Abstract
Membrane trafficking plays a fundamental role in eukaryotic cell biology. Of the numerous known or predicted protein components of the plant cell trafficking system, only a relatively small subset have been characterized with respect to their biological roles in plant growth, development, and response to stresses. In this study, we investigated the subcellular localization and function of an Arabidopsis (Arabidopsis thaliana) small GTPase belonging to the RabE family. RabE proteins are phylogenetically related to well-characterized regulators of polarized vesicle transport from the Golgi apparatus to the plasma membrane in animal and yeast cells. The RabE family of GTPases has also been proposed to be a putative host target of AvrPto, an effector protein produced by the plant pathogen Pseudomonas syringae, based on yeast two-hybrid analysis. We generated transgenic Arabidopsis plants that constitutively expressed one of the five RabE proteins (RabE1d) fused to green fluorescent protein (GFP). GFP-RabE1d and endogenous RabE proteins were found to be associated with the Golgi apparatus and the plasma membrane in Arabidopsis leaf cells. RabE down-regulation, due to cosuppression in transgenic plants, resulted in drastically altered leaf morphology and reduced plant size, providing experimental evidence for an important role of RabE GTPases in regulating plant growth. RabE down-regulation did not affect plant susceptibility to pathogenic P. syringae bacteria; conversely, expression of the constitutively active RabE1d-Q74L enhanced plant defenses, conferring resistance to P. syringae infection.
Eukaryotic cells are compartmentalized by membranes that surround organelles having specific functions. Communication and transport between these membrane-bound compartments are vital to the cell and are accomplished through complex and tightly regulated pathways. Trafficking pathways and their players have been extensively described in yeast and mammalian cells but are still poorly characterized in plants. The main effectors and regulators of these pathways appear to be shared between all eukaryotes. Among these, Rabs are a group of small monomeric GTPases that act as molecular switches to mediate vesicle transport between membrane-bound cellular compartments (Segev, 2001). Rab GTPases participate in vesicle budding from a donor compartment, transport along the cytoskeleton toward a target compartment, and, eventually, tethering and fusion of the vesicles with the target membrane. Like other small GTPases, Rabs alternate between a GTP-bound active form and a GDP-bound inactive form. Their functional specificity is determined, in part, by their unique subcellular distribution (Stenmark and Olkkonen, 2001; Zerial and McBride, 2001). The Arabidopsis (Arabidopsis thaliana) genome encodes 57 Rab proteins, divided into eight subfamilies (RabA to RabH) based on sequence similarities (Rutherford and Moore, 2002; Vernoud et al., 2003). Recent studies have indicated that members of various plant Rab subfamilies participate in secretion and recycling of cell wall components (Nielsen et al., 2008), apical growth of pollen tubes (de Graaf et al., 2005; Cheung and de Vries, 2008; Cheung and Wu, 2008) and root hair (Preuss et al., 2004, 2006), formation of the phragmoplast in dividing cells (Chow et al., 2008), endocytosis (Ueda et al., 2004), and potentially osmotic stress tolerance (Mazel et al., 2004).
Fungal and bacterial infections in plants are often associated with the activation (or suppression) of extracellular defense responses, including secretion of defense-related proteins and antimicrobial phytoalexins into the apoplast, and the formation of callose-rich cell wall appositions, known as papillae (Snyder and Nicholson, 1990; Snyder et al., 1991; Brown et al., 1995; Soylu et al., 2005; Field et al., 2006). Deployment of such defenses is expected to be dependent on vesicle trafficking and secretion (Huckelhoven, 2007; Lipka et al., 2007; Robatzek, 2007). Delivery of innate immune receptors to the plasma membrane and ligand-induced immune receptor endocytosis also play a role in plant response to pathogen-derived elicitors (Robatzek et al., 2006). Activation and integrity of the secretory pathway have been shown to be critical for resistance against potential pathogens (Collins et al., 2003; Assaad et al., 2004; Wang et al., 2005; Kalde et al., 2007). However, successful pathogens have evolved virulence effectors that inactivate vesicle traffic-associated regulators (Nomura et al., 2006; Robatzek, 2007). Detailed molecular mechanisms underlying vesicle trafficking leading to defense, and the specific cargos transported by these vesicles, have yet to be elucidated.
Pseudomonas syringae pv tomato strain DC3000 (Pst DC3000) is a Gram-negative bacterium that causes bacterial speck of tomato (Solanum lycopersicum) and Arabidopsis. Like other Gram-negative bacterial pathogens of plants and animals, Pst DC3000 delivers virulence effector proteins directly into the host cell via the type III secretion system (TTSS; Buttner and Bonas, 2003; Alfano and Collmer, 2004; He et al., 2004). These proteins, collectively called TTSS effectors, alter host cellular processes to ultimately favor pathogen growth and promote disease. To carry out virulence functions, bacterial effectors of plant and animal pathogens interact with and often biochemically modify key regulatory components of basic host cellular functions. A major virulence activity of TTSS effectors of P. syringae seems to be suppression of host defenses (Nomura et al., 2005; Abramovitch et al., 2006; Desveaux et al., 2006; Bray Speth et al., 2007; Block et al., 2008). For example, the Pst DC3000 TTSS effector AvrPto compromises Arabidopsis basal defenses and mitogen-activated protein kinase signaling and promotes Arabidopsis susceptibility to nonpathogenic bacteria such as TTSS-defective mutants and to nonhost pathogens such as P. syringae pv phaseolicola (Hauck et al., 2003; He et al., 2006). Subsequent studies show that AvrPto physically interacts with the flagellin receptor FLS2 (Xiang et al., 2008) and with its signaling partner BAK1 (Chinchilla et al., 2007; Shan et al., 2008). These results suggest that AvrPto could directly interfere with microbe-associated molecular pattern signaling, thereby blocking downstream plant basal defense responses.
A previous yeast two-hybrid (Y2H) screening of a tomato cDNA library for AvrPto-interacting proteins yielded two small GTPases, named Api2 and Api3 (Bogdanove and Martin, 2000), which are similar to the mammalian protein Rab8 (Huber et al., 1993). Rab8 and its yeast orthologues, Ypt2 of Schizosaccharomyces pombe (Craighead et al., 1993) and Sec4p of Saccharomyces cerevisiae (Goud et al., 1988), are extensively characterized regulators of polarized vesicle transport from the trans-Golgi network to specific regions of the plasma membrane (PM). The closest homologues of Rab8 in Arabidopsis are the five members of the RabE family of GTPases (Rutherford and Moore, 2002; Vernoud et al., 2003; Supplemental Fig. S1). As described in “Results,” we independently isolated a RabE GTPase in a Y2H screen for AvrPto-interacting Arabidopsis proteins.
The identification of Rab GTPases as AvrPto interactors in both tomato and Arabidopsis suggested that, as part of its virulence mechanism, this effector may perturb intracellular vesicle trafficking in the plant (Bogdanove and Martin, 2000). Interestingly, small GTPases regulating cytoskeleton dynamics and membrane trafficking are among the most common host targets of TTSS effectors produced by bacterial pathogens of animals (Harrison et al., 2004; Machner and Isberg, 2006; Murata et al., 2006; Rzomp et al., 2006; Smith et al., 2007). The biological roles of tomato Api2 and Api3 and Arabidopsis RabE GTPases in plant development and defense are unknown. Localization and function of RabE proteins have only recently begun to be investigated. Arabidopsis yellow fluorescent protein-RabE1d, transiently expressed in heterologous tobacco (Nicotiana tabacum) epidermal cells, was detected in the Golgi apparatus and in the cytoplasm, and functional analyses indicated that this protein participates in promoting post-Golgi secretory trafficking (Zheng et al., 2005). To gain further insights into the RabE GTPase subcellular localization in native Arabidopsis cells and their function in growth, development, and/or defense at the whole plant level, we generated transgenic plants constitutively expressing GFP-fused wild-type RabE1d and RabE1d-Q74L (predicted to be preferentially GTP bound and therefore in an active state) and examined these plants for morphological and developmental phenotypes and response to P. syringae infection.
RESULTS
Identification of Arabidopsis RabE Proteins as Y2H Interactors of AvrPto
We conducted a Y2H screening of two separate Arabidopsis cDNA libraries, using AvrPto as bait. Several AvrPto-interacting Arabidopsis proteins were identified, including a member of the RabE family of small GTPases (At5g59840), a putative cytoplasmic kinase (At4g11890), an auxin signaling repressor, IAA7 (At3g23050), two hypothetical proteins (At3g26600 and At5g16840), and several putatively chloroplast- or mitochondria-targeted proteins. The interaction with the small GTPase RabE was particularly interesting because RabE was predicted to be membrane localized, as is AvrPto (Shan et al., 2000; He et al., 2006), and, as mentioned above, because screening of a tomato cDNA library had also yielded two AvrPto-interacting proteins homologous to Rab8 (Bogdanove and Martin, 2000), the mammalian counterpart of Arabidopsis RabE.
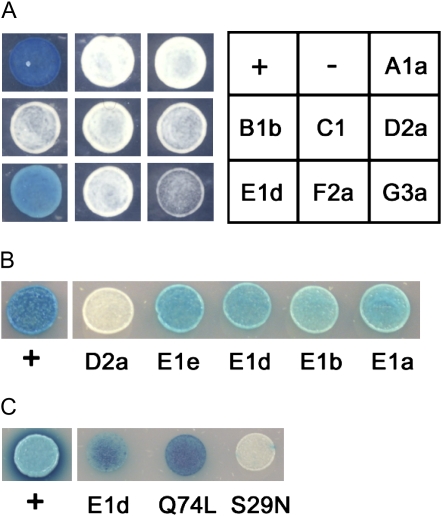
To characterize the specificity of the AvrPto-RabE interaction, we investigated whether AvrPto interacts with other members of the RabE family and with other Rab proteins. Of the other Arabidopsis Rab protein families (i.e. RabA to -D, -F, and -G), we cloned and expressed in yeast RabA1a, -B1b, -C1, -D2a, -F2a, and -G3a; none of these representatives interacted with AvrPto in the Y2H system (Fig. 1A). Of the five RabE genes, all but RabE1c were successfully cloned and expressed in yeast. All four RabE proteins tested (RabE1a, -b, -d, and -e) interacted with AvrPto (Fig. 1B). Thus, it appears that AvrPto interacts specifically with the Arabidopsis RabE family of GTPases.
Figure 1.
Arabidopsis RabE proteins interact with P. syringae AvrPto in the Y2H system. Interaction is visualized by development of blue color on medium containing X-Gal. Negative control (−) = pNLexA (bait) and pB42AD (prey) vectors; positive control (+) = pLexA-A53 and pB42AD-T. A, AvrPto (in pNLexA) interacts with Arabidopsis RabE1d but not with other members of the Rab superfamily (in pB42AD). B, AvrPto (in pNLexA) interacts with all four tested Arabidopsis RabE proteins (in pB42AD). Yeast expressing AvrPto in pNLexA and RabD2a in pB42AD is shown as a negative control. C, AvrPto (in pB42AD) interacts with RabE1d or RabE1d-Q74L but not with RabE1d-S29N. For this experiment only, wild-type RabE1d and the mutated RabE1d proteins were cloned in the bait vector pGILDA and modified by replacing the two C-terminal conserved Cys residues (sites of geranylgeranylation [Supplemental Fig. S1]) with Gly and Ser to prevent prenylation and membrane association.
Small GTPases normally cycle between the active GTP-bound and the inactive GDP-bound states. Mutation of highly conserved residues can be used to alter the nucleotide binding and hydrolysis activities of Rab proteins (Nielsen et al., 2008; Supplemental Fig. S1). The mutant forms RabE1d-S29N (predicted to be GDP bound) and RabE1d-Q74L (predicted to be GTP bound) were engineered. In the Y2H system, AvrPto interacted only with wild-type RabE1d or with GTP-restricted RabE1d-Q74L but not with GDP-restricted RabE1d-S29N (Fig. 1C), suggesting that AvrPto preferably binds to the active form.
RabE Gene Expression in Arabidopsis Leaves
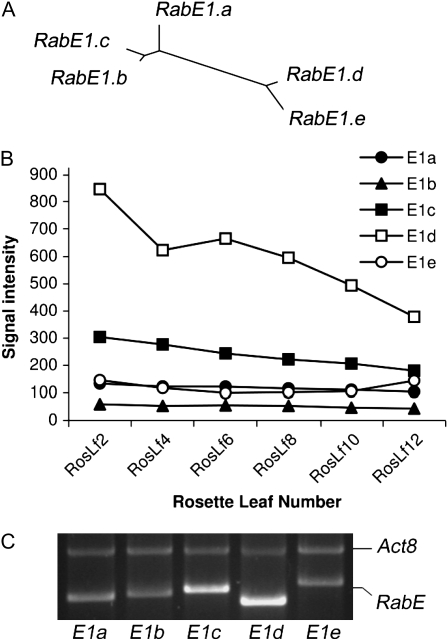
Analysis of the Arabidopsis genome showed that 44 of the 57 Rab genes reside in duplicated regions. Of the five RabE genes, RabE1d and -E1e appear to be derived from a major duplication event between chromosomes III and V, and the same holds true for RabE1b and -E1c (Rutherford and Moore, 2002; Fig. 2A). The high degree of sequence identity among the five RabE proteins, equal to or higher than 86%, suggests functional redundancy. However, members of a gene family could be preferentially expressed in different tissues, at specific developmental stages, or in response to stresses. We used the Genevestigator Gene Atlas Tool (Zimmermann et al., 2004) to investigate whether this is the case with the five RabE genes. The in silico analysis revealed that all five genes are expressed in all Arabidopsis tissues and developmental stages. RabE1d and -E1e (encoding 94% identical proteins) are the only two gene family members whose expression is much lower in pollen than in all other tissues. Based on publicly available microarray data (Genevestigator; Zimmermann et al., 2004), RabE1d appeared to be the most highly expressed RabE gene in Arabidopsis ecotype Columbia (Col-0) rosette leaves, followed by RabE1c. The RabE1a, -E1e, and -E1b genes had the lowest expression levels (Fig. 2B). We conducted reverse transcription (RT)-PCR analysis on rosette leaf total RNA and confirmed a high expression level for RabE1d (Fig. 2C).
Figure 2.
A, Phylogenetic relationship among RabE family members; E1d and E1e, as well as E1b and E1c, are predicted to be the result of genome duplication events. B, E-northern of RabE gene expression in Arabidopsis Col-0 rosette leaves at different developmental stages. Data were obtained from AtGenExpress at the Genevestigator site (https://www.genevestigator.ethz.ch/); signal intensities were averaged across three biological replicates. C, RT-PCR showing RabE gene expression in rosette leaves. RT-PCR products represent the five RabE genes, reverse transcribed and amplified from Arabidopsis Col-0 gl1 rosette leaf total RNA. A single RT reaction was performed, and equal amounts of the resulting cDNA were used as template in PCR. Each PCR sample contained primers for the Actin8 gene in addition to primers for one of the RabE genes (primers are listed in Supplemental Table S2). Equal volumes of each PCR product were loaded on a 1% agarose gel.
To investigate potential up- or down-regulation of the RabE family members in response to pathogens or other stresses, we analyzed the expression patterns of the RabE genes with the AtGenExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp; Schmid et al., 2005) across several publicly available microarray data sets. None of the five genes appeared to be significantly (more than 2.5-fold) up- or down-regulated in response to pathogens or elicitors. In the absence of any expression-based indication of whether the five RabE proteins have redundant or distinct functions, we chose the highly expressed RabE1d (At5g03520) for further study.
RabE1d Is Associated Not Only with the Golgi Apparatus But Also with the Plasma Membrane in Arabidopsis Leaf Cells
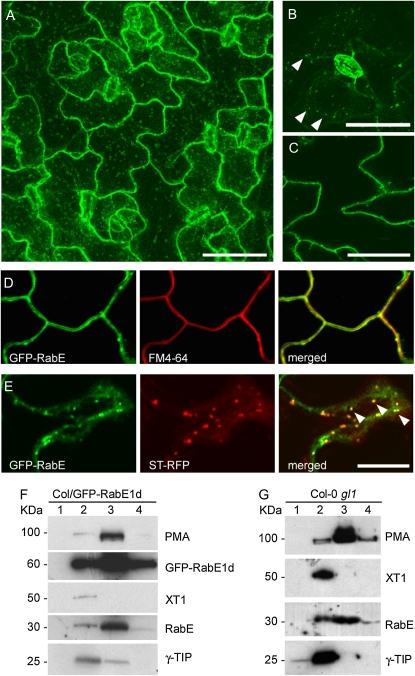
All Rab proteins are normally present in cells in two pools, one cytoplasmic and the other membrane associated (Novick and Brennwald, 1993). Nucleotide-binding state and interaction with accessory proteins determine whether a Rab is in the cytosol or the membrane at any given time. A hallmark feature of Rab proteins is that they localize to the specific membrane compartments in which they function. It was previously reported that Arabidopsis RabE1d, when transiently and heterologously expressed in tobacco epidermal cells as a fusion with yellow fluorescent protein, was detected in the Golgi apparatus and in the cytoplasm (Zheng et al., 2005). To determine RabE localization in native Arabidopsis cells, we created stable transgenic plants that express RabE1d fused with enhanced GFP under the control of the cauliflower mosaic virus 35S promoter. By convention, GFP was fused to the N terminus of RabE1d to preserve the C-terminal CAAX geranylgeranylation site that is critical for membrane association and function (Calero et al., 2003; Supplemental Fig. S1). Several independent transgenic lines were analyzed by confocal laser scanning microscopy (CLSM). GFP fluorescence was observed not only in intracellular punctate structures, consistent with the Golgi apparatus, as detected in heterologous tobacco cells (Zheng et al., 2005), but also prominently at the cell periphery (Fig. 3, A–E).
Figure 3.
GFP-RabE1d localization in Arabidopsis leaf cells. Confocal microscopy images of the leaf epidermis of transgenic Arabidopsis plants expressing GFP-RabE1d are shown. A, Projection along the z axis of several optical planes intersecting the leaf epidermal cell layer. GFP-RabE1d is visible in intracellular punctate structures and at the cell periphery. B, Single focal plane intersecting a stomate and the cortical layer of epidermal cells. Arrowheads point at intracellular punctate structures. C, Single focal plane intersecting epidermal cells. The same pattern of distribution of GFP-RabE1d was observed in at least five independent transgenic lines. Bars = 50 μm. D and E, Confocal microscopy images illustrating GFP-RabE1d colocalization with the FM4-64 dye at the plasma membrane (D) and with sialyl transferase (ST-RFP) at the Golgi apparatus (E). D, Single focal plane crossing adjacent epidermal cells (40× oil-immersion objective, 4× zoom). Left, GFP-RabE1d fluorescence, green; center, FM4-64 fluorescence, red; right, merged image, in which the yellow color results from the overlap of red and green. E, Single focal plane crossing the cytoplasm of a cell (40× oil-immersion objective, 4× zoom). Left, GFP-RabE1d fluorescence, green; center, ST-RFP fluorescence, red; right, merged image, in which the yellow color results from the overlap of red and green. Arrowheads point at some of the colabeled Golgi stacks in the merged image. Bars = 10 μm. F and G, Western blots of cellular membrane fractions separated by ultracentrifugation on a Suc step gradient. Two different samples are shown: transgenic Arabidopsis overexpressing GFP-RabE1d (F) and nontransgenic Arabidopsis Col-0 gl1 (G). Lanes 1 through 4 represent the four membrane fractions collected at the interfaces between layers of different Suc concentrations: 18% to 25% (lane 1), 25% to 34% (lane 2), 34% to 40% (lane 3), and 40% to 50% (lane 4). Equal volumes of each fraction were loaded on SDS-PAGE gels. PM-ATPase (PMA) is a plasma membrane marker, XT1 is a trans-Golgi resident protein, and γ-TIP is a marker for the tonoplast. GFP-RabE1d and endogenous RabE proteins were detected with a polyclonal anti-RabE antibody (courtesy of Dr. K. Nomura).
Leaf epidermal cells typically contain a very large vacuole that accounts for most of the cell volume. Fluorescence detected at the cell periphery may represent the PM, the vacuolar membrane (tonoplast), or the thin layer of cytoplasm that is between the PM and the tonoplast. To more precisely determine whether GFP-RabE1d was also localized at the PM, we stained live leaf tissue with the lipophylic dye FM4-64 (Fischer-Parton et al., 2000; Bolte et al., 2004). FM4-64 produces a bright red fluorescence in membranes but not in aqueous solutions and is rapidly incorporated into the PM. It is often used in microscopy as an endocytic tracker, because it is retained in the portions of PM that are internalized by endocytosis (Ueda et al., 2001). Within the first 15 to 30 min after incorporation (depending on the system used), FM4-64 selectively stains the PM. CLSM analysis revealed a precise overlap of GFP-RabE1d fluorescence with FM4-64 fluorescence immediately after staining, indicating that RabE1d is associated with the plasma membrane (Fig. 3D).
To investigate whether the punctate structures labeled by GFP-RabE1d correspond to the Golgi apparatus, we examined colocalization with rat sialyl transferase, a Golgi marker protein (Wee et al., 1998), fused to DsRed (ST-RFP). ST-RFP was transiently expressed in the GFP-RabE1d transgenic leaves via particle bombardment. Observation of cells coexpressing GFP-RabE1d and ST-RFP revealed overlapping fluorescence signals, confirming RabE1d's association with the Golgi apparatus (Fig. 3E). Taken together, our analysis of GFP-RabE1d localization revealed that, in the native Arabidopsis leaf cells, membrane-associated RabE1d is found not only in the Golgi apparatus, as reported previously in heterologous tobacco cells, but also in the PM.
Endogenous RabE Proteins Cofractionate with PM and Golgi Markers
GFP-RabE1d expression in the transgenic plants was driven by the cauliflower mosaic virus 35S constitutive promoter. To exclude the possibility that the observed RabE localization reflects patterns of only the transgenically expressed protein, we conducted additional analyses to determine the localization of endogenous RabE in transgenic as well as wild-type Arabidopsis plants. We performed subcellular membrane fractionation by centrifugation of clarified plant extracts on Suc step gradients (Zeng and Keegstra, 2008). This method allowed separation of the PM from fractions containing lighter membranes (Golgi, tonoplast). Western-blot analysis with an anti-RabE polyclonal antibody that reacts with all RabE family members revealed that the bulk of membrane-associated endogenous RabE proteins, as well as transgenically expressed GFP-RabE1d, were in the same fraction as the PM marker H+-ATPase (Morsomme et al., 1998). A lower amount of endogenous RabE, as well as GFP-RabE1d, cofractionated with xylosyl transferase (XT1), a Golgi apparatus resident protein (Faik et al., 2002; Cavalier and Keegstra, 2006; Zeng and Keegstra, 2008; Fig. 3, F and G). A tonoplast marker, γ-tonoplast integral protein (γ-TIP), was found predominantly in the same fraction as the Golgi marker. Independent membrane fractionation experiments yielded similar results, although minor differences in protein levels and distribution among fractions were occasionally observed across western blots as a result of interexperiment variability. Overall, the membrane fractionation experiments complemented the live cell imaging results; together, they indicate that endogenous and ectopically expressed RabE proteins are not only localized at the Golgi apparatus but that the majority of membrane-associated RabE is in the PM in Arabidopsis leaf cells.
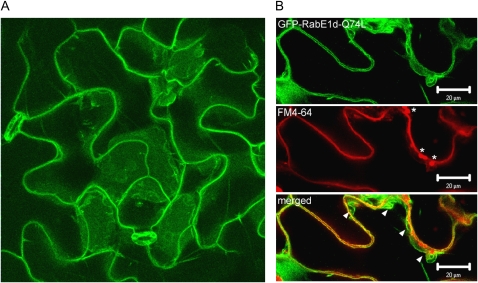
GTP-Restricted RabE1d-Q74L Displays Altered Subcellular Localization
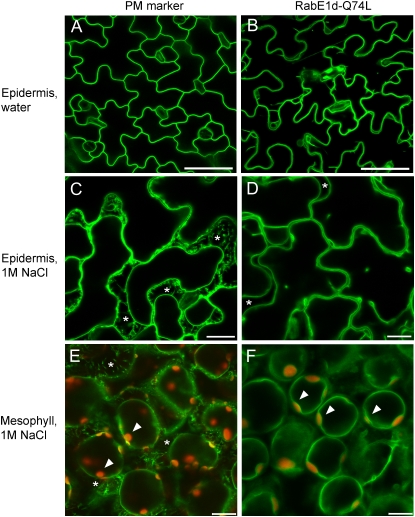
We also produced transgenic plants expressing RabE1d-Q74L, which is predicted to be constitutively active, as a GFP fusion. Unlike wild-type GFP-RabE1d, the GFP-RabE1d-Q74L fusion was not detected in intracellular punctate structures (i.e. Golgi) but was primarily found at the cell periphery (Fig. 4A). PM staining with FM4-64 revealed that the bulk of GFP-RabE1d-Q74L fluorescence did not overlap with the PM but was most likely localized in the tonoplast (Fig. 4B). We obtained from the Arabidopsis Biological Resource Center Arabidopsis transgenic lines expressing various endomembrane markers fused to GFP (Cutler et al., 2000). Comparison of GFP-RabE1d-Q74L fluorescence with that of either a PM-localized channel protein (line Q8) or a tonoplast marker (line Q5) revealed a localization pattern similar to that of the tonoplast, both in intact and in plasmolyzed leaf tissues (Supplemental Fig. S3). Plasmolysis in line Q8 resulted in the characteristic Hechtian strands (Oparka, 1994), typical of plasma membrane retracting from the cell wall; in mesophyll cells, the chloroplasts were clearly located on the inside of the GFP-labeled PM (Fig. 5). In contrast, plasmolyzed cells expressing GFP-RabE1d-Q74L did not display Hechtian strands, and chloroplasts in the mesophyll cells were distinctly on the outside of the GFP-labeled membrane (Fig. 5), further supporting RabE1d-Q74L localization in the tonoplast.
Figure 4.
Intracellular localization of the mutant GFP- RabE1d-Q74L protein. A, Confocal microscopy image of a representative Arabidopsis leaf expressing GFP-RabE1d-Q74L. Projection along the z axis of several focal planes crossing the epidermal cell layer. Several independent transgenic lines were analyzed with similar results. B, GFP-RabE1d-Q74L is mostly localized in the tonoplast. Leaves were stained with FM4-64, to visualize the plasma membrane, and immediately observed. The image represents a single focal plane (40× oil-immersion objective). Top, GFP fluorescence; middle, FM4-64 fluorescence (asterisks indicate autofluorescent chloroplasts in the mesophyll layer, below the epidermis); bottom, merged image (arrowheads indicate where the tonoplast is most clearly distinct from the plasma membrane). Invaginations and the formation of membranous structures are typical of the highly dynamic vacuolar membrane. Even in the areas where the plasma membrane and tonoplast are closest, green and red fluorescence are visibly distinct. Bars = 20 μm.
Figure 5.
Localization pattern of GFP-RabE1d-Q74L compared with the localization of a PM marker protein. A, C, and E, Arabidopsis expressing a GFP fusion to the PM integral protein PIP2A (line Q8). B, D, and F, Arabidopsis expressing GFP-RabE1d-Q74L. In C to F, samples were plasmolyzed in 1 m NaCl. Asterisks indicate areas between plasmolyzed cells; Hechtian strands are noticeable in C and E (PM) but not in D and F (RabE-Q74L). Arrowheads indicate chloroplast autofluorescence (red); chloroplasts are on the inside of the PM (E), while GFP-RabE1d-Q74L fluorescence is on the cytoplasmic side of the chloroplasts. All images are single focal planes. Bars = 50 μm (A and B), 20 μm (C), and 10 μm (D–F).
Challenge with P. syringae Bacteria Promotes Focal Accumulation of GFP-RabE1d
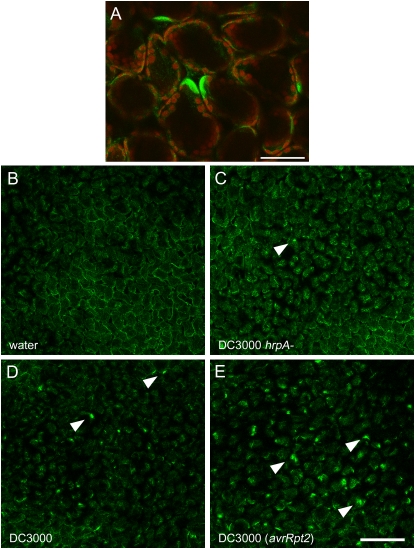
To investigate whether intracellular distribution of the RabE1d protein was perturbed in the presence of bacteria, we performed microscopic observation of transgenic GFP-RabE1d plants challenged with various strains of Pst DC3000. Leaves were syringe inoculated with bacteria at 1 × 108 colony-forming units (cfu) mL−1 and analyzed by CLSM at 5 to 6 h after inoculation. We observed that bacterial inoculation caused polarized GFP-RabE1d accumulation in mesophyll cells (Fig. 6A). Interestingly, whereas such a phenomenon was restricted to a few sparse cells in leaves challenged with wild-type virulent Pst DC3000 (Fig. 6D) or with the nonpathogenic TTSS-defective mutant hrpA− (Fig. 6C), focal accumulation of GFP-RabE1d was widespread in leaves inoculated with the avirulent strain Pst DC3000 (avrRpt2; Fig. 6E). These results suggest that the GFP-RabE1d subcellular localization is dynamic and can respond to bacterial infection, especially in a gene-for-gene interaction.
Figure 6.
Focal accumulation of GFP-RabE1d in response to bacteria. Leaves of transgenic GFP-RabE1d Arabidopsis were syringe inoculated with various strains of Pst DC3000 at a density of 1 × 108 cfu mL−1. A, Confocal microscopy observation at 6 h after inoculation revealed focal accumulation of the fluorescent GFP-RabE1d in mesophyll cells. Such accumulation is limited to a few cells in response to virulent and nonpathogenic bacteria, whereas it is widespread in response to avirulent bacteria, which trigger gene-for-gene resistance. Bar = 50 μm. B, No bacteria. C, Pst DC3000 hrpA− mutant bacteria, nonpathogenic. D, Wild-type Pst DC3000, virulent. E, Pst DC3000 (avrRpt2), avirulent. Three independent transgenic lines (A1, A5, and A14) yielded similar results. For B to E, all panels represent Z stacks of the same number of optical planes intersecting the leaf mesophyll (arrowheads indicate some of the points of accumulation). A second, independent set of images is shown in Supplemental Figure S4. Bar = 200 μm.
RabE1d Cosuppression in Transgenic Plants Resulted in Altered Plant Growth and Morphology
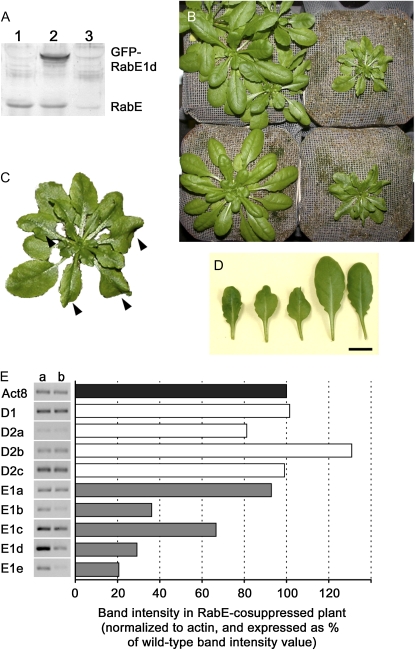
A large percentage of GFP-RabE1d transgenic Arabidopsis plants generated in this study showed cosuppression of the transgene and of endogenous RabE, as demonstrated by western-blot analysis (Fig. 7A). Severe reduction of the endogenous RabE protein level in transgenic plants invariably correlated with a distinct morphological phenotype (Fig. 7, B and C). Rosette leaves developed normally for the first 3 weeks, when Arabidopsis development is usually slower, and plants were indistinguishable from Col-0 plants. In the following 2 weeks, when Arabidopsis size increases more rapidly, the leaves of RabE-cosuppressed plants did not fully elongate (Fig. 7D); midribs remained short, while the leaf lamina continued to expand, producing a characteristic wavy phenotype. Mature (6- to 7-week-old) RabE-cosuppressed plants were significantly smaller than wild-type plants and had short midribs and petioles. RabE-cosuppressed plants flowered at the same time as wild-type Arabidopsis, although their stems were much shorter than wild-type stems and the plants produced fertile seeds. The progeny of selected silenced lines (B8, B11, and B13) maintained silencing and had the same phenotype as the parental plants.
Figure 7.
RabE cosuppression adversely affects Arabidopsis plant size and leaf morphology. A, Western blot (with anti-RabE antibody) illustrating how RabE-cosuppressed plants (lane 3) have a considerably lower amount of endogenous RabE protein compared with both the wild type (lane 1) and GFP-RabE1d overexpressors (lane 2). B, RabE-cosuppressed plants (top and bottom right) have smaller size and altered morphology compared with wild-type Arabidopsis (top left) and RabE1d-overexpressing plants (bottom left). C, Enlarged image of the RabE-cosuppressed plant in B (top right); arrowheads point to the wavy leaves. D, Size comparison of rosette leaves from 5-week-old RabE-cosuppressed (three left) and wild-type (two right) Arabidopsis plants. Bar = 10 mm. E, RT-PCR analysis of expression of the five RabE and four RabD genes in wild-type (lane a) and in RabE-cosuppressed (lane b) Arabidopsis plants. Equal volumes of the PCR samples were loaded on a 1% agarose gel. The gel was photographed with a Bio-Rad imager, and Quantity One software was used to quantify the bands. Intensity values, normalized to those of Actin8, are represented in the chart as percentages of the wild-type value.
Expression of the RabE gene family was analyzed by RT-PCR in the cosuppressed lines and compared with expression in wild-type plants. RT-PCR demonstrated that not all RabE gene family members are equally affected by cosuppression. RabE1d and RabE1e were knocked down most severely, followed by RabE1b and RabE1c. RabE1a showed only slight down-regulation (Fig. 7E). Given the high degree of sequence similarity among small GTPases of the Arabidopsis Rab superfamily, we tested whether other closely related Rabs were affected by silencing. The closest relatives of the RabE clade in Arabidopsis are the four RabD proteins (RabD1, -D2a, -D2b, and -D2c). RabD was previously characterized as a regulator of the early secretory pathway, being involved in transport from the endoplasmic reticulum to the Golgi (Batoko et al., 2000; Zheng et al., 2005). RT-PCR revealed that transcripts of all four RabD genes were present at similar levels in RabE1d-silenced plants and in wild-type Arabidopsis (Fig. 7E). Silencing in the transgenic plants, therefore, is specific and limited to the RabE genes, primarily RabE1d and -E1e.
RabE-Cosuppressed Plants Are Not Compromised in Resistance to Pst DC3000
We performed pathogenesis assays with Pst DC3000 to establish whether the partially RabE-cosuppressed plants are impaired in their defenses against P. syringae. Pst DC3000 consistently caused disease symptoms and multiplied on RabE-cosuppressed plants, reaching similar population levels as on wild-type Arabidopsis (Supplemental Fig. S2). In several instances, however, we observed that older (6- to 7-week-old) or environmentally stressed RabE-cosuppressed plants displayed some degree of basal resistance, possibly due to stress caused by RabE down-regulation (Supplemental Fig. S2). We did not observe a consistent and reproducible difference between wild-type Col-0 plants and RabE-cosuppressed plants in the multiplication of Pst DC3000 (avrRpt2) or a TTSS-defective mutant of Pst DC3000, the hrpA− mutant (Supplemental Fig. S2).
Transgenic Expression of RabE1d-Q74L Confers Resistance against Pst DC3000
GFP-RabE-Q74L plants did not exhibit significant alterations in growth and development, other than the appearance of minute sparse indentations in mature rosette leaves about 2 weeks prior to bolting. The origin and significance of these indentations are unclear; however, we have been unable to show, using trypan blue staining, that they develop as a consequence of cell death.
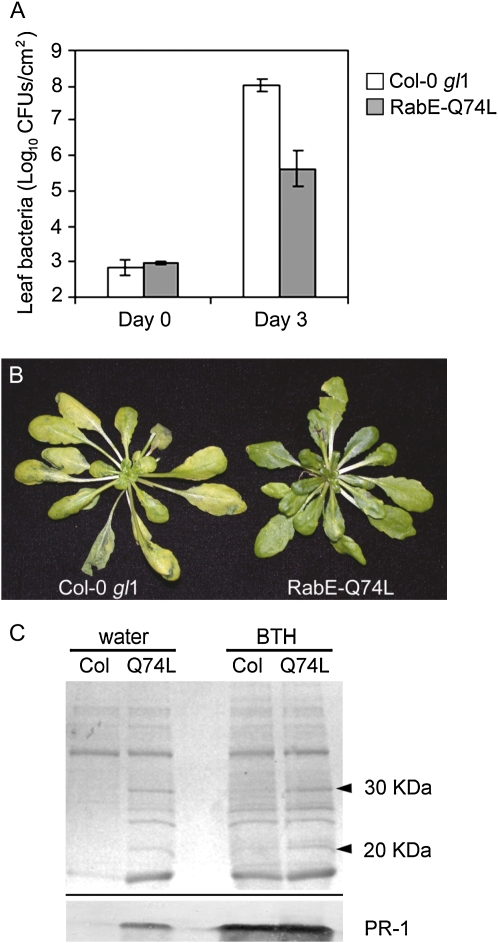
Although transgenic expression of constitutively active GFP-RabE1d-Q74L did not globally affect plant growth or development (as did RabE cosuppression), it had a remarkable effect on plant responses to P. syringae infection. Upon challenge with Pst DC3000, the GFP-RabE1d-Q74L-expressing plants displayed a considerable degree of resistance, reflected by bacterial multiplication being consistently restricted (10- to 100-fold) compared with multiplication on wild-type Arabidopsis (Fig. 8A). This observation was consistent across several experiments on different transgenic lines. Visible disease symptoms, namely chlorosis and necrosis, were also markedly reduced (Fig. 8B). This enhanced resistance phenotype apparently requires the constitutively active form of RabE1d, as transgenic expression of GFP-RabE1d did not result in enhanced resistance (Supplemental Fig. S2).
Figure 8.
Constitutive expression of RabE-Q74L confers resistance to Pst DC3000. A, Bacterial multiplication in GFP-RabE1d-Q74L-expressing plants (gray bars) compared with that in wild-type Arabidopsis (white bars). Pst DC3000 was syringe infiltrated at a density of 1 × 105 cfu mL−1. B, Disease symptoms 3 d after vacuum inoculation with Pst DC3000 at a density of 1 × 106 cfu mL−1. Left, Arabidopsis Col-0 gl1 (wild type); right, Arabidopsis expressing GFP-RabE-Q74L. C, Accumulation of extracellular proteins in plants expressing GFP-RabE1d-Q74L. Proteins in the IWF from wild-type (Col) and RabE1d-Q74L-expressing (Q74L) plants were separated by SDS-PAGE. Top, Coomassie Brilliant Blue-stained gel representing total proteins; the arrowheads indicate bands that seem to be exclusive to the Q74L plants. Bottom, Western blot with the anti-PR-1 antibody (a gift of Dr. X. Dong).
Constitutive Secretion of Extracellular Proteins in GFP-RabE1d-Q74L-Expressing Plants
Up-regulation of the secretory pathway was recently demonstrated in systemic acquired resistance (SAR; Wang et al., 2005), and plants undergoing SAR accumulate secreted proteins in the apoplast, including antimicrobial polypeptides (Uknes et al., 1992). Because RabE proteins are predicted to be involved in regulating secretory vesicle trafficking, the enhanced resistance to Pst DC3000 in GFP-RabE1d-Q74L-expressing plants could be caused by constitutive stimulation of defense-associated secretion. To test this possibility, Arabidopsis wild-type and GFP-RabE1d-Q74L-expressing plants were sprayed with benzothiadiazole (BTH), a synthetic activator known to trigger SAR in plants (Lawton et al., 1996), or with water as a control. Three days later, total protein secretion in the apoplast and secretion of the extracellular defense protein PR1 (for pathogenesis-related protein 1) were monitored. Intercellular wash fluid (IWF) collected from water-treated GFP-RabE1d-Q74L-expressing plants contained PR1 and several unknown proteins that were absent from the water-treated Arabidopsis wild-type IWF, indicating a constitutive activation of secretory and defense pathways. BTH application resulted in similar levels of secreted PR1 and several other proteins in the apoplast of both wild-type and transgenic plants (Fig. 8C). Interestingly, some protein bands were exclusively detected in the IWF of water- and BTH-treated RabE1d-Q74L-expressing plants but not in the BTH-treated wild-type plant IWF (Fig. 8C, arrowheads). These unique extracellular proteins associated with expression of RabE1d-Q74L suggest that other secretory pathways, in addition to the SAR pathway, are activated in these plants.
DISCUSSION
Plant RabE GTPases are predicted to be involved in mediating secretory vesicle traffic from the Golgi to the PM. Therefore, they could play fundamental roles in the secretion of extracellular matrix molecules, thereby influencing plant growth and development, as well as in the secretion of extracellular defense molecules in response to pathogen infections. RabE proteins from both tomato (Bogdanove and Martin, 2000) and Arabidopsis (this study) interact with a pathogen effector protein, AvrPto, in Y2H assays. Because a number of studies have implicated vesicle trafficking in pathogen defense (Collins et al., 2003; Assaad et al., 2004; Wang et al., 2005; Nomura et al., 2006; Kalde et al., 2007), physical interaction between AvrPto and RabE GTPases could indicate a potential role of RabE GTPases in plant defense. However, to date, only one report has documented subcellular localization of Arabidopsis GFP-RabE1d transiently expressed in heterologous tobacco epidermal cells (Zheng et al., 2005). Subcellular localization of RabE GTPases in native Arabidopsis cells and the biological role of RabE GTPases in regulating plant growth or defense at the whole-plant level remained unknown.
Using a combination of confocal microscopy and cell membrane fractionation on transgenic plants expressing GFP-RabE1d and on wild-type plants, we show not only that RabE proteins are localized in the Golgi, as reported by Zheng and colleagues (2005), but also that a prominent fraction is associated with the PM of Arabidopsis leaf cells. Because a hallmark feature of Rab proteins is that they localize to the specific membrane compartments in which they function, association of RabE with both the Golgi and the PM provides strong support for a role in mediating the traffic of secretory vesicles from the Golgi to the PM (i.e. instead of other post-Golgi target membranes), as suggested previously by Zheng et al. (2005).
Interestingly, the RabE-Q74L protein expressed in Arabidopsis as a GFP fusion displayed an unanticipated localization pattern. The bulk of fluorescent protein was localized at the vacuolar membrane, and we could not detect, by CLSM fluorescence in the Golgi or in the PM, the predicted destination of the active form of RabE. After delivery of vesicles to the target membrane, Rab proteins are usually recycled back to the cytoplasm and then incorporated again into the donor membrane for multiple rounds of vesicle transport. Such recycling is dependent on hydrolysis of GTP into GDP, because inactivated GDP-bound Rabs are extracted from the target membrane by accessory proteins that mediate their recycling (Segev, 2001). The Q74L mutation is predicted to inactivate the GTP hydrolysis activity of RabE. Presumably, RabE-Q74L cannot be dephosphorylated and, thus, cannot be recycled to the cytoplasm. It is possible that a default nonspecific endocytosis pathway, such as that observed for the FM4-64 dye (Ueda et al., 2001), could carry the GTP-bound RabE-Q74L protein from its target location, the PM, to the tonoplast, where it accumulates.
Analysis of RabE-cosuppressed plants clearly indicates that RabE GTPases play a role in plant growth, as evidenced by dwarf plant sizes and altered leaf morphology in RabE-cosuppressed plants. Rosette leaves of RabE-cosuppressed lines developed relatively normally for the first 3 weeks, when Arabidopsis development is usually slower. However, when Arabidopsis size increases more rapidly, in the 4th and 5th weeks, the leaves of RabE-cosuppressed plants were not able to fully elongate; leaf midribs and petioles remained short, yielding characteristically wavy leaves. Inflorescences emerged at the same time as in the wild type, but the overall stature of the RabE-cosuppressed plants remained much shorter. These observations lead us to suggest that RabE-mediated vesicle traffic from the Golgi to the PM is required for the rapid elongation of certain tissues (e.g. leaf midribs and stems) associated with rapid plant growth. The dwarf phenotype of RabE-cosuppressed plants was likely caused by simultaneous down-regulation of more than one RabE gene, as suggested by reduced expression of RabE1d and -E1e and, to a lesser extent, RabE1a, -E1b, and -E1c (Fig. 6E). Consistent with this speculation, knockout mutants of RabE1d, caused by T-DNA insertions, did not exhibit any defect in growth and development (data not shown). Notably, cosuppression did not affect the expression of RabD genes, the closest homologues of RabE genes; therefore, the observed phenotypes can be ascribed to partial silencing of multiple RabE gene family members.
Besides our interest in determining the subcellular localization of RabE GTPases in native Arabidopsis cells and a possible role of RabE GTPases in growth and development at the whole plant level, another major motivation for this work was to investigate a potential role of RabE GTPases in plant defense. Bogdanove and Martin (2000) first identified RabE GTPases as AvrPto interactors in tomato and discussed the possibility that these GTPases could be virulence targets of AvrPto. In an independent Y2H screen for AvrPto-interacting Arabidopsis proteins, we also isolated a RabE GTPase. We found that AvrPto interacted with all tested members of the RabE family but not with members of other Rab families of Arabidopsis (Fig. 1). Moreover, AvrPto interacted with a RabE mutant (RabE1d-Q74L) predicted to be GTP bound but not with a RabE mutant (RabE1d-S29N) predicted to be GDP bound. Finally, we found that the majority of membrane-associated RabE1d is localized at the PM, where AvrPto is known to be located in tomato and Arabidopsis cells (Shan et al., 2000; He et al., 2006). Despite the intriguing specificity in Y2H assays and the apparent colocalization of AvrPto and RabE1d to the same host membrane, we have been unable to detect AvrPto-RabE1d interaction using in vivo coimmunoprecipitation or in vitro pull-down methods. Physical interaction of AvrPto with confirmed virulence targets FLS2 and BAK1 was demonstrated using both in vitro and in vivo methods (Shan et al., 2008; Xiang et al., 2008). Thus, our results suggest that RabE proteins may not be host targets of AvrPto. Alternatively, RabE GTPases may interact with AvrPto in the plant cell only transiently and/or may require special conditions.
Interestingly, GFP-RabE1d plants challenged with the avirulent strain Pst DC3000 (avrRpt2) manifested dramatic focal accumulation of GFP-RabE1d in mesophyll cells (Fig. 5). GFP-RabE1d focal accumulation was limited to a few sporadic cells when plants were inoculated with wild-type virulent Pst DC3000 (i.e. in a susceptible interaction) or with the hrpA− mutant (which activates plant basal defenses). Polarized accumulation of RabE GTPase during a gene-for-gene interaction could be part of the host cell response to direct defense-associated vesicle trafficking toward invading pathogens. If so, the GFP-RabE1d fusion may be a useful cellular marker for future study of vesicle trafficking in the gene-for-gene response in Arabidopsis. Accumulation of other vesicle traffic-associated regulators (such as t-SNAREs and v-SNAREs) at the site of pathogen penetration has also been observed in fungal infections (Assaad et al., 2004; Huckelhoven, 2007; Kwon et al., 2008).
RabE cosuppression in Arabidopsis, under the conditions reported in this study, did not result in increased susceptibility to P. syringae bacterial strains. RabE proteins, therefore, may not be required for establishing defenses against this pathogen. Alternatively, partial down-regulation of RabE proteins may be insufficient to confer a discernible defense phenotype. A complete knockout of all five RabE genes would be necessary to resolve this question. However, the dwarf phenotype of partially RabE-cosuppressed plants suggests that it is unlikely that completely RabE-deficient plants would be viable and/or suitable for bacterial infection assays.
Transgenic expression of the RabE1d-Q74L variant, on the other hand, conferred in Arabidopsis a significant degree of resistance to Pst DC3000. It remains to be determined whether this resistance is caused by a direct effect of RabE1d-Q74L, due to enhancement of defense-related vesicle traffic, or rather is triggered by an indirect effect, due to overall perturbation of cellular vesicle traffic. We found that the IWF collected from water-treated GFP-RabE1d-Q74L plants contained PR1 and several unknown proteins that were absent from the IWF of water-treated wild-type Arabidopsis, indicating constitutive activation of secretory and defense pathways in these plants. BTH application resulted in similar levels of secreted PR1 and other proteins in the apoplast in both wild-type and RabE1d-Q74L transgenic plants (Fig. 8C). However, some of these extracellular proteins are detected only in the IWF of water- and BTH-treated RabE1d-Q74L-expressing plants but not in the IWF of BTH-treated wild-type plants. These unique extracellular proteins associated with the expression of RabE1d-Q74L suggest the activation of other secretory pathways in these plants in addition to the SAR pathway. Interestingly, activation of SAR and resistance to Pst DC3000 in the GFP-RabE1d-Q74L transgenic plants did not correlate with a dwarf phenotype, a common phenotype of Arabidopsis mutants that are constitutively resistant to pathogens (Lorrain et al., 2003). Further analysis of the RabE-Q74L transgenic plants may provide novel insights into plant defense and indicate possible avenues for engineering resistance in crops.
MATERIALS AND METHODS
Y2H Assay
We identified Arabidopsis (Arabidopsis thaliana) proteins that interacted with AvrPto of Pseudomonas syringae pv tomato strain DC3000 using the Matchmaker LexA-based Y2H system (Clontech Laboratories). Two Arabidopsis cDNA libraries, constructed from infected and uninfected Landsberg erecta plants (kindly provided by J. Jones), were screened. The AvrPto coding sequence was amplified from Pst DC3000 genomic DNA by PCR (sense primer, 5′-GCGAATTCCGAACCATGGGAAATATATGTGTC-3′; antisense primer, 5′-GCCTCGAGATTGCCAGTTACGGTA-3′) and cloned into pNLexA to serve as bait in the Y2H screen.
RabE Cloning and Mutagenesis
We amplified the RabE1d (At5g03520) coding sequence from Arabidopsis Col-0 cDNA using the rabE-5′ and rabE-3′ primers (Supplemental Table S1), containing the EcoRI and BamHI restriction sites, respectively. The PCR product was ligated into a TOPO vector (pCR2.1; Invitrogen) and sequenced. Single nucleotide changes were introduced in the RabE1d sequence by two-step overlapping PCR, to generate the RabE1d-S29N and RabE1d-Q74L mutant derivatives. RabE1d-S29N was obtained through a G→A substitution; in the first PCR step, two overlapping RabE1d fragments were amplified using the primer combinations rabE-5′/S29N-rev and rabE-3′/S29N-for (Supplemental Table S1). The products were purified from agarose gel, mixed, and used as template for a second PCR amplification step, with the rabE-5′ and rabE-3′ primers. The presence of an overlapping region allowed annealing of the two gene fragments and amplification of the full-length coding sequence. A similar procedure was used to introduce the Q74L mutation through an A→T substitution. In this case, the following primer combinations were used in the first PCR step: rabE-5′/Q74L-rev and rabE-3′/Q74L-for (Supplemental Table S1). RabE1d-S29N and RabE1d-Q74L amplification products were introduced into pCR2.1-TOPO and sequenced.
GFP-RabE1d Transgenic Plants
RabE1d and the mutant variant RabE1d-Q74L were subcloned in the EcoRI and BamHI sites of the binary expression vector pEGAD (Cutler et al., 2000), downstream of the enhanced GFP sequence, to create translational fusions. The binary vector was introduced in Agrobacterium tumefaciens strain GV3850 via triparental mating for plant transformation. Arabidopsis Col-0 glabrous (Col-0 gl1) plants were transformed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected based on resistance to the herbicide Basta (glufosinate). A solution containing 0.012% glufosinate (Finale concentrate; AgrEvo Environmental Health) and 0.025% Silwet L-77 was sprayed on 2-week-old seedlings growing in soil. Surviving T1 plants were screened for GFP fluorescence with a Zeiss Axiophot microscope, and expression of the correctly sized GFP-RabE fusion proteins was verified by western blotting with an anti-RabE polyclonal antibody (described below).
Protein Extraction and Immunoblotting
Total proteins were extracted as follows: approximately 20 mg (fresh weight) of fresh or frozen leaf tissue was ground with a pestle in a microfuge tube in the presence of 100 μL of 1× SDS-PAGE loading buffer (90 mm Tris-HCl, pH 8.0, 100 mm dithiothreitol, 3% SDS, 22.5% Suc, 10 μL mL−1 Protease Inhibitor Cocktail for plant cell extracts [Sigma], and bromphenol blue [to saturation]). Extracts were immediately heated at 80°C for 10 min and then frozen at −20°C. Before loading, extracts were thawed at room temperature and centrifuged at 20,000g for 2 min to pellet debris. An equal volume of each sample was used for SDS-PAGE. Total proteins were separated on precast gradient gels (4%–20%; ISC BioExpress), then transferred onto Immobilon-P membranes (Millipore) using a semidry transfer apparatus (SEMI PHOR; Hoefer Scientific Instruments). Protein detection was carried out with the following primary antibodies: anti-RabE (raised in chicken against recombinant RabE protein expressed in Escherichia coli; Cocalico Biological), anti-XT1 (Faik et al., 2002; Cavalier and Keegstra, 2006), anti-PM ATPase (Morsomme et al., 1998), and anti-γ-TIP (a gift of Dr. N. Raikhel).
Cell Membrane Fractionation
Leaves were harvested and weighed immediately prior to extraction. Leaf tissue (2.5 g) was ground with a cold mortar and pestle in the presence of 5 mL of ice-cold extraction buffer (50 mm HEPES, pH 7.5, 100 mm KCl, 10 mm EDTA, 1 mm dithiothreitol, and 10 μL mL−1 Protease Inhibitor Cocktail for plant cell extracts [Sigma]) containing 34% Suc (w/v). The extract was homogenized with a Polytron immersion blender (three pulses of 10 s each), filtered through a single layer of Miracloth, and centrifuged for 10 min at 10,000g to remove most unbroken chloroplasts and nuclei. The supernatant was adjusted to 40% Suc in about a 10-mL final volume (concentration was determined with a refractometer) and layered on a 5-mL cushion of 50% Suc in clear ultracentrifugation tubes. The homogenate was subsequently layered with 10 mL of 34% Suc, 8 mL of 25% Suc, and 8 mL of 18% Suc (w/v). All Suc solutions were prepared in the same buffer used for extraction. Gradients were centrifuged at 100,000g for 3 h, at 4°C, in an SW28 rotor (Beckman). After centrifugation, the membrane-containing interphases were collected and diluted with Suc-free extraction buffer, and membranes were collected by ultracentrifugation (1 h at 100,000g). Membrane pellets were resuspended in equal volumes of SDS-PAGE loading buffer and heated at 80°C for 10 min. Equal volumes were loaded on SDS-PAGE gels. Protein electrophoresis and western blotting were performed as described above.
Confocal Microscopy Analysis and Imaging
Pieces of leaves were sampled randomly and mounted in water. Imaging was performed using an LSM510 META inverted confocal laser scanning microscope (Zeiss) and either a 20× or a 40× oil-immersion objective. For GFP-RabE fluorescence analysis, the 488-nm excitation line of an argon ion laser was used, with a 505- to 530-nm band-pass filter, in the single-track facility of the microscope. Images were processed with the LSM Image Browser version 3.1 (Zeiss) and with the Adobe Photoshop Elements version 5.0 software (Adobe Systems). For FM4-64 staining, detached Arabidopsis leaves were submerged in 8.2 μm FM4-64 (Molecular Probes) in water for 15 min. Leaves were rinsed in distilled water and observed immediately. For imaging GFP-RabE1d and FM4-64 fluorescence, the 488-nm excitation line was used; GFP fluorescence was collected with a 505- to 530-nm band-pass filter, and FM4-64 fluorescence was collected with a 615-nm long-pass filter.
Biolistic Transformation
ST-RFP transient expression in Arabidopsis leaves was achieved by biolistic transformation. The binary vector was a gift of Dr. F. Brandizzi (Saint-Jore et al., 2002). Gold particles (1.0 μm; Bio-Rad) were coated with the ST-RFP plasmid DNA as described by Zhang et al. (2001). Arabidopsis leaves were harvested and arranged on Murashige and Skoog agar plates (4.3 g L−1 Murashige and Skoog salts, 0.8% agar, pH 5.7). The DNA-coated particles were delivered into the lower leaf epidermis with a particle gun (DuPont), using 1,100-psi rupture discs under a vacuum of 25 in mercury. After bombardment, leaves were incubated on the sealed plates at room temperature, and fluorescence was observed at 24 h after transformation. For coimaging GFP-RabE1d and ST-RFP, the argon ion laser excitation lines of 488 nm (for GFP) and 543 nm (for DsRed) were used. GFP fluorescence was collected with a 505- to 530-nm band-pass filter, and DsRed fluorescence was collected with a 615-nm long-pass filter.
Plant Growth and Bacterial Multiplication Assay
Arabidopsis plants were grown in soil, in growth chambers, under a 12-h-dark/12-h-light cycle. The light intensity averaged 100 μE m−2 s−1, and the temperature was kept constant at 20°C. Pst DC3000 bacteria were cultured in low-salt Luria-Bertani medium (10 g L−1 tryptone, 5 g L−1 yeast extract, and 5 g L−1 NaCl) supplemented with 100 μg mL−1 rifampicin. For multiplication assays in plants, bacterial liquid cultures were incubated at 30°C to the mid to late logarithmic phase. Bacteria were collected by centrifugation and resuspended in sterile water with the addition of 0.004% Silwet L-77 (OSI Specialties). Titer of the bacterial inoculum was 1 × 105 cfu mL−1, unless otherwise indicated. Arabidopsis leaves were inoculated by syringe infiltration, and bacteria enumeration in leaves was conducted as described previously (Katagiri et al., 2002).
RNA Extraction and RT-PCR Analysis
Total RNA was extracted from 100 mg of Arabidopsis leaf tissue with the RNeasy Plant Mini Kit (Qiagen), according to the manufacturer's specifications. RNA concentration in samples was determined with a NanoDrop ND-1000 Spectrophotometer (NanoDrop). RNA RT and target gene amplification (RT-PCR) were performed using the RNA LA PCR Kit (AMV), version 1.1 (TaKaRa). RT reaction mixture was prepared according to the manufacturer's protocol (5 mm MgCl2, 1× RNA PCR buffer, 1 mm deoxynucleoside triphosphate mixture, 1 unit μL−1 RNase inhibitor, 0.25 units μL−1 AMV reverse transcriptase, 0.125 μm oligo[dT] Adaptor Primer, RNase-free water, and 1 μg of total RNA). For amplification of the RabE and RabD transcripts, a single RT reaction was carried out in a total volume of 50 μL and incubated in a thermal cycler for 30 min at 45°C, followed by 5 min at 99°C and 5 min at 5°C. Five microliters of reverse-transcribed cDNA was used as template in each of 10 PCRs with gene-specific primer pairs designed to amplify the five RabE gene family members, the four RabD genes, and the Actin8 gene as a control. Primer sequences are listed in Supplemental Table S2. Each PCR sample contained 2.5 mm MgCl2, 1× LA PCR buffer II, 0.2 μm forward primer and 0.2 μm reverse primer, sterilized distilled water, and 5 μL of the RT reaction described above in a final volume of 25 μL. The reactions were placed in a thermal cycler, and amplification was performed under the following conditions: 94°C for 2 min (one cycle); 94°C for 30 s, 54°C for 30 s, and 72°C for 1 min (22 or 25 cycles); 72°C for 1 min (one cycle); and then 4°C. Ten microliters of the PCR samples was loaded on a 1% agarose gel. Gels were photographed with a Bio-Rad Gel Documentation System, and band intensity was analyzed with the Quantity One software (Bio-Rad).
BTH Treatment
BTH (Actigard) was prepared at a final concentration of 300 μm in water and sprayed onto potted GFP-RabE1d-Q74L and Col-0 gl1 Arabidopsis plants. A separate set of plants was sprayed with water as a control. Plants were covered with a tight-fitting clear plastic dome and assayed for responses at 3 d after BTH application.
IWF Collection and Analysis
BTH-treated and control plants were harvested 3 d after treatment. Whole plants were vacuum infiltrated for 2 min with distilled water containing 0.002% Silwet L-77 (OSI Specialties). The plants were placed in conical centrifuge tubes (Nalgene) containing a mesh septum placed about 2 cm above the bottom. IWF was collected by centrifuging the infiltrated plants at 400g for 20 min at 4°C. The IWF volume was measured with a micropipette, and the appropriate volume of 5× SDS-PAGE loading buffer was immediately added. Samples were heated at 85°C for 5 min, then frozen or loaded onto an acrylamide gel.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ClustalW alignment of the five Arabidopsis RabE proteins and their closest homologues in other organisms.
Supplemental Figure S2. Bacterial multiplication assays.
Supplemental Figure S3. Localization pattern of GFP-RabE1d-Q74L.
Supplemental Figure S4. Focal accumulation of GFP-RabE1d in response to bacteria.
Supplemental Table S1. Primers for RabE cloning and mutagenesis.
Supplemental Table S2. Gene-specific primers for RT-PCR.
Supplementary Material
Acknowledgments
We thank colleagues who kindly shared the antibodies used in this study: Dr. Marc Boutry (anti-PM ATPase), Dr. Xinnian Dong (anti-PR1), Dr. Ken Keegstra (anti-XT1), Dr. Kinya Nomura (anti-RabE), and Dr. Natasha Raikhel (anti-γ-TIP). We are thankful to Dr. Federica Brandizzi and Dr. Melinda Frame for assistance with confocal microscopy, Dr. Shuo Cheng Zhang for help with particle bombardment, and Beth Rzendzian and Zach Ferguson for plant care. The ST-RFP construct was a kind gift of Dr. Federica Brandizzi. Dr. Jonathan Jones provided the Arabidopsis cDNA libraries that we used in our Y2H screen. Thanks to members of the He laboratory for helpful comments and to Karen Bird for editorial review of the manuscript.
This work was supported by the U.S. Department of Energy (grant no. DEFG02–91ER20021) and the National Institutes of Health (grant no. R01AI060761).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sheng Yang He (hes@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42 385–414 [DOI] [PubMed] [Google Scholar]
- Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Li G, Fu ZQ, Alfano JR (2008) Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol 11 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Martin GB (2000) AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci USA 97 8836–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214 159–173 [DOI] [PubMed] [Google Scholar]
- Bray Speth E, Lee YN, He SY (2007) Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr Opin Plant Biol 10 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I, Mansfield J, Bonas U (1995) Hrp genes in Xanthomonas campestris pv vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol Plant Microbe Interact 8 825–836 [Google Scholar]
- Buttner D, Bonas U (2003) Common infection strategies of plant and animal pathogenic bacteria. Curr Opin Plant Biol 6 312–319 [DOI] [PubMed] [Google Scholar]
- Calero M, Chen CZ, Zhu W, Winand N, Havas KA, Gilbert PM, Burd CG, Collins RN (2003) Dual prenylation is required for Rab protein localization and function. Mol Biol Cell 14 1852–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem 281 34197–34207 [DOI] [PubMed] [Google Scholar]
- Cheung AY, de Vries SC (2008) Membrane trafficking: intracellular highways and country roads. Plant Physiol 147 1451–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu Rev Plant Biol 59 547–572 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448 497–500 [DOI] [PubMed] [Google Scholar]
- Chow CM, Neto H, Foucart C, Moore I (2008) Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Huckelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977 [DOI] [PubMed] [Google Scholar]
- Craighead MW, Bowden S, Watson R, Armstrong J (1993) Function of the Ypt2 gene in the exocytic pathway of Schizosaccharomyces pombe. Mol Biol Cell 4 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf BHJ, Cheung AY, Andreyeva T, Levasseur K, Kieliszewski M, Wu HM (2005) Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell 17 2564–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Dangl JL (2006) Type III effector proteins: doppelgangers of bacterial virulence. Curr Opin Plant Biol 9 376–382 [DOI] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Jordan F, Osbourn A (2006) First encounters: deployment of defence-related natural products by plants. New Phytol 172 193–207 [DOI] [PubMed] [Google Scholar]
- Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinson HA, Read ND (2000) Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J Microsc 198 246–259 [DOI] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, Novick PJ (1988) A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell 53 753–768 [DOI] [PubMed] [Google Scholar]
- Harrison RE, Brumell JH, Khandani A, Bucci C, Scott CC, Jiang X, Finlay BB, Grinstein S (2004) Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell 15 3146–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125 563–575 [DOI] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta 1694 181–206 [DOI] [PubMed] [Google Scholar]
- Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K (1993) Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol 123 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckelhoven R (2007) Transport and secretion in plant-microbe interactions. Curr Opin Plant Biol 10 573–579 [DOI] [PubMed] [Google Scholar]
- Kalde M, Nuhse TS, Findlay K, Peck SC (2007) The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci USA 104 11850–11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0039, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451 835–840 [DOI] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10 71–82 [DOI] [PubMed] [Google Scholar]
- Lipka V, Kwon C, Panstruga R (2007) SNARE-Ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol 23 147–174 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8 263–271 [DOI] [PubMed] [Google Scholar]
- Machner MP, Isberg RR (2006) Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11 47–56 [DOI] [PubMed] [Google Scholar]
- Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P, Dambly S, Maudoux O, Boutry M (1998) Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J Biol Chem 273 34837–34842 [DOI] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8 971–977 [DOI] [PubMed] [Google Scholar]
- Nielsen E, Cheung AY, Ueda T (2008) The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol 147 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, DebRoy S, Lee YH, Pumplin N, Jones J, He SY (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313 220–223 [DOI] [PubMed] [Google Scholar]
- Nomura K, Melotto M, He SY (2005) Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol 8 361–368 [DOI] [PubMed] [Google Scholar]
- Novick P, Brennwald P (1993) Friends and family: the role of the rab GTPases in vesicular traffic. Cell 75 597–601 [DOI] [PubMed] [Google Scholar]
- Oparka KJ (1994) Plasmolysis: new insights into an old process. New Phytol 126 571–591 [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JM, Bonner HKS, Otegui MS, Nielsen E (2006) A role for the RabA4b effector protein PI-4Kβ1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol 172 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S (2007) Vesicle trafficking in plant immune responses. Cell Microbiol 9 1–8 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Moore I (2002) The Arabidopsis Rab GTPase family: another enigma variation. Curr Opin Plant Biol 5 518–528 [DOI] [PubMed] [Google Scholar]
- Rzomp KA, Moorhead AR, Scidmore MA (2006) The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun 74 5362–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J 29 661–678 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Segev N (2001) Ypt/Rab GTPases: regulators of protein trafficking. Sci STKE 100 re11. [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan LB, Thara VK, Martin GB, Zhou JM, Tang XY (2000) The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12 2323–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Heo WD, Braun V, Jiang X, Macrae C, Casanova JE, Scidmore MA, Grinstein S, Meyer T, Brumell JH (2007) A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol 176 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder BA, Leite B, Hipskind J, Butler LG, Nicholson RL (1991) Accumulation of sorghum phytoalexins induced by Colletotrichum graminicola at the infection site. Physiol Mol Plant Pathol 39 463–470 [Google Scholar]
- Snyder BA, Nicholson RL (1990) Synthesis of phytoalexins in Sorghum as a site-specific response to fungal ingress. Science 248 1637–1639 [DOI] [PubMed] [Google Scholar]
- Soylu S, Brown I, Mansfield JW (2005) Cellular reactions in Arabidopsis following challenge by strains of Pseudomonas syringae: from basal resistance to compatibility. Physiol Mol Plant Pathol 66 232–243 [Google Scholar]
- Stenmark H, Olkkonen V (2001) The Rab GTPase family. Genome Biol 2 reviews3007.1–reviews3007.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J 40 783–789 [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauchmani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wee EGT, Sherrier DJ, Prime TA, Dupree P (1998) Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18 74–80 [DOI] [PubMed] [Google Scholar]
- Zeng W, Keegstra K (2008) AtCSLD2 is an integral Golgi membrane protein with its N-terminus facing the cytosol. Planta 228 823–838 [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2 107–117 [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wege C, Jeske H (2001) Movement proteins (BC1 and BV1) of Abutilon mosaic geminivirus are cotransported in and between cells of sink but not of source leaves as detected by green fluorescent protein tagging. Virology 290 249–260 [DOI] [PubMed] [Google Scholar]
- Zheng HQ, Camacho L, Wee E, Henri BA, Legen J, Leaver CJ, Malho R, Hussey PJ, Moore I (2005) A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17 2020–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.