Abstract
DNA-mediated charge transport (CT) is exquisitely sensitive to the integrity of the bridging π-stack and is characterized by a shallow distance dependence. These properties are obscured by poor coupling between the donor/acceptor pair and the DNA bridge, or by convolution with other processes. Previously, we found a surprising periodic length dependence for the rate of DNA-mediated CT across adenine tracts monitored by 2-aminopurine fluorescence. Here we report a similar periodicity by monitoring N2-cyclopropylguanosine decomposition by rhodium and anthraquinone photooxidants. Furthermore, we find that this periodicity is attenuated by consequent back electron transfer (BET), as observed by direct comparison between sequences that allow and suppress BET. Thus, the periodicity can be controlled by engineering the extent of BET across the bridge. The periodic length dependence is not consistent with a periodicity predicted by molecular wire theory but is consistent with a model where multiples of four to five base pairs form an ideal CT-active length of a bridging adenine domain.
INTRODUCTION
The DNA π-stack has the inherent ability to act as an efficient medium for charge transport (CT).1 Long range DNA-mediated CT is exquisitely sensitive both to the coupling of donors and acceptors into the π-stack,2 and to the presence of lesions, mismatches, protein-induced distortions, and other defects in the integrity of base stacking.3 This sensitivity has been exploited in the development of novel classes of DNA-based sensing technologies4 and might be utilized in vivo by transcriptional activation and DNA repair pathways.5 To realize fully the potential of this technology, it is necessary to understand the mechanistic underpinnings of DNA-mediated CT.
Recently, a periodic dependence on adenine tract length was observed for the fluorescence quenching of photoexcited 2-aminopurine (Ap*) by DNA-mediated CT to guanine across the adenine tract.6 By standardizing to a system containing the redox-inactive base inosine, the contribution to quenching solely due to CT between Ap* and guanine was isolated. The amplitudes associated with this periodicity are substantial and greater than the observed associated errors. Non-monotonicity of CT rate versus distance has since been observed between gold and ferrocene across methyl-substituted oligophenyleneethynylene, but that result was attributed to substantial torsional variations between polymers of different lengths, an explanation that is not adaptable to these adenine tracts7. Instead, we interpreted our surprising result in the context of four or five base pairs being conducive to forming a CT-active domain, leading to higher CT over an adenine tract that is an integer multiple of this number. This interpretation is consistent with the conformationally gated character of DNA-mediated CT over long distances,8 with evidence for delocalization of the injected hole,9 and with evidence for a similar delocalization length in the formation of excimers along adenine tracts.10 A similar argument has been made to explain this result in the context of a polaron hopping model,11 and non-monotonicity was also observed in calculations that permitted delocalization.12
Importantly, Ap* fluorescence quenching is insensitive to processes that occur after the CT event, including radical trapping, incoherent hopping or back electron transfer (BET). For hole acceptors in DNA, product yields for different photooxidants scale inversely to the propensity for BET,13 and attenuating BET, both between the hole donor and the oxidized bridge and between the hole donor and oxidized acceptor, extends the lifetime of the charge separated state.14 While other spectroscopic investigations of CT across adenine tracts have not revealed a similar periodicity, these other studies have been performed on systems for which BET is known to be substantial15,16 or where slow trapping allows charge equilibration after the initial CT step.17,18 We have recently shown that for both hole and electron transport, CT efficiency is dictated in the same manner by the dynamics and structure of the intervening DNA bases.19 If the periodicity is the result of CT-active states that serve as more efficient pathways for forward CT, then they will also mediate more efficient BET. Hence, we propose that conformations that promote forward CT also promote BET, and this BET will serve to suppress the apparent periodicity.
To test this hypothesis and determine whether this periodicity is a general property of long range DNA-mediated CT, in the present work we consider disparate donor-acceptor systems with varying extents of BET (Figure 1). Previously, by measuring quantum yield of damage at double guanine sites, we ranked a series of photooxidants by propensity for charge recombination between the guanine cation radical and the reduced hole donor.13 Two photooxidants that are subject to only moderate BET are Rh(phi)2(bpy’)3+ (Rh) and anthraquinone (AQ), while BET is highly efficient for Ap. Although these and other photooxidants typically induce oxidation of native guanine sites to 8-oxoguanine and other base-labile damage products,18,20 facile BET between guanine cation radical and aminopurine anion radical renders Ap photooxidation of guanine only observable with the CPG trap. Furthermore, to limit post-injection charge equilibration, we assay for arrival using N2-cyclopropylguanine (CPG) instead of guanine as a hole acceptor.21 This fast22 trap for cation and anion radicals allows detection of pre-equilibrium CT processes that are obscured by the slow trapping of guanine radical by water or oxygen. By modulating the extent of BET for a series of CPG-containing duplexes, we demonstrate that the periodic length dependence is inherent to adenine tracts but is attenuated with increasing BET.
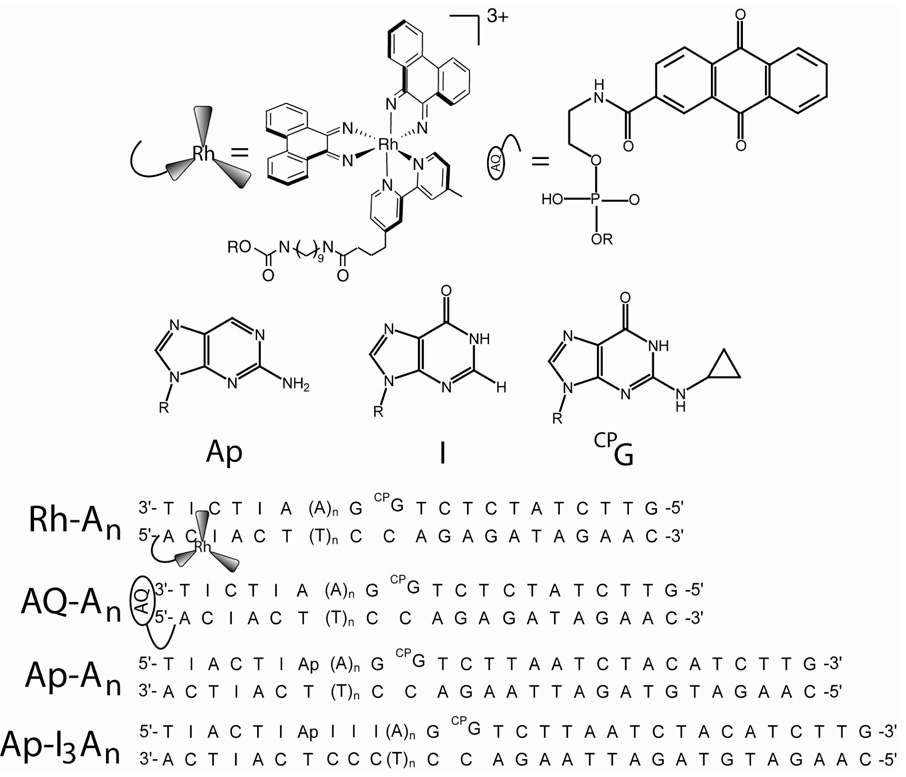
Figure 1.
Photooxidants, modified bases, and assemblies used to probe CT events in DNA. At top are the structures of the rhodium and anthraquinone complexes utilized, and structures of aminopurine, inosine, and CPG. The rhodium complex is tethered to the 5’ end of amino modified DNA by a nine carbon linker as represented in the center left, and the anthraquinone is capped on the 5’ end through the phosphate. Representative assemblies, indicating position of photooxidants, are shown on the bottom. Duplex length is conserved across individual series by removing base-pairs distal to the hole trap (see text and supplemental information).
EXPERIMENTAL
Oligonucleotide Synthesis
DNA oligonucleotides were synthesized trityl-on using standard phosphoramidite chemistry on an ABI DNA synthesizer with Glen Research reagents. 2-aminopurine was incorporated as the N2-dimethylaminomethylidene protected phosphoramidite (Glen Research). CPG-modified oligonucleotides were prepared by incorporating the precursor base, 2-fluoroinosine-O6-paraphenylethyl-2’-deoxyinosine (Glen Research), as a phosphoramidite at the desired position. The resin was then reacted with 1 M diaza(1,3)bicyclo[5.4.0]undecane (DBU, Aldrich) in acetonitrile to effectively remove the O6 protecting group. The oligonucleotides were subsequently incubated overnight in 6 M aqueous cyclopropylamine (Aldrich) at 60 °C resulting in substitution, base deprotection, and simultaneous cleavage from the resin. The cleaved strands were dried in vacuo and purified by reversed-phase HPLC, detritylated by 80% acetic acid for 15 min, and repurified by reversed-phase HPLC. Oligonucleotides were characterized by MALDI-TOF mass spectrometry.
Rhodium-modified oligonucleotides were synthesized as described previously.26 Briefly, the detritylated resin-bound oligonucleotides were first modified with a nine carbon amine linker by reaction with carbonyldiimidazole and diaminononane in dioxane. The amine-modified strands were then reacted with [Rh(phi)2(bpy’)]Cl3 (bpy’ = 4-(4’-methyl-2,2’-bipyridyl) valerate) in 1:1:1 methanol:acetonitrile:isopropanol using O-(N-succinimidyl)-1,1,3,3-tetramethyl uranium tetrafluoroborate (TSTU) as the coupling reagent. Cleavage from the resin was accomplished by incubation in NH4OH at 60 °C for 6 hours. Strands were HPLC-purified using a Varian C4 reversed-phase column. The two diasteromeric conjugates, differing in configuration at the metal center, have different retention times. However, both isomers were collected together and used for subsequent experiments. MALDI-TOF mass spectrometry was used to characterize the metallated DNA conjugates.
Anthraquinone (AQ)-tethered oligonucleotides were synthesized as described previously by incorporating an anthraquinone phosphoramidite at the 5’-end of the oligonucleotides.27 The DNA was deprotected in NH4OH at 60 °C overnight. The resulting oligonucleotides were purified once by reversed-phase HPLC and characterized by MALDI-TOF mass spectrometry.
All oligonucleotides were suspended in a buffer containing 50 mM NaCl, 20 mM or 5 mM sodium phosphate, pH 7 and quantified using UV-visible spectroscopy. Duplexes were prepared by heating equal concentrations of complementary strands to 90 °C for 5 min and slow cooling to ambient temperature. Melting temperatures (Tm) were obtained for all duplexes. All duplexes melted between 50 – 60 °C at a 1.5 µM concentration in phosphate buffer (PBS, 20 mM sodium phosphate, 50 mM NaCl, pH 7).
Photooxidation Experiments
Photooxidations of Rh-tethered oligonucleotides were carried out by irradiating 30 µL aliquots of 10 µM duplex in PBS for 30 sec at 365 nm on a 1000 W Hg/Xe lamp equipped with a 320 nm long pass filter and monochromator. AQ-containing duplexes in PBS (30 µL, 10 µM) were irradiated at 350 nm using the same apparatus for 5 min. Irradiation times were varied and the decomposition was linear over the times used (supplementary information). Samples were irradiated at various temperatures ranging from 20 to 80 °C. Ap-containing duplexes (30 µL, 10 µM) in PBS were irradiated as above at 325 nm without the long pass filter for 30 sec or 30 min.
To analyze for CPG decomposition following irradiation, samples were digested to the component nucleosides by phosphodiesterase I (USB) and alkaline phosphatase (Roche) at 37 °C, to completion. The resulting deoxynucleosides were analyzed by reversed-phase HPLC using a Chemcobond 5-ODS-H, 4.6 mm × 100 mm column. The amount of CPG per duplex was determined by taking the ratio of the area of the HPLC peak for dCPG to the area of the peak for dT. For 30 minute irradiations, a small amount of thymine decomposition was observed, as has been described previously.28 Hence, redox-inactive inosine was used as the internal standard for these experiments. The decomposition yield is taken as the percent loss of CPG between an irradiated sample and the dark control. Dark control HPLC traces were confirmed to yield the correct relative amounts of dA, dC, dG, dI, dT, and dCPG based on duplex sequence. Irradiations were performed at least three times and the results averaged. Due to the long irradiation times used for the Ap-I3AnCPG strands, actinometry was performed using a 6 mM ferrioxalate standard29 to allow comparison between experiments performed on separate days. The given quantum yield is for the efficiency from the Ap* state to the ring-opened product. Fluorescence quenching for the Ap-I3An was not expected to be observable based on the quantum yield of CPG damage, and hence was not explored.
Errors are presented at 90% standard error of the mean, using the Student’s t-distribution at the appropriate degrees of freedom to determine confidence intervals.
RESULTS
Experimental Design
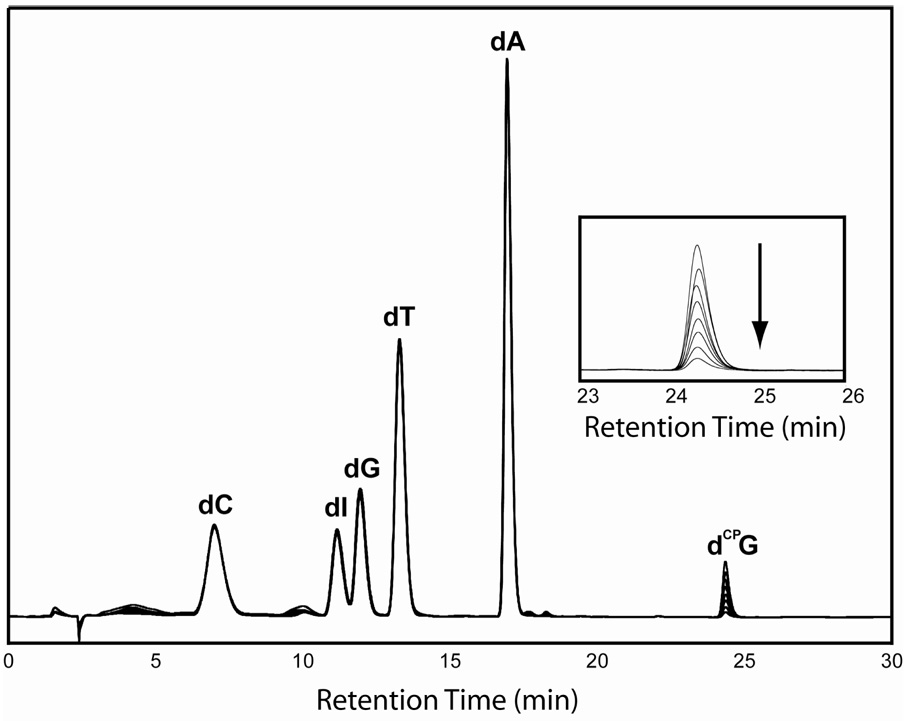
Figure 1 illustrates typical DNA-photooxidant assemblies. The Rh-An, AQ-An and Ap-An series contain rhodium, anthraquinone, or 2-aminopurine separated from CPG by a bridge containing increasing numbers of adenines. For all Rh-modified assemblies there is a four base pair segment surrounding the rhodium binding site to provide optimum intercalation of the photooxidant. In Figure 1, the rhodium is shown intercalated two base pairs from the terminus, but likely a mixture of binding sites (one and two bases in) are available to the diastereomers.26 On the side distal to the hole trap, there is a constant three base sequence so that end effects are minimized. Guanine can serve as a thermodynamic well if placed near the rhodium intercalation site and, although the trapping rate is slow, BET to rhodium is comparably fast at short distance.13 Therefore, inosine was employed as a substitute for guanine near the rhodium binding site to enhance CPG decomposition.9,19 Note that the first four adenine tract sequences, Rh-A2 through Rh-A8 are composed of 20 base pairs, while that of Rh-A8’ through Rh-A14 are slightly longer, with 26 base pairs (supplementary information). Rh-A8 and Rh-A8’, both containing the 8 base pair long adenine tract but differing in length, yield equivalent decomposition profiles with both time and temperature, and in subsequent results and figures, the data from Rh-A8’ are presented. A series of HPLC traces from the time-course of AQ-A2 degradation shows the well-resolved peaks corresponding to the six different natural and unnatural nucleosides (Figure 2).
Figure 2.
Overlaid HPLC traces at 260 nm for digested nucleosides from AQA2 irradiated at 350 nm for 0, 1, 2, 3, 5, 7, 10, and 15 min. Traces are normalized to the height of the dT peak, and the inset demonstrates that the peak corresponding to dCPG steadily degrades with respect to increased irradiation time. Conditions are as described in Experimental.
DNA-mediated Oxidative Decomposition of CPG by Rh and AQ
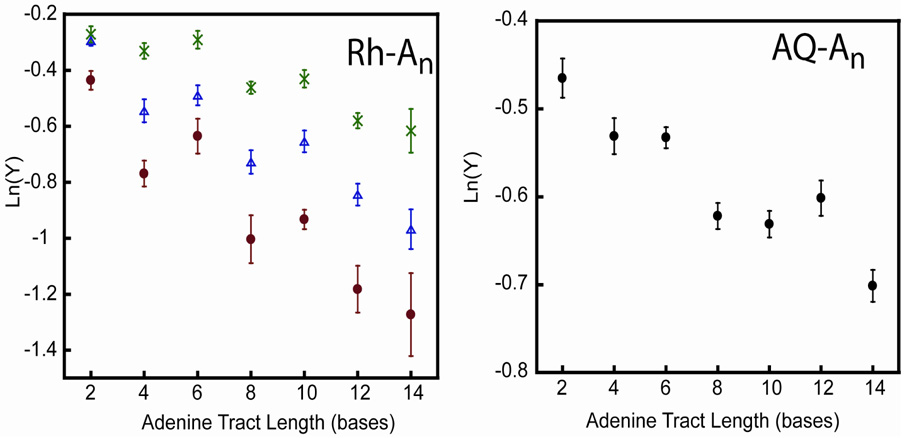
Figure 3 shows the variation in the decomposition yield (Y) as a function of bridge length for the Rh-An and AQ-An series. Notably, the same non-monotonic, apparently periodic decay is observed for the Rh-An series as was seen for the Ap* fluorescence quenching.6 The apparent period of about five base pairs is similar as well, as is the temperature dependence for the Rh-An sequences. Below the Tm of the duplex, increasing temperature leads to increased CPG decomposition, but the amplitude of the periodicity is suppressed. Once the duplexes begin to melt, unstacking the base pairs, the decomposition efficiencies sharply drop to zero (supplementary information). This decrease in decomposition occurs between 50 – 60 °C.
Figure 3.
CT yields (Y) as a function of bridge length for the Rh-An series and AQ-An series. Results at three temperatures are shown for the Rh-An series: 20 °C (red circles), 30 °C (blue triangles), and 40 °C (green x’s); AQ-An experiments are at ambient temperature. Duplexes (10 µM) were irradiated at 365 nm in 20 mM sodium phosphate, 50 mM NaCl, pH 7.0 as described in the text. The bridge length is defined as the number of adenines between the photooxidant and the trap. The experiments were repeated at least three times, the results averaged, and the error is expressed as 90% confidence intervals of the mean.
Although the apparent periodicity is dampened, a similar profile is apparent with anthraquinone as the pendant photooxidant (Figure 3). As with the Rh-An series, photooxidation of the AQ-An assemblies show a shallow, non-monotonic periodic length dependence in yield. Decay parameters and apparent period are comparable.
DNA-mediated Oxidative Decomposition of CPG by Ap
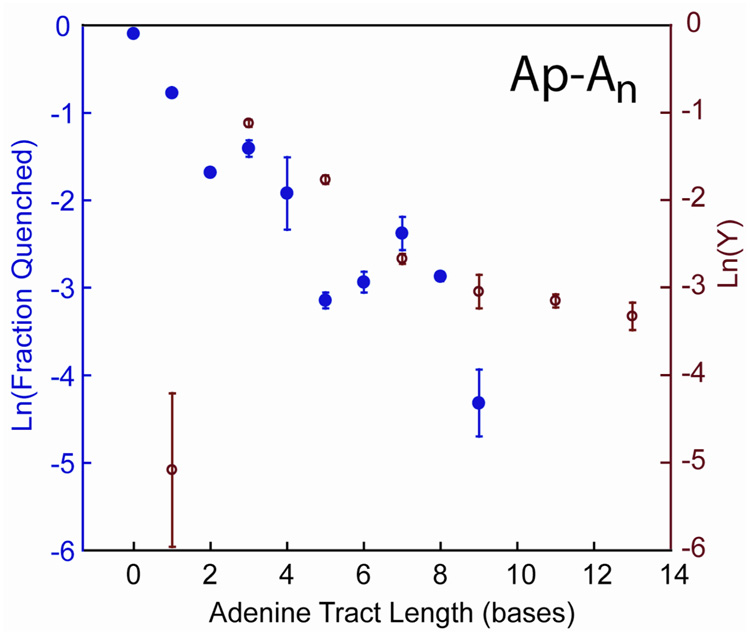
To determine if periodicities could be observed in the presence of facile BET, we prepared the series of duplexes ApAn. Figure 4 directly compares the CT yield for CPG decomposition and Ap* fluorescence quenching. Although oxidative damage to CPG is observed, CPG immediately neighboring Ap does not allow a sufficiently long-lived charge-separated state, and BET depletes the oxidized base faster than ring-opening.13 This initial low yield for a single intervening adenine, and much higher yield for three intervening adenines, is characteristic of a system with rapid charge recombination.14,15 Notably, although the length dependence in CPG ring opening is comparable to the fluorescence quenching result, the corresponding periodicity is completely suppressed.
Figure 4.
CT yields (y) as a function of bridge length for the Ap-An series (red, open circles), as determined by ring-opening of CPG. Duplexes (10 µM) were irradiated at ambient temperature for 30 sec at 325 nm in 5 mM sodium phosphate, 50 mM NaCl, pH 7.0 as described in the text. The experiments were repeated at least three times, the results averaged, and the error is expressed as 90% confidence intervals of the mean. On the same plot, fluorescence quenching from reference (6) is shown for comparison (blue, closed circles).
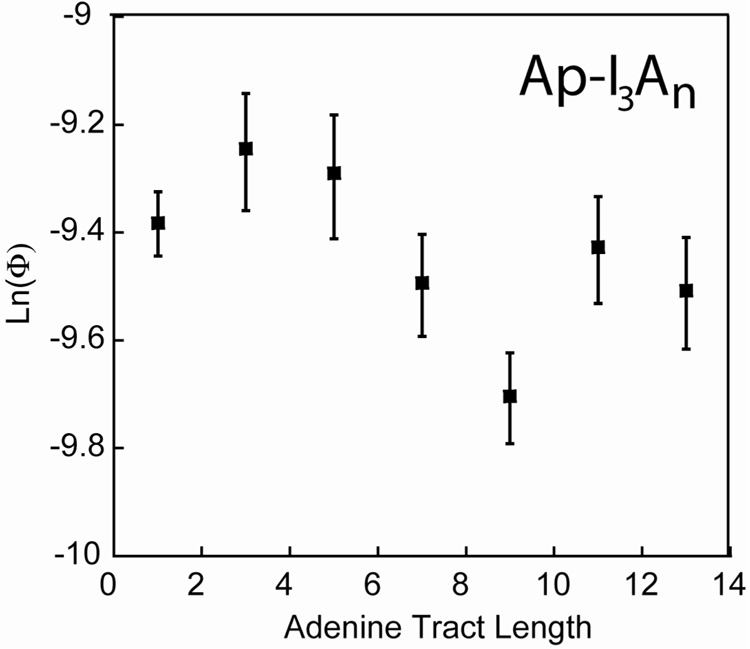
For the Ap-I3An sequences (Figure 5), there is substantially less damage, such that 30 min of irradiation is necessary to achieve significant decomposition of CPG. Nonetheless, BET is suppressed, as only slightly more decomposition is observed for the Ap-I3A3 sequence versus the Ap-I3A1 sequence. Importantly, the non-monotonicity is now recovered and is qualitatively similar to that observed for the Ap* fluorescence quenching and Rh-An systems.
Figure 5.
CT quantum yields (Φ) as a function of bridge length for the Ap-I3An series, as determined by ring-opening of CPG. Duplexes (10 µM) were irradiated at ambient temperature for 30 min at 325 nm in 5 mM sodium phosphate, 50 mM NaCl, pH 7.0 as described in the text. The experiments were repeated at least eight times, the results averaged, and the error is expressed as 90% confidence intervals of the mean. Quantum yields were determined using actinometry on 6 mM ferrioxalate.
DISCUSSION
Observation of Periodicities in Length Dependence of CPG Decomposition
The dependence of CPG oxidation by Rh or AQ on the length of the intervening adenine tract is periodic. It is striking that this result is so similar to that seen with the Ap* fluorescence quenching assay and that the periods are identical. The driving forces for photooxidation by Ap*, Rh*, and AQ* vary over a range of 700 mV.2,30,31 The fluorescence quenching assay measures direct hole injection from Ap* into an orbital that includes the acceptor guanine, while the CPG assays directly measure the total CT yield to the hole acceptor, regardless of mechanism. Nevertheless, despite these fundamental differences between the experiments, a periodic length dependence is observed for all three cases and approximately the same apparent period is observed. Importantly, when the slow, unmodified guanine trap is used, no periodicity is observed, indicating the importance of assaying pre-equilibrium states in CT experiments. Although the CPG decomposition is a chemical event, the fast timescale of ring-opening defines a fast clock where CT is rate-limiting, in contrast to biochemical experiments measuring guanine decomposition.
For the Rh-An series, with increasing temperature, the overall yield of CT increases, the length dependence becomes shallower, and the periodicity is attenuated. For a direct CT event between a donor and acceptor in contact, in which the donor and acceptor orbitals are already aligned, higher temperatures are likely to decrease the probability that the orbitals will remain aligned, and decreased CT results. In contrast, when the donor and acceptor are separated by a dynamic bridge of base pairs, increasing the temperature allows a greater fraction of these duplexes to access a CT-active domain, resulting in enhanced CT. Increased temperature has a more prominent effect on CT through longer adenine bridges because there is a lower initial probability of each bridging base being aligned in a CT-active conformation. This effect is identical to that observed for Ap* fluorescence quenching.6 Furthermore, for both cases, the apparent periodicity is suppressed with increasing temperature, implying that the underlying cause of the periodicity is the same. Periodicity is not as evident for the AQ-An system as for the Rh-An sequences. This apparent decrease in amplitude could be because the AQ is separated from the adenine tract by five bases, introducing dephasing processes. Furthermore, anionic AQ radical can equilibrate between singlet and triplet states, the former being competent to reduce oxygen,33 generating a persistent hole in the DNA that can equilibrate over a long timescale and damage CPG independently of the bridging sequence; previous work13 has, however, shown only a modest effect of oxygen on CPG ring-opening rates by AQ. Nevertheless, there is clear deviation from monotonicity that is greater than experimental error, and a period equivalent in length to that observed for the RhAn is evident.
In a sense, the examination of Ap-An-CPG sequences should represent an intermediate system between studies of Ap* fluorescence quenching by guanine and assays of CPG decomposition by Rh photooxidants. The photooxidant is the same as in the fluorescence quenching study, and CPG decomposition is used as a proxy for charge separation, as with the Rh-(A)n and AQ-(A)n series. Remarkably, the decay is monotonic (Figure 4), with a decreasing slope similar to that observed in a system using stilbene as a photooxidant.16 This could be due to a higher proportion of initial CT-active conformations for short lengths8 or to changing distribution of yield with length between superexchange, localized hopping, and delocalized hopping mechanisms. Nevertheless, the only consistent difference between the Ap-An system and the other three is the presence of efficient BET. Clearly, we can control this non-monotonic effect by changing the extent of BET.
We next considered the effect of eliminating BET while still assaying for ring-opening. The timescale required for efficient charge injection is the nanosecond lifetime of Ap*, while BET must compete with the faster ring-opening. Hence, we speculated that a bridge modification that sufficiently decreased the rate of CT in both directions could eliminate BET while still maintaining some efficiency for forward transfer.14,34 Ap* does not oxidize inosine, and the introduction of inosine into an adenine bridge substantially affects the CT yield. We introduced three inosines between the aminopurine and the adenine tract (Figure 5). As expected, the total CT efficiency dropped substantially, but the Ap-I3A1 sequence has equivalent damage yield to the Ap-I3A3 sequence, indicating that BET has been mostly excluded from the system. Importantly, the non-monotonicity is now restored, supporting the hypothesis that BET was responsible for suppressing the periodicity.
These results are straightforward to reconcile with two recent studies on CT across adenine tracts. In one system, transient absorption spectroscopy was used to measure the production of NDI radical, with PTZ across an A tract participating as the hole acceptor.15 No periodicity was observed, but it was found that BET substantially depletes the charge separated state. Similarly, another series of experiments considered CT across an adenine tract between two capping stilbenes.16 The length dependence found in this study is identical to that for Ap-An-CPG, and no periodicity was observed. Furthermore, BET of the injected hole is rapid in this system as well. Notably, although a recent theoretical treatment of three-adenine tracts implied that the stiffness introduced by the bridging stilbene used in this study does not profoundly influence local coupling constants35, this environment might well affect formation of delocalized domains.
Conformational gating through delocalized CT-active domains
Previously, we interpreted the periodic length dependence in the context of a certain number of bases being ideal for forming a CT-active domain.6 When an integer number of CT-active domains can readily form between the acceptor and donor, CT is accelerated, either coherently through two mutually CT-active domains or incoherently by hopping between such domains. For a non-integer number of domains, dephasing processes, such as domain drift, are required. These processes are slower and decrease the probability of CT to the acceptor before charge recombination. A similar argument has been made in the context of polaron hopping.11 The experiments described here do not distinguish between the two mechanistic arguments. Nevertheless, the fact that BET suppresses the periodicity supports the notion that increased CT across certain bridge lengths is the inherent source of the periodicity.
Since the conformationally gated domain hopping model ascribes the periodicity to the change in A-tract length, it is interesting to compare distance dependences to a system in which the A-tract length is fixed. This was accomplished by monitoring decomposition of cyclopropyladenine (CPA) serially substituted at each position within a 14 base pair adenine tract.2 In contrast to the CPG trapping situation, there is no periodic variation of the yield with CPA position for a given A-tract length. This result is consistent with our domain hopping model, as a given length A-tract will accommodate a similar domain structure regardless of the placement of the trap.
Other Theoretical Predictions of Periodic Distance Dependences
There have been theoretical predictions of a periodic length dependence of CT. In particular, when the energies of the donor, bridge, and acceptor are similar, on-resonance CT has been calculated to have a periodic length dependence.36–38 In these theoretical studies of molecular wires, though an exponential distance dependence was found for off-resonance CT, smooth, bounded periodicities were predicted for on-resonance coupling; energetic inhomogeneities along the bridge could attenuate the periodicities.37 Although these studies modeled the wire between metals, the same analyses could apply to a sufficiently gated charge transfer system, such that the donor can be excited independently of the bridge. It is possible that DNA fulfills that requirement based on the apparent conformational gating. A separate novel approach to determine the coupling across a molecular bridge formulated the lengthening of the bridge as iterative perturbations. Here, too, a non-monotonicity was predicted for on-resonant transfer, but was aperiodic and unstable with respect to the coupling parameters.38
Interestingly, Renger and Marcus have calculated a periodic length dependence for CT across an A-tract DNA bridge using a model that allowed delocalization of the electron hole over several bases.12 These periodicities were eliminated by incorporation of a static disorder term.
The periodic length dependence found here does not appear to be related to on-resonance CT. The periods are the same for the different photooxidants, Ap, Rh, AQ, with different oxidation potentials; this similarity argues that the periodicity is not electronic in nature. More importantly, these theoretical periodicities are all with regard to donor-acceptor separation, not adenine tract length. Only the CT-active domain model predicts that serially inserting a CPA trap along a constant A-tract will eliminate the periodicity; a quantum or symmetry effect would be, if anything, more pronounced in such a system.
It is remarkable that we are able to observe these periodicities in DNA CT using disparate assays so long as the experiments probe events on a fast timescale and isolate convoluting processes such as BET and trapping events. The observations here underscore the utility of applying cyclopropylamine-modified bases as fast traps for CT. More importantly, it is clear that engineering differing extents of BET allows control over the extent of length-dependent periodic behavior.
Supplementary Material
Sequences for Rh-An, AQ-An, Ap-An, Ap-I3An. Time-courses of CPG decomposition for Rh-A2 and AQ-A2. Temperature dependence of CPG decomposition for Rh-A2 through Rh-A8 from 20 °C to 80 °C. This material is available free of charge over the Internet at http://pubs.acs.org.
Acknowledgment
We are grateful to the NIH (GM49216) for their support. We thank also the Caltech SURF program for a summer undergraduate fellowship (M.L.D.). In addition, we are grateful for the helpful comments provided by our reviewers.
References
- 1.(a) Schuster GB, editor. Long-Range Charge Transfer in DNA, I and II. Vols 236, 237. New York: Springer; 2004. [Google Scholar]; (b) Wagenknecht HA, editor. Charge Transfer in DNA. Weinheim: Wiley-VCH; 2005. [Google Scholar]; (c) Guo X, Gorodetsky AA, Hone J, Barton JK, Nuckolls C. Nat. Nanotech. 2008;3:163–167. doi: 10.1038/nnano.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Kelley SO, Barton JK. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]; (b) Augustyn KE, Genereux JC, Barton JK. Angew. Chem. Int. Ed. 2007;46:5731–5733. doi: 10.1002/anie.200701522. [DOI] [PubMed] [Google Scholar]; (c) Delaney S, Pascaly M, Bhattacharya PK, Han K, Barton JK. Inorg. Chem. 2002;41:1966–1974. doi: 10.1021/ic0111738. [DOI] [PubMed] [Google Scholar]
- 3.(a) Kelley SO, Holmlin ER, Stemp EDA, Barton JK. J. Am. Chem. Soc. 1997;199:9861–9870. [Google Scholar]; (b) Bhattacharya PK, Barton JK. J. Am. Chem. Soc. 2001;123:8649–8656. doi: 10.1021/ja010996t. [DOI] [PubMed] [Google Scholar]; (c) Takada T, Fujitsuka M, Majima T. Proc. Natl. Acad. Sci. USA. 2007;104:11179–11183. doi: 10.1073/pnas.0700795104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Boal AK, Barton JK. Bioconj. Chem. 2005;16:312–321. doi: 10.1021/bc0497362. [DOI] [PubMed] [Google Scholar]; (e) Rajski SR, Barton JK. Biochem. 2001;40:5556–5564. doi: 10.1021/bi002684t. [DOI] [PubMed] [Google Scholar]
- 4.(a) Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Nat. Biotech. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]; (b) Boon EM, Salas JE, Barton JK. Nat. Biotech. 2002;20:282–286. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]; (c) Kelley SO, Jackson NM, Hill MG, Barton JK. Angew. Chem., Int. Ed. 1999;38:941–945. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]; (d) Inouye M, Ikeda R, Takase M, Tsuri T, Chiba J. Proc. Natl. Acad. Sci. U.S.A. 2005:11606–11610. doi: 10.1073/pnas.0502078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Merino EJ, Boal AK, Barton JK. Curr. Op. Chem. Biol. 2008;12:1–9. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, Barton JK. Biochem. 2005;44:8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]; (c) Augustyn KE, Merino EJ, Barton JK. Proc. Natl. Acad. Sci. USA. 2007;104:18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gorodetsky AA, Dietrich LEP, Lee PE, Demple B, Newman DK, Barton JK. Proc. Natl. Acad. Sci. USA. 2008;105:3684–3689. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Merino EJ, Barton JK. Biochem. 2008;47:1511–1517. doi: 10.1021/bi701775s. [DOI] [PubMed] [Google Scholar]
- 6.O’Neill MA, Barton JK. J. Am. Chem. soc. 2004;126:11471–11486. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 7.Smalley JF, Sachs SB, Chidsey CED, Dudek SP, Sikes HD, Creager SE, Yu CJ, Feldberg SW, Newton MD. J. Am. Chem. Soc. 2004;126:14620–14630. doi: 10.1021/ja047458b. [DOI] [PubMed] [Google Scholar]
- 8.(a) O’Neill MA, Barton JK. J. Am. Chem. Soc. 2004;126:13234–13235. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]; (b) Wan CZ, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. USA. 1999;96:6014–6019. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) O’Neill MA, Becker HC, Wan C, Barton JK, Zewail AH. Angew. Chem. Int. Ed. 2003;42:5896–5900. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 9.(a) Shao F, Augustyn KE, Barton JK. J. Am. Chem. Soc. 2005;127:17445–17452. doi: 10.1021/ja0563399. [DOI] [PubMed] [Google Scholar]; (b) Conwell EM, Rakhmanova SV. Proc. Natl. Acad. Sci. USA. 2000;97:4556–4560. doi: 10.1073/pnas.050074497. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kendrick T, Giese B. Chem. Comm. 2002:2016–2017. doi: 10.1039/b205928b. [DOI] [PubMed] [Google Scholar]; (d) Barnett RN, Cleveland CL, Joy A, Landman U, Schuster GB. Science. 2001;294:567–571. doi: 10.1126/science.1062864. [DOI] [PubMed] [Google Scholar]
- 10.(a) Buchvarov I, Wang Q, Raytchev M, Trifonov I, Fiebig T. Proc. Natl. Acad. Sci. USA. 2007;104:4794–4797. doi: 10.1073/pnas.0606757104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crespo-Hernández CE, Cohen B, Kohler B. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]; (c) Kadhane U, Holm AIS, Hoffmann SV, Nielsen S. Phys. Rev. E. 2008;77:021901. doi: 10.1103/PhysRevE.77.021901. [DOI] [PubMed] [Google Scholar]
- 11.Conwell EM, Bloch SM. J. Phys Chem. B. 2006;110:5801–5806. doi: 10.1021/jp0553986. [DOI] [PubMed] [Google Scholar]
- 12.Renger T, Marcus RA. J. Phys Chem. A. 2003;107:8404–8419. [Google Scholar]
- 13.(a) Williams TT, Dohno C, Stemp EDA, Barton JK. J. Am. Chem. Soc. 2004;126:8148–8158. doi: 10.1021/ja049869y. [DOI] [PubMed] [Google Scholar]; (b) O’Neill MA, Dohno C, Barton JK. J. Am. Chem. Soc. 2004;126:1316–1317. doi: 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]; (c) Dohno C, Stemp EDA, Barton JK. J. Am. Chem. Soc. 2003;125:9586–9587. doi: 10.1021/ja036397z. [DOI] [PubMed] [Google Scholar]; (d) Yoo J, Delaney S, Stemp EDA, Barton JK. J. Am. Chem. Soc. 2003;125:6640–6641. doi: 10.1021/ja034326u. [DOI] [PubMed] [Google Scholar]
- 14.(a) Kawai K, Osakada Y, Fujitsuka M, Majima T. J. Phys. Chem. B. 2008;112:2144–2149. doi: 10.1021/jp075326+. [DOI] [PubMed] [Google Scholar]; (b) Takada T, Lin C, Majima T. Angew. Chem. Int. Ed. 2007;46:6681–6683. doi: 10.1002/anie.200701525. [DOI] [PubMed] [Google Scholar]; (c) Kawai K, Osakada Y, Fujitsuka M, Majima T. Chem. Eur. J. 2008;14:3721–3726. doi: 10.1002/chem.200701835. [DOI] [PubMed] [Google Scholar]; (d) Sanii L, Schuster GB. J. Am. Chem. Soc. 2000;122:11545–11546. [Google Scholar]
- 15.(a) Takada T, Kawai K, Cai X, Sugimoto A, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2004;126:1125–1129. doi: 10.1021/ja035730w. [DOI] [PubMed] [Google Scholar]; (b) Kawai K, Takada T, Nagai T, Cai X, Sugimoto A, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2003;125:16198–16199. doi: 10.1021/ja038309g. [DOI] [PubMed] [Google Scholar]
- 16.(a) Lewis FD, Zhu H, Daublain P, Fiebig T, Raytchev M, Wang Q, Shafirovich V. J. Am. Chem. Soc. 2006;128:791–800. doi: 10.1021/ja0540831. [DOI] [PubMed] [Google Scholar]; (b) Lewis FD, Zhu H, Daublain P, Cohen B, Wasielewski MR. Angew. Chem. Int. Ed. 2006;45:7982–7985. doi: 10.1002/anie.200603455. [DOI] [PubMed] [Google Scholar]; (c) Lewis FD, Zhu H, Daublain P, Sigmund K, Fiebig T, Raytchev M, Wang Q, Shafirovich V. Photochem. Photobiol. Sciences. 2008;7:534–539. doi: 10.1039/b719715b. [DOI] [PubMed] [Google Scholar]
- 17.Giese B, Amaudrut, Köhler A-K, Sporman M, Wessely S. Nature. 2001;412:318–320. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 18.(a) Stemp EDA, Arkin M, Barton JK. J. Am. Chem. Soc. 1997;119:2921–2925. [Google Scholar]; (b) Burrows CJ, Muller JG. Chem. Rev. 1998;93:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 19.Elias B, Shao F, Barton JK. J. Am. Chem. Soc. 2008;130:1152–1153. doi: 10.1021/ja710358p. [DOI] [PubMed] [Google Scholar]
- 20.(a) Kino K, Sugiyama H. Chem. Biol. 2001;8:369–378. doi: 10.1016/s1074-5521(01)00019-9. [DOI] [PubMed] [Google Scholar]; (b) Cadet J, Douki T, Ravanat J-L. Acc. Chem. Res. 2008;41:1075–10783. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 21.(a) Nakatani K, Dohno C, Saito I. J. Am. Chem. Soc. 2001;123:9681–9682. doi: 10.1021/ja010479a. [DOI] [PubMed] [Google Scholar]; (b) Shao F, O’Neill MA, Barton JK. Proc. Natl. Acad. Sci. USA. 2004;101:17914–17919. doi: 10.1073/pnas.0408128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The rate of ring-opening for CPG has not been measured directly, although indirect results from experiments in DNA suggest a rate of ≥ 109 s−1. BET from guanine cation radical to thionine anion radical bound non-covalently to DNA has been measured as subpicosecond,23 although photolysis of thionine bound to DNA yields no detectable base-labile guanine damage despite clear evidence by transient absorption spectroscopy that photooxidation occurs.13 In contrast, CPG is facilely decomposed by DNA-tethered thionine, indicating that the CPG ring-opening rate is at least nanosecond. Similar results are obtained with Ap photooxidation, where forward transport to guanine is 200 ps over a three adenine tract,24 and no damage is observed to guanine due to facile BET, but CPG ring-opening is nonetheless observed.13 Thus CPG ring-opening appears to be much faster than charge trapping at unmodified guanine. Model studies have been less illuminating.25 A neutral N-alkylcyclopropylaminyl radical25a was observed to ring-open with a rate of at least 7.2 × 1011 s−1. However, this rate is most likely accelerated by phenyl substitution on the cyclopropyl group, though attenuated by virtue of being the neutral, rather than cation radical. A model system closer to CPG has not yet been examined.
- 23.Reid GD, Whittaker DJ, Day MA, Turton DA, Kayser V, Kelly JM, Beddard GS. J. Am. Chem. Soc. 2002;124:5518–5527. doi: 10.1021/ja0172363. [DOI] [PubMed] [Google Scholar]
- 24.Wan CZ, Fiebig T, Schiemann O, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. USA. 2000;97:14052–14055. doi: 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Musa OM, Horner JH, Shahin HE, Newcomb M. J. Am. Chem. Soc. 1996;118:3862–3868. [Google Scholar]; (b) Horner JH, Martinez FN, Musa OM, Newcomb M, Shahin NE. J. Am. Chem. Soc. 1995;117:11124–11133. [Google Scholar]; (c) Li X, Grimm ML, Igarashi K, Castagnoli N, Jr, Tanko JM. Chem. Comm. 2007:2648–2650. doi: 10.1039/b702157g. [DOI] [PubMed] [Google Scholar]
- 26.Holmlin RE, Dandliker PJ, Barton JK. Biconj. Chem. 1999;10:1122–1130. doi: 10.1021/bc9900791. [DOI] [PubMed] [Google Scholar]
- 27.Gasper SM, Schuster GB. J. Am. Chem. Soc. 1997;119:12762–12771. [Google Scholar]
- 28.(a) Joy A, Ghosh AK, Schuster GB. J. Am. Chem. Soc. 2006;128:5346–5347. doi: 10.1021/ja058758b. [DOI] [PubMed] [Google Scholar]; (b) Ghosh A, Joy A, Schuster GB, Douki T, Cadet J. Org. Biomolec. Chem. 2008;6:916–928. doi: 10.1039/b717437c. [DOI] [PubMed] [Google Scholar]
- 29.Hatchard CG, Parker CA. Proc. R. Soc. Lon. Ser. A. 1956;235:518–536. [Google Scholar]
- 30.(a) Turro C, Hall DB, Chen W, Zuilhof H, Barton JK, Turro NJ. J. Phys Chem. A. 1998;102:5708–5715. [Google Scholar]; (b) Dotse AK, Boone EK, Schuster GB. J. Am. Chem. Soc. 2000;122:6825–6833. [Google Scholar]; (c) Ly D, Sanii L, Schuster GB. J. Am. Chem. Soc. 1999;121:9400–9410. [Google Scholar]
- 31.The currently accepted oxidation potential for guanosine is ~1.29 V32, although it is notable that this value is based on irreversible electrochemistry of the isolated nucleoside in acetonitrile.
- 32.Steenken S, Jovanovic SV. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 33.Armitage B, Yu C, Devadoss C, Schuster GB. J. Am. Chem. Soc. 1994;116:9847–9859. [Google Scholar]
- 34.O’Neill MA, Barton JK. Proc. Natl. Acad. Sci USA. 2002;99:16543–16550. doi: 10.1073/pnas.012669599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siriwong K, Voityuk AA. J. Phys. Chem. B. 2008;112:8181–8187. doi: 10.1021/jp802222e. [DOI] [PubMed] [Google Scholar]
- 36.Reimers JR, Hush NS. Chem. Phys. 1990;146:89–103. [Google Scholar]
- 37.(a) Mujica V, Kemp M, Ratner MA. J. Chem. Phys. 1994;101:6856–6864. [Google Scholar]; (b) Kemp M, Mujica V, Ratner MA. J. Chem. Phys. 1994;101:5172–5178. [Google Scholar]
- 38.Hsu CP, Marcus RA. J. Chem. Phys. 1997;106:584–598. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences for Rh-An, AQ-An, Ap-An, Ap-I3An. Time-courses of CPG decomposition for Rh-A2 and AQ-A2. Temperature dependence of CPG decomposition for Rh-A2 through Rh-A8 from 20 °C to 80 °C. This material is available free of charge over the Internet at http://pubs.acs.org.