Abstract
Pairs of fluorophores in close proximity often show self-quenching of fluorescence by the well-known H-dimer mechanism. We use a pair of fluorophores in the new dicyanomethylenedihydrofuran (DCDHF) dye family in the design and characterization of a new fluorescent probe for nucleic acid detection, which we refer to as a self-quenched intramolecular dimer (SQuID) molecular beacon (MB). We obtain a quenching efficiency of 97.2%, higher than the only other reported value for a MB employing fluorophore self-quenching by H-dimer formation. Furthermore, the excellent single-molecule (SM) emitter characteristics of the DCDHF dyes allow observation of individual SQuID MB—target complexes immobilized on a surface, where the doubled SM emission intensity of our target-bound beacon ensures a higher signal-to-background ratio than conventional fluorophore-quencher MBs. Additional advantages of the SQuID MB are single-pot labeling, visible colorimetric detection of the target, and intrinsic single-molecule two-step photobleaching behavior, which offers a specific means of discriminating between functional MBs and spurious fluorescence.
Molecular beacons (MBs)1 are single-stranded, “stem—loop” (i.e., hairpin) oligonucleotide probes that are employed for the detection of specific target nucleic acid sequences in solution. Conventional MBs contain a fluorophore and a quencher at opposite ends of the oligonucleotide that are held in close proximity by the complementary stem region; fluorescence from the fluorophore is quenched in this closed hairpin conformation. The loop region of the MB is designed so that it is complementary to a target oligonucleotide sequence of interest. Upon binding, the target opens the MB hairpin, and the separation distance between the quencher and fluorophore is increased, thereby allowing fluorescence emission to occur. MBs are attractive probes for nucleic acids because their response is highly specific to the target sequence, the fluorescence intensity is proportional to the amount of target present when MB is present in excess, and it is unnecessary to remove unhybridized probes. For these reasons, MBs have found numerous applications in the sensitive and quantitative detection of biomolecules, including real-time PCR monitoring,2 in vivo mRNA detection,3 nucleic acid biosensing,4 and protein recognition.5
Here we report the preparation and characterization of a self-quenched intramolecular dimer (SQuID) MB that is homo-doubly labeled with the N-hydroxy succinimidyl (NHS) ester-functionalized dicyanomethylenedihydrofuran (DCDHF) fluorophore 1, shown in Figure 1. In this system, quenching occurs in the closed hairpin conformation due to an excitonic interaction between the two fluorophores, which form a non-emissive H-dimer.6
Figure 1.
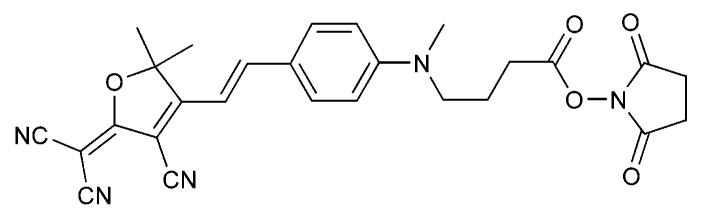
Structure of NHS ester-functionalized DCDHF fluorophore 1.
Although homo-doubly labeled MBs have been described previously,7 only one case employed H-dimer quenching, and the quenching efficiency (EQu) of that system (80%)7a is substantially lower than that for our design. With our improved SQuID MB, we are now able to achieve an EQu value comparable to widely used fluorophore—quencher pairs8 employed in conventional MBs while also realizing the inherent advantages of SQuID MBs: single-pot labeling, colorimetric detection of target, and a twofold “on” signal compared to single fluorophore-quencher MBs. Moreover, two-step photobleaching provides a powerful signature of binding to target at the single-molecule level.
SQuID MB 2 (see Table 1) was prepared by reacting the corresponding diamine-functionalized oligonucleotide with an excess of amine-reactive DCDHF 1.
TABLE 1.
Oligonucleotide Sequences for MBs (Stem Regions Are Underlined for 2 and 4a) and Targets (Mismatch Is Underlined for 3b)
| oligonucleotide sequence (5′ to 3′) | |
|---|---|
| 2 | 1-GCTCGTCCATGCCCAGGAAGGAACGAGC-1 |
| 3a | GCTCGTTCCTTCCTGGGCATGGA |
| 3b | GCTCGTTCCTTGCTGGGCATGGA |
| 4a |
1-GCTCGTCCATGCCCAGGAAGGAACGAGCTA CTTGCATTTCCATCGATTAC-biotin |
| 4b | GTAATCGATGGAAATGCAAGTA-1 |
The absorption and fluorescence emission spectra of SQuID MB 2, with and without target oligonucleotide 3a present, are shown in Figure 2. The closed beacon (no target present, blue curves) exhibited a 56 nm blue-shifted fluorophore absorption maximum and 97.2% quenched emission compared to the open beacon (opened by adding target 3a, red curves). According to molecular exciton theory,6a this spectral behavior is consistent with H-dimer formation in the closed beacon. The blue shift in the absorption spectrum of an H-dimer is the result of a splitting of the electronic excited-state into two energy levels, of which only the transition to the higher level is dipole-allowed. Electronic excitation to the higher energy level is followed by rapid internal conversion to the lower energy level, from which fluorescence is forbidden. As a result, fluorescence is quenched and intersystem crossing is likely the primary mode of relaxation.6
Figure 2.
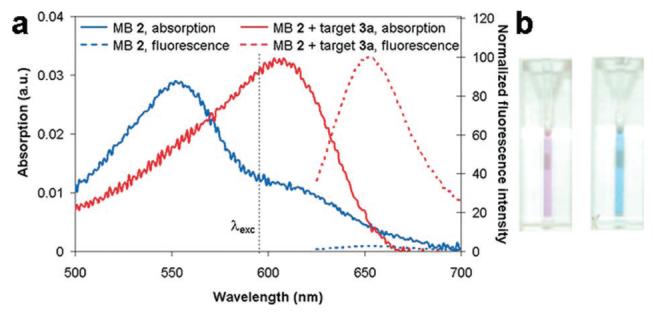
(a) Absorption (solid) and fluorescence emission (dashed, λexc) = 605 nm) spectra of SQuID MB 2 (1.49 μM) with (red) and without (blue) oligonucleotide 3a (19× excess) present. In the absence of target, the shoulder absorption at 605 nm is due to a fraction of molecules not in the H-dimer configuration. (b) Photograph of SQuID MB 2 solutions before (left) and after (right) target 3a addition showing obvious color change.
Preliminary hybridization kinetics of SQuID MB 2 were recorded by monitoring the increase in fluorescence emission with time after addition of target 3a (see Figure 3). One equivalent of target was sufficient to produce the maximum attainable fluorescence signal. As expected, the time required to achieve this maximum fluorescence intensity was strongly dependent on the target concentration and ranged from approximately 6 min with 10 equivalents of target to nearly an hour with 1 equiv of target. Further kinetic analysis will be the subject of future work.4h,i
Figure 3.
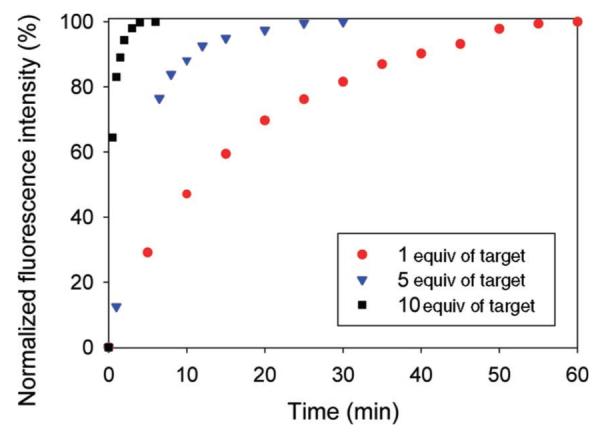
Time course of fluorescence emission (λexc = 605 nm, λcollect = 653 nm) from a solution of SQuID MB 2 (1.49 × 10-7 M) after addition of target 3a (1, 5, or 10 equiv). One equivalent of target 3a was sufficient to achieve the maximum attainable fluorescence signal. The “off” fluorescence intensity (in the absence of target) is normalized to 0%, and the maximum “on” fluorescence intensity is normalized to 100%.
The excellent single-molecule (SM) emitter characteristics of molecules in the DCDHF class (e.g., large fluorescence quantum yield, photostability comparable to R6G)9 allowed us to study the behavior of our SQuID MB system on a surface at ultralow concentration by SM fluorescence imaging.10 This allows comparison to the only previous SM study of DNA detection with surface-bound MBs.11 Accordingly, a 100 pM solution of the biotinylated two-piece hairpin 4, comprised of hybridized 4a and 4b (see Table 1 and Figure 4), was prepared and deposited on a glass coverslip coated with polyethylene glycol and NeutrAvidin, a scheme designed to minimize nonspecific adsorption of DNA to the sample surface.12 After addition of either high-salt buffer (control) or a 600 μM solution of target 3a (experimental) to the sample chamber, single molecules were imaged using a total-internal-reflection wide-field microscope with laser excitation at 532 nm, and fluorescence images were captured on a cooled EMCCD with 100-ms time resolution.
Figure 4.
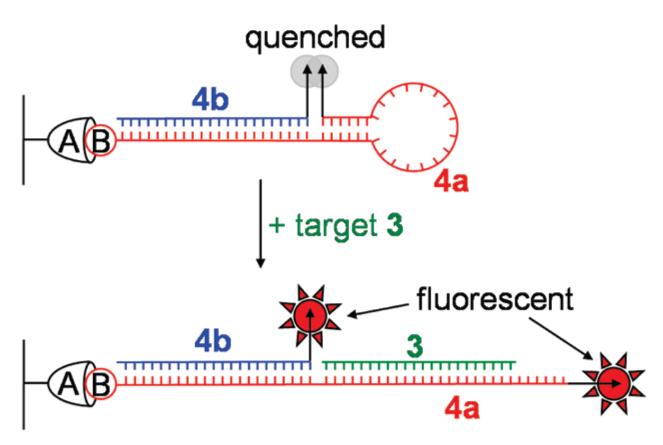
Schematic illustrating closed and open conformations of the biotin(B)-NeutrAvidin(A) surface-immobilized two-piece SQuID MB 4 used for SM imaging.
The results of our SM imaging experiments highlight the advantages of using a SQuID MB in ultrasensitive target detection. Fluorescence images of surface-immobilized SQuID MB 4 after addition of buffer and target 3a are shown in Figure 5a and b, respectively. In the control, addition of ultraclean buffer results in the appearance of a negligible number of fluorescent spots in the viewing region. In stark contrast, when target 3a was added to the sample chamber (Figure 5b), a distinct increase in the number and intensity of SM spots was observed. Time traces of individual spots were recorded; representative time traces are shown in Figure 5c, and two-step SM bleaching events are easily seen in 74% of all molecules (video in SI). Of the 26% of the molecules that did not show two discrete intensity steps, a large fraction (65%) was found to have a peak intensity corresponding to two DCDHF fluorophores (60 au or greater) and thus might also have exhibited two-step photobleaching if the data had been acquired with higher time resolution. To characterize the two-step events, average fluorescence intensities were extracted from both the time interval of high emission and the time interval of lower emission (after one dye photobleached). The resulting values are shown in the two histograms in Figure 5d. Despite slight differences in illumination uniformity and in the brightness of individual molecules, the experimental data clearly fall into two populations, one with mean intensity approximately twice (71 au) that of a typical single-molecule DCDHF fluorophore (35 au), similar to the intensity distributions reported for a carboxytetramethylrhodamine H-dimer system,6b under the stated experimental conditions. This intensity doubling is the result of fluorescence emission from two DCDHF fluorophores 1 appearing as a single diffraction-limited spot, as would be expected for target-bound SQuID MB complex 3:4. It is the unique combination of the strong single-molecule emission characteristics of the DCDHF fluorophores and their self-quenching ability via H-dimer formation that provide for robust detection of DNA at the single-molecule level by SQuID beacons.
Figure 5.
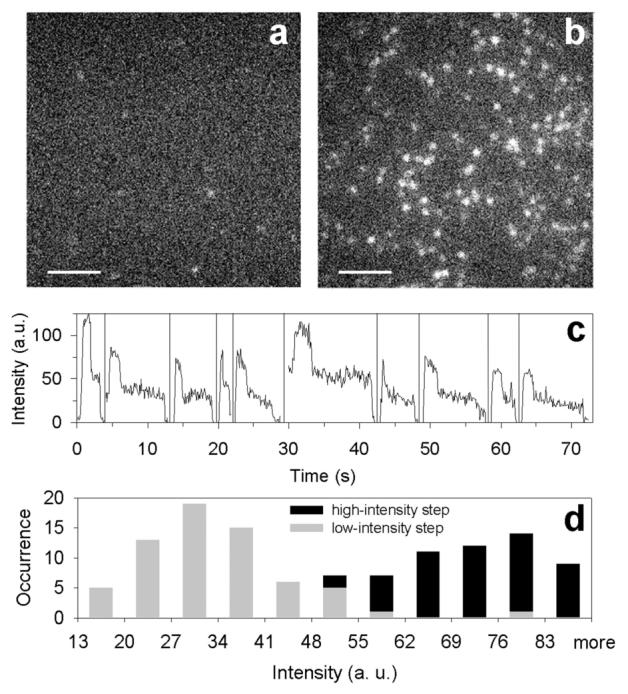
Single-molecule fluorescence images (256 × 256 pixels2; scale bar, 5 μm; 100 ms integration) of immobilized SQuID MB 4 (a) after buffer addition (control) and (b) 60 s after addition of 600 μM target DNA 3a. A similar experiment using a target with a single G mismatch (sequence 3b in Table 1) produced a sixfold smaller increase in the number of spots after target addition (data not shown). (c) Representative background-subtracted time traces for single molecules displaying two-step photobleaching, where the observation time scale is correct for the first molecule and the zero-time position for all other molecules has been horizontally offset for clarity. All traces show an initial increase due to focusing. (d) Histogram of the background-subtracted average intensity levels for molecules exhibiting two-step photobleaching after target addition (each molecule contributes two data points). The width of the distributions arises primarily from polarized, nonuniform illumination of the sample.
In summary, we have shown that SQuID molecular beacons based on H-dimer excitonic quenching of DCDHF fluorophores offer a number of advantages over conventional fluorophore-quencher MBs, including ease of synthesis and a twofold on signal. SQuID MB 2 not only has an EQu value that is similar to that of other fluorophore—quencher pairs,8 but it is also substantially improved over the EQu value of the only other reported H-dimer MB.7a Furthermore, we have demonstrated that the SQuID design has added utility in single-molecule imaging as a result of its twofold on signal and two-step photobleaching behavior. These properties offer a means, only available in a SM analysis, of discriminating between functional molecular beacons and spurious fluorescence signals. Although a number of published studies have described SM detection of MBs in flow-13 or diffusion-based14 schemes, our ability to detect DNA target binding in a surface-immobilized format holds potential for the use of SQuID probes in gene chip technology.15 Furthermore, because single DCDHF dyes can be imaged and tracked in cells,16 one can envision a SQuID aptamer beacon17 that binds selectively to specific proteins on a cell surface or a SQuID beacon that detects subcellular RNA transcripts.18 The inherent advantages of SQuID MBs warrant further development of this probe design (e.g., linker length, fluorophore quenching, orientation control of the fluorophores, etc.) for specific applications in biomolecular detection.
Acknowledgment
This work was supported in part by the National Institutes of Health Grant No. 1P20-HG003638. NRC and AKP acknowledge support from National Science Foundation Graduate Research Fellowships, and AKP acknowledges support from a G. J. Lieberman Fellowship.
Footnotes
Supporting Information Available: Experimental procedures, analyses, spectroscopic data for 1–4, quenching efficiencies for other fluorophores, and SM video. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- (2).Manganelli R, Tyagi S, Smith I. In: Methods in Molecular Medicine. Parish T, Stoker NG, editors. Vol. 54. Humana; Totowa, NY: 2001. pp. 295–310. [DOI] [PubMed] [Google Scholar]
- (3) (a).Matsuo T. Biochim. Biophys. Acta. 1998;1379:178–184. doi: 10.1016/s0304-4165(97)00090-1. [DOI] [PubMed] [Google Scholar]; (b) Sokol DL, Zhang X, Lu P, Gerwirtz AM. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei-lida Y, Kondo S, Ishibashi K. Biophys. J. 2000;78:3260–3274. doi: 10.1016/S0006-3495(00)76862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Perlette J, Tan W. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]; (e) Bratu DP, Cha B, Mhlanga MM, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Tyagi S, Alsmadi O. Biophys. J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Drake TJ, Medley CD, Sen A, Rogers RJ, Tan W. ChemBioChem. 2005;6:2041–2047. doi: 10.1002/cbic.200500046. [DOI] [PubMed] [Google Scholar]; (h) Vargas DJ, Raj A, Marras SAE, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4) (a).Broude NE. Trends Biotechnol. 2002;20:249–256. doi: 10.1016/s0167-7799(02)01942-x. [DOI] [PubMed] [Google Scholar]; (b) Lou HJ, Tan W. Instrum. Sci. Technol. 2002;30:465–476. [Google Scholar]; (c) Epstein JR, Leung APK, Lee KH, Walt DR. Biosens. Bioelectron. 2003;18:541–546. doi: 10.1016/s0956-5663(03)00021-6. [DOI] [PubMed] [Google Scholar]; (d) Zhao X, Tapec-Dytioco R, Wang K, Tan W. Anal. Chem. 2003;75:3476–3483. doi: 10.1021/ac034330o. [DOI] [PubMed] [Google Scholar]; (e) Du H, Disney MD, Miller BL, Krauss TD. J. Am. Chem. Soc. 2003;125:4012–4013. doi: 10.1021/ja0290781. [DOI] [PubMed] [Google Scholar]; (f) Yao G, Tan W. Anal. Biochem. 2004;331:216–223. doi: 10.1016/j.ab.2003.12.005. [DOI] [PubMed] [Google Scholar]; (g) Du H, Strohsahl CM, Camera J, Miller BL, Krauss TD. J. Am. Chem. Soc. 2005;127:7932–7940. doi: 10.1021/ja042482a. [DOI] [PubMed] [Google Scholar]; (h) Tsourkas A, Behlke MA, Rose SD, Bao G. Nucleic Acid Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Tsourkas A, Behlke MA, Bao G. Nucleic Acid Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5) (a).Li J, Schuster S, Tan W. Angew. Chem., Int. Ed. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]; (b) Li JJ, Fang X, Tan W. Biochem. Biophys. Res. Commun. 2002;292:31–40. doi: 10.1006/bbrc.2002.6581. [DOI] [PubMed] [Google Scholar]; (c) Vicens MC, Sen A, Vanderlaan A, Drake TJ, Tan W. ChemBioChem. 2005;6:900–907. doi: 10.1002/cbic.200400308. [DOI] [PubMed] [Google Scholar]
- (6) (a).McRae EG, Kasha M. In: Physical Processes in Radiation Biology. Augenstein L, Rosenberg B, Mason R, editors. Academic; New York: 1964. pp. 23–42. [Google Scholar]; (b) Hernando J, van der Schaaf M, van Dijk EMHP, Sauer M, García-Parajó MF, van Hulst NF. J. Phys. Chem. A. 2003;107:43–52. [Google Scholar]
- (7) (a).Bernacchi S, Mély Y. Nucleic Acid Res. 2001;29:e62. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fujimoto K, Shimizu H, Inouye M. J. Org. Chem. 2004;69:3271–3275. doi: 10.1021/jo049824f. [DOI] [PubMed] [Google Scholar]; (c) Seo YJ, Hwang GT, Kim BH. Tet. Lett. 2006;47:4037–4039. [Google Scholar]
- (8).Marras SAE, Kramer FR, Tyagi S. Nucleic Acid Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9) (a).Willets KA, Ostroverkhova O, He M, Twieg RJ, Moerner WE. J. Am. Chem. Soc. 2003;125:1174–1175. doi: 10.1021/ja029100q. [DOI] [PubMed] [Google Scholar]; (b) Willets KA, Nishimura SY, Schuck PJ, Twieg RJ, Moerner WE. Acc. Chem. Res. 2005;38:549–556. doi: 10.1021/ar0401294. [DOI] [PubMed] [Google Scholar]
- (10).Moerner WE, Fromm DP. Rev. Sci. Instrum. 2003;74:3597–3619. [Google Scholar]
- (11).Yao G, Xiaohong F, Yokota H, Yanagida T, Tan W. Chem.—Eur. J. 2003;9:5686–5692. doi: 10.1002/chem.200304977. [DOI] [PubMed] [Google Scholar]
- (12).Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- (13) (a).Wabuyele MB, Farquar H, Stryjewski W, Hammer RP, Soper SA, Cheng Y-W, Barany F. J. Am. Chem. Soc. 2003;125:6937–6945. doi: 10.1021/ja034716g. [DOI] [PubMed] [Google Scholar]; (b) Wang T-H, Peng Y, Zhang C, Wong PK, Ho C-M. J. Am. Chem. Soc. 2005;127:5354–5359. doi: 10.1021/ja042642i. [DOI] [PubMed] [Google Scholar]
- (14).Grunwell JR, Glass JL, Lacoste TD, Deniz AA, Chemla DS, Schultz PG. J. Am. Chem. Soc. 2001;123:4295–4303. doi: 10.1021/ja0027620. [DOI] [PubMed] [Google Scholar]
- (15).Culha M, Stokes DL, Griffin GD, Vo-Dinh T. Biosens. Bioelectron. 2004;19:1007–1012. doi: 10.1016/j.bios.2003.09.006. [DOI] [PubMed] [Google Scholar]
- (16).Nishimura SY, Lord SJ, Klein LO, Willets KA, He M, Lu Z, Twieg RJ, Moerner WE. J. Phys. Chem. B. 2006;110:8151–8157. doi: 10.1021/jp0574145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hamaguchi N, Ellington A, Stanton M. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- (18).Santangelo PJ, Nitin N, Bao G. J. Biomed. Opt. 2005;10:044025. doi: 10.1117/1.2011402. [DOI] [PubMed] [Google Scholar]


