Abstract
The Ellman group has been one of the most influential in the development and widespread adoption of combinatorial chemistry techniques for biomedical research. Their work has included substantial methodological development for library synthesis with a particular focus on new scaffolds rationally targeted to biomolecules of interest and biologically relevant natural products. Herein we analyze a representative set of libraries from this group with respect to their biological and biomedical relevance in comparison to existing drugs and probe compounds. This analysis reveals that the Ellman group has not only provided new methodologies to the community but also provided libraries with unique potential for further biological study.
Introduction
The field of combinatorial chemistry came into being as a discipline in 1990-1991 with the publication of a series of papers on peptide libraries.1, 2 From those seminal papers, and the work of several industrial groups reported in the patent literature, grew a substantial interest in the synthesis and use of chemical libraries in early phase drug discovery. In the early 1990’s, one of the major focuses in the field was the application of these methods to non-peptide organic syntheses. This period was co-incident with Dr. Jon Ellman establishing his independent laboratory at UC-Berkeley and this problem was one to which he devoted the majority of his attention. Subsequently, his group would become one of the first to successfully produce small molecule parallel libraries, with their 1994 report of a route to benzodiazepine libraries.3
In the ensuing decade, the field would grow rapidly, initially with unbridled enthusiasm, and later with more tempered and realistic views. Dr. Ellman’s work played a crucial role in the development of parallel (non-mixture) methods and in maintaining the course of the field. Most workers in the field of combinatorial and parallel chemistry focused their efforts in one of three areas: parallel synthesis and optimization of “drug like” molecules;4 combinatorial synthesis of libraries of highly diverse molecules, without constraint to “drug likeness;”5 and development of methodologies for library synthesis.6 Ellman’s body of work is unusual in that it contains a substantial amount of effort devoted to molecules falling in between these areas, particularly in targeting bioactive natural products and in developing rationally targeted libraries for “difficult” molecular targets or libraries targeting known targets with new scaffolds. Below we discuss a number of libraries produced within the last decade by the Ellman group that stand out in terms of the physical properties of their molecules from the landscape traversed by most other groups.
The Ellman Libraries Chosen for Review
The Ellman group has published extensively in the last decade with a wide range of libraries. Rather than exhaustively reviewing this work, we instead have chosen libraries of particular interest from a biological perspective especially with respect to bringing new chemistry to bear on difficult targets. We analyze these libraries with respect to their “drug likeness” and relevance to chemical biology from a statistical perspective.
In 1999, Dragoli et al published a manuscript describing the synthesis of a small library of prostaglandin E analogs, the penultimate in a series from the laboratory.7 The synthesis of the library was rationalized by the fact that prostaglandins have diverse physiological activities and a large set of receptors but few receptor subtype selective analogs are known. The authors understood that a flexible synthesis method was needed to allow rapid access to a wide range of analogs. The reported route uses a combination of a Suzuki coupling to introduce diversity to the α chain and cuprate addition to introduce diversity to the β chain. This solid phase route gave good access to the targeted compounds and for the first time clear controlled synthesis of prostaglandins substituted on both chains from the same modular route.
In 1996, Boojamra et al published one of the more interesting papers in a series dealing with the synthesis of 1,4-benzodiazepines.8 The scaffold was of interest because it is a privileged scaffold that binds to a wide range of potential targets. This manuscript described a route providing general access to the 2,5-dione sub-series of the benzodiazepines that afforded roughly 2,500 highly diverse compounds. The solid phase route utilized amino acid methyl esters to provide access to 3-substituents, followed by conjugation with anthranilic acids to allow substitution of the aromatic ring, followed by base induced cyclization with the lithium salt of acetanilide and subsequent trapping of the resulting anion with alkyl halides to provide for substitutions on the 1-position. This route gave the widest range of this sub-series of benzodiazepines that had been reported to date.
In 2000, Maly et al reported one of the first general fragment based approaches to finding small molecules that bind to proteins of interest that did not require prior structural knowlege.9 The process involved selection of a series of chemically diverse monomers with good solubility that could be screened for weak activity, ensuring that a synthetic route existed that could link those monomers through a flexible linker, and then assembly and testing of the combinatorial set of dimers arising from linking of binding monomers. The method was applied to rapidly generate a novel and fairly potent inhibitor of SRC kinase. While the method has not seen wide adoption itself, it clearly influences a substantial portion of the fragment based work including combinations with computational methods and with tethering approaches.
In 1998, Souers et al published the synthesis of a library of 5,600 β-turn mimetics based on a previously developed constrained amino acid scaffold.10 The approach depended upon solid phase disulfide tethering of a nascent amine that was elaborated initially with an alkyl substituent, then an amino acid, and finally a substituted α-bromocarboxylate. Upon reductive cleavage of the disulfide, the materials auto condensed to form the nine-membered ring by thiol displacement of the bromide. The route gave good yield for a wide range of substituents at each variant position. Initial screening of the library afforded inhibitors of α4β1 integrin interactions with potency in the low micromolar range. The resulting molecules were one of a handful of such inhibitors.
In 2003, Wood et al published the synthesis of a library of roughly 2,000 mecaptomethyl ketone inhibitors targeted to mechanism based inhibition of cysteine proteases.11 The route involved the polymer immobilization of chloromethyl α-aminoketones, followed by displacement of the chloride by variant thiols, elaboration with an amino acid and capping of the resulting amino terminus by acylation. Overall the route gave four variable positions on the scaffold and significant process improvements over prior work by the group, particularly with respect to removing the need for inert atmospheric conditions. Initial screening of the library revealed potent inhibitors of cathepsin B.
In 1999, Haque et al reported the iterative synthesis of series of libraries of aspartyl protease inhibitors aimed at identification of potent inhibitors of the malarial protease plasmepsin II.12 As an initial starting point, the group screened a library of hydroxyethylamine inhibitors of cathepsin D that were previously prepared. Based upon the hits from that screen they used six iterative libraries to optimize this scaffold to the plasmepsin. The initial screen turned up inhibitors with sub-micromolar potency against PM II but roughly 15 fold selective for CatD. During the optimization process, the authors were able to push the potency down to 2-5 nM against the plasmepsin and invert the selectivity to gain roughly 15 fold selectivity against the malarial enzyme. At the time these were the most potent and most drug-like inhibitors of the plasmepsins.
In 2007, Patterson et al published the synthesis of a library of tubulysin D analogs, which are mitotic spindle poisons that block polymerization of tubulin to microtubules in mammalian cells.13 The work was predicated on a prior total synthesis of tubulysin D by the group.14 While the work is modest in scope for the Ellman group, reporting only ten analogs, it afforded the first glimpse at the molecular mechanism of action of this important class of natural products. A key finding in the work was the ability to reduce the structural complexity at both ends of the peptide.
In 1999, Xu et al disclosed the synthesis of a library of roughly 40,000 analogs to the cyclic peptide antibiotic vancomycin.15 The work is unusual in this group’s portfolio in that it used an on-bead screening scheme and only deconvoluted and confirmed structures for roughly 200 of these compounds. The library itself dramatically simplified the structure of vancomycin, which contains two fused cyclic peptides. The authors included in the library one of these cyclic peptides, with a fixed structure, and then appended varied linear peptides to replace the second macrocycle. In screening the library, they were able to identify members with binding potencies for the native target of vancomycin that had affinities within one order of magnitude of the native drug. This is truly remarkable as prior to this work it was believed that both macrocycles needed to be intact to afford active molecules.
The Ellman Libraries in Chemical Space
Visualizing the distribution of synthesized molecules in the context of a biologically relevant chemical space is a powerful means to explore the relationship between the Ellman libraries and chemical biology. A chemical space is generically defined as the set of all possible molecules that satisfy some constraint, and is analogous to the range of a mathematical function. Based upon our previous work,16 the current analysis was biased towards biologically relevant chemical space by employing a reference set including known drugs and five exemplar screening collections: Bioactive (molecules with well-characterized biologically activities), Natural Products (NP, compounds extracted and purified from organisms), Fragment (compounds designed primarily for structure-based screening), Rule of Five (RO5, the bulk of commercially-available screening collections designed for compliance with Lipinski’s Rule of Five17) and Diversity-Oriented Synthesis (DOS, natural product-like compounds designed to incorporate novel chemotypes with high complexity5) (see Table 1).
Table 1.
Details for the compound collections used in this study. Library preparation and standardization is described elsewhere.18
| Library | Unique Compounds | Sources |
|---|---|---|
| Drugs | 8,152 | CMC, DrugBank, MDDR |
| Bioactive | 4,501 | Biomol, LOPAC (Sigma), Microsource, Prestwick Chemical Library, Tocris |
| Diversity-Oriented Synthesis (DOS) | 15,060 | Porco_A19, Porco_B20, Schreiber21, Shair_A22, Shair_B23 |
| Fragments | 32,220 | ACD “Rule of 3” compliant24, Enamine, Life Chemicals, Maybridge |
| Natural Products (NP) | 3,267 | Ambinter, Biomol, Interbio, Microsource, NIH MLSMR, NCI, Specs, TimTec |
| Rule of Five (RO5) | 2,133,796 | Asinex, ChemDiv, ChemBridge, Enamine, Life Chemicals, Maybridge, Specs, |
| Ellman | 53,535 | Tripos Dragoli (26), Boojamra (2,508), Maly (3,515), Souers (5,589), Wood (2,016), Haque (566), Patterson (11), Xu (39,304) |
As before, eleven commonly used computationally-derived molecular descriptors were selected as chemical space metrics: Lipinski-type (molecular weight [mw], number of hydrogen bond acceptors [hacc], number of hydrogen bond donors [hdon], log(octanol/water partition coefficient) [logP]),17 medicinal chemistry (log(aqueous solubility) [logs] and polar surface area [psa]), and topological complexity (minimum and maximum partial charge-based GCUT25 [gcut0 and gcut3], Oprea complexity26 [oprea], Kier and Hall first-order atomic valence connectivity and first kappa shape indices27 [kier1and chi1v]).
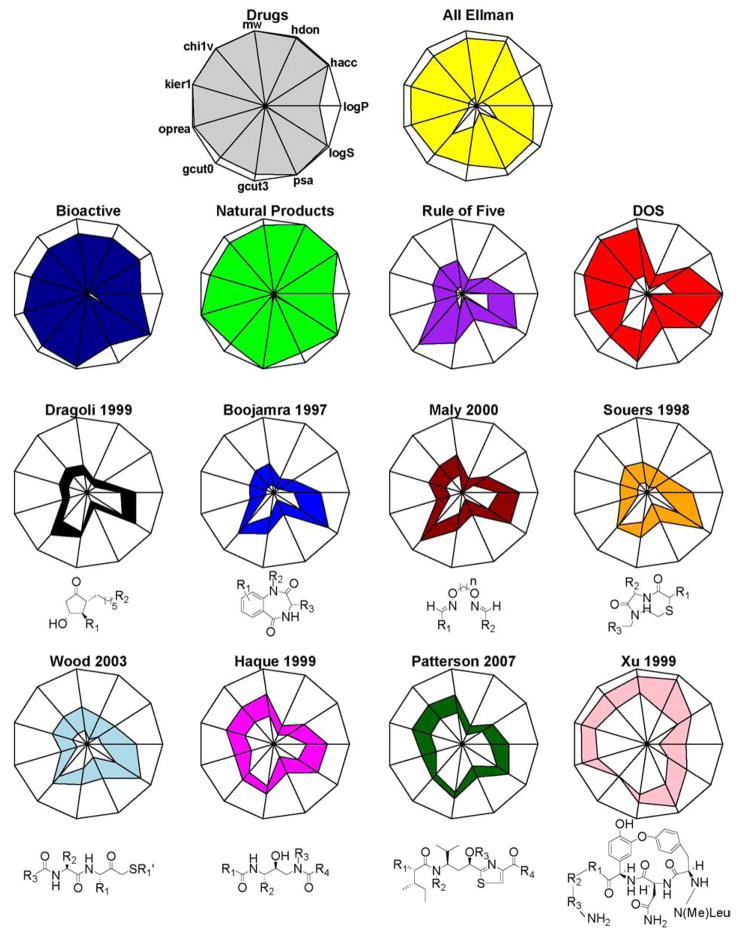
Two complimentary methods were employed to visualize the resulting 11-dimensional chemical space: radar plots and principal component analysis (PCA). Radar plots simultaneous render n dimensional data for a compound collection using a polygon with n-vertices. Each radius extending from the polygon center to a vertex is an independent axis representing the full range of a single variable calculated from all compounds in the study. The 1st and 99th percentiles of descriptor values are plotted on each axis; points on adjacent spokes are joined, yielding an enclosed area that effectively summarizes the multivariate distribution. Differences in the distribution of properties among compound libraries can then be visually assessed easily by comparing the shapes of the radar plots. PCA is a dimension reduction technique that calculates n eigenvectors from the covariance matrix generated from n molecular descriptors. Each eigenvector, or principal component, identifies an orthogonal direction of variation within the data. Often, a small subset of the eigenvectors can account for much of the variability, such that re-plotting with m < n eigenvectors yields a lower dimensional space that still faithfully represents the original data set. Whereas radar plots provide statistical summaries of the descriptors, PCA reveals information about the joint distribution of molecular properties, and can be used to identify underlying structures such as outliers and clustering that are difficult to perceive in higher dimensions.
Chemical space analysis shows that the Ellman libraries broadly sample biologically relevant chemical spaces, including regions that are not well sampled by commercially-available libraries and areas that are relatively unexplored. In the radar plots (Figure 1), the Drugs and NP collections cover the widest range of descriptor values; however, the eight Ellman libraries sample the available space well in aggregate. Four distinct shape classes are apparent. The Dragoli, Boojamra, and Maly collections tend to match the RO5 shape well, displaying relatively higher values for gcut0 compared to gcut3, moderate values for logS and logP, and low values for all the other parameters. It is of interest to note that the Dragoli library, comprised of molecules that most would intuitively view as non-druglike, fall neatly within the R05 space. The Souers and Wood collections display a noticeable departure from the characteristic RO5 shape, marked by modest increases in psa, hdon, and hacc and a contraction in gcut0. Although peptidomimetic in character, these libaries do not carry a significant liability in terms of increased logP as is generally believed to be the case when increasing hdon/hacc numbers. This is probably due to compensation in psa and/or limiting mw increase. Haque and Patterson appear more DOS-like, as indicated by the characteristic bulge in the complexity parameters oprea, kier1, and chi1v, and molecular weight, in addition to a larger gcut3 relative to gcut0. However, unlike the classical DOS libraries, there is not a strong increase in logP and ratio of hacc to hdon numbers. Finally, the Xu collection matches DOS well for parameters on the left side of the radar plot, but displays uniquely large values for hdon, hacc, and psa, and a marked contraction in logP and logS. This library samples a high complexity area of chemical space that is quite distinct from the majority of existing DOS libraries.
Figure 1.
Radar plots based on 11 commonly used molecular descriptors calculated from the chemical libraries in Table 1. Each radius of the 11-sided polygon represents the full range of a single descriptor over all compounds in the study. The 1st and 99th percentiles of descriptor values attained by a particular library are plotted on each spoke; the bounds of the enclosed colored areas are formed by connecting points between adjacent spokes. Plotted descriptors include molecular weight [mw], number of hydrogen-bond donors [hdon], number of hydrogen-bond acceptors [hacc], predicted log(octanol/water partition coefficient) [logP], predicted log(aqueous solubility) [logS], polar surface area in χ2 [psa], minimum and maximum partial charged-based GCUT [gcut0 and gcut3], Oprea complexity [oprea], and two graph theory based shape descriptors [kier1 and chi1v]. The Drugs, Bioactive, NP, RO5, and DOS libraries are included for reference. Markush structures define the contents of each Ellman library. The “All Ellman” plot aggregates the eight Ellman libraries by weighting each component’s contribution equally.
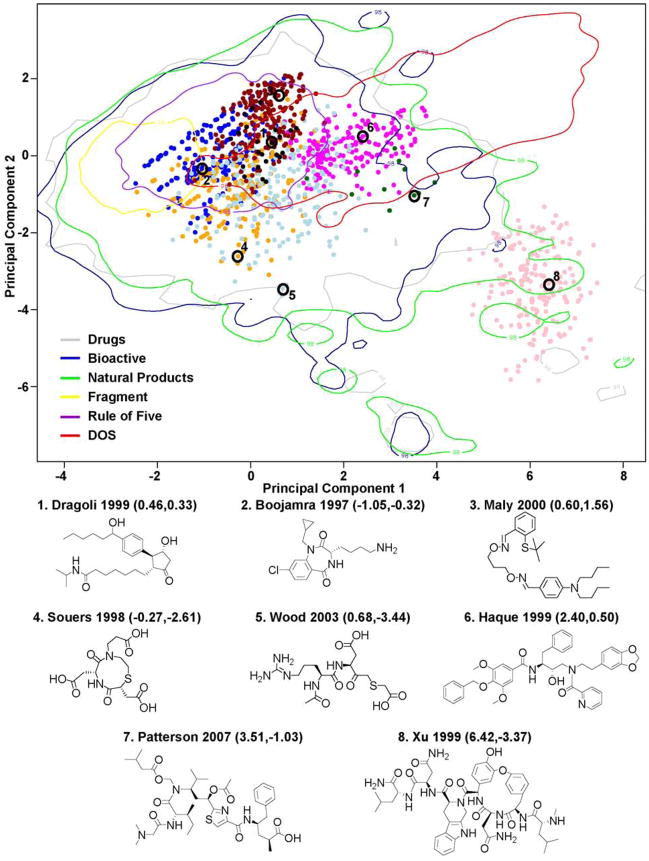
The distribution of the Ellman libraries in the PCA graph follows the patterns observed in the radar plots well (Figure 2). The Dragoli, Boojamra, and Maly libraries almost completely fall within the RO5 98% contour. The Wood and Souers libraries begin to segregate out of the RO5 bounds and into an area of chemical space mostly sampled by Drugs, Bioactive, and NP. It is likely that the molecules in these libraries might be of general interest in less target oriented screens, particularly with high content methods. Nearly all of the Haque compounds and most of the Patterson library reside in or near the DOS space. These are areas that have been poorly sampled in commercially available collections but that are also sampled well by a number of other academically sourced libraries. Finally, the Xu library exists in an extreme portion of chemical space, lying at the outer edge of the Drug and NP distributions and also in a relatively unexplored region of chemical space. This is not unexpected from a library that is essentially comprised of a fusion of a short peptide library with a fixed cyclic peptide core. Screening this library in more diverse contexts, particularly cellular adhesion, might be quite interesting as there is a possibility of gain-of-function relative to the originating peptide antibiotic.
Figure 2.
Plot of the Ellman libraries (points colored identically to Figure 1) within a chemical space described by the first two principal components (PCs) derived from the 11 commonly used descriptors calculated from the Drugs and exemplar libraries. The two PCs account for 82.7% of the total variance in the reference set (63.8% and 18.9% in the first and second PCs, respectively). The first PC is dominated by gcut3, followed by gcut0, hdon, hacc, and −logS. The second PC is largely comprised of gcut0, then gcut3, −hdon, and logP. A maximum 200 molecules per Ellman library is plotted. The Drugs, Bioactive, NP, Fragment, RO5, and DOS libraries are included for reference, and are depicted as colored contour lines enclosing 98% of each population. The chemical structure and PC coordinate for a representative from each Ellman library is also included.
Conclusion
In slightly more than a decade, the Ellman group has made profound contributions to both combinatorial chemistry and chemical biology. The eight Ellman libraries described here not only highlight technical achievement in synthetic chemistry, but have also yielded valuable chemical tools to probe biological function and have inspired potential drug leads. A statistical analysis of these compounds reveals a broad sampling of biologically relevant chemical space, including significant incursions into relatively pristine regions that have yet to be exploited by both conventional screening collections and other efforts at diversity oriented synthesis. Our analysis implies that much broader screening of these libraries, particularly outside the context of the originating hypotheses, would be well justified. As such, we eagerly look forward to the next decade and beyond of Ellman chemistry and to greater utilization of the materials produced by this dynamic group.
Acknowledgments
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC) and St. Jude Children’s Research Hospital.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Nature. 1991;354:84. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 2.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 3.Bunin BA, Plunkett MJ, Ellman JA. Proc Natl Acad Sci U S A. 1994;91:4708. doi: 10.1073/pnas.91.11.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards PJ, Allart B, Andrews MJ, Clase JA, Menet C. Curr Opin Drug Discov Devel. 2006;9:425. [PubMed] [Google Scholar]
- 5.Tan DS. Nat Chem Biol. 2005;1:74. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 6.Dolle RE, Le Bourdonnec B, Morales GA, Moriarty KJ, Salvino JM. J Comb Chem. 2006;8:597. doi: 10.1021/cc060095m. [DOI] [PubMed] [Google Scholar]
- 7.Dragoli DR, Thompson LA, O'Brien J, Ellman JA. J Comb Chem. 1999;1:534. doi: 10.1021/cc990033e. [DOI] [PubMed] [Google Scholar]
- 8.Boojamra CG, Burow KM, Thompson LA, Ellman JA. Journal of Organic Chemistry. 1997;62:1240. [Google Scholar]
- 9.Maly DJ, Choong IC, Ellman JA. Proc Natl Acad Sci U S A. 2000;97:2419. doi: 10.1073/pnas.97.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Souers AJ, Virgilio AA, Schurer SS, Ellman JA, Kogan TP, West HE, Ankener W, Vanderslice P. Bioorg Med Chem Lett. 1998;8:2297. doi: 10.1016/s0960-894x(98)00416-8. [DOI] [PubMed] [Google Scholar]
- 11.Wood WJ, Huang L, Ellman JA. J Comb Chem. 2003;5:869. doi: 10.1021/cc034008r. [DOI] [PubMed] [Google Scholar]
- 12.Haque TS, Skillman AG, Lee CE, Habashita H, Gluzman IY, Ewing TJ, Goldberg DE, Kuntz ID, Ellman JA. J Med Chem. 1999;42:1428. doi: 10.1021/jm980641t. [DOI] [PubMed] [Google Scholar]
- 13.Patterson AW, Peltier HM, Sasse F, Ellman JA. Chemistry. 2007;13:9534. doi: 10.1002/chem.200701057. [DOI] [PubMed] [Google Scholar]
- 14.Peltier HM, McMahon JP, Patterson AW, Ellman JA. J Am Chem Soc. 2006;128:16018. doi: 10.1021/ja067177z. [DOI] [PubMed] [Google Scholar]
- 15.Xu R, Greiveldinger G, Marenus LE, Cooper A, Ellman JA. J Am Chem Soc. 1999;121:4898. [Google Scholar]
- 16.Shelat AA, Guy RK. Curr Opin Chem Biol. 2007;11:244. doi: 10.1016/j.cbpa.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Deliv Rev. 2001;46:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 18.Shelat AA, Guy RK. Nat Chem Biol. 2007;3:442. doi: 10.1038/nchembio0807-442. [DOI] [PubMed] [Google Scholar]
- 19.Beeler AB, Acquilano DE, Su Q, Yan F, Roth BL, Panek JS, Porco JA., Jr Journal of combinatorial chemistry. 2005;7:673. doi: 10.1021/cc050064b. [DOI] [PubMed] [Google Scholar]
- 20.Lei X, Zaarur N, Sherman MY, Porco JA., Jr The Journal of organic chemistry. 2005;70:6474. doi: 10.1021/jo050956y. [DOI] [PubMed] [Google Scholar]
- 21.Burke MD, Berger EM, Schreiber SL. J Am Chem Soc. 2004;126:14095. doi: 10.1021/ja0457415. [DOI] [PubMed] [Google Scholar]
- 22.Pelish HE, Westwood NJ, Feng Y, Kirchhausen T, Shair MD. J Am Chem Soc. 2001;123:6740. doi: 10.1021/ja016093h. [DOI] [PubMed] [Google Scholar]
- 23.Goess BC, Hannoush RN, Chan LK, Kirchhausen T, Shair MD. Journal of the American Chemical Society. 2006;128:5391. doi: 10.1021/ja056338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Congreve M, Carr R, Murray C, Jhoti H. Drug Discov Today. 2003;8:876. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 25.Pearlman R, Smith K. J Chem Inf Comput Sci. 1999;39:28. [Google Scholar]
- 26.Allu TK, Oprea TI. J Chem Inf Model. 2005;45:1237. doi: 10.1021/ci0501387. [DOI] [PubMed] [Google Scholar]
- 27.Hall L, Kier L, editors. The molecular connectivity chi indexes and kappa shape indexes in structure-property modeling. VCH Publishers; 1991. [Google Scholar]