Abstract
Cleavage and encapsidation of newly replicated herpes simplex virus type 1 (HSV-1) DNA requires several essential viral gene products that are conserved in sequence within the Herpesviridae. However, conservation of function has not been analyzed in greater detail. For functional characterization of the UL6, UL15, UL28, UL32, and UL33 gene products of pseudorabies virus (PrV), the respective deletion mutants were generated by mutagenesis of the virus genome cloned as a bacterial artificial chromosome (BAC) in Escherichia coli and propagated in transgenic rabbit kidney cells lines expressing the deleted genes. Neither of the PrV mutants was able to produce plaques or infectious progeny in noncomplementing cells. DNA analyses revealed that the viral genomes were replicated but not cleaved into monomers. By electron microscopy, only scaffold-containing immature but not DNA-containing mature capsids were detected in the nuclei of noncomplementing cells infected with either of the mutants. Remarkably, primary envelopment of empty capsids at the nuclear membrane occasionally occurred, and enveloped tegument-containing light particles were formed in the cytoplasm and released into the extracellular space. Immunofluorescence analyses with monospecific antisera of cells transfected with the respective expression plasmids indicated that pUL6, pUL15, and pUL32 were able to enter the nucleus. In contrast, pUL28 and pUL33 were predominantly found in the cytoplasm. Only pUL6 could be unequivocally identified and localized in PrV-infected cells and in purified virions, whereas the low abundance or immunogenicity of the other proteins hampered similar studies. Yeast two-hybrid analyses revealed physical interactions between the PrV pUL15, pUL28, and pUL33 proteins, indicating that, as in HSV-1, a tripartite protein complex might catalyze cleavage and encapsidation of viral DNA. Whereas the pUL6 protein is supposed to form the portal for DNA entry into the capsid, the precise role of the UL32 gene product during this process remains to be elucidated. Interestingly, the defect of UL32-negative PrV could be completely corrected in trans by the homologous protein of HSV-1, demonstrating similar functions. However, trans-complementation of UL32-negative HSV-1 by the PrV protein was not observed.
DNA replication of herpesviruses presumably is initiated by circularization of the linear double-stranded virus genome in the host cell nucleus, followed by a complex amplification process that includes theta and rolling-circle mechanisms, as well as recombination, resulting in huge concatemeric molecules (7, 64). Viral capsids also assemble in the nucleus around a proteinaceous scaffold (61). Subsequently, the concatemeric virus genomes are simultaneously cleaved into monomers at defined positions and packaged into the preformed capsids, coinciding with removal of at least major portions of scaffold (25, 56). Cleavage-encapsidation of herpesvirus genomes exhibits considerable similarities to the corresponding process in large bacteriophages, like T4 or lambda (6), and the responsible herpesvirus enzymes have also been designated terminases.
In the prototypic alphaherpesvirus herpes simplex virus type 1 (HSV-1), seven proteins encoded by the open reading frames (ORFs) UL6, UL15, UL17, UL25, UL28, UL32, and UL33 are relevant for cleavage and encapsidation of the virus genome. Homologues of these genes were found in all subfamilies of mammalian and avian herpesviruses (52, 56). However, as shown for HSV-1 and the related alphaherpesvirus pseudorabies virus (PrV), the UL25 protein (pUL25) is obviously not absolutely required for DNA packaging but seems to function as a structural protein in nucleocapsid stabilization, nuclear egress, and/or early steps of tegumentation (15, 16, 32, 42, 57). Like pUL25, pUL17 of HSV-1 and PrV has also been identified as an essential capsid-associated virion protein (31, 58, 60), and pUL6 of HSV-1 forms a dodecameric ring at one vertex of the capsid, which serves as a portal for DNA entry (49).
The enzyme complex directly performing HSV-1 DNA cleavage and encapsidation is considered to consist of pUL15, pUL28, and pUL33. These proteins have been shown to interact with each other (5, 35), and pUL15 and pUL28 bind the portal protein pUL6 (63). There is also strong evidence that pUL15 contains a functional nuclear localization signal and promotes nuclear localization of pUL28 (34) and pUL33 (67) after binding to these proteins in the cytoplasm. In HSV-2, nuclear localization of pUL33 is enhanced by coexpression of pUL14 (65), which, however, is nonessential for replication of the closely related HSV-1 (18). The terminase subunit pUL15 possesses a predicted ATPase motif and might provide the energy required for DNA encapsidation (69). With its functional domains and overall structure, it represents one of the best-conserved herpesvirus proteins and also possesses detectable homologues in the phylogenetically distant herpesviruses of lower vertebrates and molluscs (41). Except in ostreid herpesvirus 1, the pUL15 homologues belong to the relatively rare intron-containing herpesvirus genes identified. In all investigated mammalian and avian herpesviruses, they consist of two exons (52). For PrV, this has also been suggested from sequence homology but not yet proven experimentally (30). The terminase subunit pUL28 is able to bind specifically to the packaging signals (pac sites) in the viral DNA (1), whereas the precise role of pUL33 remains to be elucidated.
This also applies to pUL32, which is indispensable for cleavage and packaging of HSV-1 DNA (38) but up to now has not been shown to interact with any of the other cleavage-encapsidation proteins. However, capsid protein distribution in the nuclei of cells infected with UL32-negative HSV-1 suggested a putative role for this protein in the transport of nucleocapsids to DNA replication compartments (38). Also not understood is the molecular linkage between cleavage-encapsidation of viral DNA and the nuclear egress of mature C capsids (52) by budding at the inner nuclear membrane, which seems to be initiated by interactions between the conserved pUL31 and pUL34 homologues of alpha-, beta-, and gammaherpesviruses (24, 37, 47, 55).
For the betaherpesvirus human cytomegalovirus (HCMV), detailed functional investigations of several components of the DNA cleavage-encapsidation machinery have also been performed (8, 10, 21). In contrast, in PrV, only the pac sequences and cleavage sites at the genome level (53) and the UL17, UL25, and UL28 genes and gene products are known (31, 32, 34, 44). Like pUL17 and pUL25, pUL28 of PrV is essential for virus replication, and unlike pUL25, also for cleavage-encapsidation (44).
In our current approach to investigate the functions of all predicted PrV genes in an isogenic viral background and cell culture system, we generated and analyzed virus mutants lacking essential parts of the homologues of the HSV-1 UL6, UL15, UL32, or UL33 genes. Monospecific rabbit antisera against bacterial fusion proteins were prepared and used for identification and subcellular localization of the investigated gene products of PrV, and yeast two-hybrid studies were performed to detect possible interactions between these proteins. Furthermore, in heterologous trans-complementation studies for pUL32, we started to investigate whether and to what extent the highly conserved cleavage-encapsidation proteins of PrV and HSV-1 can substitute for each other.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants generated in this study were derived from the bacterial artificial chromosome (BAC) pPrV-ΔgB (33), which contains the genome of PrV strain Kaplan (PrV-Ka) (28). HSV-1 recombinants were generated by mutagenesis of the BAC pHSV-1ΔgJ (39), encompassing the genome of HSV-1 strain KOS (54). Porcine (PSEK) or monkey (Vero) kidney cells were used for propagation of wild-type PrV or HSV-1, whereas defective virus mutants were propagated in stably transfected rabbit kidney (RK13) cell lines providing the deleted viral gene products in trans. RK13 cells were also used for one-step growth and plaque analyses of all investigated viruses. Cells were grown at 37°C in minimum essential medium supplemented with 10% fetal calf serum (Invitrogen). After infection, the medium was replaced by minimum essential medium that contained only 5% fetal calf serum and, for plaque assays, 6 g/liter methyl cellulose (Sigma).
Plasmid construction.
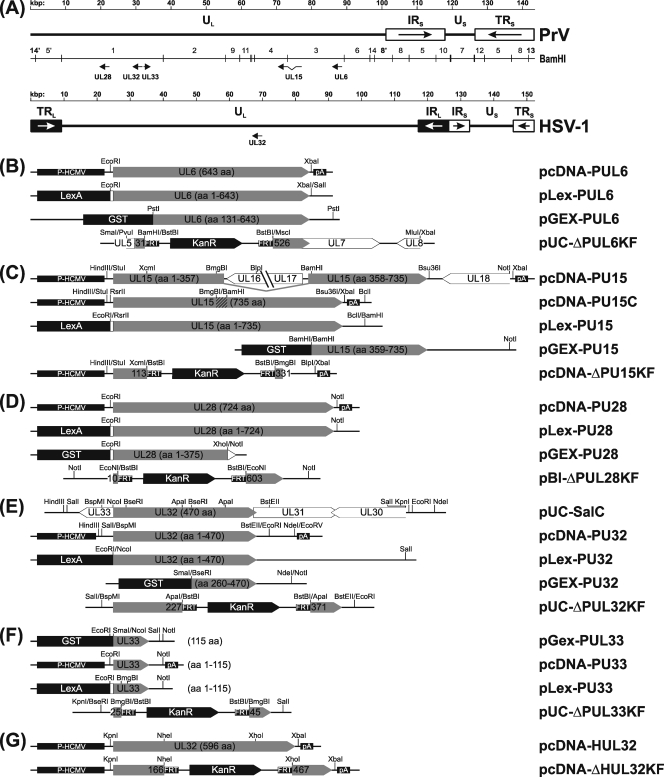
For eukaryotic expression, the UL6 ORF was amplified from genomic DNA of PrV-Ka by PCR using Platinum Pfx DNA polymerase (Invitrogen) and primers PUL6-F (5′-CACAGAATTCGCCATGTCGGCTGCGACGG-3′) and PUL6-R (CACATCTAGAGCGCGCGTCAGTCACCGAG-3′), which are complementary to nucleotides (nt) 89286 to 89304 and correspond to nt 87363 to 87381 of the PrV genome sequence (GenBank accession no. BK001744) (30), respectively. The primers include the start and stop codons of UL6 (boldface) and contain artificial EcoRI and XbaI cleavage sites (underlined), which facilitated cloning into the appropriately digested vector pcDNA3 (Invitrogen). For yeast two-hybrid studies, the 1,948-bp EcoRI/XbaI insert of pcDNA-PUL6 (Fig. 1B) was recloned in the EcoRI/SalI-digested vector pLexA (Clontech), resulting in pLex-PUL6 (Fig. 1B), to express UL6 fused to the DNA-binding LexA protein. Plasmid pGEX-PUL6 (Fig. 1B) was obtained after recloning of a 1,773-bp PstI subfragment of the genomic BamHI fragment 3 of PrV-Ka (Fig. 1A) into the SmaI-digested vector pGEX-4T-1 (GE Healthcare) and was used for expression of a fusion protein of pUL6 amino acids 131 to 643 with glutathione S-transferase (GST) in Escherichia coli. To generate a UL6-negative virus mutant, a 2,889-bp PvuI/MluI subfragment of a genomic 7,451-bp SalI fragment of PrV-Ka was recloned in pUC19 (New England Biolabs), which had been doubly digested with SmaI and XbaI. Subsequently, pUC-ΔPUL6KF (Fig. 1B) was constructed by digestion with MscI and BamHI, which led to deletion of 1,480 bp comprising UL6 codons 32 to 525 and insertion of a 1,258-bp BstBI fragment of pKD13 (19), which contained a kanamycin resistance (KanR) gene flanked by Flp recombinase recognition target (FRT) sites.
FIG. 1.
Plasmids and virus mutants. (A) Maps of the PrV and HSV-1 genomes containing long (UL) and short (US) unique regions, which are partly flanked by inverted-repeat sequences (IRL, TRL, IRS, and TRS). The investigated virus genes and BamHI fragments of PrV DNA are indicated. Shown below are the plasmids used for characterization of UL6 (B), UL15 (C), UL28 (D), UL32 (E), and UL33 (F) of PrV or of UL32 of HSV-1 (G). ORFs are shown as pointed rectangles, and relevant restriction sites, as well as expressed amino acids (aa) or retained codon ranges, are indicated. For constitutive expression in mammalian cells, the ORFs were cloned between the HCMV immediate-early promoter (P-HCMV) and a polyadenylation signal (pA). In pcDNA-PUL15C, a genomic BmgBI/BamHI fragment was replaced by a cDNA fragment (hatched). Plasmids permitting the expression of LexA fusion proteins in yeast were used for two-hybrid studies, and rabbits were immunized with bacterial GST fusion proteins to obtain monospecific antisera. For mutagenesis of the cloned virus genomes in E. coli, parts of the investigated genes were replaced by a kanamycin resistance (KanR) gene flanked by FRT sites.
For expression of PrV UL15, a 1,980-bp BamHI/NotI subfragment of genomic BamHI fragment 4 (Fig. 1A) was recloned into the appropriately cleaved vector pGEX-4T-1 or pcDNA3. By linearization with BamHI, treatment with Klenow polymerase, and religation, the GST gene was fused in frame with the 3′ part of UL15 (codons 359 to 736) in pGEX-PUL15 (Fig. 1C). The pcDNA3 construct was digested with BamHI and HindIII, and a 3,998-bp BamHI/StuI subfragment of genomic BamHI fragment 3 was additionally inserted to complete the UL15 gene of PrV. The resulting plasmid, pcDNA-PUL15 (Fig. 1C), was further modified by deletion of an 863-bp XbaI/Bsu36I fragment, followed by double digestion with BamHI and BmgBI and replacement of the released fragments, encompassing 2,964 bp, by a corresponding 85-bp cDNA fragment. The 213-bp cDNA utilized had been amplified from total RNA prepared (14) 6 h after infection of PSEK cells with PrV-Ka at a multiplicity of infection (MOI) of 10 using primers PUL15-R (5′-GGCATCGGGGCGTATAAAG-3′; nt 73050 to 73068 of BK001744) and PUL15-F (5′-GAGCCCGTGTTCGAGGAG-3′; complementary to nt 76124 to 76141 of BK001744), SuperScript II reverse transcriptase (Invitrogen), and Platinum Pfx DNA polymerase. Thus, plasmid pcDNA-PUL15C (Fig. 1C) contained the UL15 ORF of PrV, as it results from mRNA splicing. To obtain pLex-PUL15 (Fig. 1C), a 2,269-bp RsrII/BclI fragment of pcDNA-PUL15C was recloned in pLexA, which had been digested with EcoRI and BamHI. For generation of deletion plasmid pcDNA-ΔPUL15KF (Fig. 1C), pcDNA-PUL15 was shortened by the removal of a 4,429-bp BlpI/XbaI fragment, and a 649-bp XcmI/BmgBI fragment containing UL15 codons 114 to 330 was replaced by the 1,258-bp BstBI fragment of pKD13.
The UL28 ORF was amplified from virion DNA of PrV-Ka using primers PUL28-F (5′-CACAGAATTCCCGGCCATGGCGGAGCG-3′; complementary to nt 21630 to 21647 of BK001744) and PUL28-R (CACAGCGGCCGCCTAGCGCGACGGCGCCCC-3′; nt 19466 to 19483 of BK001744). The primers contained the start and stop codons of UL28 (boldface) and artificial EcoRI and NotI sites (underlined), which were utilized for cloning of the PCR product into correspondingly cleaved pcDNA3 and pLexA to obtain pcDNA-PUL28 or pLex-PUL28, respectively (Fig. 1D). The EcoRI/NotI-digested PCR product was also recloned into pGEX-4T-1 but subsequently had to be truncated to 1,133 bp by XhoI/NotI double digestion and religation to achieve sufficient expression of a GST fusion protein containing pUL28 amino acids 1 to 375 from pGEX-PUL28 (Fig. 1D). To generate the deletion plasmid pBl-ΔPUL28KF (Fig. 1D), a 2,759-bp NotI subfragment of genomic BamHI fragment 1 of PrV-Ka was recloned in pBluescript SK(−) (Stratagene), and two 1,191- and 585-bp EcoNI fragments spanning UL28 codons 11 to 602 were replaced by the 1,258-bp BstBI fragment of pKD13.
For investigation of UL32, a 3,320-bp SalI subfragment of genomic BamHI fragment 1 of PrV-Ka (Fig. 1A) was cloned into pUC19, resulting in pUC-SalC (Fig. 1E). Plasmid pLex-PUL32 (Fig. 1E) was generated by recloning of a 2,874-bp NcoI/SalI fragment of pUC-SalC into EcoRI/SalI-digested pLexA. To obtain deletion plasmid pUC-ΔPUL32KF (Fig. 1E), the insert of pUC-SalC was shortened by subsequent EcoRI/BstEII and BspMI/SalI digestions and religations, and two 230-bp and 192-bp ApaI fragments containing UL32 codons 228 to 370 were replaced by the 1,258-bp BstBI fragment of pKD13. Recloning of a 1,881-bp HindIII/NdeI fragment from the shortened pUC-SalC into HindIII/EcoRV-digested pcDNA3 resulted in pcDNA-PUL32 (Fig. 1E). For prokaryotic expression of the UL32 protein, pUC-SalC was truncated by subsequent EcoRI/BstEII and NcoI/SalI digestions, and a 1,755-bp SalI/NdeI fragment was recloned in SalI/NotI-digested pGEX-4T-3. Since protein expression from the original construct was unsatisfying, it was shortened by BseRI/SmaI double digestion and religation. The remaining 974-bp insert of pGEX-PUL32 (Fig. 1E) contained UL32 codons 260 to 471.
A 450-bp NcoI/SalI fragment containing UL33 of PrV was excised from pUC-SalC (Fig. 1E) and inserted into pGEX-4T-1, which had been digested with SmaI and Sal. From the resulting plasmid pGEX-PUL33 (Fig. 1F), the insert was recloned as a 466-bp EcoRI/NotI fragment into the appropriately digested vectors pcDNA3 and pLexA to obtain pcDNA-PUL33 and pLex-PUL33, respectively (Fig. 1F). For construction of deletion plasmid pUC-ΔPUL33KF (Fig. 1F), the insert of pUC-SalC was first shortened to 578 bp by double digestion with KpnI and BseRI and religation, and then a 57-bp BmgBI fragment containing UL33 codons 26 to 44 was replaced by the 1,258-bp BstBI fragment of pKD13.
The UL32 ORF of HSV-1 was amplified from genomic DNA using primers HUL32-F (5′-CAGGTACCATGGCAACTTCGCCC-3′), which was complementary to nt 69148 to 69164 of the genome sequence (GenBank accession no. X14112) (40), and HUL32-R (5′-CATCTAGAGGGGGTCGGTGTCATAC-3′; nt 67361 to 67377 of X14112). The primers contained the start and stop codons of UL32 (boldface), as well as artificial KpnI and XbaI restriction sites (underlined), which facilitated cloning of the PCR product in the appropriately digested vector pcDNA3. The resulting plasmid, pcDNA-HUL32 (Fig. 1G), was utilized for heterologous trans-complementation of UL32-negative PrV. For construction of an HSV-1 deletion mutant, pcDNA-ΔHUL32KF (Fig. 1G) was generated by replacement of an 896-bp NheI/XhoI fragment representing UL32 codons 167 to 466 by the 1,258-bp BstBI fragment of pKD13.
In all cloning experiments, noncompatible fragment ends were blunted by treatment with Klenow polymerase prior to ligation.
Antiserum preparation.
After transformation of E. coli strain XL1-Blue MRF′ (Stratagene) with pGEX-PUL6, -PUL15, -PUL28, -PUL32, or -PUL33 (Fig. 1B to F), expression of GST fusion proteins was induced as recommended by the manufacturer of the vector (GE Healthcare). The proteins were purified by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (24), and rabbits were immunized by four intramuscular applications of 100 μg protein emulsified in mineral oil each at 4-week intervals. Sera collected before the first and 3 weeks after the last immunization were analyzed.
Western blot analyses and indirect immunofluorescence tests.
For Western blot analyses, purified PrV particles and infected cell lysates were prepared as described previously (20). Lysates of bacteria expressing the GST fusion proteins and of RK13 cells transfected with pcDNA-PUL6, -PUL15C, -PUL28, -PUL32, or -PUL33 (Fig. 1B to F) were also investigated. The blots were probed with monospecific rabbit antisera against pUL6, pUL15, pUL28, pUL32, or pUL33 of PrV at dilutions of 1:10,000 to 1:100,000 (data not shown).
For immunofluorescence tests, RK13 cells were grown on coverslips and transfected with pcDNA3-derived expression plasmids as described above. After 24 h, the cells were fixed with acetone for 20 min at −20°C, dried, and subsequently incubated with the rabbit antisera at dilutions of 1:200 in phosphate-buffered saline (PBS) and diluted Alexa 488-conjugated anti-rabbit antibodies (Invitrogen). After each step, the slides were washed repeatedly with PBS and finally preserved with a 9:1 mixture of glycerol and PBS containing 25 mg/ml 1,4-diazabicyclooctane and 1 μg/ml propidium iodide. Green and red fluorescence was excited at 488 nm and 543 nm, respectively, and recorded in a confocal laser scanning microscope (LSM 510; Zeiss).
Yeast two-hybrid studies.
The Matchmaker LexA two-hybrid system (Clontech) was used to detect possible interactions between the investigated PrV proteins or their interactions with other viral gene products. For that purpose, Saccharomyces cerevisiae cells containing the reporter plasmid p8op-lacZ were subsequently transformed with pLex-PUL6, -PUL15, -PUL28, -PUL32, -or -PUL33 (Fig. 1B to F) and an expression library of ca. 2 × 105 random fragments of the PrV genome cloned in pB42AD (24). Yeast clones were selected on agar plates lacking amino acids that required plasmid-encoded genes for synthesis and containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for detection of LacZ expression induced by interactions between promoter-binding LexA and transcription-activating B42 fusion proteins. The insert fragments of positive library plasmids were identified by PCR amplification and DNA sequencing with vector-specific primers. Several representative library plasmids were recloned in E. coli and used for cotransformation of yeast cells, together with the respective pLexA constructs. Empty vectors (pLexA and pB42AD) and pLexA-Pos were included as negative and positive controls, respectively, and liquid cultures of three clones per plasmid combination were assayed for reporter gene expression using o-nitrophenyl-β-d-galactopyranoside as a substrate. β-Galactosidase activities were determined as described previously (46), and standard deviations were calculated.
Isolation of trans-complementing cell lines.
Subconfluent monolayers of RK13 cells were transfected (FuGene HD reagent; Roche) with plasmid pcDNA-PUL6, pcDNA-PUL15C, pcDNA-PUL28, pcDNA-PUL32, pcDNA-PUL33, or pcDNA-HUL32 (Fig. 1B to G). After 48 h, the cells were trypsinized, serially diluted in medium containing 500 μg/ml Geneticin (Invitrogen), seeded into 96-well tissue culture plates, and incubated at 37°C until resistant cell clones appeared. These clones were tested for trans-complementation of the replication defects of the respective gene deletion mutants of PrV or HSV-1, and single positive cell lines designated RK13-PUL6, -PUL15C, -PUL28, -PUL32, -PUL33, and -HUL32 were further propagated.
Generation and DNA analysis of virus recombinants.
All virus mutants were generated by Red recombinase-mediated mutagenesis of the BAC pPrV-ΔgB or pHSV-1ΔgJ in E. coli as described previously (19, 33, 39). To this end, the insert fragments of pUC-ΔPUL6KF, pBl-ΔPUL28KF, pUC-ΔPUL32KF, and pUC-ΔPUL33KF (Fig. 1B, D, E, and F) were amplified by PCR using M13/pUC (−47) and M13/pUC reverse (−48) primers (New England Biolabs), whereas T7 and SP6 primers (New England Biolabs) were used for amplification of the inserts of pcDNA-ΔPUL15KF and pcDNA-ΔHUL32KF (Fig. 1C and G). The PCR products contained a KanR gene, which permitted selection of the desired mutants resulting from homologous recombination with the cloned virus genomes. Whereas the obtained recombinant pHSV-1ΔUL32KF was directly used for replication studies, the PrV mutants were further modified in E. coli by removal of the KanR gene using Flp recombinase expressed from plasmid pCP20 (12) and replacement of the mini-F plasmid vector by the authentic PrV gB gene after recombination of the BAC with plasmid pUC-B1BclI in eukaryotic cells (33). The generated recombinants PrV-ΔUL6F, -ΔUL15F, -ΔUL28F, -ΔUL32F, and -ΔUL33F, as well as pHSV-1ΔUL32KF, were propagated in the respective trans-complementing RK13 cell lines, and the correctness of the mutations was confirmed by restriction and Southern blot analyses of genomic virus DNA.
For investigation of the cleavage of concatemeric virus genomes after DNA replication, noncomplementing RK13 cells were harvested 24 h after infection with the PrV mutants or PrV-Ka at an MOI of 5. Infected cell DNA was prepared, digested with BamHI, separated in 0.7% agarose gels, transferred to nylon membranes, and further incubated as described previously (22, 23). Southern blots were hybridized for 16 h at 65°C with the plasmid-cloned BamHI fragment 13 of the PrV-Ka genome (Fig. 1A), which had been labeled with [alpha-32P]dCTP (Rediprime II random-prime labeling system; GE Healthcare). After repeated washing at 72°C, specific hybridization reactions were detected by radioluminography of the blots (FLA-3000; Fuji).
In vitro growth studies.
The cell-to-cell spread and replication kinetics of PrV-Ka, pHSV-1ΔgJ, and the derived deletion mutants were investigated in nontransgenic RK13 cells and in stably transformed RK13 cell lines expressing the respective deleted virus genes. Replication of PrV-ΔUL32F and pHSV-1ΔUL32KF was also analyzed in cells expressing the UL32 gene of the other virus. For determination of plaque sizes, confluent monolayers in six-well plates were infected with serial virus dilutions and incubated under semisolid medium for 48 h at 37°C. PrV plaques were measured after fixation of the cells with 2% formaldehyde for 1 h, followed by staining with 1% crystal violet in 50% ethanol for 15 min. Plaques of the enhanced green fluorescent protein-expressing HSV-1 recombinants could be detected by fluorescence microscopy. The mean diameters of 30 plaques per virus and cell line were calculated. For determination of one-step growth kinetics, cell monolayers were infected at an MOI of 5 and kept on ice for 1 h to synchronize virus adsorption. Then, prewarmed medium was added, and incubation was continued at 37°C. One hour after the temperature shift, nonpenetrated virus was inactivated by low-pH treatment (43), and at different times between 2 and 72 h postinfection (p.i.), cells were scraped into the medium and lysed by freezing (−70°C) and thawing (37°C). Progeny virus titers were determined by plaque assays in the appropriate trans-complementing cell lines, and the mean titers of three or four kinetic studies per virus and cell were calculated.
Electron microscopy.
RK13 cells and the respective trans-complementing cells were infected at an MOI of 1 with PrV-ΔUL6F, PrV-ΔUL15F, PrV-ΔUL28F, PrV-ΔUL32F, or PrV-ΔUL33F. After 1 h on ice and an additional hour at 37°C, the inoculum was replaced by fresh medium, and incubation was continued for 13 h at 37°C. Fixation and embedding were performed as described previously (29), and counterstained ultrathin sections were analyzed in an electron microscope (Tecnai 12; Philips).
RESULTS
Identification of the UL6, UL15, UL28, UL32, and UL33 gene products of PrV.
In most herpesviruses, the UL15 gene consists of two exons that are linked by mRNA splicing (52). Because of sequence homologies, it has been suggested that in PrV this splicing event should occur between the reverses of nt 75995 and 73115 of the genome sequence (BK001744) to connect codons 357 and 358 of the mature ORF (30). In the present study, this could be confirmed experimentally by reverse transcription, cloning, and sequencing of the part of the viral UL15 mRNA that spans the exon boundary (Fig. 1C).
Monospecific rabbit antisera prepared against bacterial GST fusion proteins that contained parts of the deduced translation products of the UL6, UL15, UL28, and UL32 ORFs or the complete predicted UL33 protein of PrV (Fig. 1B to F) detected the respective fusion proteins in Western blot analyses (data not shown), but only the anti-pUL6 serum showed strong specific reactions with a ca. 70-kDa protein in PrV-infected cells (32) and PrV virions. In contrast, anti-pUL28, anti-pUL32, and anti-pUL33 sera reacted with ca. 80-kDa, 50-kDa, and 11-kDa proteins only in lysates of cells transfected with the respective pcDNA expression plasmids (data not shown), indicating low expression levels of these proteins during infection under the control of their own promoters.
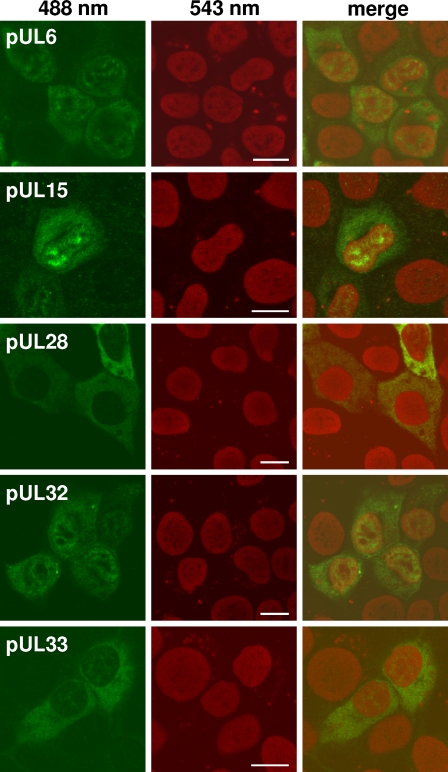
However, all antisera reacted specifically in indirect immunofluorescence tests of cells transfected with the respective pcDNA3 expression plasmids (Fig. 2). All five proteins could be detected in the cytoplasm, and pUL6, pUL15, and pUL32 additionally accumulated in distinct nuclear domains (Fig. 2). Unlike the other proteins, pUL6 was also clearly detectable in PrV-infected cells, where it also exhibited a predominantly nuclear, speckled fluorescence (not shown). The specificities of all reactions could be confirmed by the absence of signals in uninfected cells or in cells transfected with control plasmids (not shown).
FIG. 2.
Indirect immunofluorescence reactions of antisera. RK13 cells transfected with pcDNA3 expression plasmids for the indicated PrV proteins were fixed with methanol and acetone (1:1) after 24 h. Binding of the corresponding monospecific antisera was detected by Alexa Fluor 488-conjugated secondary antibodies (488 nm; green), and nuclear chromatin was stained with propidium iodide (543 nm; red). Fluorescence was analyzed by confocal laser scanning microscopy. Bars, 10 μm.
The UL6, UL15, UL28, UL32, and UL33 genes of PrV are essential for virus replication.
To determine the functional relevance of the five investigated PrV genes, parts of the individual ORFs were deleted by mutagenesis of the BAC pPrV-ΔgB (33) in E. coli. By this procedure, codons 32 to 525 of UL6, codons 114 to 330 of UL15, codons 11 to 602 of UL28, codons 228 to 370 of UL32, or codons 26 to 44 of UL33 were replaced by the 36-bp FRT site (Fig. 1B to F). It remained unclear whether any of the created virus mutants was capable of expressing truncated products of the mutated genes. However, if expressed at all, these proteins were obviously nonfunctional, since neither of the mutated BAC DNAs yielded infectious progeny virus after transfection of noncomplementing eukaryotic cells. In contrast, all virus mutants could be propagated in RK13 cell lines obtained after stable transfection with pcDNA-PUL6, -PUL15C, -PUL28, -PUL32, or -PUL33 (Fig. 1B to F), which constitutively expressed the deleted PrV genes under the control of the HCMV immediate-early promoter-enhancer complex (P-HCMV). RK13-PUL15C cells did not contain a genomic-DNA fragment of PrV, but the spliced UL15 gene, which was partly derived from cDNA (Fig. 1C). Productive replication of PrV-ΔUL15F in these cells indicated that the resulting protein was functional and that no other UL15 gene products of unspliced or alternatively spliced mRNAs are required for in vitro replication of PrV.
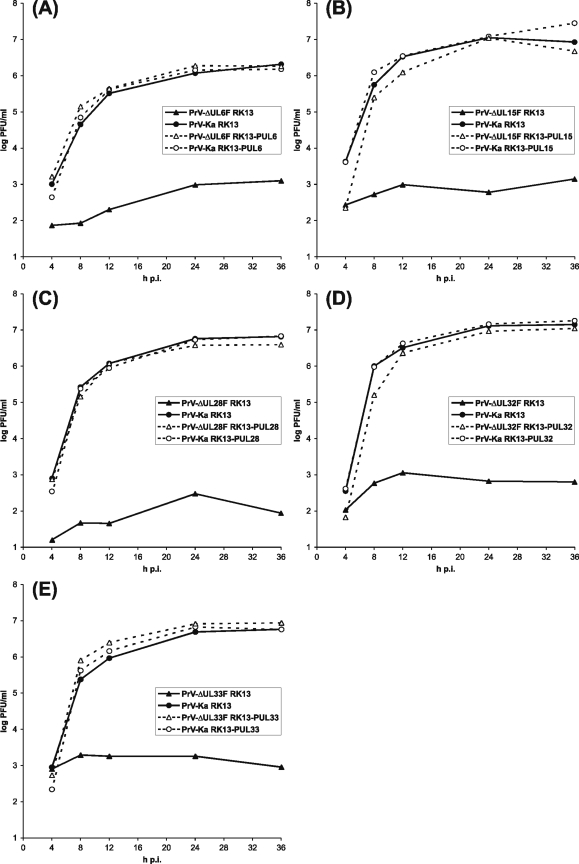
Plaque assays and one-step growth analyses revealed that all PrV mutants, including PrV-ΔUL15F, exhibited wild-type-like growth properties on the proper trans-complementing cell line (Fig. 3), whereas virus progeny isolated from these cells were unable to productively replicate in noncomplementing RK13 cells, where maximum titers did not exceed 103 PFU/ml (Fig. 3). The observed slight titer increases might be explained by the input of small amounts of the deleted proteins in virions or cell culture supernatants used for infection. None of the five deletion mutants produced plaques in RK13 cells, but indirect immunofluorescence tests with antibodies against the late envelope glycoprotein gC of PrV revealed single positive cells, and infection at a high MOI led to lysis of the cell monolayers (not shown). Thus, pUL6, pUL15, pUL28, pUL32, and pUL33 of PrV are essential for the formation and spread of infectious virus particles but presumably not required for expression of other virus genes of the lytic replication cycle.
FIG. 3.
One-step growth kinetics of PrV mutants. RK13 cells (continuous lines) or trans-complementing cells (dotted lines) were infected with PrV-Ka (circles) and the deletion mutant (triangles) PrV-ΔUL6F (A), PrV-ΔUL15F (B), PrV-ΔUL28F (C), PrV-ΔUL32F (D), or PrV-ΔUL33F (E) at an MOI of 5, harvested together with the supernatant at the indicated times, and lysed by freeze-thawing. Progeny virus titers were determined by plaque assays on RK13-PUL6 (A), RK13-PUL15 (B), RK13-PUL28 (C), RK13-PUL32 (D), or RK13-PUL33 (E) cells. The mean progeny virus titers (log PFU/ml) of four experiments are shown.
The UL6, UL15, UL28, UL32, and UL33 proteins are required for cleavage and encapsidation of viral DNA.
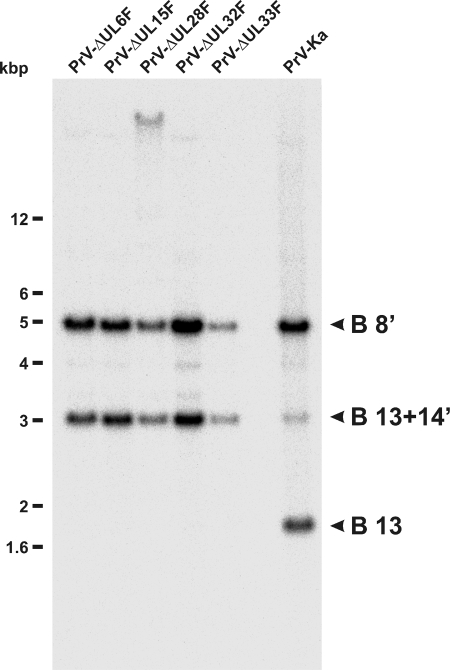
In RK13 cells harvested 24 h after infection with either of the deletion mutants at an MOI of 5, amounts of viral DNA similar to those in cells infected with PrV-Ka were found (Fig. 4), demonstrating that pUL6, pUL15, pUL28, pUL32, and pUL33 are not required for DNA replication. However, investigation of the genome termini by Southern blot hybridization of BamHI-digested DNA from infected cells with a labeled probe for one of the terminal fragments of linear virion DNA, e.g., the 1.8-kbp BamHI fragment 13 (Fig. 1A), showed its presence only in cells infected with PrV-Ka (Fig. 4). In contrast, RK13 cells infected with the deletion mutants exclusively showed the 3.1-kbp fusion fragment of BamHI 13 and 14′ (Fig. 1A and 4), as it is present in circular and concatemeric PrV DNA. In addition, the probe derived from the terminal inverted-repeat sequences of the PrV genome reacted with the 5-kbp BamHI fragment 8′ containing the corresponding part of the internal inverted-repeat sequences (Fig. 1A and 4). The absence of proper genome ends of PrV-ΔUL6F, -ΔUL15F, -ΔUL28F, -ΔUL32F, or -ΔUL33F in RK13 cells could be confirmed by Southern blot hybridization with a BamHI 14′-specific probe, and it could be also demonstrated that this defect was corrected in cells providing the product of the deleted gene in trans (not shown). The slowly migrating band observed in PrV-ΔUL28F-infected cells most likely represented partially digested DNA, as can occasionally be observed.
FIG. 4.
DNA replication of PrV mutants. RK13 cells were infected with PrV-ΔUL6F, PrV-ΔUL15F, PrV-ΔUL28F, PrV-ΔUL32F, PrV-ΔUL33F, or wild-type PrV-Ka at an MOI of 5, and total DNA was prepared 24 h p.i. After BamHI cleavage Southern blots were hybridized with the labeled terminal BamHI fragment 13 of the PrV genome, which permits differentiation of packaged unit size DNA (B 13) and the concatemeric primary replication products (B 13+14′). A fragment (B 8′) from the internal inverted-repeat sequences was also detected by the probe (Fig. 1A). The sizes of marker DNAs are indicated.
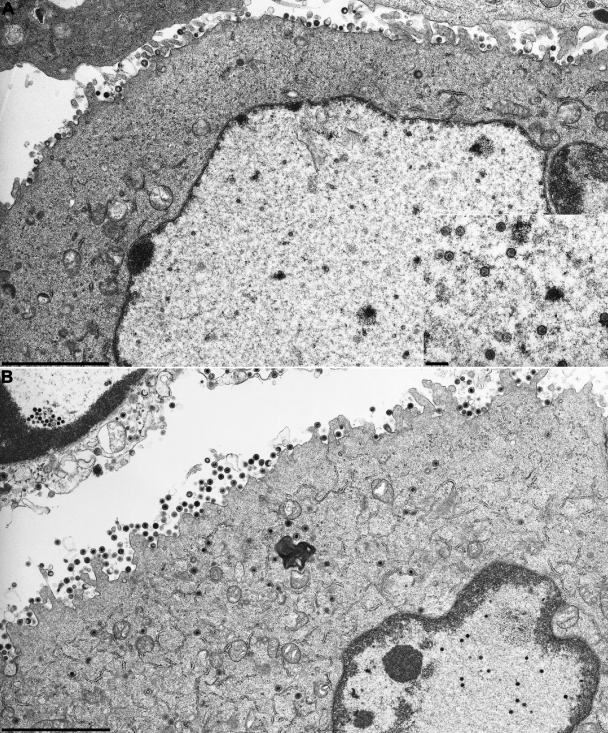
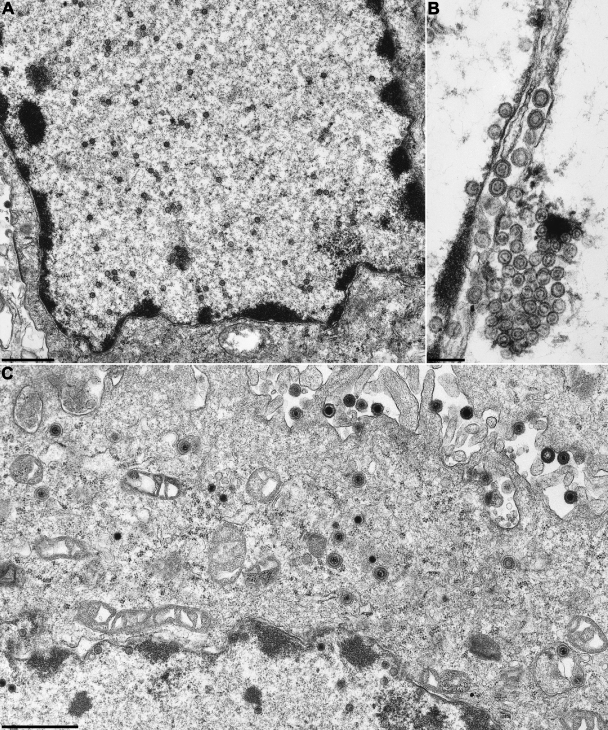
In accordance with the absence of unit-size virus genomes in RK13 cells infected with any of the five PrV mutants, no nucleocapsids could be detected by electron microscopy, as shown for PrV-ΔUL15F (Fig. 5A) and PrV-ΔUL32F (Fig. 6A). However, capsid assembly was apparently not affected, since 14 h p.i., numerous scaffold-containing B capsids (52) were found in the nuclei of infected cells (Fig. 5A, inset). Occasionally, these capsids formed semicrystalline aggregates in the vicinity of the nuclear membrane, and enveloped B capsids were also detected in the perinuclear space (Fig. 6B). Primary envelopment was found in RK13 cells infected with PrV-ΔUL6F, PrV-ΔUL32F, and PrV-ΔUL33F, but not with PrV-ΔUL15F or PrV-ΔUL28F. However, more extensive studies will be required to verify this observation. Capsids were largely absent from the cytoplasm of noncomplementing cells, and only capsidless light particles consisting of enveloped tegument proteins were released (Fig. 5A and 6A).
FIG. 5.
Electron microscopy of cells infected with PrV-ΔUL15F. RK13 (A) or RK13-PUL15 (B) cells were fixed 14 h p.i. at an MOI of 1, and uranyl-acetate-stained ultrathin sections were analyzed. The inset in panel A shows numerous scaffold-containing B capsids in the nuclei of infected cells. Bars, 3 μm or 250 nm (inset in panel A).
FIG. 6.
Electron microscopy of cells infected with PrV-ΔUL32F. RK13 (A and B) or RK13-PUL32 (C) cells were fixed 14 h p.i. at an MOI of 1, and uranyl-acetate-stained ultrathin sections were analyzed. Bars, 3 μm (A and C) or 250 nm (B).
In trans-complementing RK13-PUL15 or RK13-PUL32 cells, the respective gene deletion mutants exhibited all stages of PrV virion morphogenesis (26). DNA-containing nucleocapsids were found in the nucleus and in the cytoplasm, and complete virions were released in the extracellular space (Fig. 5B and 6C). Similar results were obtained on cell lines complementing the replication defects of PrV-ΔUL6F, PrV-ΔUL28F, and PrV-ΔUL33F (not shown). Taken together, these results demonstrate that pUL6, pUL15, pUL28, pUL32, and pUL33 are required for cleavage and packaging of genomic DNA into preformed PrV capsids.
Interactions of the investigated PrV proteins.
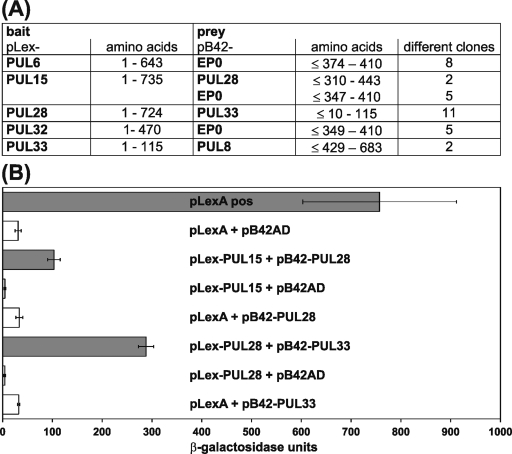
Two-hybrid studies were performed to identify possible interactions of pUL6, pUL15, pUL28, pUL32, and pUL33 of PrV with other viral proteins or with each other. The five ORFs were expressed as DNA-binding LexA fusion proteins (Fig. 1B to F) in S. cerevisiae cells and used to screen a PrV expression library (24). Interactions between LexA bait and B42 prey proteins permitted autotrophic growth on minimal medium and induced a LacZ reporter gene. For each bait construct, ca. 20 positive clones were characterized by DNA sequencing of the prey plasmids, and PrV genes represented by at least two different overlapping fragments were listed (Fig. 7A). Interestingly, pUL6, pUL15, and pUL32 are apparently able to bind to the C-terminal part of the early protein EP0 of PrV, which is an activator of viral gene expression homologous to ICP0 of HSV-1 (13, 62). In contrast, pUL33 interacted with the C-terminal part of pUL8 (Fig. 7A), which is predicted to be a subunit of the helicase-primase complex of PrV (30). Reciprocal interactions of EP0 and pUL8 could not be detected after recloning of the identified gene fragments in pLexA.
FIG. 7.
Physical interactions of DNA cleavage-encapsidation proteins of PrV. (A) DNA-binding fusion proteins were expressed from pLex-PUL6, pLex-PUL15, pLex-PUL28, pLex-PUL32, and pLex-PUL33 in transformed yeast cells and used as baits for two-hybrid screening of an expression library of PrV gene products cloned in pB42AD, which provides a trans-activation domain. The minimum amino acid ranges of reproducibly detected viral prey proteins and the numbers of positive clones are indicated. (B) Induction of β-galactosidase reporter gene expression by the fusion protein complexes expressed by pLex-UL15 and pB42-PUL28 or pLex-PUL28 and pB42-PUL33 was quantified and compared to positive (pLexA pos) and negative (empty expression vectors pLexA and pB42AD) controls. The mean results of three experiments and standard deviations are shown.
Furthermore, pUL15 reacted with proteins representing the central part of pUL28, and the pUL28 bait detected numerous clones encoding pUL33, but not vice versa (Fig. 7A). The putative interactions between pUL15, pUL28, and pUL33 of PrV correlate with similar biological functions (see above), and a functional complex consisting of the HSV-1 homologues of these three proteins has been postulated (67). Therefore, the results were verified by retransformation of yeast cells with representative plasmid pairs, including the empty vectors expressing native LexA or the B42 protein as negative controls, and quantified by determination of the β-galactosidase activity in liquid cultures (Fig. 7B). Although lower than in cultures expressing a transactivating LexA fusion protein, the activity in cells coexpressing LexA-pUL15 and B42-pUL28 or LexA-pUL28 and B42-pUL32 was still significantly higher than after replacement of either of the proteins by the respective negative control (Fig. 7B). Thus, our yeast two-hybrid studies support the hypothesis that pUL15, pUL28, and pUL33 of PrV form a tripartite terminase complex, which is required for cleavage and encapsidation of viral DNA.
The UL32 gene product of HSV-1 can substitute for the homologous PrV protein, but not vice versa.
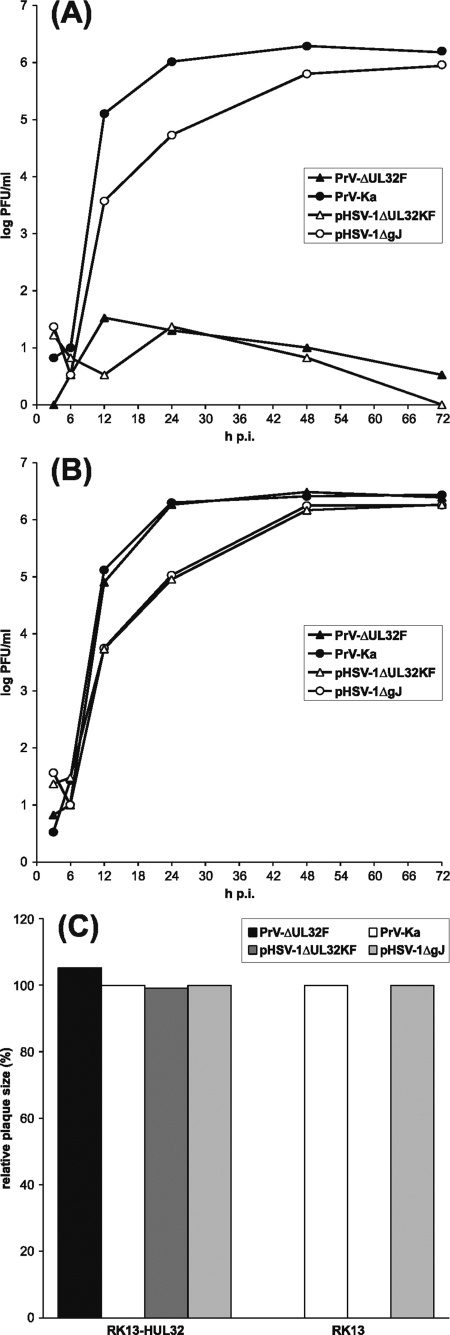
Since all PrV genes investigated in this study are highly conserved between herpesviruses, we started to investigate whether the homologous proteins of different viruses could compensate for each other. In a first experiment, an HSV-1 recombinant lacking codons 167 to 466 of UL32 was generated by mutagenesis of the infectious BAC clone pHSV-1ΔgJ (39) in E. coli, and a corresponding RK13 cell line expressing pUL32 of HSV-1 under the control of P-HCMV was also isolated (Fig. 1G). In these cells, pHSV-1ΔUL32KF exhibited plaque sizes and growth kinetics similar to those of the parental BAC clone (Fig. 8B and C), whereas neither productive replication nor cell-to-cell spread of the mutant was observed in normal RK13 cells (Fig. 8A and C). UL32-negative HSV-1 also did not replicate in RK13-PUL32 cells expressing the UL32 gene of PrV (not shown). In contrast, replication of the UL32-negative mutant PrV-ΔUL32F was supported, not only by the authentic pUL32 (Fig. 3D), but also by the homologous HSV-1 protein expressed in RK13-HUL32 cells (Fig. 8B). trans-Complementation by the HSV-1 protein was apparently efficient, since plaque sizes and virus titers of the PrV mutant in RK13-HUL32 cells were comparable to those of wild-type PrV-Ka (Fig. 8B and C). Whereas unidirectional heterologous complementation could be shown for pUL32 of PrV and HSV-1, similar experiments including a UL33-negative HSV-1 mutant and a corresponding cell line proved to be negative in both directions (not shown), while pUL6, pUL15, and pUL28 remain to be tested.
FIG. 8.
trans-Complementation of UL32-negative PrV by the homologous HSV-1 protein. One-step growth kinetics were determined after infection of RK13 (A) or RK13-HUL32 (B) cells with PrV-Ka, PrV-ΔUL32F, pHSV-1ΔgJ, or pHSV-1ΔUL32KF at an MOI of 5. At the indicated times, progeny virus titers were determined by plaque assays on RK13-HUL32 cells. Shown are the mean results of three experiments. (C) Cell-to-cell spread was investigated 48 h after infection of RK13 or RK13-HUL32 cells at low MOI. The average diameters of 30 plaques each were calculated, and the plaque sizes of PrV-ΔUL32F and pHSV-1ΔUL32 were displayed as percentages of those of PrV-Ka and pHSV-1ΔgJ on the same cell line, which were set at 100%.
DISCUSSION
In the present study, five components of the DNA cleavage-encapsidation machinery of PrV were identified and analyzed in a standardized isogenic system. The salient findings were as follows. (i) The UL6, UL15, UL28, UL32, and UL33 gene products of PrV are essential for productive virus replication in cell culture. (ii) Their absence correlates with a defect in cleavage of replicated viral DNA into monomers and packaging into preformed capsids. (iii) The defects of UL6-, UL15-, UL28-, UL32-, or UL33-negative PrV mutants were complemented in trans in cells expressing the corresponding protein. (iv) The defect of UL32-negative PrV was corrected by pUL32 of HSV-1, but not vice versa. (v) Yeast two-hybrid studies revealed physical interactions of pUL28 with pUL15 and with pUL33, indicating the existence of a functional complex.
For UL28-negative PrV, the present results confirm an earlier study (44), and the phenotypes of all generated deletion mutants are similar to those of temperature-sensitive insertion or deletion mutations of the homologous genes in HSV-1 (2, 4, 38, 50, 59). However, since all PrV mutants described here have been derived from the same parental virus clone by mutagenesis in E. coli, the close functional relationships between pUL6, pUL15, pUL28, pUL32, and pUL33 become more evident than before. In contrast, functional investigations of two other proteins involved in DNA cleavage and stable encapsidation, pUL17 and pUL25, using the same PrV clone revealed different functions (31, 32).
pUL6 of PrV is a component of mature virions, and immunoelectron microscopy indicated an association with nucleocapsids (results not shown). Therefore, it can be assumed that pUL6 of PrV, like its homologues in HSV-1 and HCMV (21, 49), forms the portal for DNA incorporation into capsids. Transient-expression studies showed that pUL6 of PrV, like the HSV-1 protein (50), is able to enter the cell nucleus independently of other viral gene products, but unlike in HSV-1, direct interactions with terminase subunits (63, 67) were not detected.
As deduced from other herpesviruses, the UL15 gene of PrV should consist of two exons (30), and removal of an intron ranging from nt 73116 to 75994 was experimentally proven in this study. Moreover, the protein translated from the spliced mRNA fully corrected the replication defect of UL15-negative PrV in trans. Similar observations were made in HSV-1 (3), but nevertheless, additional, smaller gene products presumably representing the 3′-terminal part of the ORF have been identified (4, 68). Unfortunately, in PrV, the weak reactivity of the UL15-specific antiserum prohibited identification of the gene product or products by Western blotting or immunoprecipitation. However, immunofluorescence analyses of cells transfected with a UL15 expression plasmid showed that pUL15 exhibited a speckled nuclear localization similar to that of pUL6. This is again in agreement with previous observations in HSV-1, although the nuclear localization signal at amino acid positions 183 to 189 of HSV-1 pUL15 (67) is not conserved in the predicted PrV homologue (30). However, two other putative nuclear localization signals could be identified at amino acid positions 16 to 19 and 305 to 321 of PrV pUL15 using PSORTII programs (48).
Unlike pUL15, the transiently expressed UL28 and UL33 gene products of PrV did not accumulate in the cell nucleus. Previous studies showed that pUL28 exhibits nuclear localization in infected cells, whereas it remains cytoplasmic in the absence of other viral proteins (34, 51). Furthermore, it has been demonstrated that pUL15 of HSV-1 mediates nuclear localization, not only of the autologous UL28 gene product, but also of its PrV homologue (34, 35), and transient-coexpression studies indicated that the portal protein pUL6 can also relocalize pUL28 to the nucleus (63). Presumably, pUL33 of HSV-1 is also translocated to nuclear sites of viral DNA replication by interactions with the UL15 protein, either directly (27) or together with pUL28 (66, 67). Furthermore, it has been reported that in HSV-2 nuclear localization of pUL33 is mediated by pUL14 (65), which, however, is nonessential for HSV-1 replication (18). For PrV, it remains to be tested which other viral proteins may influence the subcellular localization of pUL33.
The yeast two-hybrid studies presented here revealed direct interactions of PrV pUL28 with pUL15 and pUL33, whereas direct interactions between pUL15 and pUL33 were not detected. This indicates that the tripartite terminase complex of PrV might be organized as proposed for HSV-1, with pUL28 as the central component (66). Interactions between terminase subunits and the portal protein, as found by coimmunoprecipitation of pUL6 with pUL15 and pUL28 of HSV-1 (63), were not detected in two-hybrid experiments with the homologous PrV proteins. However, pUL6, as well as pUL15 and pUL32, of PrV interacted with the C-terminal part of the ICP0 homologue EP0, which is a regulator of viral transcription (62). Thus, a second function of EP0 might be the linkage of proteins involved in cleavage-encapsidation of viral DNA. However, like pUL14 of HSV-1, EP0 of PrV is also nonessential for virus replication (9), and therefore, its putative role in DNA packaging cannot be crucial. Nevertheless, EP0 and the predicted helicase-primase subunit pUL8 (17, 30), which was identified as an interaction partner of pUL33 of PrV, function in the cell nucleus via direct or indirect interaction with viral DNA, suggesting the possibility of accessory roles in DNA cleavage and packaging.
Up to now, no direct interactions of the pUL32 homologues of any herpesvirus with other essential components of the DNA cleavage and packaging machinery have been described, and the HSV-1 protein was predominantly detected in the cytoplasm of infected cells (11, 38), whereas pUL32 of PrV, in addition, showed a speckled intranuclear distribution similar to those of pUL6 and pUL15. Thus, at least in PrV, a direct involvement of pUL32 in DNA cleavage and encapsidation is conceivable, as is a role in the localization of capsids to DNA replication compartments, which was proposed for HSV-1 (38).
Despite these differences, the biological functions of PrV and HSV-1 pUL32 are obviously very similar, since the replication defect of UL32-negative PrV could be fully corrected in trans by the homologous HSV-1 protein. In contrast, a UL32 deletion mutant of HSV-1 was not complemented by the corresponding protein of PrV. It is conceivable that pUL32 of HSV-1 executes an additional function(s) that cannot be fulfilled by the PrV protein. More likely, the function of pUL32 might be dependent on interactions with other viral gene products, which are impaired, e.g., for sterical reasons, between PrV pUL32 and the HSV-1 proteins. Heterologous complementation between PrV and HSV-1 has already been described for several other conserved viral proteins, which reflects the relatively close phylogenetic relationship of the two alphaherpesviruses (41). However, complementation was always only unidirectional: pUL25 of HSV-1 partly corrected the replication defect of a UL25-negative PrV mutant (36), whereas gB and pUL11 of PrV rescued corresponding HSV-1 mutants (39, 45), but not vice versa. In all these cases, including pUL32, construction of chimeric proteins consisting of PrV and HSV-1 portions could contribute to the identification of domains relevant for the function of the respective protein and to deeper insights into the biochemical basis for protein function.
Remarkably, for the conserved subunit pUL33 of the proposed terminase complexes of PrV and HSV-1 (67), heterologous trans-complementation was not observed in either direction (results not shown). This might indicate that a precise interplay between the three different components of the enzyme complex performing cleavage and packaging of herpesvirus DNA is required for functionality and that substitutions of single subunits are not tolerated. Thus, it will be interesting to test whether cells concomitantly expressing pUL15, pUL28, and pUL33 of either HSV-1 or PrV permit replication of deletion mutants of the other virus. In this way, heterologous trans-complementation studies might not only support evolutionary investigations, but also help to identify protein interactions required for herpesvirus replication.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/8-2).
We thank G. A. Smith and B. Wanner for providing plasmid vectors used for BAC cloning and mutagenesis and E. Mundt and J. Veits for rabbit immunization. The technical help of C. Ehrlich, M. Jörn, P. Meyer, and D. Werner is greatly appreciated.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 983086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180380-388. [DOI] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The cDNA of UL15, a highly conserved herpes simplex virus 1 gene, effectively replaces the two exons of the wild-type virus. J. Virol. 665621-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., A. P. Poon, J. Rovnak, and B. Roizman. 1994. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 688118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 764785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, L. W. 1995. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays 171025-1030. [DOI] [PubMed] [Google Scholar]
- 7.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66347-384. [DOI] [PubMed] [Google Scholar]
- 8.Bogner, E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 12115-127. [DOI] [PubMed] [Google Scholar]
- 9.Boldogköi, Z., A. Braun, and I. Fodor. 2000. Replication and virulence of early protein 0 and long latency transcript deficient mutants of the Aujeszky's disease (pseudorabies) virus. Microbes Infect. 21321-1328. [DOI] [PubMed] [Google Scholar]
- 10.Borst, E. M., K. Wagner, A. Binz, B. Sodeik, and M. Messerle. 2008. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J. Virol. 822065-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. E., A. P. Poon, and B. Roizman. 1995. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J. Virol. 703938-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 13.Cheung, A. K. 1991. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J. Virol. 655260-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 15.Cockrell, S. K., M. E. Sanchez, A. Erazo, and F. L. Homa. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 8347-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coller, K. E., J. I. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 8111790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crute, J. J., T. Tsurumi, L. A. Zhu, S. K. Weller, P. D. Olivo, M. D. Challberg, E. S. Mocarski, and I. R. Lehman. 1989. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc. Natl. Acad. Sci. USA 862186-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham, C., A. J. Davison, A. R. MacLean, N. S. Taus, and J. D. Baines. 2000. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 7433-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 745083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dittmer, A., J. C. Drach, L. B. Townsend, A. Fischer, and E. Bogner. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-d-ribonucleosides. J. Virol. 7914660-14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, W., and T. C. Mettenleiter. 1996. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 772221-2229. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs, W., and T. C. Mettenleiter. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J. Gen. Virol. 802173-2182. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson, W., and B. Roizman. 1974. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 101044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 753675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgs, M. R., V. G. Preston, and N. D. Stow. 2008. The UL15 protein of herpes simplex virus type 1 is necessary for the localization of the UL28 and UL33 proteins to viral DNA replication centres. J. Gen. Virol. 891709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7394-407. [DOI] [PubMed] [Google Scholar]
- 29.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 7410063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klupp, B. G., H. Granzow, A. Karger, and T. C. Mettenleiter. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J. Virol. 7913442-13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 806235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 775339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koslowski, K. M., P. R. Shaver, X. Y. Wang, D. J. Tenney, and N. E. Pederson. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 719118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 731704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn, J., T. Leege, B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 825725-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 32099-106. [DOI] [PubMed] [Google Scholar]
- 38.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 722463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leege, T., W. Fuchs, H. Granzow, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus 1. J. Virol. 83896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 41.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 11790-104. [DOI] [PubMed] [Google Scholar]
- 42.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 721060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171623-625. [DOI] [PubMed] [Google Scholar]
- 44.Mettenleiter, T. C., A. Saalmüller, and F. Weiland. 1993. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J. Virol. 671236-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mettenleiter, T. C., and P. G. Spear. 1994. Glycoprotein gB (gII) of pseudorabies virus can functionally substitute for glycoprotein gB in herpes simplex virus type 1. J. Virol. 68500-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 48.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 2434-36. [DOI] [PubMed] [Google Scholar]
- 49.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217111-123. [DOI] [PubMed] [Google Scholar]
- 51.Pederson, N. E., and L. W. Enquist. 1991. Overexpression in bacterial and identification in infected cells of the pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5. J. Virol. 653746-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pellet, P. E., and B. Roizman. 2007. The family Herpesviridae: a brief introduction, p. 2479-2499. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 53.Rall, G. F., S. Kupershmidt, N. Sugg, R. A. Veach, and T. Ben-Porat. 1992. Functions of the sequences at the ends of the inverted repeats of pseudorabies virus. J. Virol. 661506-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rawls, W. E., D. Laurel, J. L. Melnick, J. M. Glicksman, and R. H. Kaufman. 1968. A search for viruses in smegma, premalignant and early malignant cervical tissues. The isolation of herpesviruses with distinct antigenic properties. Am. J. Epidemiol. 87647-655. [DOI] [PubMed] [Google Scholar]
- 55.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roizman, B., D. M. Knipe, and R. J. Whitney. 2007. Herpes simplex viruses, p. 2501-2601. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 57.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252115-125. [DOI] [PubMed] [Google Scholar]
- 59.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 673470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 802118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263447-462. [DOI] [PubMed] [Google Scholar]
- 62.Watanabe, S., E. Ono, Y. Shimizu, and H. Kida. 1995. Pseudorabies virus early protein 0 transactivates the viral gene promoters. J. Gen. Virol. 762881-2885. [DOI] [PubMed] [Google Scholar]
- 63.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 776351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilkinson, D. E., and S. K. Weller. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55451-458. [DOI] [PubMed] [Google Scholar]
- 65.Yamauchi, Y., K. Wada, F. Goshima, H. Takakuwa, T. Daikoku, M. Yamada, and Y. Nishiyama. 2001. The UL14 protein of herpes simplex virus type 2 translocates the minor capsid protein VP26 and the DNA cleavage and packaging UL33 protein into the nucleus of coexpressing cells. J. Gen. Virol. 82321-330. [DOI] [PubMed] [Google Scholar]
- 66.Yang, K., and J. D. Baines. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 805733-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, K., F. Homa, and J. D. Baines. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 816419-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, D., A. K. Sheaffer, D. J. Tenney, and S. K. Weller. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 712656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 24332-44. [DOI] [PubMed] [Google Scholar]