Abstract
Amplification artifacts introduced during library preparation for the Illumina Genome Analyzer increase the likelihood that an appreciable proportion of these sequences will be duplicates, and cause an uneven distribution of read coverage across the targeted sequencing regions. As a consequence, these unfavorable features result in difficulties in genome assembly and variation analysis from the short reads, particularly when the sequences are from genomes with base compositions at the extremes of high or low GC content. Here we present an amplification-free method of library preparation, in which the cluster amplification step, rather than the polymerase chain reaction, enriches for fully ligated template strands, reducing the incidence of duplicate sequences, improving read mapping and SNP calling and aiding de novo assembly. We illustrate this by generating and analysing DNA sequences from extremely GC-poor (Plasmodium falciparum), GC-neutral (Escherichia coli) and high GC (Bordetella pertussis) genomes.
Introduction
Sequencing genomes with biased nucleotide compositions poses great technical challenges to the currently available sequencing platforms, most notably the highly GC-poor genomes of Plasmodium species, which are difficult even for the traditional BAC to BAC Sanger method 1-4. In several malaria species, including Plasmodium falciparum, the mean exonic AT content is > 75 %, and in intergenic and intronic regions can be close to 100 % 5,6.
One lane of an Illumina Genome Analyzer (GA) flowcell 7 can yield 700 × 106 bases of purity filtered (PF) sequence data in a 2 × 36-base paired end (PE) run, which would represent > 30 × coverage of the genome of the 23 Mb reference P. falciparum clone, 3D7 6. To make the most of the sequencing capacity of a GA, it is essential to obtain as broad a representation of the genome as possible, but amplification and sampling biases during library preparation result in libraries that are lower in complexity than the genomic DNA from which they were derived. Further sequencing runs of the same library are often not sufficient to improve coverage of regions that are poorly represented, and it becomes necessary to prepare and sequence additional libraries.
The standard Illumina library preparation pipeline is a multi-step process ending with a PCR amplification step prior to loading the sample into the flowcell. For the last 20 years, PCR has been used ubiquitously to amplify specific sections of DNA exponentially 8, but it is an inherently biased procedure 9-12. To help overcome amplification biases, and to reduce the formation of primer dimers, the Illumina library preparation protocol uses universal PCR primers, which allow simultaneous amplification of all loci in complex template pools 7. There is a narrow range of conditions in the PCR that will give clean libraries with adequate representation 13, but even when performed under optimal conditions, the PCR step is still sensitive to biases, particularly when the template to be amplified has a high AT content such as P. falciparum.
The malaria sequencing programme at the Wellcome Trust Sanger Institute aims to sequence hundreds of cell lines, including clinical isolates. As a pilot study, we started with GA sequencing runs of P. falciparum 3D7, the reference genome sequenced by the Sanger dideoxy method 6, with the intention of correcting base errors in the reference. This was followed by several more sequencing runs for a variety of malaria strains. Quality of read mapping against the reference was very poor, with a high proportion of duplicate reads, and uneven coverage. This increases sequencing costs, as only a portion of the reads are useful. For the GC-neutral Escherichia coli and GC-rich Bordetella pertussis genomes, the coverage bias is far less pronounced.
Here we report an alternative method of Illumina library preparation that omits the PCR step entirely. For the extremely GC-poor malaria genomes, datasets obtained from these libraries not only improve SNP detection, but also facilitate de novo assemblies using short read assemblers.
Results
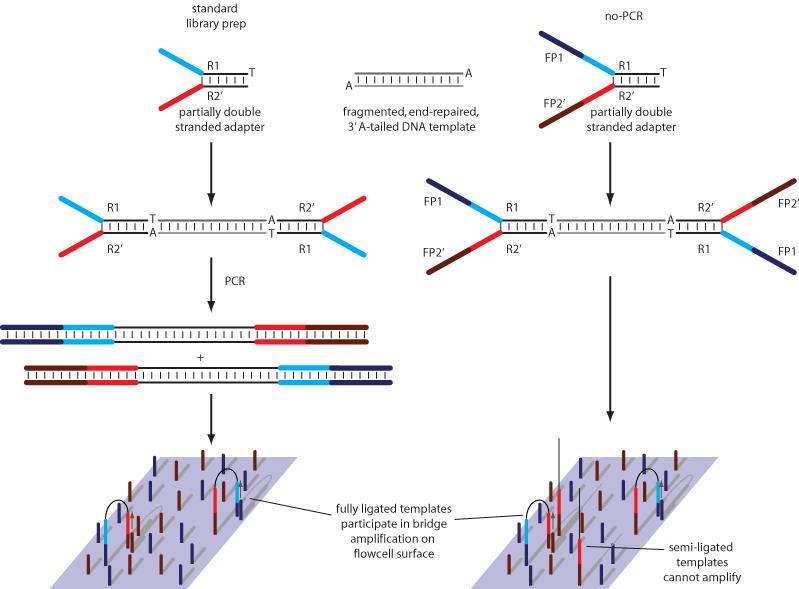
In the course of Illumina library preparation, sample DNA is fragmented, end-repaired and A-tailed. Adapters, essentially consisting of the sequencing primer annealing sequences, are then ligated via a 3′ T-overhang. The structure of the adapters ensures that each strand receives a unique adapter sequence at either end. Finally, ligated fragments are amplified by PCR 7. Amplification is needed to generate sufficient quantities of template DNA to allow accurate quantification and to enrich for successfully ligated fragments. The PCR also adds additional adapter sequence, using tailed primers, resulting in template molecules which are capable of hybridization to oligonucleotides on the flowcell surface. Even though the number of cycles of PCR amplification is kept low (10-12 cycles for PE libraries) 7, the PCR is still a source of duplicate sequences, and amplification bias, and struggles with AT-rich base compositions 14. Runs therefore become less efficient, and assembly, mapping and SNP detection are made more complicated than necessary.
In our no-PCR protocol, partially double stranded adapters are also added to end-repaired, template DNA with a 3′ A overhang, by ligation (Figure 1). Unlike the standard Illumina adapters, no-PCR adapters contain additional sequences that allow hybridization of templates directly to the flowcell surface. Incompletely ligated fragments are inert in the cluster amplification step. Thus it is not necessary to retain the PCR step to enrich for properly ligated fragments, but in order to obtain an optimal cluster density, it is necessary to accurately quantify only those template fragments with an adapter at either end. We achieve this by quantitative PCR, using primers that target the adapter regions 13.
Figure 1. No-PCR library preparation.
In both standard and no-PCR library preps, partially complementary (‘Y-shaped’) adapters with a 3′ T overhang are ligated onto fragmented, end-repaired, 3′ A-tailed DNA. Whereas standard adapters consist only of sections to which read 1 and read 2 sequencing primers hybridize (R1 and R2′), no-PCR adapters also contain sequences that facilitate hybridization to oligonucleotides attached to the flowcell surface (FP1 and FP2′). The standard library prep uses PCR to add these sections, and to enrich for fully ligated templates which then amplify on the flow cell surface. With the no-PCR approach, the flowcell itself is used to select for fully ligated template molecules. All no-PCR templates hybridise to the flowcell in the same orientation, because only the FP2′ sequence is reverse complementary to a flowcell oligonucleotide.
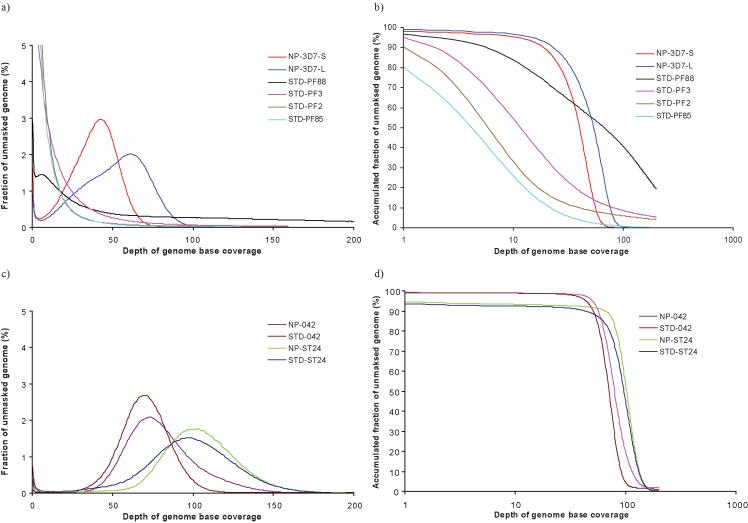
To investigate differences between standard and no-PCR library preparations, we produced four sets of 2 × 35 and 36-base PE P. falciparum 3D7 data from standard libraries STD-PF88, STD-PF2, STD-PF3 and STD-PF85, corresponding to read coverage of 174x, 114x, 96x and 21x respectively (Supplementary Table 1). We also produced two sets of PE 3D7 data from one no-PCR library: 2 × 36-bases (NP-3D7-S) and 2 × 76-bases (NP-3D7-L), corresponding to read coverage of 44x and 65x respectively. We mapped reads to the 3D7 reference sequence using a modified version of SSAHA 15. Data from the standard 3D7 libraries (STD-PF2, STD-PF3 and STD-PF85) and one run of a standard library made from a clinical isolate (STD-PF88) all failed to show a typical Poisson distribution with the peak around the average read depth._In contrast, the peak for both 36-base and 76-base no-PCR data agree closely with read depth (Figure 2a).
Figure 2. Distribution of genome sequence coverage.
The distribution of sequence coverage across the unmasked genomes are shown for various datasets with (STD) or without (NP) the PCR step. (a) % of unmasked genome against depth of genome base coverage and (b) accumulated % of unmasked genome against depth of genome base coverage for malaria strains (P. falciparum (PF) 2,3,88 and 85 and 3D7) with either long (L) or short (S) reads. (c) % of unmasked genome against depth of genome base coverage and (d) accumulated % of unmasked genome against depth of genome base coverage for E. coli 042 and B. pertussis ST24
Sequence coverage and SNP calling
A plot of accumulated fractions of unmasked genome - in which Repeatmasker ( Smith A.F.A., Hubley R. & Green P. RepeatMasker Open-3.0. http://www.repeatmasker.org 1996-2004) was not run to mask repetitive elements - against depth of base coverage (Figure 2b) reveals that for STD-PF2 only 30 % bases are covered by the mapped reads at 10 times or higher, whereas the 2.2 Gb of raw data should cover the 23 Mb genome 96 times on average. Between 4.8 and 19.9 % of bases in the reference sequence have no coverage (Supplementary Table 1). A highly uneven coverage distribution makes it difficult to identify duplicated regions. In such regions, variant nucleotides could be misinterpreted as SNPs, when in reality they are paralogous sequence variants. Assuming that at least 10-fold coverage is required to call SNPs reliably, the no-PCR data perform substantially better than the other four datasets, with 97 % of bases covered 10 times or more (Figure 2b). In contrast, for the GC-neutral Escherichia coli and GC-rich Bordetella pertussis genomes, this situation is less pronounced, with data generated from libraries produced in the standard way showing a Poisson-like distribution (Figure 2c) and a high proportion of bases being covered by mapped reads at 30x or higher (Figure 2d; Supplementary Table 1).
We aligned no-PCR reads using SSAHA_pileup and identified 2059 SNPs. To estimate the accuracy of our SNP calls in the 3D7 data, we determined a baseline of accuracy from E. coli data, for which a high quality finished sequence is available. Short read Illumina sequence data, generated from the no-PCR library preparation, only differed at 5 positions in the entire 5.3 Mb genome. We confirmed these by de novo assembly to be finishing errors (data not shown). Assuming that read accuracy is similar between the E. coli and P. falciparum Illumina datasets, virtually all of the SNPs called in the P. falciparum dataset are actually base errors in our Sanger sequence data.
Amplification bias
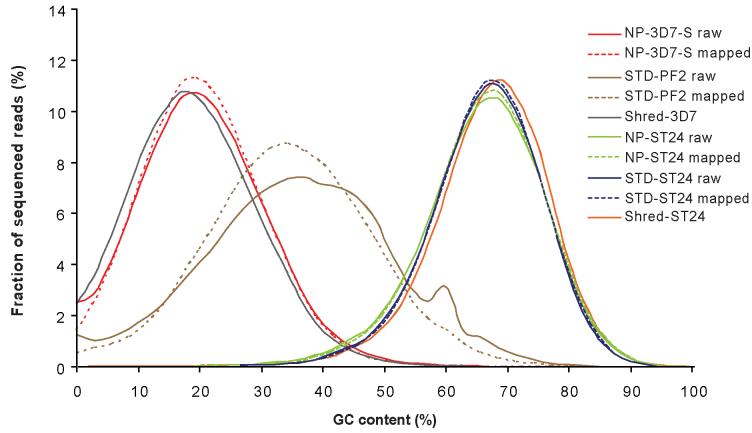
To assess systematic biases in base composition introduced during the library preparation and sequencing procedures, it is necessary to evaluate how closely the sequence data represent the base composition of the original genome, and to identify any major shifts in GC content by comparison with a reference sequence. We divided reference sequences of P. falciparum 3D7, E. coli 042 and B. pertussis ST24 into tiled fragments, corresponding to the read length used in the different sequencing runs. For each fragment, we calculated % GC content, and used this information to plot theoretical GC profiles for these genomes, with which we compared the sequence data (Figure 3). The coverage for this ‘shredded’ data for 3D7 and ST24 were both 70x, though as read fraction is independent of read depth, we should not see any changes in curve patterns at different depths.
Figure 3. Distribution of sequenced reads for different values of GC content.
GC profiles for raw and mapped sequence data for the malaria strains NP-3D7-S and STD-PF2 are shown alongside simulated data (‘Shred-3D7′) for comparison. GC levels are calculated in a window size of read length and therefore the peak of fraction reads is dependent upon read length. A shift away from the simulated data curve, towards a more balanced GC composition is evident for the STD-PF2 sequence data, indicating severe bias.
For both the raw and mapped datasets of the 3D7 STD-PF2 sequence data, there is an appreciable shift away from the theoretical shredded data towards higher GC content, indicating severe anti AT bias in the sequences. Although both mapped and raw data are shifted in this way, the curve of mapped STD-PF2 data is smoother, suggesting some low quality and GC-biased reads in the raw sequencing data could not be aligned against the reference. A similar pattern can be seen for the other standard P. falciparum libraries (Supplementary Figure 1). In each case, a shift towards higher GC content indicates poor performance of these standard libraries. In contrast, for both the raw and mapped datasets of the 2 × 36 base no-PCR library are in good agreement with the shredded curve, showing that the base composition of the sequence data represent that of the original genome (Figure 3). For B. pertussis, GC profiles of both standard and no-PCR libraries correlate well with the simulated data (Figure 3). For E. coli, the standard library agrees closely with the simulated data, whereas the no-PCR curve is shifted to the left, indicating a slight bias in this library (Supplementary Figure 1).
Duplicate sequences
Duplicate sequences are a major concern in Illumina sequencing. We defined duplicate reads as those sharing exactly the same start and end locations, and counted these reads to determine the extent of duplication. In addition to PCR duplicates, duplicate sequences arise from adapter dimers created during the library prep, sequencing artifacts such as poly-A and poly-N reads, noise in the cluster detection and analysis software (data not shown), and potentially from genomic DNA shearing at the same position in different molecules. Reducing PCR duplicates would be beneficial, both in lowering costs and allowing improved read mapping. We assessed the frequency of duplicate sequences in our no-PCR libraries by mapping to the reference sequence.
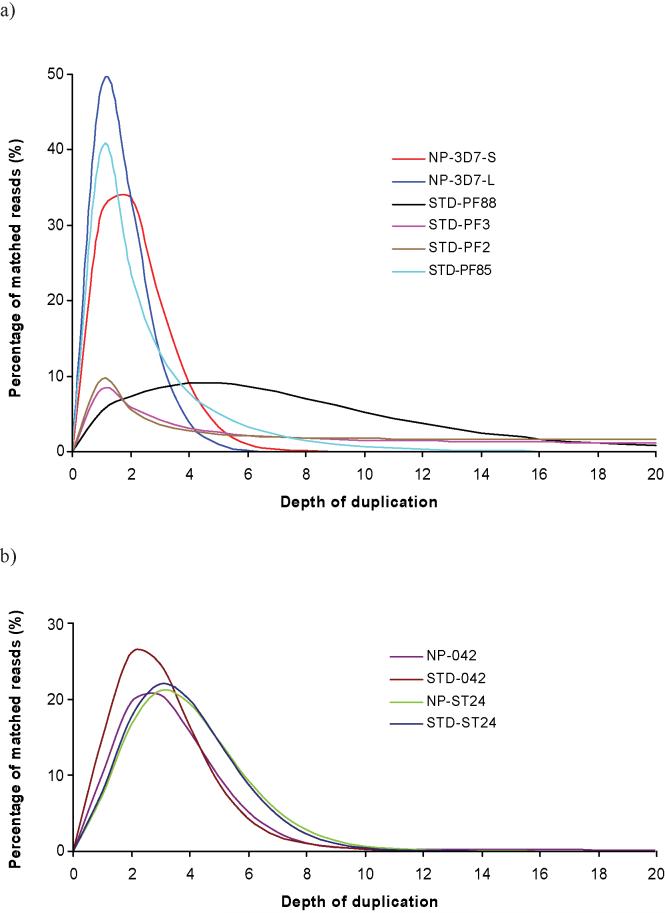
The frequency of duplicate sequences is high for STD-PF3 and STD-PF2 (Figure 4a): tails on the STD curves extend far, indicating that for such an AT-rich genome, PCR duplicates comprise the major source of duplicate sequences. It is interesting to note that the duplication rate looks high for STD-PF88, a P. falciparum clinical isolate, but is, in fact, normal judged from the distribution of read duplication. The duplication distribution has a relatively short tail and a peak value at ~5.0, which is close to the theoretical value of 3.4, obtained by dividing mean coverage by read length. In contrast, the abundance of duplicate sequences in the no-PCR and standard libraries of E. coli and B. pertussis did not differ appreciably, suggesting that a greater proportion of these two genomes are able to amplify in the PCR (Figure 4b and Supplementary Table 1). The duplication rate is low for the 36-base NP-3D7-S dataset, but not the 76-base NP-3D7-L dataset, in spite of the fact that the same library was used for both sequencing runs. Trimming the 76-base sequence data back to 36 bases revealed that although the base composition of mapped sequences agrees well with the theoretical data, the raw data shows a tail shifting away from the theoretical predictions (Supplementary Figure 2), showing that the data from the 76 bp run has a higher level of bias than the 36 bp run, indicating a problem with the longer sequencing run itself.
Figure 4. Frequencies of duplicate sequences.
Percentage of matched reads against duplication depth for sequence data derived from libraries prepared both with and without a PCR step. (a) Duplication frequencies for Plasmodium libraries; (b) duplication frequencies for E. coli and B. pertussis libraries.
De novo assembly
The low bias of the no-PCR P. falciparum datasets makes de novo assembly possible, whereas standard libraries do not permit this, due to uneven coverage and inadequate representation of the genome (Table 1). From the 2 × 36-base dataset with approximately 14 million paired end reads, we obtained an assembly of 15.5 Mb with N50 = 1.38 kb (i.e. 50 % of all bases are contained within contigs of 1.38 kb or longer). Using the 2 × 76-base data, we produced an assembly of 20.8 Mb with N50 = 1.28 kb from 9.8 million paired end reads.
Table 1. Summary of sequence data for the no-PCR and standard libraries.
No-PCR libraries have the prefix ‘NP’, whereas standard libraries have the prefix ‘STD’. Suffixes ‘L’ and ‘S’ indicate long and short different sequencing runs performed on the same library. No assembly was possible on data generated from the standard P. falciparum libraries
| Library | Organism | Genome Size (Mb) |
Insert Size (bp) |
Read Length (bp) |
Number of Reads |
Raw Read Coverage |
Assembled Bases |
Contig Coverage % |
Number of contigs >100 bp |
Contig N50 |
|---|---|---|---|---|---|---|---|---|---|---|
| NP-3D7-S | P. falciparum 3D7 | 23 | 200 | 36 | 28,009,122 | 43 | 19,025,823 | 82.7 | 26,920 | 1456 |
| NP-3D7-L | P. falciparum 3D7 | 23 | 200 | 76 | 19,556,224 | 64 | 21,092,855 | 91.7 | 22,839 | 1621 |
| STD-PF88 | P. falciparum 3D7 | 23 | 200 | 37 | 110,939,984 | 174 | N/A | N/A | N/A | N/A |
| STD-PF3 | P. falciparum 3D7 | 23 | 200 | 37 | 75,083,768 | 114 | N/A | N/A | N/A | N/A |
| STD-PF2 | P. falciparum 3D7 | 23 | 200 | 37 | 62,802,164 | 96 | N/A | N/A | N/A | N/A |
| STD-PF85 | P. falciparum 3D7 | 23 | 200 | 37 | 13,530,194 | 21 | N/A | N/A | N/A | N/A |
| NP-042 | E. coli 042 | 5.3 | 200 | 36 | 14,110,696 | 95 | 5,362,633 | 99.9 | 186 | 91605 |
| STD-042 | E. coli 042 | 5.3 | 200 | 37 | 10,719,672 | 75 | 5,309,673 | 99.9 | 177 | 95860 |
| NP-ST24 | B. pertussis ST24 | 4.0* | 200 | 36 | 12,549,138 | 113 | 3,821,094 | 95.5 | 306 | 17808 |
| STD-ST24 | B. pertussis ST24 | 4.0* | 200 | 37 | 11,756,654 | 109 | 3,763,213 | 94 | 386 | 14200 |
indicates the approximate size of the ST24 genome - in the absence of a finished assembly, it is only possible to estimate this.
The standard library of Bordetella pertussis yielded an assembly with N50 = 10.6 kb from 6 million 36-base PE reads, whereas an N50 of 20.5 kb was obtained from the no-PCR library, also from 6 million PE reads (Table 1). No finished quality reference sequence is available from this organism - the genome has a very high mean GC content (68 %) which, coupled with a complicated repeat structure, makes assembly more difficult than for GC neutral genomes such as E. coli.
For E. coli strain 042, a 5.35 Mb genome with 50.5 % GC, a finished sequence obtained from Sanger sequencing is available for comparison (http://www.sanger.ac.uk/Projects/Escherichia_Shigella/). Using 7 million PE reads of 2 × 36 bases from a standard library, we assembled the genome into contigs with N50 = 146 kb, compared to N50 of 71.7 kb with reads from a no-PCR library. The poorer assembly of the no-PCR library of the E. coli compared to the standard library is presumably due to variation in read quality, combined with a very limited effect of the no-PCR library prep on this GC neutral genome.
Discussion
PCR amplification introduces artifacts into sequencing libraries 9-12. In addition to nucleotide misincorporation, amplification tends to be uneven, so that some sequence species become overrepresented in the resulting library. This situation is exacerbated by templates with GC-biased compositions.
By ligating adapters which consist of all sections required for sequencing primer annealing and attachment to the flowcell surface, we can avoid the requirement of a PCR step. The quantity of template DNA generated in this way is lower than when PCR is employed, but library quantification by qPCR 13 showed that from 5 μg starting DNA sufficient 200 bp no-PCR library can be obtained for > 400 high density GA lanes, more than enough for most sequencing purposes. Starting with lower quantities, e.g. 500ng of genomic DNA we can obtain sufficient library with a200 bp insert size for ~ 12 lanes. Inserts of 500 bp result in a lower yield than shorter fragment libraries, presumably due to the lower number of fragments present in the same mass of DNA.
As with standard Illumina adapters, the structure of no-PCR adapters ensures that all fully ligated template strands receive the unique adapter sequence complementary to the flow-cell adapters at their 5′ and 3′ end (Figure 1). Because the efficiency of ligation is not 100 %, many template strands will receive no adapters, or will only be partially ligated. However, Illumina cluster amplification can only amplify template strands that have a different adapter at either end and thus the cluster amplification step performs the enrichment that is otherwise provided in the PCR.
We have demonstrated that for genomes of extreme GC composition, the sequence coverage provided by the no-PCR approach is more even than the standard, PCR-based Illumina library preparation, contains very few duplicates, aids mapping and SNP calling, and makes assembly more straightforward. This is best illustrated by the P. falciparum genome, which until now has resisted attempts at de novo assembly from short read data. The differences between the short- and long-read malaria assemblies are not large as the average fragment size for the no-PCR 3D7 library is only 170 bp, close to the long paired read length of 152 bp (2 × 76 bases).
It is important to note that P. falciparum genomes are extremely difficult to assemble even using 600-700 base Sanger sequence reads: assembly of clinical isolates from 6-fold Sanger coverage, yielded a contig N50 of only 7 kb (data not shown). Although it seems unlikely that assemblies from short read data alone will ever generate N50 values in the 7 kb range, we believe that we will be able to increase our malaria N50 beyond this by combining short read data with Sanger reads.
Approximately 2 % of the P. falciparum 3D7 reference sequence is not covered by the NP-3D7-S sequence data, since reads were only placed to their best location, while repetitive reads were not placed. In contrast, between 4.8 and 19.9 % of bases were not covered by mapping for the standard P. falciparum libraries. Using an alternative alignment tool, MAQ 17, which places repetitive reads to a random location, the uncovered regions were reduced to just 5585 bp for NP-3D7-S, indicating that 99.98 % of the 3D7 genome is represented in the sequence data.
Anecdotally, sequences with a GC content exceeding 80 % are difficult to sequence on a GA. The genome of B. pertussis has a mean GC content of 68 %, and only a small proportion of sequence reads would have > 80 % GC. Nevertheless, both standard and no-PCR B. pertussis libraries revealed GC profiles that were almost identical to simulated data, with no loss towards higher GC contents (Figure 3), indicating that the standard library preparation protocol finds no difficulty with GC contents within this range. If there are difficulties in sequencing organisms with a higher GC content than B. pertussis on a GA, our data indicates that these are not the result of PCR artefacts, though it is conceivable that biases are introduced at other stages in the sequencing process, such as cluster growth 7. However, the high GC content of ST24 has hindered the generation of a finished-standard reference sequence by Sanger sequencing: the assembly still contains 115 contigs, of which some are vector contamination, and this prevents a more thorough analysis.
Because of the absence of the PCR step, the method is quicker to perform than the standard Illumina library prep 7, and we feel that it should be employed routinely in the preparation of libraries for Illumina sequencing.
Methods
Adapter preparation
We obtained two HPLC-purified oligonucleotides (Sigma): A_adapter_t and A_adapter_b. We phosphorylated 40 μM oligos at the 5′ end by 1 unit / μl T4 polynucleotide kinase in 1x T4 ligase buffer (both New England Biolabs) for 30 minutes at 37 °C in a thermocycler (MJ Research). We then denatured the kinase by heating, and annealed the oligos by cooling to 20 °C by 0.1 °C every 2 seconds. We divided adapter oligos into single-use aliquots and stored them at -20 °C.
For all other methods, see Supplementary Methods
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Wellcome Trust [grant number WT079643]
Footnotes
URLs. Alignment software can be found at: http://www.sanger.ac.uk/Software/analysis/SSAHA2/.
All computer codes on the detection of SNPs and short indels can be found at: ftp://ftp.sanger.ac.uk/pub/zn1/ssaha_pileup/
All raw Illumina reads and assemblies can be found at: ftp://ftp.sanger.ac.uk/pub/zn1/PCR_free/
See also: ftp://ftp.sanger.ac.uk/pub/zn1/PCR_free/readme and ftp://ftp.sanger.ac.uk/pub/zn1/PCR_free/data_info.xls
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Goman M, et al. The establishment of genomic DNA libraries for the human malaria parasite Plasmodium falciparum and identification of individual clones by hybridisation. Mol Biochem Parasitol. 1982;5:391–400. doi: 10.1016/0166-6851(82)90012-3. [DOI] [PubMed] [Google Scholar]
- 2.Camargo AA, Fischer K, Lanzer M, del Portillo HA. Construction and characterization of a Plasmodium vivax genomic library in yeast artificial chromosomes. Genomics. 1997;42:467–473. doi: 10.1006/geno.1997.4758. [DOI] [PubMed] [Google Scholar]
- 3.de Bruin D, Lanzer M, Ravetch JV. Characterization of yeast artificial chromosomes from Plasmodium falciparum: construction of a stable, representative library and cloning of telomeric DNA fragments. Genomics. 1992;14:332–339. doi: 10.1016/s0888-7543(05)80223-x. [DOI] [PubMed] [Google Scholar]
- 4.Triglia T, Kemp DJ. Large fragments of Plasmodium falciparum DNA can be stable when cloned in yeast artificial chromosomes. Mol Biochem Parasitol. 1991;44:207–211. doi: 10.1016/0166-6851(91)90006-r. [DOI] [PubMed] [Google Scholar]
- 5.Pollack Y, Katzen AL, Spira DT, Golenser J. The genome of Plasmodium falciparum. I: DNA base composition. Nucleic Acids Res. 1982;10:539–546. doi: 10.1093/nar/10.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saiki RK, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 9.Day DJ, et al. Identification of non-amplifying CYP21 genes when using PCR-based diagnosis of 21-hydroxylase deficiency in congenital adrenal hyperplasia (CAH) affected pedigrees. Human Molecular Genetics. 1996;5:2039–2048. doi: 10.1093/hmg/5.12.2039. [DOI] [PubMed] [Google Scholar]
- 10.Barnard R, Futo V, Pecheniuk N, Slattery M, Walsh T. PCR bias toward the wild-type k-ras and p53 sequences: implications for PCR detection of mutations and cancer diagnosis. Biotechniques. 1998;25:684–691. doi: 10.2144/98254dt08. [DOI] [PubMed] [Google Scholar]
- 11.Hahn S, Garvin AM, Di Naro E, Holzgreve W. Allele drop-out can occur in alleles differing by a single nucleotide and is not alleviated by preamplification or minor template increments. Genet Test. 1998;2:351–355. doi: 10.1089/gte.1998.2.351. [DOI] [PubMed] [Google Scholar]
- 12.Ogino S, Wilson RB. Quantification of PCR bias caused by a single nucleotide polymorphism in SMN gene dosage analysis. J Mol Diagn. 2002;4:185–190. doi: 10.1016/S1525-1578(10)60702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quail MA, et al. A large genome centre’s improvements to the Illumina sequencing system. Nature Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.