Abstract
We investigated the clinical implications of lung developmental transcription factors (TTF-1, NKX2–8, and PAX9) that we recently discovered as cooperating oncogenes activated by way of gene amplification at chromosome 14q13 in lung cancer. Using stable transfectants of human bronchial epithelial cells, RNA expression profiles (signatures) representing activation of the biological pathways defined by each of the 3 genes were determined and used to risk stratify a non-small-cell lung cancer (NSCLC) clinical data set consisting of 91 early stage tumors. Coactivation of the TTF-1 and NKX2–8 pathways identified a cluster of patients with poor survival, representing ≈20% of patients with early stage NSCLC, whereas activation of individual pathways did not reveal significant prognostic power. Importantly, the poor prognosis associated with coactivation of TTF-1 and NKX2–8 was validated in 2 other independent clinical data sets. Furthermore, lung cancer cell lines showing coactivation of the TTF-1 and NKX2–8 pathways were shown to exhibit resistance to cisplatin, the standard of care for the treatment of NSCLC. This suggests that the cohort of patients with coactivation of TTF-1 and NKX2–8 pathways appears to be resistant to standard cisplatin therapy, suggesting the need for alternative therapies in this cohort of high-risk patients.
Keywords: gene expression, lung cancer, transcription factors
The complexity of the oncogenic process involving the somatic acquisition of large numbers of mutations, coupled with the varied host genetic constitution, produces a disease of enormous complexity. The ability to dissect this complexity by better understanding models of carcinogenesis, lineage dependence, and oncogene addiction is critical for developing new effective therapeutic strategies.
There is strong evidence for the involvement of developmental and cell lineage-dependent processes in the development of human tumors. For example, multiple genes (e.g., AML1, PML-RAR fusion gene, and PAX5) that are critical to hematopoietic progenitor cell development are frequently altered in hematologic malignancies (1). Similar examples exist in cell proliferation and differentiation events that lead to melanoma (2). In contrast to the oncogene addiction model that invokes a tumor-specific gain-of-function event (3), the lineage-survival oncogene model (4) proposes that tumors may become highly dependent on survival mechanisms that are built into the lineage precursor cells during development and that alteration of the genes involved in these survival mechanisms could provide selective advantage for evolving tumor cells.
Despite the large number of transcription factors involved in mammalian lung development, the role of these factors in lung cancer progression, particularly non-small-cell lung cancer (NSCLC), and their clinical relevance have not been comprehensively evaluated. Recently, we discovered and functionally characterized a novel, recurrent amplicon located at 14q13.3 that appears to harbor lung cell lineage-specific oncogenes due to its preferential occurrence in lung cancer and the role that these 3 genes play in lung development (5). High-resolution quantitative PCR mapping of the commonly amplified region revealed genes encoding 3 transcription factors in the 413-kb core region of the amplicon: TTF-1, NKX2–8, and PAX9. Both TTF-1 and NKX2–8 are known to mediate lung development and maturation (5, 6). Similarly, PAX9 is also likely involved in lung development as evident by its expression in fetal lung tissue and the diverse negative impact on organogenesis in knockout animals (7). Whereas overexpression of individual members of this 3-gene set in the core region of the amplicon was ineffective in stimulating proliferation, combinatorial overexpression of the genes led to significant proliferation of premalignant human bronchial epithelial cells, implicating oncogene cooperation for the 3 coamplified lung development regulators. Furthermore, our reported RNAi knockdown experiments in concert with other reports establish that each of these 3 genes can have essential tumor maintenance function (5, 8, 9).
To investigate the impact of the 3 collaborating lung cell lineage oncogenes on clinical outcome, we adapted a validated strategy that utilizes gene expression profiles as a measure of the consequence of an activated oncogenic pathway, irrespective of how the signaling pathway might have been altered. Thus, even if the known oncogene is not mutated, but rather another component of the pathway is altered, the expression profile will likely still detect the biological alteration. Building upon this approach, here we describe a significant correlation of lung tumors with dual activation of the biological pathways directed by both TTF-1 and NKX2–8 with poor survival. Furthermore, we demonstrate that lung cancer cells possessing coactivated TTF-1 and NKX2–8 pathways exhibit cisplatin resistance, suggesting the need for alternative therapies in this cohort of high-risk patients.
Results
Gene Expression Signatures That Reflect the Activity of TTF-1, NKX2–8, and PAX9.
Using gene expression levels of TTF-1 (Probe IDs 207771_at, 207772_s_at), NKX2–8 (Probe ID 207451_at), and PAX9 (207059_at), we first explored the possibility that individual gene expression levels of the developmental transcription factors may reveal correlations with clinical parameters. To test this hypothesis, we chose survival in patients with surgically resected stage I NSCLC. Supporting information (SI) Fig. S1 and Fig. S2 depict an analysis of transcription factor status in a cohort of NSCLC tumors (n = 91) that included comparable numbers of squamous cell carcinoma and adenocarcinoma (GSE3141). Importantly, none of these patients had received adjuvant chemotherapy. Kaplan–Meier survival analysis shows that no statistically significant difference was detected between high or low expression level of NKX2–8, TTF-1, and PAX9 (Fig. S1) or when combinations of high or low expression level of NKX2–8/TTF-1 were examined (Fig. S2), suggesting that gene expression levels of these transcription factors hold no definitive prognostic implications.
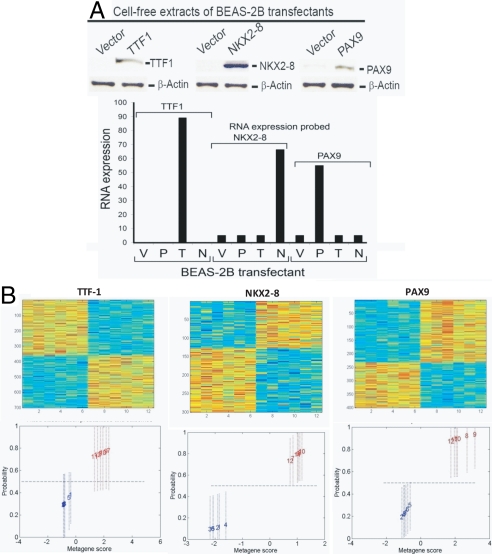
As gene expression levels of NKX2–8, TTF-1, and PAX9 could not be used to identify a clinically relevant phenotype, we hypothesized that signatures representative of coordinated activation of NKX2–8, TTF-1, and PAX9 pathways may be more informative. Using oncogene-immortalized, premalignant lung epithelial cells (BEAS-2B), pathway signatures were developed that embodied the activation of TTF-1, NKX2–8, or PAX9. Individual genes were retrovirally transduced into BEAS-2B cells for stable expression and uninfected cells were eliminated by drug selection. Quantitative assays evaluating overexpression of TTF-1, NKX2–8, and PAX9 confirmed the biochemical activation of each gene (Fig. 1A).
Fig. 1.
(A) Individual genes (NKX2–8, PAX9, and TTF-1) were retrovirally transfected into BEAS-2B cells for stable expression and uninfected cells were eliminated by drug selection. Quantitative assays evaluating TTF-1, NKX2–8, and PAX9 activation confirmed the overexpression of each gene by Western analysis (Upper) and RT-PCR (Lower). V, vector; P, PAX9; T, TTF-1; N, NKX2–8. (B) (Upper) An image intensity display of the expression levels of genes most highly weighted in a profile differentiating control cell lines from cell lines representative of activated TTF-1, NKX2–8, and PAX9. (Lower) The fitted classification probabilities of the samples used to develop the predictor. Each cell line in the training set is indicated by a sample index number and is color coded, with blue indicating control cell lines and red indicating transfected cell lines for a given signature.
Gene expression signatures represented by metagenes, which represent groups of genes that together exhibit a consistent pattern of expression in a collection of samples, were identified and correlated to the observable phenotype of activation of a transcription factor (e.g., TTF-1, NKX2–8, or PAX9), as described previously (10). This analysis selects a set of genes whose expression levels are most highly correlated with the classification of cell lines into oncogene activated/deregulated vs. control (Fig. 1B Upper). The dominant principal component from such a set of genes then defines a relevant phenotype-related metagene, and regression models assign the probability of TTF-1, NKX2–8, or PAX9 activation in an independent validation set of tumor or cell line samples.
Fig. 1B (Upper) illustrates the gene expression patterns that reflect the activity of the 3 transcription factors (TTF-1, NKX2–8, and PAX9) coded for by 3 coamplified genes in the 14q13.3 amplicon. Each individual signature distinguishes cells expressing the transcription factors from control cells (Fig. 1B Lower). As such, this result suggests that these gene expression signatures represent tools to accurately profile the biological pathway status defined by each transcription factor. Importantly, gene annotation of the individual gene expression profiles using GATHER (11) further demonstrated that the TTF-1, NKX2–8, and PAX9 signatures are composed of genes/pathways that have previously been shown to be critical in both embryonal tissue development (JAK/STAT, Wnt, BMP, and Hedgehog) (12–15) and more specifically lung development (MAPkinase, PI3kinase, and JAK/STAT) (Table S1) (15–17).
TTF-1, NKX2–8, and PAX9 Activation Correlates with Molecular and Pathologic Phenotypes of NSCLC.
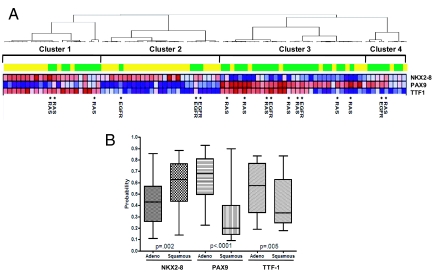
Using robust predictors of activation of TTF-1, NKX2–8, and PAX9, we explored the possibility that signatures representative of deregulated developmental pathways may further dissect lung cancer phenotypes. Fig. 2A depicts an analysis of pathway status of the 3 transcription factors in a cohort of 91 NSCLC tumors described above (GSE3141). Following prediction of transcription factor activation probabilities, the resulting probabilities of a tumor having activation of TTF-1, NKX2–8, or PAX9 pathways in the NSCLC cohort were grouped using hierarchical clustering. Three distinct clusters were observed, with cluster 1 consisting of NKX2–8/TTF-1 coactivation, cluster 2 consisting of NKX2–8 activation, and cluster 3 consisting of PAX9/TTF-1 coactivation (Fig. 2A). Interestingly, PAX9, TTF-1, and NKX2–8 activation status correlates with the tissue subtype of non-small-cell lung cancer; the majority of the adenocarcinoma samples exhibit a high probability of PAX9 and TTF-1 activation (Fig. 2B) relative to the squamous cell carcinoma samples, in line with published reports that TTF-1 protein expression is higher in adenocarcinoma than in squamous cell carcinoma (18–21). In contrast to TTF-1 and PAX9, NKX2–8 deregulation is more prominent in squamous cell carcinomas. (Fig. 2B). Furthermore, an examination of KRAS and epidermal growth factor receptor (EGFR) mutational status in these tumors identified 11 of 91 (12%) samples with KRAS (codon 12) mutations (11/45, 24.4% of adenocarcinomas) and only a small number of samples, 6 of 91 (6.5%), contained EGFR mutations (exons 18, 19, and 21), all restricted to adenocarcinomas (Fig. 2A), consistent with current literature (22, 23). Interestingly, among the 2 largest clusters of patients, only 3 of 11 (27%) of KRAS mutations were confined to tumors with NKX2–8 and TTF-1 coactivation while 5 of 11 (45%) were confined to tumors with coactivated PAX9 and TTF-1, suggesting that the specific patterns of coactivation of developmental transcription factors (TTF-1, NKX2–8, and PAX9) are independent of either KRAS or EGFR mutational events.
Fig. 2.
(A) Unsupervised hierarchical clustering of patients with clinical stage I NSCLC identifies unique patterns of TTF-1, NKX-2, and PAX9 coactivation (red, high level of activation; blue, low level of activation) in our cohort of 91 NSCLC (GSE3141) tumors that includes comparable numbers of squamous cell carcinoma (yellow) and adenocarcinoma (green). *, KRAS and EGFR mutations. (B) Prediction of PAX9, TTF-1, and NKX2–8 activation in NSCLC and correlation with the histologic subtype of NSCLC. The majority of the adenocarcinoma samples exhibit a high probability of PAX9 (P < 0.001, Mann–Whitney) and TTF-1 (P = 0.005, Mann–Whitney) activation relative to the squamous cell carcinoma samples. This is in contrast to NKX2–8 deregulation, which is prominent in squamous cell carcinomas (P = 0.002, Mann–Whitney).
We further explored the pathway coactivation of NKX2–8 and TTF-1 coactivation in a series of tumors in which KRAS was spontaneously activated by homologous recombination in adult mice (24). In these animal lung tumors, TTF-1 and NKX2–8 coactivation was observed in a subgroup, similar to the pattern of coactivation seen in human lung tumors. Importantly, not all of the KRAS-dependent murine tumors had evidence of TTF-1/NKX2–8 coactivation (Fig. S3). Taken together, these results further support the hypothesis that coactivation of the developmental pathways TTF-1 and NKX2–8 represents an important, potentially aggressive phenotype of NSCLC that is not defined just by the RAS pathway.
TTF-1 and NKX2–8 Coactivation and Survival Patterns in NSCLC.
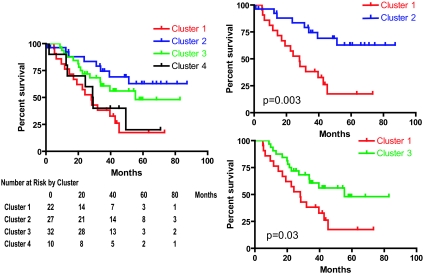
Using survival as a clinically relevant phenotype, we investigated the extent to which analysis of the activation status of the developmental pathways delineates prognosis in lung cancer. Kaplan–Meier survival curves of the 3 independent clusters identified in Fig. 2A were analyzed independent of tumor histology (squamous vs. adenocarcinoma); concerted activation of TTF-1 and NKX2–8 target genes (cluster 1) identified a population of patients (22/91, 24%) with poor survival when compared to cluster 2 (activation of NKX2–8 only) (P = 0.002) or cluster 3 (activation of PAX9 and TTF-1) (P = 0.02) (Fig. 3). When subgroup analyses involving only activation of any one transcription factor were performed, no statistically significant survival difference was observed (P > 0.3) (Fig. S4). Importantly, the poor prognosis cluster of patients represented by coactivation of both TTF-1 and NKX2–8 had a survival pattern similar to that of patients with metastatic NSCLC (Fig. S5, n = 37), suggesting that the coactivation of developmental pathways TTF-1 and NKX2–8 in a group of early stage patients portends a poor overall survival that phenotypically mimics metastatic disease.
Fig. 3.
Kaplan–Meier survival analysis of the different clusters showing significant differences (P = 0.01, log rank). Cluster 1, represented by coactivation of TTF-1 and NKX2–8 had the worst prognosis compared to the survival patterns of those in clusters 2 and 3 (P = 0.002 and P = 0.02, log rank, respectively). The number of patients at risk by clusters at 0, 20, 40, 60, and 80 months is shown for clarity (Left).
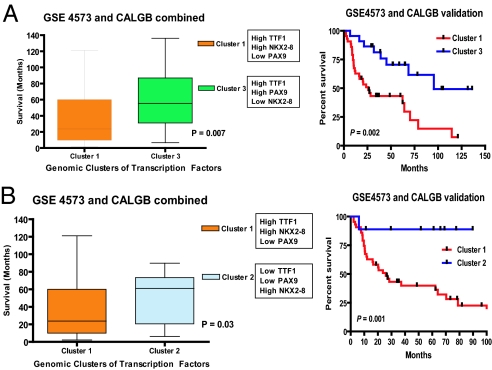
Further validation of the prognostic importance of cluster 1 (TTF-1, NKX2–8 coactivation) was performed in an independent pooled analysis based on tumor samples derived from 2 large independent data sets, 84 adenocarcinoma samples (GSE3593) and 130 squamous carcinoma samples (GSE4573) (25, 26). In this large validation cohort (n = 214), coderegulation of TTF-1 and NKX2–8 pathways was also associated with the worst outcome, when compared to other combinations of transcription factor activation (clusters 2 and 3) (Fig. 4 A and B). Importantly, a similar trend was seen when we repeated the analysis using either the GSE3593 or the GSE4573 data set alone (Fig. S6 and Fig. S7). As further confirmation, univariate and multivariate analyses (Table 1) revealed a statistically significant (univariate Cox proportional hazards model, P = 0.009) prognostic implication of NKX2–8/TTF-1 coactivation (cluster 1), independent of traditional prognostic criteria such as tumor size, vascular invasion, lymph node status, or stage of disease (multivariate Cox proportional hazards model, P = 0.03).
Fig. 4.
(A) Independent validation (n = 214) of the poor prognosis associated with coactivation of TTF-1 and NKX2–8. Box and whisker plots of survival are plotted against the probability of samples being classified as having high expression of TTF-1 and NKX2–8. Patients represented by cluster 1 (high TTF-1 and NKX2–8) have a significantly poorer outcome when compared to cluster 3 (high TTF-1 and PAX9, Left) and confirmed in independent Kaplan–Meier survival analysis (Right). (B) Box and whisker plots of survival plotted against the probability of samples being classified as having high expression of TTF-1 and NKX2–8. Patients represented by cluster 1 (high TTF-1 and NKX2–8) have a significantly poorer outcome when compared cluster 2 (high NKX2–8 alone, Left) and confirmed in independent Kaplan–Meier survival analysis (Right).
Table 1.
Univariate and multivariate Cox proportional hazards models of clinical, pathologic, and genomic variables
| Hazard ratio | Lower 95% CI | Upper 95% CI | P-value | |
|---|---|---|---|---|
| Univariate Cox proportional hazards model | ||||
| Age | 1.02 | 0.99 | 1.05 | 0.2175 |
| Tumor size | 1.24 | 1.07 | 1.44 | 0.0041 |
| Gender | 1.05 | 0.57 | 1.92 | 0.8797 |
| Histologic subtype | 1.02 | 0.57 | 1.83 | 0.9499 |
| Vascular invasion | 2.89 | 1.55 | 5.39 | 0.005 |
| Lymphatic invasion | 2.49 | 1.21 | 5.15 | 0.0106 |
| Lymph node status | 1.96 | 1.05 | 3.67 | 0.0311 |
| Pleural invasion | 1.69 | 0.81 | 3.52 | 0.1544 |
| Pathologic stage | ||||
| Stage I/stage II | 1.64 | 0.82 | 3.3 | 0.004 |
| Stage I/stage III | 5.62 | 2.12 | 14.86 | |
| Genomic cluster | ||||
| Cluster 2/cluster 1 | 0.28 | 0.12 | 0.66 | 0.0095 |
| Cluster 3/cluster 1 | 0.45 | 0.22 | 0.92 | |
| Cluster 4/cluster 1 | 0.83 | 0.34 | 2.02 | |
| Multivariate Cox proportional hazards model | ||||
| Hazard ratio | Lower 95%CI | Upper 95%CI | P-value | |
| Tumor size | 1.22 | 1.02 | 1.46 | 0.03 |
| Vascular invasion | 2.50 | 0.97 | 6.47 | 0.07 |
| Lymphatic invasion | 0.93 | 0.3 | 2.92 | 0.90 |
| Lymph node status | 0.78 | 0.27 | 2.3 | 0.65 |
| Pathologic stage | ||||
| Stage I/stage II | 1.42 | 0.45 | 4.42 | 0.11 |
| Stage I/stage III | 4.87 | 1.09 | 21.84 | 0.01 |
| Genomic cluster | ||||
| Cluster 2/cluster 1 | 0.33 | 0.13 | 0.86 | 0.03 |
| Cluster 3/cluster 1 | 0.64 | 0.26 | 1.54 | |
| Cluster 4/cluster 1 | 1.28 | 0.45 | 3.66 | |
Only significant factors with univariate P < 0.05 were included in the multivariate model. Likelihood-ratio test against full model: P = 0.005. The likelihood-ratio test was done for the multivariate model with and without a genomic cluster. CI, confidence interval.
As such, it appears that the ability to integrate developmental pathway analysis by identifying patterns of coderegulation does provide a mechanism to better categorize patients with lung cancer. Importantly, this analysis demonstrates the strong association of TTF-1 and NKX2–8 coactivation in non-small-cell lung cancer with an adverse clinical outcome.
TTF-1 and NKX2–8 Coactivation and Resistance to Cisplatin Therapy.
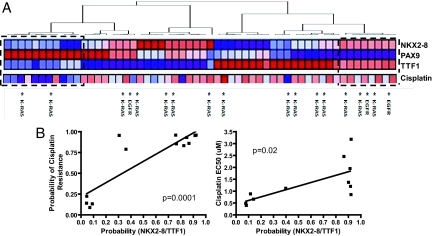
An ability to identify patients at high risk for an adverse outcome also emphasizes the need for better therapeutic strategies, particularly in NSCLC. Platinum compounds constitute the backbone of combination chemotherapy in NSCLC, although only 30–40% of patients with NSCLC respond to cisplatin-based therapy (27). Using a previously validated gene expression signature of cisplatin resistance, we compared the correlation between the likelihood of TTF-1/NKX2–8 coactivation (the worst prognosis cohort) and cisplatin resistance in a cohort of 56 lung cancer cell lines. As shown in Fig. 5A, clusters similar to those seen in patient samples were also seen in this large cohort of NSCLC cell lines. More importantly, PAX9 activation was associated with cisplatin sensitivity while coactivation of TTF-1 and NKX2–8 had a direct linear relationship with likelihood of cisplatin resistance (P = 0.0001, r = 0.87, Fig. 5B Left). This association was confirmed using drug sensitivity assays, wherein the EC50s of cisplatin in the cluster of NSCLC cell lines that showed coactivation of TTF-1 and NKX2–8 were significantly higher than in the cluster of cell lines that did not demonstrate activation of TTF-1 and NKX2–8 (P = 0.02, r = 0.80, Fig. 5B Right), and is consistent with our previous observation. In our prior work, we used the amplified NCI-H2170 cell system of the squamous cell carcinoma subtype (which lacks transcription of TTF1) to test the tumor maintenance function of NKX2–8 (and PAX9) by stably expressing small hairpin RNA (shRNA) to knock down the protein expression of NKX2–8 (or PAX9) (5). In this study, H2170 was found to have coactivation of NKX2–8 and TTF-1 pathways and the H2170 transfectant cells of knocked-down expression of NKX2–8 presented an increased sensitivity to cisplatin (data not shown), in line with our statistical prediction and experimental observation that coactivation of TTF-1/NKX2–8 positively correlates with cisplatin resistance.
Fig. 5.
(A) Unsupervised hierarchical clustering of 56 lung cancer cell lines identifies unique patterns of TTF-1, NKX2, and PAX9 coactivation (red, high level of activation; blue, low level of activation) consistent with patterns seen in tumor samples (Fig. 3). Predicted probability of cisplatin resistance in the individual lung cancer cell lines is plotted beneath the predicted probabilities of transcription factor coactivation (red, resistance to cisplatin; blue, sensitive to cisplatin). *, KRAS and EGFR mutations. (B) (Left) A linear regression analysis of the predicted probability of cisplatin resistance plotted against the predicted probability of transcription factor coactivation of NKX2–8 and TTF-1 (dashed box in A) reveals a positive correlation (P = 0.0001). (Right) A linear regression analysis demonstrates a significant relationship between the EC50 of cisplatin and the predicted probability of transcription factor coactivation of NKX2–8 and TTF-1 (dashed box in A) in lung cancer cell lines (P = 0.02).
To determine if the coactivation of TTF-1/NKX2–8 was correlated with other commonly used chemotherapy agents in the treatment of NSCLC, we examined the sensitivity of the lung cancer cell lines with coactivation of TTF-1/NKX2–8 to other cytotoxic agents commonly used as therapeutic options in patients with NSCLC (i.e., pemetrexed, paclitaxel, gemcitabine, and vinorelbine). Fig. S8 shows that coactivation of TTF-1/NKX2–8 is associated with resistance to taxanes, gemcitabine, and vinorelbine (data not shown) but was positively correlated with antifolate therapy with pemetrexed (P = 0.03, r = −0.68), suggesting that pemetrexed should be considered in the initial treatment for this cohort of patients. This finding might explain, at least in part, the increased clinical response of lung adenocarcinoma to pemetrexed.
Discussion
The capacity to understand biological complexity is often limited by the ability to define relevant phenotypes. There is perhaps no better example of this challenge than that seen in cancer, in particular lung cancer. More than 180,000 new patients are diagnosed with NSCLC each year and 150,000 lung cancer-related deaths occur annually (28), yet only minimal advances in therapeutic strategies that significantly alter patient outcome have been observed over the past 2 decades. This is likely due to the complexity of the oncogenic process that involves the somatic acquisition of large numbers of mutations, coupled with the varied host genetic constitution, which produces a disease of enormous complexity that is extremely difficult to characterize and treat effectively.
Traditional methods of characterizing tumors rely on gross visual information (size of the tumor, degree of spread, histological characteristics of the tumor) that is coupled to represent the TNM staging system. Although these tools do provide a capacity to define tumor subgroups with distinct biology, it is abundantly clear that these classifications are imprecise, creating heterogeneous groupings of tumors and patients. Numerous examples can now be found where gene expression profiles have been able to dissect tumor subtypes (29, 30). The use of gene expression signatures as surrogate phenotypes has been particularly important, linking diverse experimental systems that dissect the complexity of biological systems in a way that was not previously feasible (31, 32). However, none of these studies to date have addressed the potential impact of specific developmental pathways on defining cancer phenotypes. It is very likely that tissue-specific developmental pathways may be critical in dissecting subtypes of cancer and may complement current classification systems. Here we report on the role of biological activities defined by the developmental oncogenes TTF-1, NKX2–8, and PAX9 in lung cancer. All 3 genes are localized to a unique, recurrent lung cancer amplicon at 14q13.3. Given its essential function in lung development and relative restrictive expression in lung tissue, TTF-1 is an appealing candidate as a cell lineage-survival oncogene in lung cancer, similar to the MITF oncogene specific to melanoma (2, 8). However, our recent discovery of TTF-1, NKX2–8, and PAX9 as coamplified oncogenes with functional synergism suggests that the 14q13.3 amplicon in lung cancer provides selective advantage to tumor cells by way of activation of multiple oncogenes, not just TTF-1 (5). Intriguingly, in this present study, simultaneous activation of TTF-1 and NKX2–8 pathways was associated with poor survival, in contrast to activation of individual genes that did not have prognostic significance. This finding not only echoes our thesis of multiple collaborating driver genes at the 14q13.3 amplicon, but also potentially sheds light on the prognostic value of TTF-1 protein expression, which has thus far been controversial (9, 33, 34). We believe that the prognostic relevance of TTF-1 is best evaluated with consideration of its oncogenic collaborator, NKX2–8. Furthermore, although the prognostic significance of TTF-1 and NKX2–8 coactivation was validated in 2 large patient data sets and the impact on survival in early stage NSCLC was found to be independent of standard clinico-pathologic variables such as tumor size and stage (in the univariate and multivariate analyses shown in Table 1), we emphasize, however, that there are several approaches to refining prognosis in early stage NSCLC and the aims of this study were not to develop yet another prognostic model. Instead, in the present context, we use survival merely as a phenotype to demonstrate the biologic relevance of TTF-1 and NKX2–8 coactivation in lung tumors. To further elaborate, 3 large studies in lung cancer have been described, all of which were specifically designed to find a prognostic gene signature in early stage NSCLC using supervised classification on gene expression data from lung tumor samples with known clinical outcomes (25, 35, 36). Although the number of genes that overlap between the 3 independently derived gene sets is minimal, recent data suggest that because all 3 gene signatures could be independently validated in large patient cohorts (25, 35, 36), it is more likely that these gene signatures use different reporter genes to monitor the same biological pathways or processes. When comparing the pathways involved in the TTF-1 and NKX2–8 signatures, coactivation is associated with a higher likelihood of recurrence in our present study. In addition, several biologically relevant pathways are now identified (e.g., MAPKinase signaling, JAK/STAT signaling, Wnt signaling, etc.) that are common among the 3 previously described signatures of recurrence involved in activation of TTF-1 and NKX2–8 (Table S2). Despite this observed overlap in biologic pathways, it is important to note that none of the prognostic models described to date, including our own previous work (25), have shown predictive ability in being able to identify patients resistant to standard cytotoxic therapy or sensitive to specific targeted strategies.
The ability to identify a poor prognosis group of patients with early stage NSCLC characterized by NKX2–8 and TTF-1 coactivation whose survival pattern mimics that of patients with more advanced disease (Fig. S4) suggests that platinum-based chemotherapy, the standard of care for patients with advanced disease, may be beneficial in this “high-risk” group as well. However, in sharp contrast, we found that patients defined by the poor prognosis pattern (of TTF-1 and NKX2–8 coactivation) were in fact more likely to be resistant to cisplatin therapy. This suggests that a rational strategy to targeted therapy in the high-risk phenotype represented by NKX2–8 and TTF-1 coactivation is needed to treat this cohort of patients. We also emphasize that although these findings are encouraging, prospective validation strategies that are currently being initiated are needed before implementation into clinical practice.
In conclusion, the developmental oncogenes TTF-1, NKX2–8, and PAX9 are important biologically and clinically in lung cancer initiation and progression. Gene signatures reflecting the status of these transcription factors and patterns have prognostic and predictive value and may provide a rational approach to individualizing therapeutic options for a given patient with NSCLC.
Methods
Overexpression of NKX2–8, PAX9, and TTF-1.
Oncogene-immortalized, premalignant normal human lung epithelium (BEAS-2B) was purchased from American Type Cell Collection. Retrovirus-mediated gene transfer was performed as described previously (5) with expression vectors bearing each individual transcription factor and 1 of 3 types of selection markers: puromycin, hygromycin, or blasticidin. In each instance, cells were harvested for preparation of total RNA. Infections of each individual transcription factor were performed 10 times to generate sufficient consistent data for pattern analysis. Quantitative assays including Western analysis and RT-PCR were performed to evaluate the extent of overexpression and activation of TTF-1, NKX2–8, and PAX9 (Fig. 1A). Anti-TTF-1, anti-NKX2–8, and anti-PAX9 antibodies were purchased from abCam, Invitrogen, and GenWay, respectively.
Clinical Data.
The “discovery” data set consists of 91 early stage NSCLC tumors (Ia/Ib, IIa/IIb, and IIIa) (GSE3141) identified from the Duke Lung Cancer Prognostic Laboratory (10). This data set represents equal numbers of squamous and adenocarcinoma samples. The validation sample set included 214 samples: 84 lung adenocarcinomas (GSE3593) (25) and 130 lung squamous cell carcinomas (GSE4573) (26). Details of the relevant demographic and clinical information by data set are shown in Table S3.
Gene Expression Arrays.
Total RNA from stable transfectant cells overexpressing each individual transcription factor was extracted using the QiaShredder and Qiagen RNeasy Mini kit. RNAs were quantified using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies) and quality was assessed with an Agilent 2100 Bioanalyzer. Total RNAs were amplified by a modified Eberwine technique (37), using a Message Amp II kit (Ambion) for 2 rounds of amplification. The resulting biotin-labeled RNA samples were then hybridized with Affymetrix U133Plus2.0 GeneChip arrays according to the manufacturer's instructions. All analyses were performed in a minimal information about a microarray experiment (MIAME)-compliant fashion, as defined in the guidelines established by MGED (www.mged.org). The gene expression data used in the analyses are available at GEO (GSE9212, GSE3141, GSE3593, and GSE4573).
Gene Expression Analysis.
Gene expression data obtained from cell lines transfected with each individual transcription factor were used to populate a matrix with MATLAB software. Statistical modeling representing activation of the individual transcription factors (NKX2–8, PAX9, and TTF-1) was performed using metagene construction and binary prediction analysis, as described previously (10, 25, 38). Gene expression signatures that reflect the activity of a given transcription factor were applied to a clinically annotated data set of 91 tumor samples (GSE3141) to predict patterns of transcription factor activation. Predictions of transcription factor pathway activation within the tumor samples were evaluated to produce estimated relative probabilities and associated measures of uncertainty of transcription factor activation across the validation set. An estimated probability of 0.5 was classified as high probability of transcription factor activation and a probability of <0.5 was classified as low probability of transcription factor activation. Applications of the transcription factor predictors to other data sets (GSE4573, GSE3593) and the gene expression-based predictor of cisplatin resistance (38) to lung cancer cell lines and lung tumor data sets (GSE3141) were performed as described above. See SI Methods for complete details.
Hierarchical clustering using the gene pattern software (http://www.broad.mit/edu/cancer/software/genepatter) based on the probability of transcription activation of the tumor samples was used to generate clusters with coactivation of specific transcription factors. Specifically, the clustering was performed using a complete linkage with a Pearson correlation metric method on the preprocess data to organize all of the data elements into a single tree with the highest levels of the tree representing the discovered classes (29).
Standard Kaplan–Meier mortality curves and their significance level were generated to evaluate the prognostic role of individual transcription factors and in clusters of patients with coactivated transcription factors, using the graph pad software. The log-rank test was used to assess the differences between the survival curves and to calculate the nominal P-values between groups.
Supplementary Material
Acknowledgments.
The authors thank the Duke University microarray core facility and the Cold Spring Harbor Laboratory microarray facility for collecting the microarray data. A.P. was supported by the Emilene Brown Cancer Research Fund and by grants from the Jimmy V. Foundation, the American Cancer Society, and the Burroughs Wellcome Fund. D.M. was supported by the Joan's Legacy Foundation, a National Institutes of Health grant (1R01CA127547-01), and a Penn State University Pathology Internal Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900827106/DCSupplemental.
References
- 1.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 2.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB. Addiction to oncogenes—the Achilles heel of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 4.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 5.Kendall J, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwei KA, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 9.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 11.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 12.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 13.Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M. Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns. 2004;4:713–717. doi: 10.1016/j.modgep.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 15.Nakasato M, et al. Involvement of the STAT5 signaling pathway in the regulation of mouse preimplantation development. Biol Reprod. 2006;75:508–517. doi: 10.1095/biolreprod.105.047860. [DOI] [PubMed] [Google Scholar]
- 16.Cantrell DA. Phosphoinositide 3-kinase signalling pathways. J Cell Sci. 2001;114:1439–1445. doi: 10.1242/jcs.114.8.1439. [DOI] [PubMed] [Google Scholar]
- 17.Paliga AJ, Natale DR, Watson AJ. p38 mitogen-activated protein kinase (MAPK) first regulates filamentous actin at the 8–16-cell stage during preimplantation development. Biol Cell. 2005;97:629–640. doi: 10.1042/BC20040146. [DOI] [PubMed] [Google Scholar]
- 18.Bejarano PA, et al. Surfactant proteins and thyroid transcription factor-1 in pulmonary and breast carcinomas. Mod Pathol. 1996;9:445–452. [PubMed] [Google Scholar]
- 19.Di Loreto C, et al. Immunocytochemical expression of tissue specific transcription factor-1 in lung carcinoma. J Clin Pathol. 1997;50:30–32. doi: 10.1136/jcp.50.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabbro D, et al. TTF-1 gene expression in human lung tumours. Eur J Cancer. 1996;32A:512–517. doi: 10.1016/0959-8049(95)00560-9. [DOI] [PubMed] [Google Scholar]
- 21.Khoor A, Whitsett JA, Stahlman MT, Olson SJ, Cagle PT. Utility of surfactant protein B precursor and thyroid transcription factor 1 in differentiating adenocarcinoma of the lung from malignant mesothelioma. Hum Pathol. 1999;30:695–700. doi: 10.1016/s0046-8177(99)90096-5. [DOI] [PubMed] [Google Scholar]
- 22.Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J Clin Oncol. 1998;16:1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 23.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 24.Sweet-Cordero A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 25.Potti A, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 26.Raponi M, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 27.Schiller JH, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 28.Jemal A, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 29.Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 30.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 31.Dave SS, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 32.Huang E, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 33.Barlesi F, et al. Positive thyroid transcription factor 1 staining strongly correlates with survival of patients with adenocarcinoma of the lung. Br J Cancer. 2005;93:450–452. doi: 10.1038/sj.bjc.6602717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haque AK, Syed S, Lele SM, Freeman DH, Adegboyega PA. Immunohistochemical study of thyroid transcription factor-1 and HER2/neu in non-small cell lung cancer: strong thyroid transcription factor-1 expression predicts better survival. Appl Immunohistochem Mol Morphol. 2002;10:103–109. doi: 10.1097/00129039-200206000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Gelder RN, et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu DS, et al. Pharmacogenomic strategies provide a rational approach to the treatment of cisplatin-resistant patients with advanced cancer. J Clin Oncol. 2007;25:4350–4357. doi: 10.1200/JCO.2007.11.0593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.