 in EBA-140 tend to be more dependent on sialic acid-dependent invasion pathways. Interestingly, four out of the five detected nucleotide exchanges result in amino acid changes, arguing against a random fluctuation and raising the question of selective pressures being involved in their generation. One possibility is that selection for different EBA-140 binding affinities to GYP C could be a strategy to circumvent host red blood cell polymorphisms, such as Gerbich negativity. Gerbich negativity is highly prevalent in regions of Papua New Guinea where malaria is endemic, but this negativity is rarely found in other parts of the world. However, the presence of all observed EBA-140 polymorphisms in Papua New Guinean, Gambian, and Brazilian isolates (6, 25, 27, 29) argues against this hypothesis. In addition, no obvious association between the geographical origin of the laboratory strains and the observed mutations was seen (Table 1).
The position of the polymorphisms in the EBA molecules is similar to what is seen in related molecules: in the P. vivax Duffy binding protein, for example, polymorphic sites are separate from the receptor binding site (44) and the molecule is released from micronemes just before binding to the erythrocyte receptor (47), limiting exposure to the immune system. The polymorphic sites in the Var2CSA DBL domains are on exposed surfaces or on connecting loops, whereas the structural elements are semiconserved (9), a fact which is very much reflected in the polymorphic positions in EBA-181.
The absence of a signature of diversifying selection in EBA-140 and EBA-181 also might indicate that these proteins represent a more recently evolved invasion pathway for P. falciparum (6). The dominance and therefore the possible evolutionary age of EBA-175-GYP A make sense. GYP A is one of the most abundant proteins on the red blood cell, and it is not surprising that invasion through this receptor, via EBA-175, would be more important than, for example, GYP C, which is present at lower levels (21). Further support for a less significant role for EBA-140 and EBA-181 in merozoite invasion, in comparison to EBA-175, is the ability to disrupt the corresponding genes in most parasites so far attempted (13, 14). In contrast, it does not seem to be possible to disrupt the EBA-175 gene in D10, in which the available invasion pathways appear to be limited (13). Nevertheless, the availability of alternate ligands that are functionally homologous provides a backup for occasions when the function of the dominant pathway is blocked either from immune mechanisms or host red blood cell receptor polymorphisms and would be an advantage in avoiding host immune responses.
Supplementary Material
Acknowledgments
We thank the Red Cross Blood Service (Melbourne, Australia) for the supply of erythrocytes and serum. Our thanks go to M. Cravino, M. O'Neill, M. Brown, M. Duraisingh, J. Thompson, and T. Triglia for their support. We thank Melanie Rug, Brendan Crabb, and Matthias Marti for critical comments. We are grateful to Joyce Poole and David Anstee from the Red Cell Reference Department of IBGRL (United Kingdom), Peter Zimmerman, Sheral Patel (Case Western University), and John Reeder (PNG Institute of Medical Research) for the mutant erythrocytes and the blood donors for their participation.
A.G.M. is an ARC Australian Research Fellow, J.B. is recipient of an NHMRC Career Development Award and A.F.C. is a Howard Hughes International Scholar. This work was supported by the National Health and Medical Research Council of Australia and the Wellcome Trust.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 9 February 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Adams, J. H., P. L. Blair, O. Kaneko, and D. S. Peterson. 2001. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 17297-299. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. H., B. K. L. Sim, S. A. Dolan, X. Fang, D. C. Kaslow, and L. H. Miller. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 897085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnwell, J. W., and M. R. Galinski. 1998. Invasion of vertebrate cells: erythrocytes, p. 93-120. In I. W. Sherman (ed.), Malaria: parasite biology, pathogenesis and protection. ASM Press, Washington DC.
- 4.Baum, J., A. G. Maier, R. T. Good, K. M. Simpson, and A. F. Cowman. 2005. Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLoS Pathog. 1e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum, J., M. Pinder, and D. J. Conway. 2003. Erythrocyte invasion phenotypes of Plasmodium falciparum in The Gambia. Infect. Immun. 711856-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum, J., A. W. Thomas, and D. J. Conway. 2003. Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics 1631327-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bei, A. K., C. D. Membi, J. C. Rayner, M. Mubi, B. Ngasala, A. A. Sultan, Z. Premji, and M. T. Duraisingh. 2007. Variant merozoite protein expression is associated with erythrocyte invasion phenotypes in Plasmodium falciparum isolates from Tanzania. Mol. Biochem. Parasitol. 15366-71. [DOI] [PubMed] [Google Scholar]
- 8.Bhasin, V. K., and W. Trager. 1984. Gametocyte-forming and non-gametocyte-forming clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 33534-537. [DOI] [PubMed] [Google Scholar]
- 9.Bockhorst, J., F. Lu, J. H. Janes, J. Keebler, B. Gamain, P. Awadalla, X. Z. Su, R. Samudrala, N. Jojic, and J. D. Smith. 2007. Structural polymorphism and diversifying selection on the pregnancy malaria vaccine candidate VAR2CSA. Mol. Biochem. Parasitol. 155103-112. [DOI] [PubMed] [Google Scholar]
- 10.Breuer, W. V., H. Ginsburg, and Z. I. Cabantchik. 1983. An assay of malaria parasite invasion into human erythrocytes. The effects of chemical and enzymatic modification of erythrocyte membrane components. Biochim. Biophys. Acta 755263-271. [DOI] [PubMed] [Google Scholar]
- 11.Burkot, T. R., J. L. Williams, and I. Schneider. 1984. Infectivity to mosquitoes of Plasmodium falciparum clones grown in vitro from the same isolate. Trans. R. Soc. Trop. Med. Hyg. 78339-341. [DOI] [PubMed] [Google Scholar]
- 12.Camus, D., and T. J. Hadley. 1985. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230553-556. [DOI] [PubMed] [Google Scholar]
- 13.Cowman, A. F., D. L. Baldi, J. Healer, K. E. Mills, R. A. O'Donnell, M. B. Reed, T. Triglia, M. E. Wickham, and B. S. Crabb. 2000. Functional analysis of proteins involved in Plasmodium falciparum merozoite invasion of red blood cells. FEBS Lett. 47684-88. [DOI] [PubMed] [Google Scholar]
- 14.Duraisingh, M. T., A. G. Maier, T. Triglia, and A. F. Cowman. 2003. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and -independent pathways. Proc. Natl. Acad. Sci. USA 1004796-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felger, I., L. Tavul, and H.-P. Beck. 1993. Plasmodium falciparum: a rapid technique for genotyping the merozoite surface protein 2. Exp. Parasitol. 77372-375. [DOI] [PubMed] [Google Scholar]
- 16.Fiser, A., and A. Sali. 2003. ModLoop: automated modeling of loops in protein structures. Bioinformatics 192500-2501. [DOI] [PubMed] [Google Scholar]
- 17.Galinski, M. R., C. C. Medina, P. Ingravallo, and J. W. Barnwell. 1992. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 691213-1226. [DOI] [PubMed] [Google Scholar]
- 18.Gilberger, T. W., J. K. Thompson, M. B. Reed, R. T. Good, and A. F. Cowman. 2003. The cytoplasmic domain of the Plasmodium falciparum ligand EBA-175 is essential for invasion but not protein trafficking. J. Cell Biol. 162317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilberger, T. W., J. K. Thompson, T. Triglia, R. T. Good, M. T. Duraisingh, and A. F. Cowman. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J. Biol. Chem. 27814480-14486. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C. H., K. Johe, J. J. Moulds, P. D. Siebert, M. Fukuda, and O. O. Blumenfeld. 1987. Delta glycophorin (glycophorin B) gene deletion in two individuals homozygous for the S-s-U- blood group phenotype. Blood 701830-1835. [PubMed] [Google Scholar]
- 21.Issitt, P. D., and D. J. Anstee. 1998. Applied blood group serology, 4th ed. Montgomery Scientific Publications, Durham, NC.
- 22.Jensen, J. B. 1978. Concentration from continuous culture of erythrocytes infected with trophozoites and schizonts of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 271274-1276. [DOI] [PubMed] [Google Scholar]
- 23.Lanzillotti, R., and T. L. Coetzer. 2006. The 10 kDa domain of human erythrocyte protein 4.1 binds the Plasmodium falciparum EBA-181 protein. Malar. J. 5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauterbach, S. B., R. Lanzillotti, and T. L. Coetzer. 2003. Construction and use of Plasmodium falciparum phage display libraries to identify host parasite interactions. Malar. J. 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobo, C. A., K. de Frazao, M. Rodriguez, M. Reid, M. Zalis, and S. Lustigman. 2004. Invasion profiles of Brazilian field isolates of Plasmodium falciparum: phenotypic and genotypic analyses. Infect. Immun. 725886-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobo, C. A., M. Rodriguez, M. Reid, and S. Lustigman. 2003. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 66. [DOI] [PubMed] [Google Scholar]
- 27.Maier, A. G., M. T. Duraisingh, J. C. Reeder, S. S. Patel, J. W. Kazura, P. A. Zimmerman, and A. F. Cowman. 2003. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 987-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer, D. C., O. Kaneko, D. E. Hudson-Taylor, M. E. Reid, and L. H. Miller. 2001. Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc. Natl. Acad. Sci. USA 985222-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer, D. C., J. B. Mu, X. Feng, X. Z. Su, and L. H. Miller. 2002. Polymorphism in a Plasmodium falciparum erythrocyte-binding ligand changes its receptor specificity. J. Exp. Med. 1961523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer, D. C., J. B. Mu, O. Kaneko, J. Duan, X. Z. Su, and L. H. Miller. 2004. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc. Natl. Acad. Sci. USA 1012518-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, L. H., J. D. Haynes, F. M. McAuliffe, T. Shiroishi, and J. R. Durocher. 1977. Evidence for differences in erythrocyte surface receptors for the malarial parasites, Plasmodium falciparum and Plasmodium knowlesi. J. Exp. Med. 146277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, L. H., S. J. Mason, D. F. Clyde, and M. H. McGinniss. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295302-304. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, G. H., T. J. Hadley, M. H. McGinniss, F. W. Klotz, and L. H. Miller. 1986. Invasion of erythrocytes by Plasmodium falciparum malaria parasites: evidence for receptor heterogeneity and two receptors. Blood 671519-1521. [PubMed] [Google Scholar]
- 34.Narum, D. L., S. R. Fuhrmann, T. Luu, and B. K. Sim. 2002. A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol. Biochem. Parasitol. 119159-168. [DOI] [PubMed] [Google Scholar]
- 35.Nieper, H., and H. Muller. 1996. Susceptibility of chicken lymphoid cells to infectious bursal disease virus does not correlate with the presence of specific binding sites. J. Gen. Virol. 771229-1237. [DOI] [PubMed] [Google Scholar]
- 36.Oduola, A. M., W. K. Milhous, N. F. Weatherly, J. H. Bowdre, and R. E. Desjardins. 1988. Plasmodium falciparum: induction of resistance to mefloquine in cloned strains by continuous drug exposure in vitro. Exp. Parasitol. 67354-360. [DOI] [PubMed] [Google Scholar]
- 37.Pandey, K., S. Singh, P. Pattnaik, C. Pillai, U. Pillai, A. Lynn, S. Jain, and C. Chitnis. 2002. Bacterially expressed and refolded receptor binding domain of Plasmodium falciparum EBA-175 elicits invasion inhibitory antibodies. Mol. Biochem. Parasitol. 12323. [DOI] [PubMed] [Google Scholar]
- 38.Perkins, M. 1981. Inhibitory effects of erythrocyte membrane proteins on the in vitro invasion of the human malarial parasite (Plasmodium falciparum) into its host cell. J. Cell Biol. 90563-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins, M. E., and E. H. Holt. 1988. Erythrocyte receptor recognition varies in Plasmodium falciparum isolates. Mol. Biochem. Parasitol. 2723-34. [DOI] [PubMed] [Google Scholar]
- 40.Rayner, J. C., M. R. Galinski, P. Ingravallo, and J. W. Barnwell. 2000. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc. Natl. Acad. Sci. USA 979648-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed, M. B., S. R. Caruana, A. H. Batchelor, J. K. Thompson, B. S. Crabb, and A. F. Cowman. 2000. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid independent pathway of invasion. Proc. Natl. Acad. Sci. USA 977509-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310243-257. [DOI] [PubMed] [Google Scholar]
- 43.Sim, B. K., J. M. Carter, C. D. Deal, C. Holland, J. D. Haynes, and M. Gross. 1994. Plasmodium falciparum: further characterization of a functionally active region of the merozoite invasion ligand EBA-175. Exp. Parasitol. 78259-268. [DOI] [PubMed] [Google Scholar]
- 44.Singh, S. K., R. Hora, H. Belrhali, C. E. Chitnis, and A. Sharma. 2006. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature 439741-744. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. K., T. Triglia, M. B. Reed, and A. F. Cowman. 2001. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol. Microbiol. 4147-58. [DOI] [PubMed] [Google Scholar]
- 46.Tolia, N. H., E. J. Enemark, B. K. Sim, and L. Joshua-Tor. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122183-193. [DOI] [PubMed] [Google Scholar]
- 47.VanBuskirk, K. M., E. Sevova, and J. H. Adams. 2004. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc. Natl. Acad. Sci. USA 10115754-15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verra, F., S. D. Polley, A. W. Thomas, and D. J. Conway. 2006. Comparative analysis of molecular variation in Plasmodium falciparum and P. reichenowi maebl gene. Parassitologia 48567-572. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.