Abstract
Myotonic dystrophy type 1 (DM1) is an RNA dominant disease in which mutant transcripts containing an expanded CUG repeat (CUGexp) cause muscle dysfunction by interfering with biogenesis of other mRNAs. The toxic effects of mutant RNA are mediated partly through sequestration of splicing regulator Muscleblind-like 1 (Mbnl1), a protein that binds to CUGexp RNA. A gene that is prominently affected encodes chloride channel 1 (Clcn1), resulting in hyperexcitability of muscle (myotonia). To identify DM1-affected genes and study mechanisms for dysregulation, we performed global mRNA profiling in transgenic mice that express CUGexp RNA, when compared with Mbnl1 knockout and Clcn1 null mice. We found that the majority of changes induced by CUGexp RNA in skeletal muscle can be explained by reduced activity of Mbnl1, including many changes that are secondary to myotonia. The pathway most affected comprises genes involved in calcium signaling and homeostasis. Some effects of CUGexp RNA on gene expression are caused by abnormal alternative splicing or downregulation of Mbnl1-interacting mRNAs. However, several of the most highly dysregulated genes showed altered transcription, as indicated by parallel changes of the corresponding pre-mRNAs. These results support the idea that trans-dominant effects of CUGexp RNA on gene expression in this transgenic model may occur at the level of transcription, RNA processing and mRNA decay, and are mediated mainly but not entirely through sequestration of Mbnl1.
INTRODUCTION
Myotonic dystrophy type 1 (DM1) is the most prevalent form of muscular dystrophy. This disorder is caused by expansion of a CTG repeat in the 3′ untranslated region (UTR) of DMPK, a gene encoding a protein kinase (1). The mutant DMPK mRNA contains a highly expanded CUG repeat (CUGexp) and is retained in nuclear foci (2–4). Accumulation of this transcript in the nucleus leads to RNA dominant disease by interfering with the regulated expression of other genes (reviewed in 5). Expression of CUGexp RNA in transgenic mice, or RNA containing an interrupted CUG repeat, reproduces features of DM1, such as, repetitive action potentials (myotonia) and degenerative changes in skeletal muscle (6,7).
The mutant DMPK mRNA is believed to affect gene expression through several distinct mechanisms. The first indication of trans-dominant effects on RNA metabolism came from studies suggesting an effect of the mutant DMPK mRNA on polyadenylation (8). Splicing regulators in the Muscleblind-like (MBNL) family, such as MBNL1, were shown to bind to CUGexp RNA with high affinity and become sequestered in nuclear foci (9–11). Reduced MBNL1 activity leads to abnormal regulation of alternative splicing for several genes expressed in skeletal muscle and heart (10,12). MBNL proteins may also regulate mRNA localization (13), but it is unclear whether this activity is affected in DM1. In addition to effects on MBNL proteins, DM1 is associated with upregulation of two additional RNA binding proteins, CUGBP1 and hnRNP H (14–16). These proteins regulate alternative splicing, stability and translation for a group of muscle-expressed transcripts, some of which are also regulated by Mbnl1 (17–19). Moreover, CUGexp hairpins are processed by Dicer, resulting in short poly(CUG) RNAs that may induce posttranscriptional silencing of genes that contain CAG repeats (20). Finally, CUGexp RNA may influence transcription by activating signaling proteins, such as protein kinase C or the dsRNA-dependent protein kinase PKR, or by leaching transcription factors from chromatin (15,21,22).
A trans-effect of CUGexp RNA on RNA processing that is linked to functional impairment in DM1 involves the muscle-specific chloride channel, Clcn1. Expression of CUGexp RNA or ablation of Mbnl1 causes inclusion of an additional exon in the Clcn1 mRNA. This leads to a shift of the reading frame, truncation of the Clcn1 protein, loss of channel function, repetitive action potentials and delay of muscle relaxation (myotonia) (23–26). A similar derangement exists in human DM1 (23,27). Myotonic discharges are known to influence gene expression in skeletal muscle (28–31), but the full spectrum of genes responding to muscle hyperactivity has not been determined.
To identify genes and pathways impacted by CUGexp RNA, we used oligonucleotide microarrays to examine gene expression in human skeletal actin long repeat (HSALR) transgenic mice. These mice express skeletal actin mRNA containing ∼250 CUG repeats in the 3′-UTR (6). The CUGexp-containing transcript is only expressed in skeletal muscle, and the mice display characteristic features of DM1, such as sequestration of Mbnl1 protein in nuclear foci, structural changes in muscle and myotonia. To determine which changes in CUGexp-expressing mice may result from repetitive action potentials, results were compared with Clcn1 null mice that have severe myotonia. To determine which changes in CUGexp-expressing mice may result from Mbnl1 sequestration, results were compared with Mbnl1 knockout mice (12). We also used RNA immunoprecipitation to identify Mbnl1-interacting mRNAs whose expression may be affected by Mbnl1 sequestration. Our results indicate that gene expression abnormalities in DM1 may result from effects on transcription, processing and stability of RNA, and point to pathways impacted by this disease.
RESULTS
Effects of CUGexp RNA on gene expression in skeletal muscle
To determine global effects of CUGexp RNA on gene expression, we used Affymetrix Mouse Genome 430 microarrays to compare two founder lines of HSALR transgenic mice with WT controls (n = 6 per group). Of ∼45 000 probe sets on the expression arrays, ∼23 000 showed detectable gene expression in muscle (see Materials and Methods). Among the muscle-expressed transcripts, 269 were dysregulated either in transgenic line HSALR20b, transgenic line HSALR41, or both, based on criteria of a fold-change >2, nominal P-value <0.001 according to t-tests, and false discovery rate (FDR) <1% according to significance analysis of microarrays (SAM) (32) (Supplementary Material, Table S1). The number of differentially expressed transcripts was higher in line HSALR20b (n = 252) than in line HSALR41 (n = 75), and the fold-change was generally greater (P < 0.001 for paired t-test among transcripts differentially expressed in both lines). These results were consistent with our previous observations that the level of CUGexp expression, severity of myotonia and extent of muscle histopathology were greater in line HSALR20b than in line HSALR41 (6). Out of 75 transcripts that were differentially expressed in line HSALR41, 73 (97%) showed a similar change in line HSALR20b (change in the same direction with a nominal P-value <0.01, Supplementary Material, Table S1). Out of 252 transcripts that were differentially expressed in line HSALR20b, 175 transcripts (69%) showed a similar change in line HSALR41 (change in the same direction with a nominal P-value <0.01). These results indicated that effects of CUGexp RNA on gene expression were broadly similar in both HSALR founder lines, but the effect size was greater in line HSALR20b. We therefore focused our subsequent analyses on line HSALR20b. However, to eliminate changes that may depend on the site of transgene integration or extreme levels of CUGexp accumulation, we report only those transcripts dysregulated in line HSALR20b that also showed a similar, if less profound change in line HSALR41 (i.e. transcripts showing a fold-change >2 with nominal P-value <0.001 and FDR <1% in line HSALR20b, and a change in the same direction with nominal P < 0.01 in line HSALR41). These transcripts constitute the ‘dysregulated in HSALR’ set.
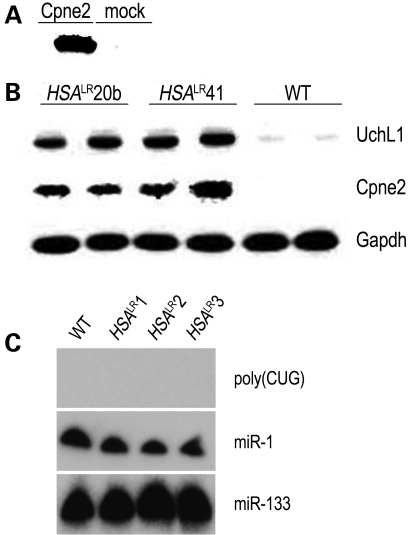
By these criteria, 175 transcripts were dysregulated in HSALR mice, comprising 110 transcripts that were upregulated and 65 that were downregulated (Supplementary Material, Table S2). The 10 most-upregulated and downregulated transcripts are shown in Table 1. Quantitative RT–PCR analysis confirmed the change in mRNA level for each of the seven transcripts that we tested (Supplementary Material, Table S3). Moreover, for two genes, Cpne2 encoding a Ca2+ binding protein involved in membrane trafficking, and Uchl1 encoding ubiquitin carboxyl-terminal hydrolase L1, we confirmed a marked upregulation of protein expression by immunoblot (Fig. 1A–B).
Table 1.
Genes most highly dysregulated in HSALR mice
| Gene | Functional category | HSALR20b |
HSALR41 |
Clcn1 null |
Mbnl1 knockout |
||||
|---|---|---|---|---|---|---|---|---|---|
| Fold-change | P(t) | Fold-change | P(t) | Fold-change | P(t) | Fold-change | P(t) | ||
| Sarcolipin | Ca2+ homeostasis | 98.3 | 2.8E−06 | 72.8 | 6.0E−07 | 15.6 | 0.010 | 42.6 | 0.176 |
| Ubiquitin carboxy terminal hydrolase L1 | Proteolysis, neuronal | 20.0 | 4.6E−12 | 25.3 | 0.003 | 1.2 | 0.102 | 16.1 | 0.002 |
| Myosin IA | Vesicle traficking | 17.9 | 1.2E−06 | 28.7 | 7.0E−05 | −1.1 | 0.362 | 1.7 | 0.007 |
| Cartilage intermediate layer protein | ECM, IGF-1 signaling | 16.9 | 3.7E−06 | 7.8 | 2.8E−04 | 3.6 | 0.008 | 5.1 | 4.4E−05 |
| Myelin protein zero-like 3 | Cell adhesion | 16.7 | 1.0E−06 | 7.7 | 1.3E−05 | −1.5 | 0.204 | 1.6 | 0.053 |
| Copine II | Ca-dependent membrane binding | 14.0 | 3.4E−06 | 10.1 | 7.2E−07 | 1.4 | 0.124 | 11.0 | 0.001 |
| Plekho1 | PI3-kinase signaling | 11.0 | 4.5E−06 | 6.3 | 5.5E−06 | 1.4 | 0.004 | 6.6 | 0.001 |
| Ectodysplasin A2 isoform receptor | Transmembrane receptor | 9.8 | 2.8E−06 | 5.2 | 1.7E−05 | −1.0 | 0.943 | 14.0 | 2.0E−04 |
| Poliovirus receptor-related 3 | Cell membrane and adhesion | 8.2 | 1.6E−04 | 2.5 | 4.3E−05 | 1.0 | 0.968 | 2.5 | 0.010 |
| Contactin associated protein-like 2 | Cell adhesion, neuronal | 7.9 | 1.4E−07 | 3.7 | 0.001 | −8.2 | 0.002 | 3.7 | 1.1E−04 |
| HtrA serine peptidase 1 | IGF signaling | −4.3 | 1.8E−05 | −2.2 | 4.7E−04 | −4.3 | 0.001 | −1.8 | 0.001 |
| Caspase 12 | Apoptosis | −5.3 | 5.8E−05 | −2.2 | 0.010 | −2.3 | 0.081 | −3.3 | 6.0E−05 |
| Solute carrier family 38, member 4 | Amino acid transporter | −5.6 | 2.6E−06 | −1.8 | 0.001 | −15.5 | 6.4E−05 | −5.4 | 0.001 |
| Dickkopf homolog 3 | Wnt signaling | −5.9 | 5.0E−05 | −3.6 | 1.6E−04 | −18.8 | 1.6E−04 | −1.8 | 0.093 |
| Mindbomb homolog 1 | Notch signaling | −7.4 | 5.6E−07 | −3.4 | 2.8E−05 | −6.0 | 0.001 | −5.0 | 0.018 |
| Gdap1 | Mitochondrial fission | −7.6 | 5.5E−05 | −2.1 | 0.007 | −123.1 | 0.021 | −3.7 | 2.5E−04 |
| Gremlin 2 homolog | BMP signaling | −7.6 | 3.2E−04 | −4.3 | 9.1E−04 | −2.6 | 0.006 | −3.1 | 4.9E−05 |
| Phospholipase A2, group VII | Inflammatory response | −9.3 | 1.5E−06 | −3.3 | 3.6E−05 | −28.5 | 0.039 | −2.8 | 0.006 |
| Tiam1 | Receptor signaling, microtubule function | −10.0 | 5.3E−05 | −3.8 | 3.4E−04 | −10.3 | 0.009 | −3.9 | 9.7E−05 |
| Kcnab1 potassium channel | Ion channel | −24.7 | 3.4E−07 | −3.6 | 2.3E−05 | −4.1 | 3.4E−04 | −21.6 | 8.4E−05 |
P(t) denotes nominal P-value for t-test.
Figure 1.
Upregulation of Cpne2 and UchL1 protein, and absence of short poly(CUG) RNA, in mice that express CUGexp RNA. (A) Specificity of anti-Cpne2 antibody. Immunoblot shows detection of appropriate 60 kDa protein in COS cells transfected with Cpne2 expression construct, but not in mock transfected cells. (B) Cpne2 protein is detected by immunoblot in muscle homogenate from HSALR20b and HSALR41 mice, but not in muscle from wild-type (WT) mice of the same strain background (FVB). Gapdh serves as loading control. A parallel blot was probed with the anti-UchL1 antibody. (C) RNA blot analysis was performed under conditions designed to detect short CUG repeats. Shown are RNAs isolated from quadriceps muscle of three HSALR transgenic mice and WT control (FVB). Probes for miR-1 and miR-133 RNAs serve as controls for RNA loading and miRNA detection. Note that short poly(CUG) RNA was undetectable even after a 20-fold longer film exposure (not shown).
Comparisons with Clcn1 and Mbnl1 null mice
Gene expression in skeletal muscle is modulated by patterns of electrical activity (33). To identify changes induced by CUGexp RNA that may result from myotonia, results in HSALR mice were compared with Clcn1 null mice that have generalized myotonia. Out of 175 transcripts dysregulated in HSALR mice, 42 showed a similar effect in the Clcn1−/− versus Clcn1+/+ comparison, defined as a change in the same direction with a nominal P-value <0.01 (Supplementary Material, Table S4, Section A). In terms of function, many genes that were dysregulated in HSALR and Clcn1−/− mice pertained to cell signaling. These results suggest that a substantial fraction (∼24%) of the major changes in gene expression in HSALR mice may result from increased electrical activity due to misregulated alternative splicing of a single Mbnl1 splicing target, Clcn1.
The set of 175 HSALR-dysregulated transcripts showed little overlap with a previous dataset from mice with muscular dystrophy due to dystrophin loss (34). Only four transcripts were dysregulated in common (Supplementary Material, Table S2). Furthermore, whereas Clcn1−/− mice showed upregulation of embryonic myosin and vimentin, two markers for muscle regeneration (35,36), the regenerative changes in HSALR20b mice were less pronounced (Supplementary Material, Table S4, Section B). These results suggest that relatively few of the changes in the HSALR-dysregulated transcript set result from a non-specific response of muscle to chronic disease or regeneration.
To identify changes induced by CUGexp RNA that depend on Mbnl1 sequestration, we compared gene expression changes in HSALR mice with Mbnl1 knockouts. We first backcrossed a disrupted allele of Mbnl1 (12) onto the same genetic background as HSALR mice (FVB strain), and then compared gene expression in Mbnl1 knockouts with their WT littermates (n = 5 per group). Among 175 transcripts in the HSALR-dysregulated set, 127 (73%) were similarly dysregulated in Mbnl1 knockout mice (fold-change >1.5, P < 0.01), and most of these were not affected in Clcn1−/− mice (70 transcripts, Supplementary Material, Table S4, Section C). However, a group of 17 HSALR-dysregulated transcripts showed no trend for altered expression either in Mbnl1 knockout or Clcn1−/− mice (nominal P-value >0.05 for both comparisons, Supplementary Material, Table S4, Section D). These results indicate that most of the major changes in gene expression in HSALR mice result directly or indirectly (via myotonia) from sequestration of Mbnl1, but that loss of function for this protein cannot account for all effects of CUGexp RNA.
Pathways dysregulated in HSALR, Mbnl1 knockout, or Clcn1−/− mice
The results presented here were based on stringent criteria for differential gene expression. However, more subtle differences in gene expression may have important consequences, particularly if several genes involved in the same pathway are affected. We used ingenuity pathway analysis (37) (IPA), gene set enrichment analysis (38) (GSEA) and expression analysis systematic explorer (39) (EASE) to test for effects CUGexp RNA, Mbnl1 loss or myotonia on functionally related genes, including effects that might not be obvious based on the largest fold differences or lowest P-values. Results of pathway analysis are summarized in Table 2, and specific findings from GSEA are shown in Supplementary Material, Table S5.
Table 2.
Pathway analysis of gene expression profiles in mouse models of DM1 and Clcn1 null mice
| HSALR20b |
HSALR41 |
Clcn1 null |
Mbnl1 knockout |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPA | GSEA | EASE | IPA | GSEA | EASE | IPA | GSEA | EASE | IPA | GSEA | EASE | |
| Signaling | ||||||||||||
| Calcium signaling and homeostasis | ↑↓ | ↑↓ | ↑↓ | ↓ | ↑↓ | |||||||
| Downregulated by TNF alpha in endothelial cells | ↓ | ↓ | ||||||||||
| Metabolism | ||||||||||||
| Glycerolipid and fatty acid metabolism | ↓ | ↑ | ↑ | ↑ | ||||||||
| Mitochondrial oxidative phosph./TCA cycle | ↑ | ↑ | ↑ | ↑ | ||||||||
| Amino acid degradation/metabolism | ↑ | ↑ | ||||||||||
| Glutathione metabolism | ↑ | |||||||||||
| Glycolysis/glycogen metabolism | ↓ | ↓ | ↓ | |||||||||
| Other | ||||||||||||
| Ribosomal proteins | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ||||||
| Translation/translation elongation | ↑ | ↑ | ||||||||||
| MyoD targets | ↑ | |||||||||||
| Microtubule binding | ↑ | |||||||||||
| Caveola | ↓ | |||||||||||
| Calcium-dependent cell adhesion molecule activity | ↓ | |||||||||||
| Purine nucleotide/GTP binding | ↓ | |||||||||||
‘↑↓’ denotes pathway in which some genes are upregulated (↑) and others are downregulated (↑).
By IPA, the pathway most prominently affected in HSALR and Mbnl1 knockout mice comprised genes involved in Ca2+ signaling and homeostasis (Supplementary Material, Table S6). These changes were complex, including upregulation of some transcripts and downregulation of others. The transcript most highly upregulated in HSALR mice encoded sarcolipin, a protein that modulates activity of the Ca2+ reuptake pump of the sarcoplasmic reticulum (Serca) (40,41). Upregulation of sarcolipin mRNA ranged from 16-fold in Clcn1−/− mice to 98-fold in HSALR20b mice by microarray, and was confirmed as 60-fold increased in both lines of HSALR mice by qRT–PCR (Supplementary Material, Table S3). Also common to Clcn1 null, Mnbl1 knockout and HSALR transgenic mice were downregulation of CaM kinase 2 alpha, CaM kinase 2 gamma, calcineurin-B and all three of the calmodulin genes expressed in skeletal muscle. However, other effects on Ca2+ signaling and homeostasis differed between Clcn1 null mice and the DM1 models (Supplementary Material, Table S6).
Clcn1−/− mice showed upregulation of genes involved in fat metabolism and oxidative phosphorylation, with a concurrent decrease in genes involved in carbohydrate catabolism, consistent with known effects of increased muscle activity on these pathways (28–30). However, these effects were generally absent in Mbnl1 knockout or HSALR mice, despite the presence of myotonia (Supplementary Material, Table S5). In contrast, a set of genes upregulated in Mbnl1 knockout and HSALR mice but not in Clcn1 null mice comprised transcripts that encode ribosomal proteins. While the statistical significance of this change was high (false detection rate <1% by GSEA and P < 10−10 by EASE), owing to coordinate upregulation of many different transcripts in this pathway, the magnitude of the effect on individual transcripts was small (1.1- to 1.3-fold increase for most transcripts).
The computer algorithms did not identify pathways that differed between HSALR and Mbnl1 knockout mice. However, inspection of gene lists did show a functional connection. Among transcripts strongly affected by inactivation of Mbnl1 (3-fold up- or downregulated in Mbnl1−/− mice with a nominal P < 0.0001, n = 39 transcripts), most were similarly dysregulated in line HSALR20b or HSALR41 (n = 31 or 79%), but eight showed no parallel change in either HSALR transgenic line. Six of these eight transcripts encoded contractile proteins of slow-twitch muscle fibers, and were strongly downregulated in Mbnl1 knockout and Clcn1−/− mice but not in HSALR mice (Supplementary Material, Table S4, Section E, see Discussion).
Mechanism of gene dysregulation in response to CUGexp RNA
Krol et al. (20) proposed that CUGexp hairpins are processed by Dicer to short CUG fragments that induce post-transcriptional silencing of transcripts that contain CAG repeats. To examine this mechanism, we performed Northern blots to detect short CUG repeat RNAs. Under conditions that readily detected miR-1 and miR-133 microRNAs, we were unable to demonstrate short poly(CUG) RNAs in skeletal muscle of HSALR mice (Fig. 1C). We also carried out BLAST analysis of mouse RefSeq mRNAs to identify potential targets for degradation by this mechanism. Out of 89 muscle-expressed transcripts that contained 19 or more nucleotides of CAG repeats, 11 showed a trend for differential expression in HSALR transgenic mice (nominal P < 0.05) that did not occur in Mbnl1 knockout or Clcn1−/− mice. Among these 11 transcripts, 8 were downregulated (−1.2- to −2.0-fold) whereas 3 were upregulated (1.3- to 2.7-fold) (Supplementary Material, Table S4, Section F). However, only one of the downregulated transcripts, Med12, satisfied stringent criteria for differential expression (see Discussion).
Changes in RNA decay may also result from misregulated alternative splicing. For example, this occurs when variant splice products contain premature termination codons (PTCs) that induce nonsense-mediated decay (NMD). In DM1 patients and mouse models, inclusion of Clcn1 exon 7a is increased, which creates a PTC that triggers NMD of the Clcn1 mRNA (23,42). Consistent with this effect, the microarray data showed reduction of Clcn1 mRNA in HSALR20b (−1.9-fold-change, P < 0.0001) and Mbnl1 knockout mice (−2.1-fold-change, P = 0.001). However, cross-referencing of other HSALR- and Mbnl1 knockout-dysregulated transcripts with an alternative splicing database (43) yielded only a single additional candidate, ninein, for dysregulation by this mechanism, and RT–PCR analysis failed to confirm a splicing change (data not shown). We also cloned and sequenced cDNAs from two genes that were highly dysregulated in HSALR20b and Mbnl1 knockout mice, ectodysplasin A2 isoform receptor (Xedar, 39 cDNA clones sequenced) and copine 2 (Cpne2, 35 clones sequenced), and found splicing patterns that were similar in WT and HSALR20b mice.
RNA processing may also influence gene expression through use of alternative 3′ ends. Microarray data suggested that this occurred for Asph/junctin/junctate, a gene encoding three functionally distinct proteins, depending on which 3′ end is utilized (44). Among seven probe sets for Asph/junctin/junctate on the arrays, only those targeting the longer isoforms, utilizing downstream 3′ ends, were upregulated 2- to 4-fold in HSALR, Mbnl1−/−, and Clcn1−/− mice. RT–PCR analysis confirmed that utilization of 3′ ends was altered (Supplementary Material, Fig. S1).
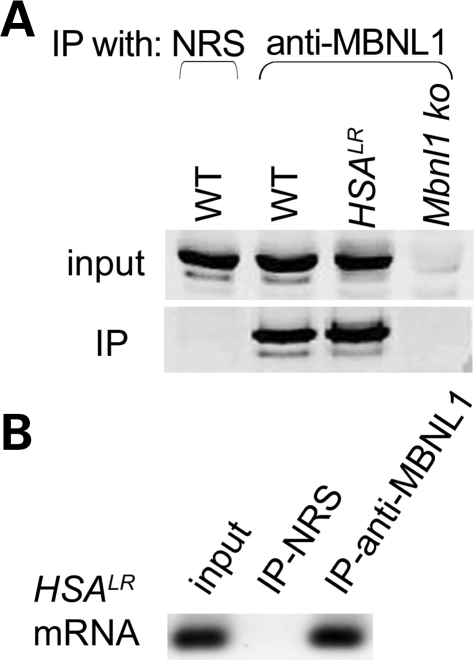
Proteins that bind to pre-mRNA to regulate splicing may also bind to mRNA to regulate stability or translation. To address this possibility for Mbnl1, we prepared muscle homogenates for immunoprecipation of ribonucleoprotein complexes using anti-Mbnl1 antibodies (Supplementary Methods). We first showed that Mbnl1 protein was highly enriched in the immunoprecipitate (IP) from WT and HSALR muscle (Fig. 2A). The HSALR (CUGexp-containing) transcript was also enriched in the IP from HSALR muscle, confirming that a known Mbnl1-interacting transcript was pulled down by this method (Fig. 2B). We then used microarrays to analyze transcripts that co-immunoprecipitated with Mbnl1 from WT muscle. A parallel IP from Mbnl1 knockout muscle served as a control for non-specific pull down of mRNA. Whereas the number of transcripts expressed in WT and Mbnl1 knockout muscle was similar (22 917 versus 23 174 probe sets detected in total cellular RNA), the number of transcripts in the IP was much higher in WT than Mbnl1−/− muscle (9144 versus 2609 probe sets detected). These findings indicate that many transcripts have direct or indirect association with Mbnl1 protein, suggesting that functions of this protein are broader than originally suspected. Notably, the spectrum of RNAs in the control (Mbnl1−/−) IP was limited, mainly comprising transcripts above the 90th percentile for expression level (Supplementary Material, Fig. S2, right panels), consistent with non-specific pull down of high abundance mRNAs. In contrast, the IP from WT mice represented a broad spectrum of expression levels, including many mRNAs of low or intermediate abundance (Supplementary Material, Fig. S2, left panels).
Figure 2.
Immunoprecipitation (IP) of ribonucleoprotein complexes containing Mbnl1 protein and Mbnl1-interacting mRNA. (A) Immunoblot shows Mbnl1 protein in homogenate prepared from WT or HSALR muscle (‘input’) but not in homogenate prepared from Mbnl1 knockout (ko) mice. Immunoblot in lower panel shows that Mbnl1 protein is Immunoprecipitated by affinity-purified anti-Mbnl1 antibody but not by normal rabbit serum (NRS). (B) RT–PCR assay showing that a known Mbnl1-interacting mRNA (HSALR mRNA) is co-Immunoprecipitated by anti-Mbnl1 antibodies by not by non-immune serum.
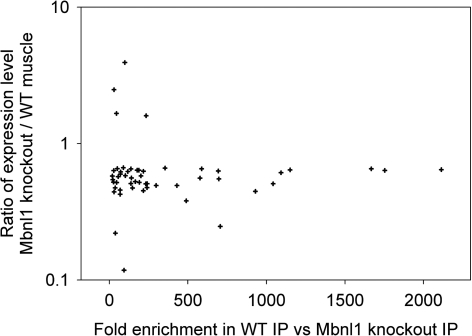
To test for effects of Mbnl1 ablation on expression of Mbnl1-interacting mRNAs, we next defined a set of transcripts that were misregulated in Mbnl1 knockout mice in a myotonia-independent fashion (fold-change >1.5 with nominal P < 0.01 in Mbnl1−/− versus Mbnl1+/+ mice, P > 0.05 in Clcn1−/− versus Clcn1+/+ mice). This set included 733 probe sets, of which 339 (46%) were downregulated and 394 (54%) were upregulated. We compared this transcript set with transcripts that were highly enriched in the anti-Mbnl1 IP (>50-fold higher signal in the WT than Mbnl1−/−-IP, n = 847, see Supplementary Methods). The intersection comprised 52 transcripts, of which 48 (92%) were downregulated in Mbnl1−/− muscle, whereas only 4 (8%) were upregulated (Fig. 3, Supplementary Material, Table S7). The overrepresentation of downregulated transcripts (P < 0.001) among Mbn1-interacting mRNAs suggests that one mechanism for gene dysregulation in Mbnl1-deficient muscle involves destabilization of transcripts that normally interact with Mbnl1 protein. However, it appears that relatively few of the major changes induced by CUGexp RNA can be accounted for by this mechanism. Only five transcripts in the HSALR-dysregulated set were highly enriched in the anti-Mbnl1 IP, all of which were downregulated (Supplementary Material, Table S7).
Figure 3.
Overrepresentation of downregulated transcripts among Mbnl1-interacting mRNAs that are dysregulated in Mbnl1 knockout mice. Vertical axis shows fold-change in expression level in Mbnl1 knockout mice, expressed as a ratio with WT mice.
Altered transcription is another putative mechanism for gene deregulation in DM1. Previously, we found that CUGexp RNA binds to and activates the double-stranded-RNA-dependent protein kinase, PKR, in vitro (21). Activation of this kinase in vivo would be expected to induce genes involved in the interferon response. However, HSALR mice did not show upregulation of interferon-responsive genes (data not shown). Mutant DMPK mRNA also is reported to cause leaching of transcription factors RARγ and Sp1 from chromatin, resulting in downregulation of several transcripts, including Clcn1, Sp1, Sp3 and RARγ in myogenic cells (22). Among these target transcripts, however, only the expression of Clcn1 was reduced in HSALR lines, an effect that is not caused by reduced transcription (42). To test for effects on transcription more directly, we used competitive RT–PCR to quantify pre-mRNA levels for several genes that showed major dysregulation in HSALR and Mbnl1 knockout mice. There was marked upregulation of Cpne2 mRNA (14-fold and 11-fold increase for HSALR and Mbnl1 knockout mice, respectively, P < 0.0001) and protein (Fig. 1) in DM1 models, but the level of pre-mRNA was not different from WT controls, supporting a post-transcriptional mechanism for dysregulation of this gene. However, levels of pre-mRNA for Uchl1, ectodysplasin A2 isoform receptor (Xedar), voltage-gated K+ channel β subunit 1 (Kcnab1), myosin 1A (Myo1A) and sarcolipin (Sln) all showed changes in HSALR and Mbnl1 knockout mice that paralleled the change in mRNA (Table 3), supporting a transcriptional mechanism. These results indicate that altered transcription is an important disease mechanism in mouse models of DM1.
Table 3.
Competitive RT–PCR analysis of pre-mRNA
| Gene |
HSALR41 versus WT |
HSALR20b versus WT |
Mbnl1−/− versus Mbnl1+/+ |
|||
|---|---|---|---|---|---|---|
| FC | P(t) | FC | P(t) | FC | P(t) | |
| Sarcolipin | 16 | 0.02 | 10 | 0.001 | 4.9 | 0.01 |
| Ubiquitin carboxy-terminal hydrolase L1 | 3.8 | 0.02 | 2.9 | 0.04 | 2.2 | 0.09 |
| Copine II | 0.77 | 0.17 | 1.1 | 0.27 | 1.0 | 1.0 |
| Ectodysplasin A2 isoform receptor | 2.5 | 0.06 | 3.1 | 0.003 | 2.0 | 0.04 |
| Kcnab1 | −1.5 | 0.03 | −4.0 | 0.002 | −2.0 | 0.13 |
| Myosin IA | 5.7 | 0.13 | 1.7 | 0.27 | 5.0 | 0.03 |
P(t) denotes P-value for t-test; FC, fold-change.
DISCUSSION
Studies of RNA dominance in DM1 have focused mainly on misregulated alternative splicing in cardiac and skeletal muscle. The present study is the first to examine global effects of CUGexp RNA on gene expression. The major findings are that CUGexp transcripts induce specific changes in gene expression, and that similar but non-identical changes occur in Mbnl1 knockout mice. Together with previous observations that splicing abnormalities in HSALR transgenic and Mbnl1 knockout mice are quite similar (10,12), these findings suggest that sequestration of Mbnl1 is the predominant mechanism for CUGexp-induced re-modeling of the muscle transcriptome in this transgenic model. While many of the expression changes can be attributed to hyperexcitability, as evidenced by similar changes in Clcn1−/− mice, it appears that most of the effects of CUGexp RNA or Mbnl1 loss are independent of myotonia. Furthermore, gene expression changes in HSALR mice show little overlap with dystrophin null mice (34), suggesting that changes in these DM1 models mainly reflect a specific response to CUGexp RNA rather than a non-specific consequence of muscle injury or repair. Taken together, our results support the hypothesis that CUGexp-Mbnl1 interaction is a critical event in DM1 pathogenesis, and strengthen the rationale for targeting this interaction as a therapeutic strategy.
A subset of changes (at least 10%) induced by CUGexp RNA did not occur in Mbnl1 knockout or Clcn1 null mice. As Mbnl2 and Mbnl3 proteins are also sequestered by CUGexp RNA (45), it is possible that these changes may result from combinatorial loss of several proteins in the muscleblind-like family. Alternatively, these changes may result from overexpression of CUG-BP1 or NKX-2.5, or other CUGexp-binding proteins that have not yet been identified.
Mouse models of DM1 used in this study have recognized limitations. Most importantly, the circumstances of RNA toxicity that exist in patients with DM1 are not fully recapitulated in HSALR mice. The (CTG)250 repeat in HSALR mice is ∼10-fold shorter than repeat tracts in muscle tissue of DM1 patients (46–48). Unlike DM1, the mutant transcript in HSALR mice is not expressed in myogenic precursor cells or subsynaptic nuclei (49), and therefore cannot replicate problems with muscle regeneration or synaptic function that may exist in DM1. Furthermore, the CUGexp element is expressed in the context of a different 3′-UTR and does not contain any other sequence from the DMPK gene. Presently, it is unclear exactly how these differences may relate to the failure of HSALR mice to replicate certain biochemical [upregulation of CUGBP1 (14,18) and NKX2-5 protein (50)] or histologic (progressive muscle wasting) features of DM1, or whether humans and mice may differ in their response to CUGexp RNA. These considerations suggest that the changes in gene expression observed in this study could reflect a subset of all changes that may occur in human DM1 muscle.
Our results point to calcium signaling and homeostasis as an aspect of muscle biology especially prone to derangements in DM1. A structure that may be particularly impacted is the sarcoplasmic reticulum (SR). The transcript showing the most extreme splicing change in DM1 is Serca1 (10,51), encoding the calcium reuptake pump of the SR. This splicing switch results in the expression of Serca1 protein with a different carboxyl terminus, but the functional impact has not been determined. Intriguingly, the gene most highly upregulated in DM1 models encodes sarcolipin, a modulator of Serca1 function. We have not determined the extent to which sarcolipin protein is overexpressed, because we were unable to generate antibodies to this protein, but the levels of sarcolipin mRNA were increased 60-fold in both founder lines of HSALR mice. In cardiac myocytes, sarcolipin overexpression caused reductions of Ca2+ re-uptake, Ca2+ transient amplitude and contractility (40,52,53), suggesting that sarcolipin overexpression may alter the function of striated muscle. In addition to Serca1 and sarcolipin, other sarcoplasmic reticulum-associated genes whose expression or splicing is altered in DM1 models include Asph, triadin and ryanodine receptor.
Increased muscle activity modulates gene expression in several pathways, but chiefly those involved in energy metabolism, contractility and calcium homeostasis. Experimentally, such effects have been induced by chronic low frequency electrical stimulation of muscle fibers, but similar changes are observed without external stimuli in Clcn1 null mice, and are driven by myotonic discharges (28–30). Our results provide a more comprehensive picture of myotonia-induced alterations of gene expression than was previously available. Consistent with previous findings, Clcn1−/− mice showed altered expression of genes involved in energy metabolism (upregulation of genes involved in fat metabolism and oxidative phosphorylation, downregulation of carbohydrate catabolism), contractility (increase of myosin heavy chain 2A) and calcium signaling (downregulation of CaM kinases and calmodulins). While all mice that we examined have myotonia, the pattern of activity-dependent gene expression in DM1 models differed from Clcn1−/− mice in several respects. In terms of genes involved in energy metabolism, the changes in Mbnl1 knockout and HSALR mice were much less apparent than in Clcn1−/− mice. Whereas changes in calcium signaling and homeostasis were conspicuous in all myotonic mice, the changes in DM1 models were qualitatively different (Supplementary Material, Table S6). These differences may reflect subtotal loss of Clcn1 channels and less severe myotonia in the DM1 models (25). Alternatively, it is possible that CUGexp RNA may have modified the transcriptional response to muscle hyperexcitability, enhancing certain aspects while repressing others.
An unexpected finding in Clcn1−/− and Mbnl1 knockout mice was the marked downregulation of genes encoding contractile proteins of type 1 muscle fibers (Supplementary Material, Table S4, section E). These data suggest that fiber-type transformations driven by muscle activity are not invariably characterized by a unidirectional shift from fast to slow, but in the particular case of myotonia there is a convergence from both ends of the spectrum (type 1 or type 2B) on an intermediate contractile phenotype. When compared with continuous low frequency stimulation, the electrical discharges in myotonic muscle are higher frequency (40–150 Hz) and intermittent, which may resemble natural firing patterns in type 2 muscle fibers. This pattern of electrical activity may repress the program of gene expression for type 1 fibers. Notably, these changes did not occur in HSALR mice. We suggest that this disparity results from decreased severity of chloride channelopathy in type 1 fibers of HSALR mice, which may reflect differences in the stoichiometry of CUGexp RNA and Mbnl1 protein in these fibers.
Effects of CUGexp RNA on the transcriptome must reflect changes in the balance of RNA synthesis and decay. Among several potential effects of CUGexp RNA on mRNA decay, our findings suggest that destabilization of Mbnl1-interacting mRNAs may impact the greatest number of genes, but the magnitude of the expression change was typically modest (−1.5- to −3.1-fold). Further work will be needed to determine whether Mbnl1 protein has a direct or indirect interaction with these mRNAs, and how the interaction may affect stability. While misregulated alternative splicing can also impact mRNA decay, as shown for Clcn1, we could not identify other candidates for dysregulation by this mechanism. However, because PTC-containing transcripts are underrepresented in the EST/mRNA database, other examples of increased turnover via NMD may have been overlooked.
Processing of CUGexp RNA to short poly(CUG) RNAs may induce posttranscriptional silencing of genes containing CAG repeats (20). Although we were not able to directly demonstrate short CUG repeat RNAs by Northern blot, our results indicate that Med12 is a candidate for this effect. Med12 encodes the transcriptional coactivator mediator, mutations of which are associated with infantile hypotonia and mental retardation (54), making it an interesting candidate for the involvement in congenital DM1. We suggest that this transcript is selectively vulnerable to short poly(CUG) because it contains five distinct CAG repeat tracts, each ≥15 nt in length. Notably, each of these CAG tracts is conserved in human MED12. However, few other CAG-repeat-containing transcripts showed downregulation. Our data indicate that posttranscriptional silencing of (CAG)n-containing transcripts is not a prevalent mechanism for gene dysregulation in CUGexp-expressing mice.
By examining six of the more extreme changes in gene expression, we found that most are accompanied by a parallel change in the level of pre-mRNA, supporting an effect of CUGexp RNA on transcription. The mechanism for this effect is presently unknown. GSEA analysis of promoter regions in genes that are dysregulated, when compared with genes not affected, did not show enrichment for known transcription factor binding sites (data not shown). Mbnl1 is not known to have DNA binding or transcriptional-regulatory activity. While we cannot eliminate the possibility that this protein may directly regulate transcription, it seems more likely that effects of Mbnl1 loss on transcription are indirect, via post-transcriptional modulation of other genes that encode transcription factors. Of note, Ebralidze et al. (22) have postulated that transcription factors are ‘leached’ from chromatin by mutant DMPK transcripts. However, this mechanism cannot account for HSALR-dysregulated genes that show similar changes in Mbnl1 knockout mice, unless Mbnl1 itself is a transcription factor.
MATERIALS AND METHODS
Mouse strains
HSALR transgenic mice in lines 20b and 41 were derived and maintained on the FVB inbred background (6). Clinical and electromyographic myotonia was verified in each mouse as previously described (42). Mbnl1 knockout mice (12) were backcrossed six generations onto the FVB background. Clcn1 null myotonic mice were obtained from Jackson Laboratory (strain adr-mto2J, BALB background). Mice were studied according to guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care.
Experimental groups
Microarray analysis was carried out on 34 mice in seven experimental groups. Homozygous HSALR transgenic mice in line 20b were compared with WT controls (FVB background, n = 6 mice per group, age 6 months). Homozygous HSALR transgenic mice in line 41 (n = 6) were compared with the same WT FVB controls. Mbnl1 knockout mice were compared with Mbnl1+/+ littermates (n = 5 per group, age 3 months). Clcn1−/− mice were compared with Clcn1+/+ littermates (n = 3 per group, age 3 months). Clcn1 and Mbnl1 knockouts were studied at an earlier age because they display increased mortality after age 5 months.
RNA isolation and microarray hybridization
Mice were euthanized and vastus (quadriceps) muscle was immediately dissected, flash frozen in liquid nitrogen and stored at −70°C. Total cellular RNA was extracted using Tri-reagent (Molecular Research Center). Biotinylated cRNA was generated and used to probe Affymetrix microarrays (Santa Clara, CA, USA) as previously described (55). HSALR20b, HSALR41, FVB WT, Clcn1−/− and Clcn1+/+ samples were processed in parallel and analyzed on Mouse Genome 430 A/B GeneChip Arrays. Because additional time was required for backcrossing Mbnl1 knockouts, the Mbnl1−/− and Mbnl1+/+ samples were processed at a later date using 430 2.0 Arrays. 430 2.0 Arrays have the same probes as the 430 A/B Arrays, but all probes are included on a single chip. Arrays were hybridized with cRNA, washed, stained with streptavidin–phycoerythrin and scanned (before and after antibody enhancement) according to standard Affymetrix protocols.
Microarray data analysis
Array data were analyzed using the gcRMA algorithm (56). Comparisons of gene expression were limited to probe sets that showed detectable expression in skeletal muscle. Expression was defined according to the Affymetrix probability of detection (Pdetection) algorithm as previously described (55). In brief, a probe set was considered non-expressed if the Pdetection was >0.1 for more than half the samples in whichever group showed the higher mean signal value. For between-group comparisons, we calculated a fold-change in mean signal intensity and a nominal P-value using an equal-variance two-tailed t-test. A nominal P < 0.001 in this study always corresponded to a FDR of <1% as defined by the SAM procedure (32). Probe sets showing off-target hybridization with CTG repeats were eliminated (see Supplementary Methods). The primary data (.CEL) files and primary analysis have been deposited in the gene expression omnibus (GEO) archive (http://www.ncbi.nlm.nih.gov/geo/).
IP of ribonucleoprotein complexes and microarray analysis
Homogenates were prepared from quadriceps muscle and then immunoprecipitated using anti-Mbnl1 antibodies (see Supplementary Methods). The co-precipitating RNA was extracted, amplified by isothermal linear amplification, converted to biotin-labeled cDNA and then analyzed on Affymetrix U430 2.0 arrays (Supplementary Methods).
Competitive RT–PCR
Microarray data were validated by RT–PCR with competitive internal standards as described (57), using n = 4 mice per each experimental group. The RT–PCR confirmations were carried out on different mice than those used in the microarray analysis. For RT–PCR analysis of pre-mRNA, RNA was treated with DNase (DNA-free Kit, Ambion) prior to synthesis of cDNA, and controls omitting reverse transcriptase were included. All primers are listed in Supplementary Methods.
Immunoblot
Mouse quadriceps muscle was pulverized under liquid nitrogen, homogenized in lysis buffer, resolved on SDS–PAGE gels and transferred to nitrocellulose (Bio-Rad Laboratories) as previously described (58). Equal protein loading was confirmed by Ponceau Red staining. After incubation with primary antibody at 4°C overnight, membranes were incubated with secondary antibody at room temperature for 1 h, washed and then imaged on the Odyssey Infrared Imaging System (Licor). Antibodies were anti-Mbnl1 polyclonal antibody A2764 (1:10,000), anti-GAPDH mouse monoclonal (1:10,000, Biogenesis, Brentwood, NH, USA) and anti-UCHL1 monoclonal ZMD.339 (1:1,000, Zymed Laboratories, South San Francisco, CA, USA). Secondary antibodies were IRDye 800- or IRDye 700DX-conjugated anti-rabbit or anti-mouse IgG (Rockland Immunochemicals). To generate primary antibodies against copine 2 (Cpne2), rabbits were immunized with peptide QYFKHKNLPPTNSEPA, the C terminus of mouse Cpne2.
Search for alternative splice isoforms that are substrates for nonsense mediated decay
For Xedar and Cpne2, cDNAs extending from the first to the final exon were amplified from WT and HSALR20b RNA through 30–31 cycles of RT–PCR. Products from five to seven PCR reactions were combined, gel-purified, cloned and sequenced. The EMBL-EBI Alternative Splicing and Transcript Diversity 1.1 database (http://www.ebi.ac.uk/astd/main.html) (43) was used to survey HSALR-dysregulated genes for alternative splice products that contained PTCs.
Small RNA northern
Twenty microgam of total RNA was resolved on 10% urea/polyacrylamide gels and transferred to Hybond N+ membranes (Amersham) at 200 mA for 3 h using a Trans-blot SD semi-dry transfer cell (Biorad). Blots were cross-linked with ultraviolet radiation using a Stratalinker. Blots were prehybridized at least 1 h at 37°C in ULTRAhyb™-Oligo hybridization buffer (Ambion) before overnight incubation at 37°C in hybridization buffer containing denatured [32P]-end-labeled probe. Probes were DNA oligonucleotides consisting of (CAG)7 or sequence complementary to the indicated miRNA. Blots were washed (2X SSC, 0.1% SDS) at 37°C for 30 min followed by two 30 min room temperature washes and exposed to Kodak BioMax film.
Analysis of repeat containing transcripts
To determine whether transcripts containing CAG repeats had abnormal expression, we used BLAST to identify 286 mouse RefSeq mRNAs having 19 nt of uninterrupted CAG repeats [i.e. (CAG)6C, (AGC)6A or (GCA)6G].
Pathway analysis
To identify functionally related genes whose expression was altered in DM1 models, we used ingenuity pathways analysis (37) (www.ingenuity.com), GSEA (38) (www.broad.mit.edu/gsea/) and EASE (39) (see Supplementary Methods).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (AR046806, U54NS48843, AR48143), the Run America Foundation and the Saunders Family Neuromuscular Research Fund.
Supplementary Material
ACKNOWLEDGEMENTS
This authors wish to thank Linda Richardson and Kirti Bhatt for technical assistance. This work comes from the Wellstone Muscular Dystrophy Cooperative Research Center of the University of Rochester.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 2.Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamshere M.G., Newman E.E., Alwazzan M., Athwal B.S., Brook J.D. Transcriptional abnormality in myotonic dystrophy affects DMPK but not neighboring genes. Proc. Natl Acad. Sci. USA. 1997;94:7394–7399. doi: 10.1073/pnas.94.14.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne R.J., Thornton C.A. RNA-dominant diseases. Hum. Mol. Genet. 2006;15:R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 6.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 7.Orengo J.P., Chambon P., Metzger D., Mosier D.R., Snipes G.J., Cooper T.A. Expanded CTG repeats within the DMPK 3′-UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Pegoraro E., Menegazzo E., Gennarelli M., Hoop R.C., Angelini C., Hoffman E.P. Myotonic dystrophy: evidence for a possible dominant-negative RNA mutation. Hum. Mol. Genet. 1995;4:599–606. doi: 10.1093/hmg/4.4.599. [DOI] [PubMed] [Google Scholar]
- 9.Miller J.W., Urbinati C.R., Teng-umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A. Failure of MBNL1-dependent postnatal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y., Compton S.A., Sobczak K., Stenberg M.G., Thornton C.A., Griffith J.D., Swanson M.S. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 13.Adereth Y., Dammai V., Kose N., Li R., Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat.Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 15.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul S., Dansithong W., Kim D., Rossi J., Webster N.J., Comai L., Reddy S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 18.Timchenko N.A., Iakova P., Cai Z.J., Smith J.R., Timchenko L.T. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho T.H., Charlet B., Poulos M.G., Singh G., Swanson M.S., Cooper T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol J., Fiszer A., Mykowska A., Sobczak K., de Mezer M., Krzyzosiak W.J. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 21.Tian B., White R.J., Xia T., Welle S., Turner D.H., Mathews M.B., Thornton C.A. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebralidze A., Wang Y., Petkova V., Ebralidse K., Junghans R.P. RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science. 2004;303:383–387. doi: 10.1126/science.1088679. [DOI] [PubMed] [Google Scholar]
- 23.Mankodi A., Takahashi M.P., Jiang H., Beck C.L., Bowers W.J., Moxley R.T., Cannon S.C., Thornton C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 24.Kanadia R.N., Shin J., Yuan Y., Beattie S.G., Wheeler T.M., Thornton C.A., Swanson M.S. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lueck J.D., Mankodi A., Swanson M.S., Thornton C.A., Dirksen R.T. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler T.M., Lueck J.D., Swanson M., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charlet-B N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 28.Silverman H., Atwood H.L. Increase in oxidative capacity of muscle fibers in dystrophic mice and correlation with overactivity in these fibers. Exp. Neurol. 1980;68:97–113. doi: 10.1016/0014-4886(80)90069-2. [DOI] [PubMed] [Google Scholar]
- 29.Soothill P.W., Kouseibati F., Watts R.L., Watts D.C. Glycolytic, pentose-phosphate shunt and transaminase enzymes in gastrocnemius muscle, liver, heart, and brain of two mouse mutants, 129J-dy and A2g-adr, with abnormal muscle function. J. Neurochem. 1981;37:506–510. doi: 10.1111/j.1471-4159.1981.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 30.Reininghaus J., Fuchtbauer E.M., Bertram K., Jockusch H. The myotonic mouse mutant ADR: physiological and histochemical properties of muscle. Muscle Nerve. 1988;11:433–439. doi: 10.1002/mus.880110504. [DOI] [PubMed] [Google Scholar]
- 31.Wu H., Olson E.N. Activation of the MEF2 transcription factor in skeletal muscles from myotonic mice. J. Clin. Invest. 2002;109:1327–1333. doi: 10.1172/JCI15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pette D., Vrbova G. Adaptation of mammalian skeletal muscle fibers to chronic electrical stimulation. Rev. Physiol. Biochem. Pharmacol. 1992;120:115–202. doi: 10.1007/BFb0036123. [DOI] [PubMed] [Google Scholar]
- 34.Haslett J.N., Kang P.B., Han M., Kho A.T., Sanoudou D., Volinski J.M., Beggs A.H., Kohane I.S., Kunkel L.M. The influence of muscle type and dystrophin deficiency on murine expression profiles. Mamm. Genome. 2005;16:739–748. doi: 10.1007/s00335-005-0053-8. [DOI] [PubMed] [Google Scholar]
- 35.Gallanti A., Prelle A., Moggio M., Ciscato P., Checcarelli N., Sciacco M., Comini A., Scarlato G. Desmin and vimentin as markers of regeneration in muscle diseases. Acta Neuropathol. 1992;85:88–92. doi: 10.1007/BF00304637. [DOI] [PubMed] [Google Scholar]
- 36.Whalen R.G., Harris J.B., Butler-Browne G.S., Sesodia S. Expression of myosin isoforms during notexin-induced regeneration of rat soleus muscles. Dev. Biol. 1990;141:24–40. doi: 10.1016/0012-1606(90)90099-5. [DOI] [PubMed] [Google Scholar]
- 37.Calvano S.E., Xiao W., Richards D.R., Felciano R.M., Baker H.V., Cho R.J., Chen R.O., Brownstein B.H., Cobb J.P., Tschoeke S.K., et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosack D.A., Dennis G., Jr, Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asahi M., Otsu K., Nakayama H., Hikoso S., Takeda T., Gramolini A.O., Trivieri M.G., Oudit G.Y., Morita T., Kusakari Y., et al. Cardiac-specific overexpression of sarcolipin inhibits sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs cardiac function in mice. Proc. Natl Acad. Sci. USA. 2004;101:9199–9204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babu G.J., Bhupathy P., Timofeyev V., Petrashevskaya N.N., Reiser P.J., Chiamvimonvat N., Periasamy M. Ablation of sarcolipin enhances sarcoplasmic reticulum calcium transport and atrial contractility. Proc. Natl Acad. Sci. USA. 2007;104:17867–17872. doi: 10.1073/pnas.0707722104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lueck J.D., Lungu C., Mankodi A., Osborne R., Welle S., Dirksen R.T., Thornton C.A. Chloride channelopathy in myotonic dystrophy resulting from loss of post-transcriptional regulation for CLCN1. Am. J. Physiol. Cell Physiol. 2007;292:C1245–C1247. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 43.Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong C.S., Kwon S.J., Kim do H. Multiple functions of junctin and junctate, two distinct isoforms of aspartyl beta-hydroxylase. Biochem. Biophys. Res. Commun. 2007;362:1–4. doi: 10.1016/j.bbrc.2007.07.166. [DOI] [PubMed] [Google Scholar]
- 45.Fardaei M., Rogers M.T., Thorpe H.M., Larkin K., Hamshere M.G., Harper P.S., Brook J.D. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 46.Ashizawa T., Dubel J.R., Harati Y. Somatic instability of CTG repeat in myotonic dystrophy. Neurology. 1993;43:2674–2678. doi: 10.1212/wnl.43.12.2674. [DOI] [PubMed] [Google Scholar]
- 47.Thornton C.A., Johnson K., Moxley R.T. Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol. 1994;35:104–107. doi: 10.1002/ana.410350116. [DOI] [PubMed] [Google Scholar]
- 48.Zatz M., Passos-Bueno M.R., Cerqueira A., Marie S.K., Vainzof M., Pavanello R.C.M. Analysis of the CTG repeat in skeletal muscle of young and adult myotonic dystrophy patients: when does the expansion occur? Hum. Mol. Genet. 1995;4:401–406. doi: 10.1093/hmg/4.3.401. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler T.M., Krym M.C., Thornton C.A. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007;17:242–247. doi: 10.1016/j.nmd.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadava R.S., Frenzel-McCardell C.D., Yu Q., Srinivasan V., Tucker A.L., Puymirat J., Thornton C.A., Prall O.W., Harvey R.P., Mahadevan M.S. RNA toxicity in myotonic muscular dystrophy induces NKX2-5 expression. Nat. Genet. 2008;40:61–68. doi: 10.1038/ng.2007.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura T., Nakamori M., Lueck J.D., Pouliquin P., Aoike F., Fujimura H., Dirksen R.T., Takahashi M.P., Dulhunty A.F., Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 52.Babu G.J., Bhupathy P., Petrashevskaya N.N., Wang H., Raman S., Wheeler D., Jagatheesan G., Wieczorek D., Schwartz A., Janssen P.M., et al. Targeted overexpression of sarcolipin in the mouse heart decreases sarcoplasmic reticulum calcium transport and cardiac contractility. J. Biol. Chem. 2006;281:3972–3979. doi: 10.1074/jbc.M508998200. [DOI] [PubMed] [Google Scholar]
- 53.Gramolini A.O., Trivieri M.G., Oudit G.Y., Kislinger T., Li W., Patel M.M., Emili A., Kranias E.G., Backx P.H., Maclennan D.H. Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proc. Natl Acad. Sci. USA. 2006;103:2446–2451. doi: 10.1073/pnas.0510883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risheg H., Graham J.M., Jr, Clark R.D., Rogers R.C., Opitz J.M., Moeschler J.B., Peiffer A.P., May M., Joseph S.M., Jones J.R., et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 55.Welle S., Brooks A.I., Thornton C.A. Computational method for reducing variance with Affymetrix microarrays. BMC Bioinformat. 2002;3:23. doi: 10.1186/1471-2105-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z., Irizarry R.A. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. [DOI] [PubMed] [Google Scholar]
- 57.Welle S., Bhatt K., Thornton C.A. Stimulation of myofibrillar synthesis by exercise is mediated by more efficient translation of mRNA. J. Appl. Physiol. 1999;86:1220–1225. doi: 10.1152/jappl.1999.86.4.1220. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler T.M., Krym M.C., Thornton C.A. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007;17:242–247. doi: 10.1016/j.nmd.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.