Stomatal development provides a model for understanding the molecular basis of how cell lineages are established and how cells differentiate into functionally mature structures. This article describes recent advancements in understanding the role of basic helix-loop-helix (bHLH) proteins in stomatal lineage choice and differentiation in Arabidopsis. The emerging picture unravels that SPEECHLESS (SPCH), MUTE, and FAMA form heterodimers with SCREAM (SCRM)/ICE1 and SCRM2 to specify the sequential steps during stomatal development. Intriguingly, both key genes and mechanisms needed for stomatal development are also required for the formation of skeletal muscle in animals.
STEPS IN STOMATAL DEVELOPMENT
Stomata are epidermal structures that occur in most aerial organs of all terrestrial plants. They consist of two guard cells that delimit a pore and play an essential role in establishing adequate gas exchange between the plant and the atmosphere. The pore opening depends on changes in the turgor of the guard cells, and this in turn is controlled by the flow of water and ions between the guard cells and their neighboring adjacent epidermal cells (Taiz and Zeiger, 2006). Loss of guard cell turgor triggers stomatal closure, resulting in cessation of gas exchange, whereas gain in turgor induces the opposite effect.
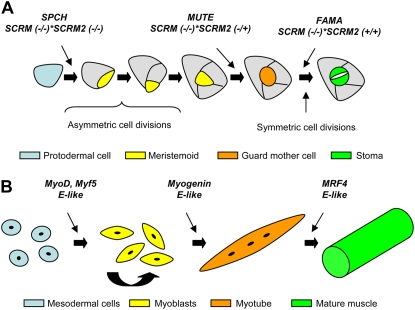
In Arabidopsis (Arabidopsis thaliana), stomatal development starts with an asymmetric cell division from an epidermal cell named the meristemoid mother cell (Bergmann and Sack, 2007; Fig. 1A). This cell division produces a small meristemoid with a triangular shape and a larger cell. Meristemoids can divide asymmetrically in an inward spiral up to three times, always yielding a larger cell and a smaller meristemoid that maintains its stem cell character. The meristemoid, after these asymmetric cell divisions, loses its stem cell activity and adopts a rounded shape, giving rise to the guard mother cell. The guard mother cell undergoes a symmetric cell division that produces the paired guard cells, which do not divide further. The larger cells that result from the asymmetric divisions and make contact with the stoma (or its precursor) can either assume meristemoid mother cell identity entering into the stomatal cell lineage or become pavement cells.
Figure 1.
The role of bHLH genes in stomatal and muscle development. A, Stomatal development. SPCH starts stomatal development by inducing the first asymmetric division, which gives rise to the first meristemoid. Two or three divisions after the formation of the first meristemoid, MUTE drives the last asymmetric cell division, producing the guard mother cell. Then, FAMA regulates the symmetric division that gives rise to the two guard cells. SCRM and SCRM2, in a dosage-dependent manner, specify the actions of SPCH, MUTE, and FAMA. (Adapted from MacAlister et al. [2007], Pillitteri et al. [2007], Serna [2007], and Kanaoka et al. [2008].) B, Muscle development. MyoD and Myf5 induce myoblast determination from mesodermal cells. Myoblasts remain in a proliferative state until myogenin instructs them to differentiate into myotubes. MRF4 acts in late differentiation events, producing mature muscle. These MyoD family members function as heterodimers with the E-like proteins. (Adapted from Weintraub [1993] and Pillitteri and Torii [2007].)
Although stomata are essential for the plant even under laboratory conditions, which makes it difficult to isolate stomata-specific mutants, a large number of genes have been cloned that enable the understanding of stomatal formation in a molecular context. These include genes that encode for extracellular molecules (Berger and Altmann, 2000; Hara et al., 2007), cell membrane components (Nadeau and Sack, 2002; Shpak et al., 2005), cytoplasmic factors (Bergmann et al., 2004; Kutter et al., 2007; Wang et al., 2007), and nuclear factors (Lai et al., 2005; Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008). This article discusses the role of genes encoding for bHLH nuclear proteins during stomatal development of Arabidopsis. Interestingly, the similarities found between stomatal and skeletal muscle development reinforce the idea of a common underlying regulatory mechanism guiding these fates.
SPCH, MUTE, AND FAMA
Three earlier studies have shown that three bHLH genes, SPCH, MUTE, and FAMA, act sequentially in stomatal development, from the early decision to enter into the stomatal cell lineage to the last step when stomata are formed (Fig. 1A; for review, see Barton, 2007; Gray, 2007; Pillitteri and Torii, 2007; Serna, 2007). Plants either lacking detectable SPCH transcripts (spch-3 and spch-4) or encoding a truncated SPCH protein without the last seven amino acids (spch-1) do not form stomata and exhibit an epidermal tissue consisting of only jigsaw-puzzle-piece-shaped pavement cells (Fig. 2; MacAlister et al., 2007). The lack of stomatal lineage cells in these mutants suggests that SPCH controls the initiation of stomatal development (Fig. 1A; MacAlister et al., 2007). Supporting such a role, the number of cells that initiate stomatal development, although not forming stomata, increases when SPCH is overexpressed (MacAlister et al., 2007; Pillitteri et al., 2007). The mutant spch-2, which encodes a protein that differs from the wild type at the C terminus, exhibits a reduced number of stomata (MacAlister et al., 2007). The presence of stomata in spch-2 enabled the unraveling of additional SPCH functions. Indeed, studies in the pedicel epidermis of this mutant showed that SPCH, in addition to controlling stomatal initiation, also maintains the stem cell activity of the meristemoids (MacAlister et al., 2007). Consistent with this dual function, the SPCH gene is broadly expressed, from undifferentiated epidermal cells to stomatal lineage cells (MacAlister et al., 2007).
Figure 2.
Stomatal phenotype of wild-type and mutant plants in genes encoding for bHLH proteins. Wild-type plants develop stomata that are spaced by intervening cells. Cells do not enter into the stomatal pathway in spch mutants. The mute mutant does not develop stomata but forms meristemoids that abort after excessive asymmetric cell divisions. fama lacks mature stomata and, instead, develops clusters of guard mother cells or young guard cells. The scrm, scrm scrm2/+, and scrm scrm2 mutants phenocopy fama, mute, and spch, respectively. (Adapted from MacAlister et al. [2007] and Serna [2007].)
MUTE also encodes a bHLH protein, which, in addition, is very similar in sequence to SPCH (Pillitteri et al., 2007). The loss-of-function mute mutant, with truncations in various positions within the bHLH domain, is completely devoid of stomata but develops meristemoids that abort after excessive asymmetric cell divisions (Fig. 2; MacAlister et al., 2007; Pillitteri et al., 2007). This suggests that MUTE represses stem cell activity of the meristemoids and induces guard mother cell formation (Fig. 1A). Consistently, the overexpression of MUTE converts all epidermal cells into stomata (MacAlister et al., 2007; Pillitteri et al., 2007). Both MUTE promoter activity and the MUTE protein localization are restricted to a subset of meristemoids, with very low activity in guard mother cells and developing stomata (MacAlister et al., 2007, Pillitteri et al., 2007). It is probable, therefore, that MUTE performs its function in those cells where it is expressed (Serna, 2007). MUTE also controls hydathode pore formation (Pillitteri et al., 2008).
FAMA was the first bHLH protein controlling stomatal development to be identified. It shares high sequence identity with SPCH and MUTE in the bHLH domain and C-terminal region (MacAlister et al., 2007; Pillitteri et al., 2007). Plants lacking detectable FAMA transcripts (fama-1) do not have mature stomata; instead, they develop groups of guard mother cells or immature guard cells (Fig. 2; Ohashi-Ito and Bergmann, 2006). These observations support the idea that FAMA induces guard mother cell division into two guard cells and that it promotes guard cell differentiation (Fig. 1A). Acknowledging its positive role in stomata formation, when FAMA is overexpressed nonstomatal cells convert directly to guard cells (Ohashi-Ito and Bergmann, 2006). Intriguingly, these guard cells are not organized into pairs; they develop from guard mother cells that become guard cells without undergoing a symmetrical division (Ohashi-Ito and Bergmann, 2006). One explanation for this finding is that FAMA levels are essential in the regulation of cell division versus differentiation, with high levels repressing cell division and forcing guard mother cells to differentiate directly into guard cells (Ohashi-Ito and Bergmann, 2006). FAMA promoter is induced in guard mother cells and young guard cells, and the FAMA protein localizes to the nucleus of these cells (Ohashi-Ito and Bergmann, 2006), which indicates that FAMA localizes in those cells where it exerts its action.
In summary, these bHLH genes have nonoverlapping but sequential roles in regulating stomatal development. At least MUTE and FAMA appear to play a major role in controlling stomata formation, because ectopic expression of any one in the nonstomatal cells results in a conversion of these cells to stomata or unpaired guard cells. Consequently, the cells must prevent their expression in the wrong cells or at inappropriate times or levels. Therefore, these genes necessarily have to be masterfully regulated.
SCRM/ICE1 AND SCRM2
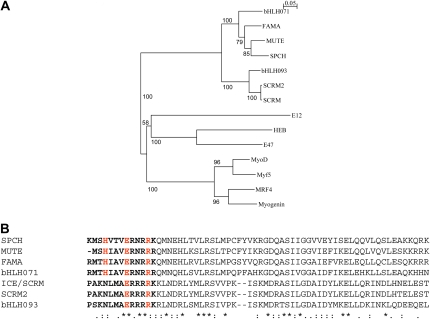
The role for broadly expressed bHLH-Leu zipper proteins in stomatal development came from the analysis of gain-of-function mutants. Homozygous scrm-D mutants develop an epidermis composed of only stomata (Kanaoka et al., 2008), whereas heterozygous scrm-D/+ plants exhibit a less severe phenotype with a high density of stomata making direct contact (Kanaoka et al., 2008). This strongly suggests that SCRM promotes stomatal development in a dose-dependent manner. Sequence analysis showed that the SCRM gene encodes the previously isolated bHLH-Leu zipper protein named ICE (Chinnusamy et al., 2003; Kanaoka et al., 2008). ICE is a regulator of freezing tolerance (Chinnusamy et al., 2003). The scrm-D phenotype is the result of a missense mutation within the KRAAM motif that replaces Arg-236 with His (Kanaoka et al., 2008). The truncated protein, similar to the wild-type one, localizes to the nucleus, which indicates that the scrm-D phenotype is not caused by a failure in the subcellular localization of the mutated SCRM protein (Kanaoka et al., 2008). Interestingly, an additional mutation destroying the DNA-binding ability restores the wild-type phenotype (Kanaoka et al., 2008). This strongly suggests that the scrm-D phenotype depends on the DNA binding. Equivalent amino acid substitution (Arg to His) within the KRAAM motif of the closely related SCRM paralog, SCRM2, gives rise to stomatal cluster formation (Kanaoka et al., 2008). Both SCRM and SCRM2 cluster in the same clade, which is distinct from the SPCH, MUTE, and FAMA clade (Fig. 3A). Similar to SCRM, SCRM2 also localizes to the cell nucleus (Kanaoka et al., 2008).
Figure 3.
Phylogeny and comparison of bHLH proteins regulating stomatal development in Arabidopsis. A, Phylogeny of the bHLH proteins that control stomatal development. Myogenic bHLH proteins serve as an outgroup. SPCH, MUTE, and FAMA cluster in the same clade. bHLH093 clusters in the SCRM/SCRM2 clade. The bHLH domains were used to calculate the neighbor-joining phylogenetic tree using ClustalX2 software. Branch lengths are proportional to sequence distance. Bootstrap values are based on 1,000 replicates. The GenBank accession numbers are as follows (in parentheses): bHLH071 (NP_568666), FAMA (Q56YJ8), MUTE (ABI74926), SPCH (ABI26170), bHLH93 (NP_001078801), SCRM2 (ACA63683), SCRM (AAP14668), E12 human (CAC14267), HEB human (NP_996923), E47 human (NP_001129611), MyoD human (CAA40000), Myf5 human (NP_005584), MRF4 human (NP_002460), and myogenin human (NP_002470). B, Sequence comparisons among the bHLH domains. The H-E-R residues are shown in red. The basic region is shown in boldface type. Asterisks indicate identical residues, colons indicate conservative changes, and periods indicate semiconservative changes. Proteins were aligned using the ClustalW2 software.
In contrast to MUTE and FAMA, both SCRM and SCRM2 are broadly expressed in stomatal cell lineages, including meristemoids, guard mother cells, and differentiating guard cells (Kanaoka et al., 2008). Although SPCH and SCRM/SCRM2 are broadly expressed in the epidermal tissue, their expression patterns are not completely overlapping. Certainly, SPCH is expressed in the entire protoderm (MacAlister et al., 2007; Pillitteri et al., 2007), being the first of the regulatory genes to be transcribed. Kanaoka et al. (2008) have proposed that SPCH just might confer the competency to enter into the stomatal pathway and that SCRM and SCRM2 might be required to initiate stomatal development.
Null alleles confirmed the positive role of these broadly expressed bHLH proteins in stomatal formation and also allowed in-depth insight into their specific roles. The loss-of-function mutation in SCRM induces the formation of groups of guard mother cells or immature guard cells instead of single and fully differentiated stomata, similar to those developing in fama (Fig. 2; Kanaoka et al., 2008). The epidermis of the scrm scrm2 double loss-of-function mutant is indistinguishable from that of the spch mutant (Fig. 2; Kanaoka et al., 2008). Finally, the scrm scrm2/+ mutant exhibits an identical phenotype to mute (Fig. 2; Kanaoka et al., 2008). Together, these findings demonstrate that the dosage of these broadly expressed genes determines the successive steps that take place during stomatal development (Fig. 1A).
BROADLY EXPRESSED BHLH PROTEINS BIND TO CELL TYPE-SPECIFIC ONES
The bHLH family is defined by two functionally distinct regions (Littlewood and Evan, 1998). The helix-loop-helix region is located at the C-terminal end of the domain and is constituted mainly of hydrophobic residues. This region adopts a helix-loop-helix conformation in which two amphipathic α-helices are separated by an intervening loop of variable length, and it is required for dimerization with a partner of the same family. The basic region, with a large number of basic residues, is located at the N-terminal end, and it provides the contact points for an appropriate DNA target. In general, outside of the conserved bHLH domain, these proteins exhibit considerable sequence divergence (Atchley et al., 1999). Dimerization is a prerequisite for binding of bHLH-containing proteins to DNA (Murre et al., 1989a, 1989b; Davis et al., 1990). Some bHLH proteins form homodimers or restrict their heterodimerization activity to related members of the family (Littlewood and Evan, 1998). The core DNA sequence motif recognized by the bHLH proteins is a consensus hexanucleotide sequence known as the E-box (5′-CANNTG-3′) and identified for the first time in the immunoglobulin enhancers (Church et al., 1985).
Kanaoka et al. (2008) have made an in-depth study into the physical interaction among the bHLH proteins that control stomatal development. Bimolecular fluorescence complementation assays have shown that MUTE and FAMA strongly heterodimerize with SCRM and SCRM2. SPCH also associates with these broadly expressed proteins, but it shows only a weak interaction. In addition, with the exception of MUTE, these proteins do not form homodimers. The yeast two-hybrid system has confirmed that MUTE, FAMA, and SPCH physically associate with SCRM. The possible interactions between SCRM2 and cell type-specific bHLH factors in yeast have not been investigated. In summary, both yeast two-hybrid and bimolecular fluorescence complementation assays have shown that, in general, the cell type-specific bHLH proteins form heterodimers with the broadly expressed ones.
Yeast two-hybrid screening allowed the identification of two broadly expressed proteins, bHLH071 and bHLH093, as possible partners of FAMA (Ohashi-Ito and Bergmann, 2006). Bimolecular fluorescence complementation confirmed that these interactions take place in planta (Ohashi-Ito and Bergmann, 2006). It is unknown whether they interact with the remaining stomatogenic bHLH proteins. Loss-of-function mutations in bHLH071 and bHLH093 genes produce no obvious phenotype; however, the overexpression of either gene produces a weak fama phenotype (Ohashi-Ito and Bergmann, 2006). Either bHLH71 or bHLH093 overexpression may have titrated out an available pool of FAMA interfering with the formation (or function; see next paragraph) of biologically functional SCRM-FAMA or SCRM2-FAMA heterodimers.
It is likely that upon dimerization, these heterodimers with their basic regions recognize and bind to specific E-boxes and lead to transcriptional regulation of their target genes. Certainly, it is known that SCRM (ICE) binds to the CANNTG motif (Chinnusamy et al., 2003). In addition, SPCH, MUTE, and FAMA all contain the conserved residues H-E-R in their putative DNA-binding domains, which are present in proteins that bind DNA at the E-box (Fig. 3B; Shimizu et al., 1997). SCRM and SCRM2 contain a variant of such residues (N-E-R; Fig. 3B), which most probably also reflects binding to this conserved sequence. Both bHLH071 and bHLH93 also contain these conserved residues (H-E-R in bHLH071 and N-E-R in bHLH93; Fig. 3B). This suggests that in plants overexpressing either bHLH71 or bHLH093, the hypothetical bHLH71-FAMA or bHLH93-FAMA heterodimers might successfully compete with SCRM-FAMA and/or SCRM2-FAMA for binding to specific E-boxes, preventing FAMA action.
PARALLELS BETWEEN STOMATAL AND MUSCLE DEVELOPMENT
Members of the bHLH family of transcription factors have also been shown to regulate the determination and differentiation of a variety of cell types, including skeletal muscle, neurons, and hematopoietic cells. Some parallels between muscle and stomatal development were previously highlighted (MacAlister et al., 2007; Pillitteri and Torii, 2007); however, the recent implication of SCRM and SCRM2 in stomatal development and their interaction with SPCH, MUTE, and FAMA reflect that a similar mechanism guides muscle and stomatal fate. In muscle development, an interplay of both tissue-specific bHLH regulators (MyoD family, which includes MyoD, myogenin, Myf5, and MRF4) and non-tissue-restricted bHLH factors (E-like proteins) acts at multiple points to establish myoblast identity and control terminal differentiation (Fig. 1B; Lassar et al., 1991; Weintraub, 1993). In concert with E-like proteins, MyoD and Myf5 specify myoblast state, myogenin initiates myotube differentiation, and MRF4 acts in a later differentiation state producing mature muscle (Fig. 1B). Similar to SPCH, MUTE, and FAMA, the members of the MyoD family exhibit sequential expression patterns (for review, see Buckingham, 1992; Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). E-like proteins regulate wide varieties of developmental processes and pathogenesis. Similarly, SCRM controls not only stomatal development but also cold tolerance (Chinnusamy et al., 2003; Kanaoka et al., 2008). The four members of the MyoD family cluster in the same clade, which is distinct from the E-like protein clade (Fig. 3A).
The formation of heterodimers between regulators with restricted expression and those that exhibit a broad transcription not only affects stomatal development. MyoD family members also function predominantly as heterodimers with the E-like proteins, which include E12, E47, and HEB (Murre et al., 1989b; Hu et al., 1992). Myogenic bHLH factors can homodimerize, but the resulting complex can neither bind to nor activate muscle-specific genes (Murre et al., 1989a; Lassar et al., 1991). MUTE also forms homodimers (Kanaoka et al., 2008); the role of such complexes is unknown.
Myogenic heterodimers recognize and bind to the E-box consensus sequence (CANNTG) in gene muscle promoters and enhancers (Olson, 1990; Weintraub et al., 1991; Rudnicki and Jaenisch, 1995). E-boxes have been identified in promoters and enhancers of many skeletal muscle-specific structural genes, where they are required for activation by myogenic bHLH factors (Wentworth et al., 1991; Bessereau et al., 1993; Li and Capetanaki, 1994). As stated previously, it is known that SCRM binds to the CANNTG motif (Chinnusamy et al., 2003). In addition, SPCH, MUTE, FAMA, and bHLH071 all contain the conserved residues H-E-R in their putative DNA-binding domains (Fig. 3B), which are present in proteins that bind DNA at the E-box (Shimizu et al., 1997). Mutations in these residues in FAMA resulted in a nonfunctional protein (Ohashi-Ito and Bergmann, 2006). It seems that both myogenic and stomatogenic heterodimers bind to similar sequences, the E-boxes.
It is known that the activity of bHLH myogenic factors is regulated by phosphorylation/dephosphorylation events. For example, p38 mitogen-activated protein kinase phosphorylates MRF4, modulating its transcriptional activity (Suelves et al., 2004). The p38 mitogen-activated protein kinase also phosphorylates E47, which promotes MyoD/E4 association and muscle-specific transcription (Lluís et al., 2005). An elegant set of experiments showed that MPK3 and MPK4, which negatively regulate stomatal development (Wang et al., 2007), phosphorylate SPCH in vitro and modulate its activity in vivo (Lampard et al., 2008). So phosphorylation events regulate the activity of both myogenic and stomatogenic bHLH regulators.
In addition, both auto-regulatory and cross-regulatory interactions among bHLH factors have been demonstrated during muscle development (Braun et al., 1989; Thayer et al., 1989; Miner and Wold, 1990). For example, MyoD positively autoregulates its own expression and myogenin and MyoD regulate each other's expression (Thayer et al., 1989). Ohashi-Ito and Bergmann (2006) demonstrated that FAMA acts as a transcriptional activator. However, it seems that FAMA is not required to activate its own expression, as shown by the fact that the FAMA promoter is activated in fama-1 cells (Ohashi-Ito and Bergmann, 2006). Reverse transcription-PCR analysis showed that MUTE is not required to activate its own transcription (Pillitteri et al., 2007). In contrast, SPCH positively autoregulates its own transcription (MacAlister et al., 2007; Pillitteri et al., 2007). Reverse transcription-PCR analysis also showed that SCRM activates its own expression (Kanaoka et al., 2008). SCRM2 also seems required to maintain its wild-type expression levels (Kanaoka et al., 2008). Interestingly, no SCRM∷GUS expression has been detected in the spch epidermis (Kanaoka et al., 2008). In addition, the scrm scrm2 double loss-of-function mutant does not express the SPCH gene (Kanaoka et al., 2008). This indicates that SPCH and SCRM/SCRM2 regulate each other's expression. SPCH is also required to maintain the wild-type expression levels of FAMA and MUTE (MacAlister et al., 2007; Pillitteri et al., 2007). However, at least MUTE is not required to activate SPCH expression (Pillitteri et al., 2007). SCRM and SCRM2 are also required to activate both MUTE and FAMA expression (Kanaoka et al., 2008).
Furthermore, the forced expression of any of four bHLH genes from the MyoD family induces muscle differentiation in the transfected cell (Emerson, 1990; Weintraub et al., 1991; Buckingham, 1992; Weintraub, 1993; Olson and Klein, 1994), much as the ectopic expression of either MUTE or FAMA induces stomata or guard cell formation (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). In contrast, ectopic expression of SPCH increases the number of cells that initiate stomatal development, although they seem to arrest their development because stomata are not formed (MacAlister et al., 2007; Pillitteri et al., 2007).
Myoblast differentiation or division is dictated by a balance of opposing signals controlled by myogenic bHLH proteins and peptide growth factors. The forced expression at high levels of the myogenic factors can induce cell cycle withdrawal and initiate myogenesis (Crescenzi et al., 1990; Sorrentino et al., 1990). This is similar to those occurring in plants overexpressing FAMA, which, in addition to activating guard cell formation, also play a role in regulating the exit from the cell cycle (Ohashi-Ito and Bergmann, 2006). It is likely then that these bHLH proteins modulate cell cycle regulatory protein activity in a similar fashion in both animal and plant cells.
In spite of these strong similarities between these two systems, some differences have been found. Although myogenic bHLH factors are functionally interchangeable in gain-of-function studies (Wang and Jaenisch, 1997; Zhu and Miller, 1997), neither the expression of FAMA nor the expression of MUTE from the SPCH promoter substituted for SPCH function.
CONCLUDING REMARKS
Several recent findings have shown that myogenesis and stomatal development share not only a set of similar bHLH genes but also a common underlying mechanism. An intriguing property of the myogenic factors is their ability to self-regulate and activate the expression of other myogenic factors. Such regulatory interactions seem to reinforce the decision to differentiate and confer stability to the phenotype. An important challenge for the future will be to complete the possible cross-regulation and auto-regulation among the bHLH factors that guide stomatal development. An understanding of how these regulators control the cell cycle machinery ultimately to establish stomatal differentiation will also be important. The study of these problems will allow the similarities between muscle and stomatal development to be extended and, most importantly, some differences to be unraveled.
This work was supported by the Communities Council of Castilla-La Mancha (grant no. PCI08–0041–1136) and the Ministry of Science and Innovation of Spain (grant no. BIO2008–02149).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Laura Serna (laura.serna@uclm.es).
References
- Atchley WR, Therhalle W, Dress A (1999) Positional dependence, cliques and predictive motifs in the bHLH protein domain. J Mol Evol 48 501–516 [DOI] [PubMed] [Google Scholar]
- Barton MK (2007) Making holes in leaves: promoting cell state transitions in stomatal development. Plant Cell 19 1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D, Altmann T (2000) A subtilisin-like protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD (2007) Stomatal development. Annu Rev Plant Biol 58 163–181 [DOI] [PubMed] [Google Scholar]
- Bessereau JL, Mendelzon D, LePoupon C, Fiszman M, Changeux JP, Piette J (1993) Muscle-specific expression of the acetylcholine receptor alpha-subunit gene requires both positive and negative interactions between myogenic factors, Sp1 and GBF factors. EMBO J 12 443–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Bober E, Buschhausen-Denker G, Kohtz S, Grzeschik KH, Arnold HH, Kotz S (1989) Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J 8 3617–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M (1992) Making muscle in mammals. Trends Genet 8 144–148 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Ephrussi A, Gilbert W, Tonegawa S (1985) Cell-type-specific contacts to immunoglobulin enhancers in nuclei. Nature 313 798–801 [DOI] [PubMed] [Google Scholar]
- Crescenzi M, Fleming TP, Lassar AB, Weintraub H, Aaronson SA (1990) MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci USA 87 8442–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Cheng PF, Lassar AB, Weintraub H (1990) The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60 733–746 [DOI] [PubMed] [Google Scholar]
- Emerson CP (1990) Myogenesis and developmental control genes. Curr Opin Cell Biol 2 1065–1075 [DOI] [PubMed] [Google Scholar]
- Gray JE (2007) Plant development: three steps for stomata. Curr Biol 17 R213–R215 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JS, Olson EN, Kingston RE (1992) HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol 12 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Schöb H, Stadler M, Meins F Jr, Si-Ammour A (2007) MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, Lee EK, Nakagawa T, Zhao L, Geisler M, Sack FD (2005) The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal lineage. Plant Cell 17 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, MacAlister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H (1991) Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66 305–315 [DOI] [PubMed] [Google Scholar]
- Li H, Capetanaki Y (1994) An E box in the desmin promoter cooperates with the E box and MEF-2 sites of a distal enhancer to direct muscle-specific transcription. EMBO J 13 3580–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Evan GI (1998) Basic Helix-Loop-Helix Transcription Factors. Oxford University Press, Oxford
- Lluís F, Ballestar E, Suelves M, Esteller M, Muñoz-Cánoves P (2005) E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J 24 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC (2007) Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445 537–540 [DOI] [PubMed] [Google Scholar]
- Miner JH, Wold B (1990) Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc Natl Acad Sci USA 87 1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D (1989. a) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56 777–783 [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al (1989. b) Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58 537–544 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 1697–1700 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN (1990) MyoD family: a paradigm for development? Genes Dev 4 1454–1461 [DOI] [PubMed] [Google Scholar]
- Olson EN, Klein WH (1994) bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev 81 1–8 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Bogenschutz NL, Torii KU (2008) The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol 49 934–943 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU (2007) Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays 29 861–870 [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Jaenisch R (1995) The MyoD family of transcription factors and skeletal myogenesis. Bioessays 17 203–209 [DOI] [PubMed] [Google Scholar]
- Serna L (2007) bHLH proteins know when to make a stoma. Trends Plant Sci 12 483–485 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T (1997) Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J 16 4689–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293 [DOI] [PubMed] [Google Scholar]
- Sorrentino V, Pepperkok R, Davis RL, Ansorge W, Philipson L (1990) Cell proliferation inhibited by MyoD1 independently of myogenic differentiation. Nature 345 813–815 [DOI] [PubMed] [Google Scholar]
- Suelves M, Lluís F, Ruiz V, Nebreda AR, Muñoz-Cánoves P (2004) Phosphorylation of MRF4 transactivation domain by p38 mediates repression of specific myogenic genes. EMBO J 23 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E (2006) Plant Physiology, Ed 4. Sinauer Associates, Sunderland, MA
- Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H (1989) Positive autoregulation of the myogenic determination gene MyoD1. Cell 58 241–248 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jaenisch R (1997) Myogenin can substitute for Myf5 in promoting myogenesis but less efficiently. Development 124 2507–2513 [DOI] [PubMed] [Google Scholar]
- Weintraub H (1993) The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75 1241–1244 [DOI] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Rapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al (1991) The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251 761–766 [DOI] [PubMed] [Google Scholar]
- Wentworth BM, Donoghue M, Engert JC, Berglund EB, Rosenthal N (1991) Paired MyoD-binding sites regulate myosin light chain gene expression. Proc Natl Acad Sci USA 88 1242–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Miller JB (1997) MRF4 can substitute for myogenin during early stages of myogenesis. Dev Dyn 209 233–241 [DOI] [PubMed] [Google Scholar]