Abstract
Knowing the geographical origin of economically important plants is important for genetic improvement and conservation, but has been slowed by uneven geographical sampling where relatives occur in remote areas of difficult access. Less biased species sampling can be achieved when herbarium collections are included as DNA sources. Here, we address the history of Cucurbitaceae, one of the most economically important families of plants, using a multigene phylogeny for 114 of the 115 genera and 25 per cent of the 960 species. Worldwide sampling was achieved by using specimens from 30 herbaria. Results reveal an Asian origin of Cucurbitaceae in the Late Cretaceous, followed by the repeated spread of lineages into the African, American and Australian continents via transoceanic long-distance dispersal (LDD). North American cucurbits stem from at least seven range expansions of Central and South American lineages; Madagascar was colonized 13 times, always from Africa; Australia was reached 12 times, apparently always from Southeast Asia. Overall, Cucurbitaceae underwent at least 43 successful LDD events over the past 60 Myr, which would translate into an average of seven LDDs every 10 Myr. These and similar findings from other angiosperms stress the need for an increased tapping of museum collections to achieve extensive geographical sampling in plant phylogenetics.
Keywords: Bayesian molecular clock, biogeography, dispersal, economic plants, museomics, test of monophyly
1. Introduction
Molecular clock analyses suggest that the majority of lineages of legumes that occur on islands are younger than 30 Myr (Lavin & Beyra Matos 2008) and that plant diaspores from source areas hundreds or thousands of kilometres away regularly reach isolated Arctic islands (Alsos et al. 2007), island-like mountains in Eastern Africa and mountain ranges in the Northern Cape, South Africa (Galley et al. 2007). Striking dispersal events have also been documented for the flora of Hawaii (Wagner et al. 1990), the montane region of New Zealand (Winkworth et al. 2005) and many other island systems. Such frequent long-distance dispersal (LDD) implies that long-established views on the origin of economically important plants may need to be re-evaluated based on drastically enlarged geographical sampling. An example is the origin of Cucumis sativus, the cucumber. Cucumber ranks among the top 10 vegetables in world production (Chen et al. 2004). Until 2006, it was thought that the genus Cucumis had 32 species and was essentially African. Only C. sativus and C. hystrix were thought to occur naturally in India, China, Burma and Thailand (Ghebretinsae & Barber 2006). However, broader geographical species sampling revealed that C. sativus is closer to 13 species from Australia, India, Yunnan and Indochina than to any African species (Renner & Schaefer 2008).
Biogeographic inference for economically important plants is complicated by human transport of seeds between continents for at least 10 000 years (Smith 1997; Sanjur et al. 2002; Dillehay et al. 2007). The extent of the anthropogenic transfer, however, is difficult to work out without comprehensive phylogenetic frameworks, which can be prohibitively expensive if worldwide collecting of material is required. In the economically important plant family Cucurbitaceae, these difficulties have led to the geography of the closest relatives of watermelon (Citrullus lanatus), cucumber (Cucumis sativus), loofah (Luffa acutangula), bitter gourd (Momordica charantia), chayote (Sechium edule), ivy gourd (Coccinia grandis), snake gourd (Trichosanthes cucumerina) and creeping cucumber (Melothria pendula) remaining ambiguous. Natural LDD of cucurbit diaspores may be frequent because many are adapted for transport by birds or wind, or they can withstand long periods in water (Cayaponia, Fevillea, Hodgsonia, Lagenaria, Luffa and Sicana; Ridley 1930; Whitaker & Carter 1954).
Here, we use worldwide sampling, based on museum specimens, to infer the biogeographic history of Cucurbitaceae, a family consisting of climbers or trailers of tropical and subtropical regions that are typically strongly seasonal, lacking aboveground parts during part of the year. These traits have caused cucurbits to be undercollected (Gentry 1991), resulting in dozens of species still known from only one or two collections even in the world's leading herbaria (e.g. De Wilde & Duyfjes 2007). Of the approximately 960 accepted species of Cucurbitaceae, approximately 40 per cent are endemic in the American continent, and the remainder occur in Africa (28%), Asia (26%), Australia (2%) and Europe (1%; Schaefer & Renner in press).
Based on chloroplast sequences from all but one of the 115 genera and 25 per cent of the 960 species, and employing specimens from 30 herbaria and up to 172 years old, we address here the following questions: did Cucurbitaceae initially diversify in Asia, in America, or in Africa and Madagascar? What are the geographical sources of the world's major Cucurbitaceae floras? Are the transoceanic geographical ranges in the genera Cayaponia, Lagenaria, Luffa and Sicyos anthropogenic or the result of natural LDD? We also use Cucurbitaceae to illustrate the still barely tapped potential of museum collections to achieve less biased geographical sampling than has traditionally been employed in tropical plant phylogenetics.
2. Material and methods
(a) Taxon sampling
We generated 126 sequences, representing 32 species from seven genera not sampled in previous studies (Anangia, Cucumeropsis, Gomphogyne, Hodgsonia, Papuasicyos, Pseudosicydium and Zanonia). GenBank accession numbers (EU436320–EU436422) and vouchers for newly sequenced taxa are listed in table 1 in the electronic supplementary material. Accession numbers and voucher information for additional Cucurbitaceae sequences from our earlier studies are given in Zhang et al. (2006), Kocyan et al. (2007), Schaefer et al. (2008a) and Nee et al. (submitted). Fourteen sequences from Genbank were included to represent Indomelothria (EF065456), Neoachmandra (EF065484–86), Urceodiscus (EF065464) and Zehneria (EF065485, EF065489, EF065491–493, EF065497, EF065499–500 and EF065502). This resulted in a sampling of 114 of the 115 genera currently recognized in Cucurbitaceae (Schaefer & Renner in press). The only genus of Cucurbitaceae not yet sequenced is Khmeriosicyos W. J. de Wilde & B. Duyfjes, which is only known from the Cambodian type collection. Judging from morphology, it is expected to group with other Asian Benincaseae. As outgroups, we used 15 species of the Cucurbitales families Anisophyllaceae, Begoniaceae, Coriariaceae, Corynocarpaceae, Datiscaceae and Tetramelaceae, based on Zhang et al. (2006). Of these, the Begoniaceae, Datiscaceae and Tetramelaceae, with the Cucurbitaceae, constitute a morphologically and molecularly well-defined clade, traditionally called the core Cucurbitales (Zhang et al. 2006).
(b) DNA extraction, amplification, sequencing and alignments
Total genomic DNA was isolated from herbarium specimens or, more rarely, silica-dried leaves with a commercial plant DNA extraction kit (NucleoSpin, MACHEREY-NAGEL, Düren, Germany), following the manufacturer's manual. We amplified the rbcL and matK genes, the trnL intron and the trnL-F and rpl20–rps12 intergenic spacers. Polymerase chain reactions (PCRs) were performed with the standard protocol and primers described in Kocyan et al. (2007), and products were purified with the Wizard SV PCR clean-up kit (Promega GmbH, Mannheim, Germany). Cycle sequencing was performed with BigDye Terminator cycle sequencing kits on an ABI Prism 3100 Avant automated sequencer (Applied Biosystems, Foster City, California, USA). Sequences were edited with Sequencher v. 4.6 (Gene Codes, Ann Arbor, Michigan, USA) and aligned by eye, using MacClade v. 4.06 (Maddison & Maddison 2003).
The data matrices comprised 245 ingroup species plus 15 outgroup species. The lengths of the individual loci were 1356 aligned nucleotides for the rbcL gene, 1195 for the matK gene, 667 for the tRNA-Leu (trnL) intron (after exclusion of a poly A run and a highly variable microsatellite region), 803 for the tRNA-Leu–tRNA-Phe (trnL-F) intergenic spacer and 1010 for the rpl20–rps12 intergenic spacer. The combined dataset comprised 5031 aligned nucleotides.
(c) Phylogenetic analysis
Maximum-likelihood (ML) tree searches and ML bootstrap searches were performed using RAxML v. 7.0.3 (Stamatakis et al. 2008; available at http://phylobench.vital-it.ch/raxml-bb/) and GARLI v. 0.951 (Zwickl 2006; available at www.bio.utexas.edu/faculty/antisense/garli/Garli.html). RAxML and GARLI searches relied on the GTR+Γ+I model (six general time-reversible substitution rates, assuming gamma rate heterogeneity and a proportion of invariable sites), with model parameters estimated over the duration of specified runs. Analyses in RAxML were run both with the combined unpartitioned data and with a model that partitioned the rbcL gene from the remaining non-coding regions. GARLI does not allow data partitioning. The data matrix and trees have been deposited in TreeBASE (www.treebase.org; study number S2210).
(d) Molecular clock analysis
Estimation of divergence times relied either on a strict clock or on a Bayesian relaxed clock with autocorrelated rates (Thorne et al. 1998). Very short (‘zero-length’) branches are known to cause problems for time estimation algorithms, and we therefore reduced their number by using the best-scoring ML tree for 147 taxa instead of the full 260-taxon tree. The clock tree was rooted on Coriariaceae and Corynocarpaceae, instead of Anisophyllaceae, because the latter are extremely rich in autapomorphies, contributing towards rate heterogeneity near the base.
For the Bayesian approach, we used baseml from the PAML package (Yang 2007) and multidivtime (Thorne et al. 1998; Thorne & Kishino 2002) in LAGOPUS, an R package written by Heibl & Cusimano (2008). LAGOPUS checks the input data for consistency, automates the assignment of constraints to nodes and connects the executables of the mentioned software packages in a pipeline. Model parameters for the 147-taxon matrix were estimated in baseml, and branch lengths and their variance then calculated in estbranches, all under the F84+G model (the only model implemented in multidivtime). Priors for multidivtime were as follows: based on outgroup fossils (below), the prior on the mean age of the root node was set to 84 Myr, with an equally large standard deviation. The prior on the substitution rate at the root was set to the value obtained by dividing the median distance between the root and the tips in the estbranches phylogram by 84 Myr. This yielded a rate of 0.0009 substitutions per site and million years [S/(S×Myr)]. The prior for the Brownian motion parameter, which controls the magnitude of autocorrelation along the descending branches of the tree, was set to 1.11, with a standard deviation of the same size. Markov chain Monte Carlo (MCMC) samples were drawn for every 100th generation up to one million generations, with a burn-in of 100 000 cycles. Confidence in node ages was assessed using the 95 per cent credibility intervals calculated by multidivtime.
To translate relative times into absolute times, the Bayesian clock relied on the following simultaneous constraints. (i) The age of core Cucurbitales (the root node) was constrained to maximally 84 Myr, based on the earliest fossils of the sister group of Cucurbitales, the Fagales (Herendeen et al. 1995). However, we also performed two runs in which the root node was unconstrained or constrained to minimally 84 Myr. This yielded age estimates for the Cucurbitaceae crown group that were older than the oldest angiosperm fossils (132 Myr). In general, relaxed molecular clocks will yield reliable ages only with at least one minimal and one maximal constraint, the latter preferentially at or near the root (Thorne et al. 1998). (ii) The age of the split between Datisca and Octomeles/Tetrameles was set to minimally 68 Myr old or, in an alternative run, to minimally 65.5 Myr old, based on the fossil wood of Tetrameleoxylon prenudiflora from the Deccan intertrappean beds at Mohgaonkalan in India (Lakhanpal & Verma 1965; Lakhanpal 1970). These beds have been dated to the Maastrichtian or Late Maastrichtian (Khajuria et al. 1994; Kar et al. 2003), and we therefore used either the midpoint of the Maastrichtian (68 Myr) or the Maastrichtian/Palaeocene border (65.5 Myr). (iii) The crown group of Cucurbitaceae was set to minimally 65 Myr or, alternatively, 55.8 Myr, based on the seeds from the Palaeocene Felpham flora (Collinson 1986; Collinson et al. 1993). These dates span the upper and lower boundary of the Palaeocene. (iv) The split between Linnaeosicyos, with tetracolpate–reticulate pollen, and the remaining New World Sicyeae (Schaefer et al. 2008a), which usually have polycolpate pollen, was set to minimally 33.9 Myr or, alternatively, 23 Myr, based on Hexacolpites echinatus pollen from the Oligocene of Cameroon, which is the oldest hexacolpate Sicyeae-type pollen (Salard-Cheboldaeff 1978; Muller 1985). Polycolpate pollen is not found in other Cucurbitaceae except the African Neoachmandra peneyana (Van der Ham & Pruesapan 2006). The Oligocene epoch ranges from 33.9 to 23 Myr, and the stratum containing Hexacolpites has not been precisely dated; therefore, in alternative analyses, we used the upper or lower boundary as minimal constraints. (v) The split between the Hispaniola endemics Anacaona and Penelopeia was set to maximally 20 or 15 Myr, based on the age of Dominican amber, which was produced by tropical trees and provides a proxy for the presence of tropical forest on that island; the amber age is estimated as 15–20 Myr (Iturralde-Vinent & MacPhee 1996).
To explore the sensitivity of the Bayesian clock to the various priors, we performed alternative MCMC runs in which we tested (a) the effect of a more clock-like Brownian motion parameter of 0.4 instead of 1.11, (b) the effect of using up to four data partitions, thereby allowing the genes and spacer regions to have different rates, and (c) the effects of varying the age constraints. For the latter exploration, we ran an analysis in which all constraints were set to the lowest age boundaries, another in which all constraints were set to the highest age boundaries, and four analyses that used the minimum age for one of the constraints and the maximum age for the remaining constraints. All other parameters, such as root rate and MCMC chain length, were constant between these six runs. Finally, we performed a run (d) with mean ages for four constraints, namely 66.8 Myr (constraint ii), 60.4 Myr (iii), 28.5 Myr (iv) and 17.5 Myr (v).
For the strict clock approach, rbcL branch lengths were calculated under a GTR+Γ+I+ clock model on the preferred ML topology. The tree was imported into PAUP, rooted on Coriaria (Zhang et al. 2006), and branch lengths were then calculated under the ‘enforce clock’ option. The distance between a calibration node and the present was divided by the age of the calibration node to obtain a substitution rate, and this rate was then used to calculate the age of divergence events of interest. As calibration nodes, we used either the age of the earliest Cucurbitaceae seeds (constraint iii) or the oldest Sicyeae-type pollen (constraint iv).
(e) Biogeographic analysis
For a dispersal-vicariance analysis, we used the 147-taxon dataset also used for the clock runs and coded the distribution ranges of all species in a binary matrix in MacClade. The species were recorded as present in one of five regions: Asia; Europe; Africa (including Madagascar); America (including Caribbean, Galapagos, and Hawaii); and Australia (including New Guinea and Polynesia). We then used the parsimony-based approach implemented in DIVA v. 1.1 (Ronquist 1996, 1997) to infer vicariance and dispersal events. The maximum number of areas simultaneously occupied by hypothetical ancestral lineages was experimentally constrained to 4, 3 or 2 because it is unlikely that an ancestral species would have ranged over several continents.
3. Results
Herbarium material yielded suitable DNA in more than 95 per cent of the cases, even for 50–100-year-old collections.
The highest-scoring ML tree obtained for the 260-taxon dataset (figure 1) shows Cucurbitaceae highly supported as monophyletic and family relationships similar to those found by Zhang et al. (2006) with a much larger amount of sequence data. Within Cucurbitaceae, there are five main clades (figure 1), namely: (i) a group of approximately 100 genera traditionally treated as subfamily Cucurbitoideae (Kosteletzky 1833) and usually subdivided into several tribes (below); (ii) a clade of Asian genera, including Alsomitra, Bayabusua and Neoalsomitra that corresponds to the tribe Gomphogyneae of Bentham & Hooker (1867); (iii) a clade of one African and five Neotropical genera, including Fevillea and Sicydium, that corresponds to the tribe Fevilleeae of Bentham & Hooker (1867); (iv) a clade of a few genera from Madagascar, continental Africa, Asia and South America corresponding to the tribe Zanonieae of Blume (1826); and (v) a clade consisting of the two Asian genera Actinostemma and Bolbostemma.
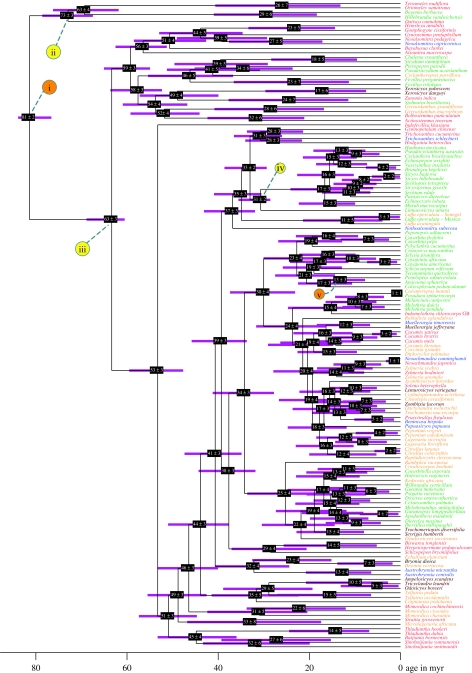
Figure 1.
Best ML tree for Cucurbitaceae and relatives found with combined chloroplast gene, spacer and intron sequences (5031 nucleotides) analysed under a GTR+Γ+I model with unlinked partitions for coding and non-coding regions. Likelihood bootstrap values greater than 60% are given at the nodes. Rooting follows Zhang et al. (2006). The geographical occurrence of genera is colour coded as follows: green, America (including Galapagos, Hawaii and the Caribbean); yellow, mainland African; brown, Madagascar; red, Asia; blue, Australia/New Guinea/Polynesia; black, Europe.
Clades (ii–v) have been treated as subfamily Nhandiroboideae (an illegitimate name) or Zanonioideae (a taxonomic synonym of Fevilleoideae), but this subfamily is not supported as monophyletic by our data. Clade (i), Cucurbitoideae, can be divided into geographically or morphologically more homogeneous groups that correspond to the traditional tribes Herpetospermeae, Bryonieae, Sicyeae, Coniandreae, Benincaseae and Cucurbiteae, plus a few clades of similar phylogenetic depth that have not traditionally been ranked as tribes, such as the Asian Thladiantha and Baijiania, the Asian/African Siraitia and Microlagenaria, the African/Asian Momordica, the African Telfairia, Cogniauxia, a group of Madagascan genera and the Himalayan Indofevillea, which is sister to all remaining Cucurbitoideae (figure 1). Well-known genera found to be poly- or paraphyletic include Citrullus (must include Acanthosicyos), Ampelosicyos (must include Tricyclandra and Odosicyos), Gomphogyne (must include Hemsleya), Xerosicyos (must include Zygosicyos), Apodanthera, Psiguria and Trichosanthes.
multidivtime dating runs with a Brownian motion parameter of 0.4 instead of 1.11, yielding barely different estimates for the ingroup nodes of interest. Runs that allowed uncoupled rates for the two genes and the spacers also yielded essentially identical estimates, and final runs therefore modelled the data under a single model. The estimates from the run in which all constraints were set to their lowest boundaries differed significantly from those obtained when all constraints were set to their highest boundaries (Wilcoxon signed-rank test, p=0.0085; see fig. 1c,d in the electronic supplementary material). Among the test runs in which one constraint was set to the minimum age and the others to the maximum age, only two yielded significantly different results: the Tetrameleoxylon fossil set to the minimum age and the Felpham flora seed set to the minimum age (see fig. 1e in the electronic supplementary material). However, all results were within the 95 per cent confidence intervals of the estimates obtained when the constraints were set to mean ages (see fig. 1a,b in the electronic supplementary material).
The substitution rates obtained under a strict clock model calibrated with either the seed or the pollen fossil (§2) were 0.00018 S/(S×Myr) (oldest Sycieae-type pollen, constraint (iv)) or 0.00030 S/(S×Myr) (earliest Cucurbitaceae seeds, constraint (iii)). An average rate of 0.00024 S/(S×Myr) yielded absolute times that for the most part were older than those obtained with the relaxed clock model (see table 2 in the electronic supplementary material that lists the ages obtained with the relaxed clock and with the strict clock model). The following discussion focuses on the relaxed clock estimates because they provide 95 per cent confidence intervals as a measure of uncertainty.
The split between the two genera of Begoniaceae, Begonia and Hillebrandia, is estimated as 29 (41–18) Myr old, roughly the age of the Hawaiian archipelago (ca 30 Myr), where Hillebrandia is endemic (Clement et al. 2004). The deepest split in the Cucurbitaceae is ca 63 (69–61) Myr old, while crown group Cucurbitoideae are 53 (60–48) Myr old, Gomphogyneae 56 (63–51) Myr, Fevilleeae 46 (55–37) Myr and the Actinostemma/Bolbostemma clade 52 (59–44) Myr. The Madagascan Xerosicyos clade (split Zanonia–Xerosicyos) appears to be 49 (57–40) Myr old; the likewise Madagascan Ampelosicyos clade is 29 (39–19) Myr old. The Madagascar/Southeast Asia disjunction between the two species of Muellerargia is only ca 12 (18–7) Myr old. The South America/Asia disjunction in the Zanonia clade (Siolmatra–Zanonia split), finally, is 24 (38–11) Myr old, and the South America/Africa disjunction in the Fevillea clade (Chalema–Cyclantheropsis split) 41 (51–31) Myr. Other estimates of specific interest (Introduction; see table 2 in the electronic supplementary material) concern Cayaponia (stem age 10 (17–5) Myr), Luffa (stem age 35 (41–31) Myr), Sicyos (stem age 8 (13–4) Myr) and the Lagenaria crown group (8 (14–3) Myr; figure 2).
Figure 2.
Chronogram obtained for Cucurbitaceae under a Bayesian autocorrelated rates relaxed clock model applied to the combined data (5031 nucleotides) and calibrated with three minimal (yellow) and two maximal (orange) constraints as in run (d) of §2(d). Age estimates with their 95% confidence ranges shown in purple. Rooting follows Zhang et al. (2006). Green, America (including Galapagos, Hawaii and the Caribbean); yellow, mainland Africa; brown, Madagascar; red, Asia; blue, Australia/New Guinea/Polynesia; black, Europe.
The DIVA analysis yielded Asia as the most likely region of origin of the Cucurbitaceae (figure 3a). From there, at least five lineages reached Africa, and 12 lineages reached Australia. No fewer than seven lineages independently reached the American continent, and no fewer than 13 lineages dispersed from Africa to Madagascar. Some disjunctions are best explained as secondary dispersals from Africa back to Asia (Coccinia and Momordica). A few lineages reached Africa via LDD from America (Cayaponia africana, Cucumeropsis, perhaps Cyclantheropsis and Kedrostis).
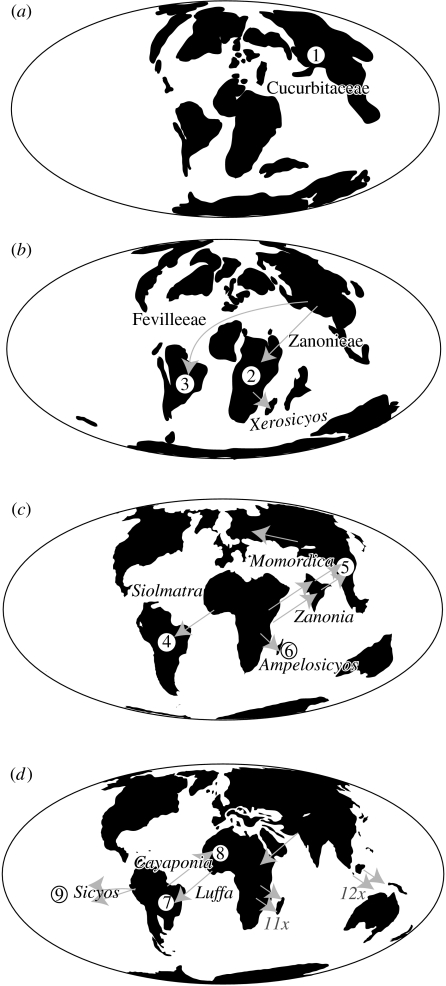
Figure 3.
The biogeographic history of Cucurbitaceae as inferred from the statistical approach described in the text (coastlines drawn after Smith et al. 1994). (a) Late Cretaceous, ca 70 Myr ago; (1) origin of the Cucurbitaceae in Asia. (b) Palaeocene/Eocene, 60–40 Myr ago; (2) the ancestor of the Zanonieae reaches Africa, the ancestor of the Xerosicyos lineage reaches Madagascar, and (3) the ancestor of Fevilleae reaches South America. (c) Oligocene, ca 30 Myr ago; (4) the ancestor of the Siolmatra lineage disperses over the Atlantic into South America, (5) the ancestors of Momordica cochinchinensis and Zanonia indica independently disperse from Africa to Southeast Asia, and (6) the ancestor of the Ampelosicyos lineage reaches Madagascar. (d) Middle Miocene, ca 10 Myr ago; (7) the ancestors of Neotropical Luffa disperse from Africa to the Americas, (8) the ancestor of Cayaponia africana disperses from South America to West Africa, and (9) ancestors of several Sicyos species groups spread from South America to Hawaii, Galapagos, New Zealand and Australia.
4. Discussion
(a) The geographical origins of the world's regional Cucurbitaceae floras
Our results suggest that Cucurbitaceae initially diversified in Asia (specifically, the region north of the Tethys) sometime in the Late Cretaceous. This fits with the observation that India contains more deeply divergent lineages of Cucurbitaceae than any other similar-sized geographical area (Chakravarty 1946, 1959; this study). Of the family's Late Cretaceous radiations, two clades (the Gomphogyneae and the Actinostemmateae) are now almost restricted to subtropical Asia (figure 1). A third clade, Fevilleeae, is mainly Neotropical except for a small African ‘extension’, Cyclantheropsis. The ancestors of Fevilleeae were probably more widely distributed in the Laurasian tropics and reached the American continent by dispersing across a still-narrow Atlantic (figure 3b; seeds of Fevilleeae are wind- and water-dispersed). Cyclantheropsis must result from a back dispersal from South America to Africa in the Middle Eocene. The ancestors of the fourth ancient clade, Zanonieae, apparently reached the African continent early and from there dispersed to Madagascar (the Early Eocene Xerosicyos lineage; figure 3b). Later, in the Oligocene, at least two LDD events brought the Siolmatra lineage to America and the Zanonia lineage back to tropical Asia (figure 3c). The fifth and last ancient clade, the Cucurbitoideae, diversified partly in Asia (e.g. Thladiantha, Siraitia, Trichosanthes), and partly in Africa (e.g. Momordica, Cucumis, Coccinia, Kedrostis). The cucumber (Cucumis sativus) and its closest relatives (not all included in the present study) evolved from a common ancestor ca 3 (6–1) Myr ago. The wax gourd Benincasa and its sister group Praecitrullus, an important vegetable in parts of India, apparently split only 5 (10–1) Myr ago. Further dispersals from Africa back to Asia are present within Momordica, Coccinia, Kedrostis and Corallocarpus (figure 3c).
The native European cucurbit flora consists only of Bryonia, with 10 species (Volz & Renner 2008), and its monotypic sister Ecballium, which probably represent a lineage that spread along the Tethys border from Asia to the Mediterranean 32 (41–24) Myr ago. The remaining cucurbit species that occur in Europe are the result of recent introductions (Echinocystis lobata, Sicyos angulatus and Thladiantha dubia) or casual escapes from cultivation (Citrullus lanatus, Cucumis melo, C. sativus and Cucurbita pepo). The closest extant relative of Bryonia and Ecballium is the Australian genus Austrobryonia (four species), which may have reached Australia from Asia ca 13 (21–6) Myr ago (Schaefer et al. 2008b).
African Cucurbitoideae (25 genera) are the result of five dispersals from Asia to Africa and two from America to Africa (in the genera Cucumeropsis and Cayaponia). The watermelon (Citrullus lanatus) and its sister species (C. colocynthis) apparently evolved from a common ancestor as recently as 2 (6–0.1) Myr ago. The lineage leading to the cucumber tree, Dendrosicyos socotranus, endemic on Socotra, some 350 km off the Arabian peninsula, is estimated as 22 (30–14) Myr old, while the Socotra archipelago is only some 10 Myr old (Ghebreab 1998). Dendrosicyos thus seems to be an island relict of a progenitor lineage that went extinct on the mainland. This example of a species that is twice as old as the island on which it occurs cautions against using geological calibrations in molecular clock dating. Another supposed example of a species being older than the island on which it lives, that of the Hawaiian Hillebrandia sandwicensis (Clement et al. 2004), is not supported by our data (see table 2 in the electronic supplementary material).
Madagascar has 16 native Cucurbitaceae genera with 50 species in total. From our data it appears that Cucurbitaceae reached Madagascar at least 13 times, apparently always from the African mainland, and that these 13 ancestors then underwent local radiations, giving rise to today's 50 species. Using Madagascar as a stepping stone, one of these clades, Peponium, later reached the Seychelles (the endemic species there has not yet been sequenced).
South America has approximately 350 species of Cucurbitoideae in 47 genera that all descend from five LDD events, mostly from Africa to South America. These involved the ancestors of Cucurbiteae, Sicyinae, a clade of Coniandreae, the Melothria clade and a subclade of Luffa. Based on the tree topology (figure 1), Luffa originated in the Old World or Australia, and one species then reached the New World by LDD from Africa across the Atlantic as suggested by Heiser & Schilling (1988). The fruit is dry with fibrous tissue and probably well adapted to floating (Ridley 1930). The Neotropical Melothria clade (figure 1) appears to have crossed the Pacific because the sister group of Melothria, Indomelothria, is endemic in Southeast Asia. Today's pumpkin and squash species (Cucurbita spp. in the Cucurbiteae) apparently originated in Central or South America, and the genus Cucurbita split from its sister clade, Peponopsis, only some 16 (23–9) Myr ago. North American Cucurbitaceae, finally, all descend from seven expansions of Central and South American lineages that occurred at widely different times (figure 2; see table 2 in the electronic supplementary material).
The indigenous Australian Cucurbitaceae flora consists of 30 species in 12 genera of which two are endemic: Nothoalsomitra, a liana species of Queensland's rainforests; and Austrobryonia, four species of trailers or creepers in the dry regions of (mostly) Central Australia. This low Australian species diversity is in marked contrast with the minimally 12 independent dispersal events into Australia (figures 1 and 3d). The largest Australian ‘radiation’ comprises only four species (Schaefer et al. 2008b), even though the ecological conditions in Australian rainforests and bushland are similar to those in Southeast Asian forests and African bushland, where cucurbit diversity is much higher.
Overall, Cucurbitaceae underwent at least 43 successful LDD events over the past 60 Myr, which would translate into an average of seven LDDs every 10 Myr. These events seem to have occurred throughout the evolutionary past of the family, rather than being clustered at a particular geological time (see table 2 in the electronic supplementary material). The most striking case of a rapid radiation following LDD is Sicyos, which reached Hawaii only ca 2 (6–0.1) Myr ago (figure 3d) and now has 15 Hawaiian species. Sicyos species often grow in seabird colonies (www.botany.hawaii.edu/gradstud/eijzenga/OIRC/lehua.htm# Vegetation), and the hooked barbs on the fruits may help external dispersal on bird feathers. Another extreme case is Muellerargia, the closest relative of Cucumis (Renner & Schaefer 2008), which comprises just two species, one endemic in Madagascar, and the other in northeast Australia and Timor.
(b) Using museomics to achieve less biased taxon sampling
Results of this study may have implications for the conservation of wild Cucurbitaceae gene pools for future crop improvement; this is almost certainly true for Luffa and Cucumis. They also show that the gourd family, which appears to be of Asian origin, has undergone numerous natural LDD events, prior to anthropogenic LDD, and that these events have played a large role in the build-up of local cucurbit floras. More generally, this study illustrates the great potential of herbarium material for generating broad phylogenetic frameworks at relatively low costs and in a time-efficient manner (no lengthy permit procedures). With rapidly improving molecular techniques for DNA isolation from tiny fragments of ancient collections, it is now possible to study the phylogeny of worldwide lineages without time-consuming and difficult field trips. And broad geographical sampling in turn is the precondition for assessing the full extent of LDD in the angiosperms at different geological times and across different ocean basins. Given that habitat destruction in the centres of cucurbit diversity (Madagascar, Southeast Asia, West Africa and Central America) is extremely high, the use of herbarium material also may soon be the only option for future research on the phylogenetics of this plant group.
Acknowledgements
We thank B. Duyfjes and W. J. de Wilde (L) for silica samples and advice; M. Nee (NY), M. Pignal (P), J. Wieringa (WAG) and the curators of the herbaria AAU, ASU, B, BKF, BR, BSC, CHR, CMU, E, EA, F, FTG, G, GIFU, K, KUN, L, LE, LISC, M, MEXU, MO, NE, RSA, ULM, US and Z for permission to sample DNA from specimens in their care; E. Vosyka for help in the laboratory; and the German science foundation for financial support (DFG RE603/3-1).
Supplementary Material
Species of Cucurbitaceae and chloroplast loci sequenced newly for this study, their sources, geographic origin, and GenBank accession numbers
Estimates (in Myr) for Cucurbitaceae divergence events of special biogeographic interest
Results of the sensitivity analysis testing the effects of uncertainty in the fossil ages used in the Bayesian relaxed clock runs: (a) age estimates from the MCMC analyses using the maximum (left orange), mean (red), and minimum (right orange) values for age constraints (ii)–(v) sorted according to age and overlaid on the chronogram from run d; (b) same as (a) but the estimated node ages from the runs that used maximum and minimum values for age constraints (ii)–(v) indicated, respectively, by open red and purple circles
Results of the sensitivity analysis testing the effects of uncertainty in the fossil ages used in the Bayesian relaxed clock runs: (c) pair-wise bar plots of estimated node ages from the MCMC analyses using maximum (red) and minimum (blue) values for age constraints (ii)–(v); (d) the same as (c) but pair-wise bar plots sorted by node age; (e–h) pair-wise bar plots comparing node ages obtained when the three of the four age constraints were set to their maximum age (red) while one was set to its minimum (blue), namely, in run e, the age of the Tetrameleoyxlon fossil wood (constraint (ii)), in run f, the age of Cucurbitaceae seeds from the Felpham flora (constraint (iii)); in run g, the split between the tetracolpate-reticulate and polycolpate pollen (constraint (iv)), and in run h, the age of Hispaniola amber (constraint v).
References
- Alsos I.G., Eidesen P.B., Ehrich D., Skrede I., Westergaard K., Jacobsen G.H., Landvik J.Y., Taberlet P., Brochmann C. Frequent long-distance plant colonization in the changing arctic. Science. 2007;316:1606–1608. doi: 10.1126/science.1139178. doi:10.1126/science.1139178 [DOI] [PubMed] [Google Scholar]
- Bentham G., Hooker J.D. Genera plantarum. vol.1. Reeve & Co; London, UK: 1867. [Google Scholar]
- Blume C. L. von. Ter Lands Drukkerij; Batavia, IL: 1826. Bijdragen tot de Flora van Nederlandsch Indie 15. [Google Scholar]
- Chakravarty H.L. Studies on Indian Cucurbitaceae with special remarks on distribution and uses of economic species. Indian J. Agr. Sci. 1946;16:1–89. [Google Scholar]
- Chakravarty H.L. Monograph on Indian Cucurbitaceae. Rec. Bot. Surv. India. 1959;17:1–234. [Google Scholar]
- Chen J.F., Zhuang F.Y., Liu X.A., Qian C.T. Reciprocal differences of morphological and DNA characters in interspecific hybridization in Cucumis. Can. J. Bot. 2004;82:16–21. doi:10.1139/b03-107 [Google Scholar]
- Clement W.L., Tebbitt M.C., Forrest L.L., Blair J.E., Brouillet L., Eriksson T., Swensen S.M. Phylogenetic position and biogeography of Hillebrandia sandwicensis (Begoniaceae): a rare Hawaiian relict. Am. J. Bot. 2004;91:905–917. doi: 10.3732/ajb.91.6.905. doi:10.3732/ajb.91.6.905 [DOI] [PubMed] [Google Scholar]
- Collinson M.E. The Felpham flora—a preliminary report. Tertiary Res. 1986;8:29–32. [Google Scholar]
- Collinson M.E., Boulter M.C., Holmes P.R. Magnoliophyta (‘Angiospermae’) In: Benton M.J., editor. The fossil record. Chapman and Hall; London, UK: 1993. pp. 809–841. [Google Scholar]
- De Wilde W.J.J.O., Duyfjes B.E.E. The wild species of Cucumis L. (Cucurbitaceae) in South-East Asia. Adansonia, sér. 3. 2007;29:239–248. [Google Scholar]
- Dillehay T.D., Rossen J., Andres T.C., Williams D.E. Preceramic adoption of peanut, squash, and cotton in Northern Peru. Science. 2007;316:1890–1893. doi: 10.1126/science.1141395. doi:10.1126/science.1141395 [DOI] [PubMed] [Google Scholar]
- Galley C., Bytebier B., Bellstedt D.U., Linder H.P. The Cape element in the Afrotemperate flora: from Cape to Cairo? Proc. R. Soc. B. 2007;274:535–543. doi: 10.1098/rspb.2006.0046. doi:10.1098/rspb.2006.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry A.H. The distribution and evolution of climbing plants. In: Putz F.E., Mooney H.A., editors. The biology of vines. Cambridge University Press; Cambridge, UK: 1991. pp. 3–42. [Google Scholar]
- Ghebreab W. Tectonics of the Red Sea region reassessed. Earth Sci. Rev. 1998;45:1–44. doi:10.1016/S0012-8252(98)00036-1 [Google Scholar]
- Ghebretinsae, A. & Barber, J. C. 2006. Phylogenetic relationships among species of Cucumis and Cucumella (Cucurbitaceae): evidence from ITS, rpl16 and trnS-G sequences. See www.2006.botanyconference.org/engine/search/index.php?func=detail&aid=498.
- Heibl, C. & Cusimano, N. 2008. LAGOPUS—divergence time estimation in R. See www.christophheibl.de/mdt/mdtinr.html.
- Heiser C.B., Schilling E.E. Phylogeny and distribution of Luffa (Cucurbitaceae) Biotropica. 1988;20:185–191. doi:10.2307/2388233 [Google Scholar]
- Herendeen P.S., Crane P.R., Drinnan A.N. Fagaceous flowers, fruits, and capsules from the Campanian (Late Cretaceous) of central Georgia, USA. Int. J. Plant Sci. 1995;156:93–116. doi:10.1086/297231 [Google Scholar]
- Iturralde-Vinent M.E., MacPhee R.D.E. Age and paleogeography of Dominican amber. Science. 1996;273:1850–1852. doi:10.1126/science.273.5283.1850 [Google Scholar]
- Kar R.K., Ambwani K., Sahni A., Sharma P. Unisexual flowers from the Deccan Intertrappean Bed of Madhya Pradesh, India. Palaeobotanist. 2003;52:73–79. [Google Scholar]
- Khajuria C.K., Prasad G.V.R., Manhas B.K. Palaeontological constraints on the age of Deccan Traps, Peninsular India. Newsletter on Stratigraphy. 1994;31:21–32. [Google Scholar]
- Kocyan A., Zhang L., Schaefer H., Renner S.S. A multi-locus chloroplast phylogeny for the Cucurbitaceae and its implications for character evolution and classification. Mol. Phyl. Evol. 2007;44:553–577. doi: 10.1016/j.ympev.2006.12.022. doi:10.1016/j.ympev.2006.12.022 [DOI] [PubMed] [Google Scholar]
- Kosteletzky, V. F. 1833. Allgemeine medizinisch-pharmazeutische Flora. vol. 6. Prague, Czech Republic: Borrosch & Andre.
- Lakhanpal R.N. Tertiary floras of India and their bearing on the historical geology of the region. Taxon. 1970;19:675–694. doi:10.2307/1219280 [Google Scholar]
- Lakhanpal R.N., Verma J.K. Fossil wood of Tetrameles from the Deccan Intertrappean beds of Mohgaonkalan, Madhya Pradesh. Palaeobotanist. 1965;14:209–213. [Google Scholar]
- Lavin M., Beyra Matos A. The impact of ecology and biogeography on legume diversity, endemism, and phylogeny in the Caribbean region: a new direction in historical biogeography. Bot. Rev. 2008;74:178–196. doi:10.1007/s12229-008-9006-8 [Google Scholar]
- Maddison W.P., Maddison D.K. Sinauer Associates; Sunderland, MA: 2003. MacClade: analysis of phylogeny and character evolution. [DOI] [PubMed] [Google Scholar]
- Muller J. Significance of fossil pollen for angiosperm history. Ann. Miss. Bot. Garden. 1985;71:419–443. doi:10.2307/2399033 [Google Scholar]
- Nee, M., Schaefer, H. & Renner, S. S. Submitted. A new, anomalous species of Fevillea (Cucurbitaceae) from Bolivia. Syst. Bot.
- Renner, S. S. & Schaefer, H. 2008. Phylogenetics of Cucumis (Cucurbitaceae) as understood in 2008. In Proc. IXth EUCARPIA meeting on genetics and breeding of Cucurbitaceae (ed. M. Pitrat), pp. 53–58. See https://w3.avignon.inra.fr/dspace/handle/2174/236.
- Ridley H.N. L. Reeve; Ashford, UK: 1930. The dispersal of plants throughout the world. [Google Scholar]
- Ronquist, F. 1996. DIVA v. 1.1. Computer program and manual available by FTP from Uppsala University. See ftp.uu.se or ftp.systbot.uu.se.
- Ronquist F. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 1997;46:195–203. doi:10.2307/2413643 [Google Scholar]
- Salard-Cheboldaeff M. Sur la palynoflore Maestrichtienne et Tertiaire du bassin sédimentaire littoral du Cameroun. Pollen et Spores. 1978;20:215–260. [Google Scholar]
- Sanjur O.I., Piperno D.R., Andres T.C., Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proc. Natl Acad. Sci. USA. 2002;99:535–540. doi: 10.1073/pnas.012577299. doi:10.1073/pnas.012577299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, H. & Renner, S. S. In press. Cucurbitaceae. In Families and genera of vascular plants. (ed. K. Kubitzki). Berlin, Germany: Springer.
- Schaefer H., Kocyan A., Renner S.S. Linnaeosicyos (Cucurbitaceae): a new genus for Trichosanthes amara, the Caribbean sister species of all Sicyeae. Syst. Bot. 2008;33:349–355. doi:10.1600/036364408784571707 [Google Scholar]
- Schaefer H., Telford I.R.H., Renner S.S. Austrobryonia (Cucurbitaceae), a new Australian endemic genus, is the closest living relative to the Eurasian and Mediterranean Bryonia and Ecballium. Syst. Bot. 2008;33:125–132. doi:10.1600/036364408783887357 [Google Scholar]
- Smith B.D. The initial domestication of Cucurbita pepo in the Americas 10,000 years ago. Science. 1997;276:932–934. doi:10.1126/science.276.5314.932 [Google Scholar]
- Smith A.G., Smith D.G., Funell M. Cambridge University Press; Cambridge, UK: 1994. Atlas of mesozoic and cenozoic coastlines. [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. doi:10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Thorne J.L., Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. doi:10.1080/10635150290102456 [DOI] [PubMed] [Google Scholar]
- Thorne J.L., Kishino H., Painter I.S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Van der Ham R., Pruesapan K. Pollen morphology of Zehneria s.l. (Cucurbitaceae) Grana. 2006;45:241–248. doi:10.1080/00173130600874834 [Google Scholar]
- Volz S.M., Renner S.S. Hybridization, polyploidy, and evolutionary transitions between monoecy and dioecy in Bryonia (Cucurbitaceae) Am. J. Bot. 2008;95:1297–1306. doi: 10.3732/ajb.0800187. doi:10.3732/ajb.0800187 [DOI] [PubMed] [Google Scholar]
- Wagner W.L., Herbst D.R., Sohmer S.H. University of Hawaii Press; Honolulu, HI: 1990. Manual of the flowering plants of Hawai'i. [Google Scholar]
- Whitaker T.W., Carter G.F. Oceanic drift of gourds—experimental observations. Am. J. Bot. 1954;41:697–700. doi:10.2307/2438952 [Google Scholar]
- Winkworth R.C., Wagstaff S.J., Glenny D., Lockhart P.J. Evolution of the New Zealand mountain flora: origins, diversification and dispersal. Org. Div. Evol. 2005;5:237–247. doi:10.1016/j.ode.2004.12.001 [Google Scholar]
- Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. doi:10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang L., Simmons M.P., Kocyan A., Renner S.S. Phylogeny of the Cucurbitales based on DNA sequences of nine loci from three genomes: implications for morphological and sexual system evolution. Mol. Phyl. Evol. 2006;39:305–322. doi: 10.1016/j.ympev.2005.10.002. doi:10.1016/j.ympev.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Zwickl, D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. PhD dissertation, The University of Texas, Austin, TX.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Species of Cucurbitaceae and chloroplast loci sequenced newly for this study, their sources, geographic origin, and GenBank accession numbers
Estimates (in Myr) for Cucurbitaceae divergence events of special biogeographic interest
Results of the sensitivity analysis testing the effects of uncertainty in the fossil ages used in the Bayesian relaxed clock runs: (a) age estimates from the MCMC analyses using the maximum (left orange), mean (red), and minimum (right orange) values for age constraints (ii)–(v) sorted according to age and overlaid on the chronogram from run d; (b) same as (a) but the estimated node ages from the runs that used maximum and minimum values for age constraints (ii)–(v) indicated, respectively, by open red and purple circles
Results of the sensitivity analysis testing the effects of uncertainty in the fossil ages used in the Bayesian relaxed clock runs: (c) pair-wise bar plots of estimated node ages from the MCMC analyses using maximum (red) and minimum (blue) values for age constraints (ii)–(v); (d) the same as (c) but pair-wise bar plots sorted by node age; (e–h) pair-wise bar plots comparing node ages obtained when the three of the four age constraints were set to their maximum age (red) while one was set to its minimum (blue), namely, in run e, the age of the Tetrameleoyxlon fossil wood (constraint (ii)), in run f, the age of Cucurbitaceae seeds from the Felpham flora (constraint (iii)); in run g, the split between the tetracolpate-reticulate and polycolpate pollen (constraint (iv)), and in run h, the age of Hispaniola amber (constraint v).