Abstract
The dynamic regulation of actin polymerisation plays crucial roles in cell morphology and endocytosis. The mechanistic details of these processes and the proteins involved are not fully understood, especially in neurons. PICK1 is a PDZ-BAR-domain protein involved in regulated AMPAR endocytosis in neurons. Here, we demonstrate that PICK1 binds F-actin and the actin-nucleating Arp2/3 complex, and potently inhibits Arp2/3-mediated actin polymerisation. RNAi knockdown of PICK1 in neurons induces a reorganisation of the actin cytoskeleton resulting in aberrant cell morphology. Wild-type PICK1 rescues this phenotype, but a mutant PICK1 (W413A) that does not bind or inhibit Arp2/3 has no effect. Furthermore, this mutant also blocks NMDA-induced AMPAR internalisation. This study identifies PICK1 as a new negative regulator of Arp2/3-mediated actin polymerisation that is critical for a specific form of vesicle trafficking and also for the development of neuronal architecture.
The dynamic actin cytoskeleton plays multiple roles in eukaryotic cells to regulate cell morphology, cell motility and vesicle trafficking by exerting mechanical forces that alter the shape of the plasma membrane. The Arp2/3 complex is the major catalyst for the formation of branched actin networks that mediate changes in membrane geometry. Proteins such as N-WASP, WAVE and cortactin bind and activate the Arp2/3 complex so that changes in cell morphology or vesicle trafficking occur at appropriate times and subcellular locations1-3.
Regulation of the actin cytoskeleton is crucial for neuronal morphogenesis1,4,5. Recent reports have suggested a role for the Arp2/3 activator N-WASP in neurite elongation and branching in neuronal development6,7. However, negative regulators of the Arp2/3 complex in these processes have not yet been identified. A role for Arp2/3-mediated actin dynamics in the regulation of endocytosis in mammalian cell lines is becoming increasingly well established. Localised alterations in actin turnover are proposed to provide mechanical forces that contribute to plasma membrane curvature, vesicle scission, and propulsion of nascent vesicles away from the membrane2,8-10. The specific molecular mechanisms for regulating actin to control endocytosis in neurons are unknown. Dendritic spines, the sites of excitatory synapses in neurons, are particularly enriched in filamentous (F-) actin11-13, which is extremely dynamic, rapidly cycling between globular (G-) actin and F-actin14. Since designated endocytic sites are found in dendritic spines15,16, it seems likely that the densely packed, dynamic actin filaments influence endocytosis. PICK1 is a PDZ and BAR-domain containing protein that binds to a number of membrane proteins including AMPA receptor (AMPAR) subunits GluR2/3. This interaction is required for AMPAR internalisation in response to Ca2+ influx via NMDAR activation in hippocampal neurons17-21. This is a major mechanism that underlies the reduction in synaptic strength in Long Term Depression (LTD), which is thought to play a role in learning and memory20,21. The precise molecular mechanisms for the action of PICK1 in receptor endocytosis are unclear.
Here, we demonstrate that PICK1 is an inhibitor of Arp2/3-mediated actin assembly. This regulation is required for the organisation of the actin cytoskeleton to establish appropriate neuronal morphology. Furthermore, we demonstrate that inhibition of Arp2/3 activity by PICK1 is required for NMDA-induced AMPAR endocytosis.
RESULTS
PICK1 binds actin
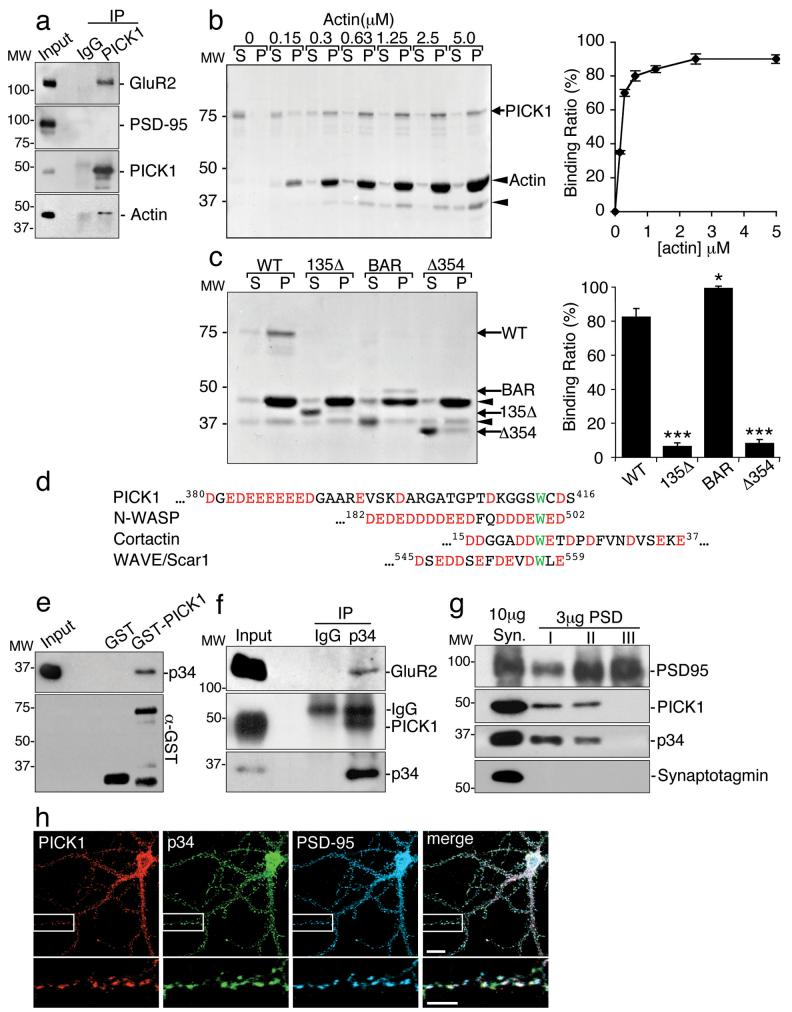
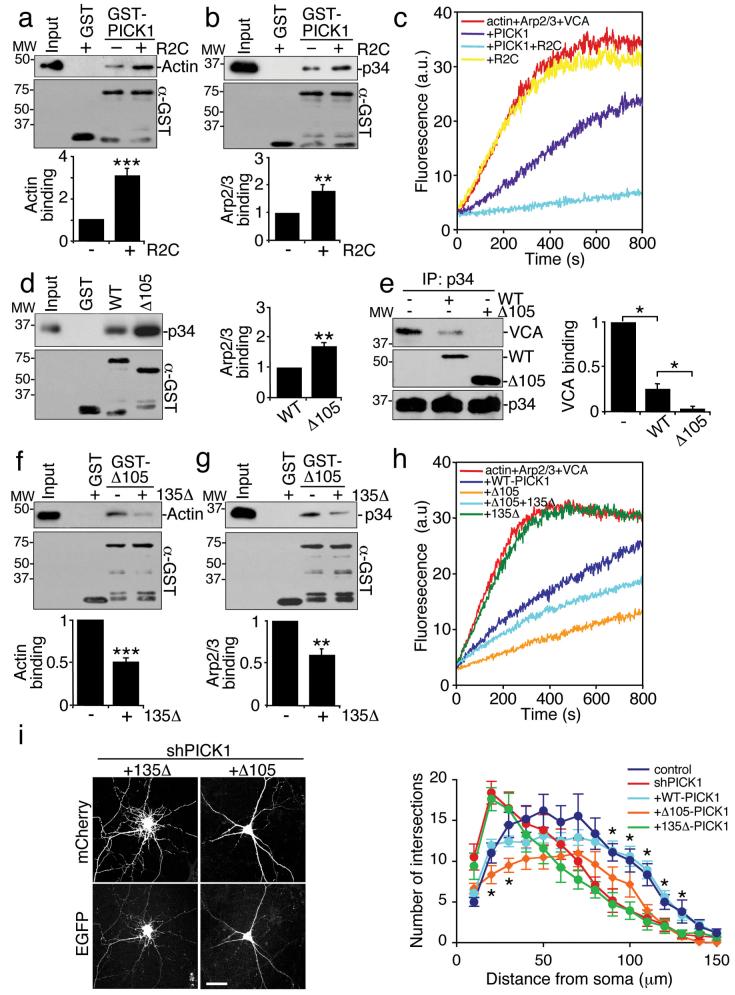
Since certain proteins with BAR-like domains are involved in actin regulation22-24, we asked whether PICK1 interacts with actin. Co-immunoprecipitations from neuronal extract demonstrate the presence of a PICK1-actin-GluR2 complex (Fig. 1a). Actin-binding proteins may associate with G-actin or F-actin, so we analysed which form binds to PICK1. In pull-down assays, GST-PICK1 does not bind G-actin (see Supplementary Information, Fig. S1 online). To investigate F-actin binding, we carried out actin sedimentation assays using actin filaments polymerised in vitro. As the concentration of actin is increased, GST-PICK1 is enriched in the F-actin pellet (Fig. 1b). GST-PICK1 binds F-actin with a KD of ~0.3 μM. To identify the actin-binding region of PICK1, we analysed truncation mutants of GST-PICK1. Figure 1c demonstrates that the BAR domain is necessary and sufficient for binding F-actin; in fact the isolated BAR domain binds actin more efficiently than full-length PICK1, suggesting that either the PDZ domain or the C-terminal region inhibit the interaction.
Figure 1.
PICK1 binds F-actin and Arp2/3 complex.
(a) PICK1 interacts with actin in neurons. Extracts from cultured neurons were immunoprecipitated with anti-PICK1 or control IgG, and bound proteins detected by immunoblotting.
(b) PICK1 binds directly to F-actin. Left panel: co-sedimentation assay using 0-5 μM rabbit muscle actin and 100nM GST-PICK1 was resolved by SDS-PAGE followed by Coomassie staining. S=supernatants, P=pellets. Arrowhead indicates actin-degradation product. Right panel: quantification of the percentage of PICK1 bound to F-actin as a function of actin concentration. n=4. Data are means +/− SEM.
(c)PICK1 binds F-actin via its BAR domain. Left panel: co-sedimentation assay using 2.5 μM rabbit muscle actin and 100 nM GST-PICK1 or truncation mutants, resolved by SDS-PAGE followed by Coomassie staining. S=supernatants, P=pellets. Arrowheads indicate actin and actin-degradation product. Right panel: quantification of the percentage of WT PICK1 and PICK1 mutants bound to F-actin. *p<0.05, ***p<0.001, n=5. Data are means +/− SEM.
(d) Sequence alignment of the C-terminus of PICK1 with known activators of the Arp2/3 complex. Conserved tryptophan residues are in green. Acidic amino acids are in red.
(e)GST-PICK1 binds directly to purified Arp2/3 complex. Immobilised GST and GST-PICK1 were incubated with 10 nM purified Arp2/3. Bound proteins were detected by immunoblotting (p34 Arc is a subunit of the Arp2/3 complex).
(f) PICK1 and GluR2 interact with the Arp2/3 complex in neurons. Extracts from cultured cortical neurons were immunoprecipitated with anti-p34 antibodies or control IgG, and bound proteins detected by immunoblotting.
(g) PICK1 and the Arp2/3 complex are similarly associated with the postsynaptic density (PSD). PSD fractions (PSD I, II and III) were prepared by extraction of synaptosomes (Syn) once (I) or twice (II) with Triton-X-100 or with Triton-X-100 and SDS (III). Proteins were detected by immunoblotting.
(h) PICK1 colocalises with Arp2/3 at synapses. Hippocampal neurons were stained for PICK1 (red), Arp2/3 subunit p34 (green) and the synaptic marker PSD-95 (blue). Colocalisation is white in the merged image. Scale bars: 10μm (top panels) and 5μm (bottom panels).
PICK1 binds the Arp2/3 complex
PICK1 contains an acidic region (D380GEDEEEEEED390) and a tryptophan residue (W413), resembling Arp2/3 activators (Fig. 1d). A pull-down assay using GST-PICK1 and purified Arp2/3 complex indicates that these proteins can interact directly (Fig. 1e), and co-immunoprecipitations from neuronal extract demonstrate an interaction between Arp2/3, PICK1 and GluR2 in native tissue (Fig. 1f). PICK1 is known to be loosely associated with the postsynaptic density (PSD25). Figure 1g indicates that both PICK1 and Arp2/3 are found in PSD fractions I and II purified from brain synaptosomes. Furthermore, PICK1 and Arp2/3 colocalise at PSD-95 immunopositive synaptic sites in a subset of hippocampal neurons (Fig. 1h).
PICK1 inhibits Arp2/3-mediated actin polymerisation
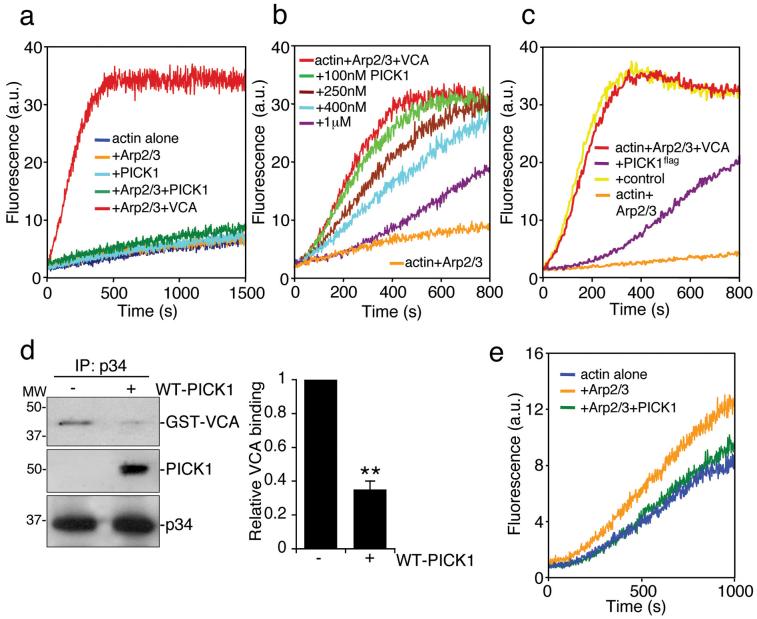
We utilised in vitro pyrene-labelled actin polymerisation assays to investigate regulation of the Arp2/3 complex by PICK1. PICK1 does not stimulate Arp2/3-mediated actin polymerisation, in contrast to the N-WASP VCA domain26 (Fig. 2a). Surprisingly, PICK1 reduces the rate of VCA-stimulated Arp2/3-mediated actin assembly in a dose-dependent manner (Fig. 2b). PICK1flag purified from neurons has similar inhibitory activity (Fig. 2c; also see Supplementary Information, Fig. S1 online). Co-immunoprecipitation from transfected COS cells shows that PICK1 and N-WASP VCA do not interact (see Supplementary Information, Fig. S1 online), indicating that PICK1 is not inhibiting VCA by direct interaction. Therefore, we asked whether PICK1 could compete with VCA for binding to the Arp2/3 complex. When PICK1 is present, VCA is displaced from the Arp2/3 complex (Fig. 2d), suggesting that PICK1 and VCA compete for the same binding site on Arp2/3. A high concentration of Arp2/3 complex has some actin nucleating activity even in the absence of VCA27. PICK1 inhibits actin assembly stimulated by 100nM Arp2/3 complex without VCA (Fig 2e), suggesting that PICK1 can directly disrupt the nucleating activity of Arp2/3.
Figure 2.
PICK1 inhibits VCA and Arp2/3-mediated actin polymerisation.
(a) PICK1 does not activate the Arp2/3 complex. In vitro polymerisation of pyrene-labelled actin (2.5 μM) was monitored using time-based fluorimetry. Additional components were: 25 nM Arp2/3 complex, 100 nM GST-VCA, 1 μM his6PICK1.
(b) PICK1 inhibits VCA and Arp2/3-stimulated actin polymerisation in a dose-dependent manner. Various concentrations of his6PICK1 (0.1 – 1 μM) were added to 2.5 μM pyrene-actin, 25 nM Arp2/3 and 100 nM GST-VCA in an in vitro polymerisation assay.
(c) PICK1 purified from neurons inhibits VCA and Arp2/3-stimulated actin polymerisation. High-density cortical neurons were transduced with PICK1flag-IRES-EGFP Sindbis virus. PICK1flag was purified by immunoprecipitation (see methods). Proteins eluted with the flag peptide were added to pyrene-actin, Arp2/3 complex and GST-VCA in an in vitro polymerisation assay. Eluates of immunoprecipitations from non-transduced neurons were used as control.
(d) PICK1 and VCA compete for binding to the Arp2/3 complex. Purified Arp2/3 was immunoprecipitated with anti-p34 antibody. Immunocomplexes were incubated with 100 nM purified GST-VCA with or without 500 nM his6PICK1. After washing of beads, proteins were detected by immunoblotting. Left panel shows representative western blots, right panel shows quantification of relative GST-VCA binding to Arp2/3 in the absence or presence of PICK1. n=3 **p<0.01. Data are means +/− SEM.
(e) PICK1 inhibits the Arp2/3 complex directly. Polymerisation of 5 μM pyrene-labelled actin in the presence of 100 nM Arp2/3 was inhibited by 400 nM his6PICK1.
These data demonstrate that PICK1 reduces the rate of actin assembly by two distinct mechanisms. First, by disrupting VCA-Arp2/3 interactions and second, by inhibiting the nucleating activity of Arp2/3 directly.
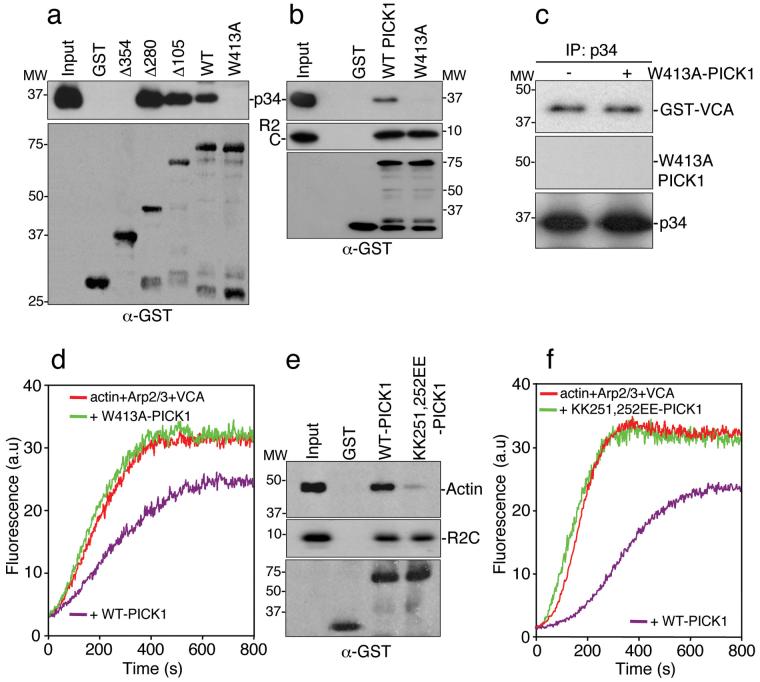
Tryptophan 413 is required for Arp2/3 binding to PICK1 and inhibition of actin polymerisation
We analysed the PICK1 sequence requirements for Arp2/3 binding using PICK1 truncation mutants. Figure 3a demonstrates that the minimal region for Arp2/3 interaction comprises amino acids 280-416 (Δ280). We mutated the C-terminal tryptophan in full-length PICK1 to alanine (W413A). Figure 3b shows that this single amino acid substitution eliminates Arp2/3 binding. As expected, GluR2 C-terminus binds equally well to both W413A-PICK1 and WT-PICK1 (Fig. 3b). Additional PICK1 binding partners are also unaffected by this mutation (see Supplementary Information, Fig. S1 online). Consistent with an inability to bind Arp2/3, W413A-PICK1 does not compete with VCA domain for Arp2/3 binding (Fig. 3c). In actin polymerisation assays W413A-PICK1 has no effect on VCA-stimulated Arp2/3-mediated actin assembly (Fig. 3d), demonstrating that Arp2/3 binding is an absolute requirement for the inhibitory effect of PICK1 on actin polymerisation.
Figure 3.
Identification of functional Arp2/3 and actin binding sites on PICK1.
(a) Amino acids 280-416 are the minimal region for Arp2/3 binding to PICK1. GST fusions corresponding to the stated regions of PICK1 (see Supplementary Information, Fig. S1 online) were immobilised on beads and incubated with 10 nM Arp2/3 complex. Bound proteins were detected by immunoblotting.
(b) Tryptophan 413 is required for Arp2/3 binding. GST-WT-PICK1 and GST-W413A-PICK1, in which the tryptophan residue is mutated to alanine, were immobilised on beads and incubated with 10 nM Arp2/3 complex or 10 nM his6-myc-tagged GluR2 C-terminus (his6mycR2C). Bound proteins were detected by immunoblotting.
(c) W413A-PICK1 does not compete with VCA for binding to Arp2/3 complex. Purified Arp2/3 was immunoprecipitated with anti-p34 antibody. Immunocomplexes were incubated with 100 nM GST-VCA with or without 500 nM his6W413A-PICK1. Bound proteins were detected by immunoblotting.
(d) W413A-PICK1 does not inhibit actin polymerisation. In vitro polymerisation of 2.5 μM pyrene-labelled actin in the presence of 25 nM Arp2/3, 100 nM GST-VCA and 400 nM his6WT-PICK1 or his6W413A-PICK1.
(e) Lysines 251, 252 are required for actin binding. GST-WT-PICK1 and GST-KK251,252EE-PICK1 were immobilised on beads and incubated with in vitro-polymerised F-actin (from 1 μM purified G-actin) or 10 nM his6-myc-tagged GluR2 C-terminus (his6mycR2C). Bound protein was detected by immunoblotting.
(f) KK251,252EE-PICK1 does not inhibit actin polymerisation. In vitro polymerisation of 2.5 μM pyrene-labelled actin in the presence of 25 nM Arp2/3, 100 nM GST-VCA and 400 nM his6WT-PICK1 or his6KK251,252EE-PICK1.
Lysines 251, 252 are required for actin binding and inhibition of actin polymerisation
We demonstrate in figure 1 that PICK1 binds actin via the BAR domain. To investigate a precise actin-binding motif, we analysed a double lysine motif (KK251,252) similar to the F-actin binding motif in pan1p28. Mutating lysines 251 and 252 in GST-PICK1 BAR domain to glutamate (KK251,252EE) abolishes F-actin binding (Fig 3e). In actin polymerisation assays, KK251,252EE-PICK1 has no effect on VCA-stimulated Arp2/3-mediated actin assembly (Figure 3f), demonstrating that actin binding is required for the inhibition of actin polymerisation by PICK1. In agreement with previous studies29,30, the KK251,252EE mutation also inhibits PICK1-lipid vesicle interactions. However, Arp2/3 binding is unaffected by this mutation (see Supplementary Information, Fig. S1 online).
Regulation of Arp2/3 complex by PICK1 is critical for maintaining the neuronal actin cytoskeleton
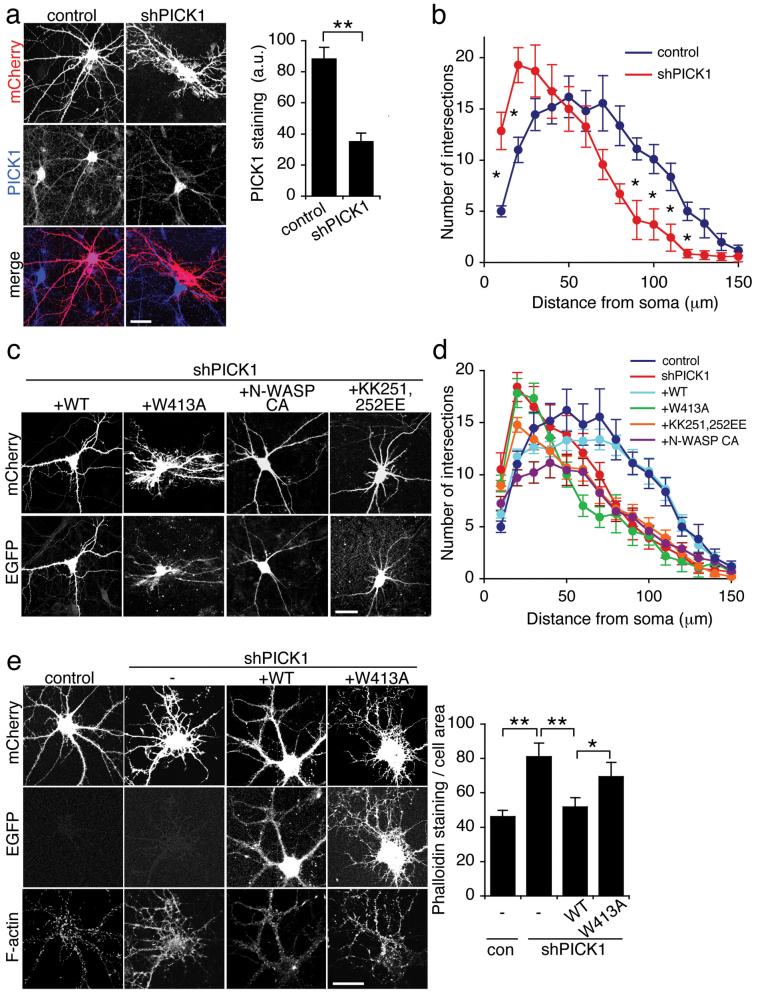
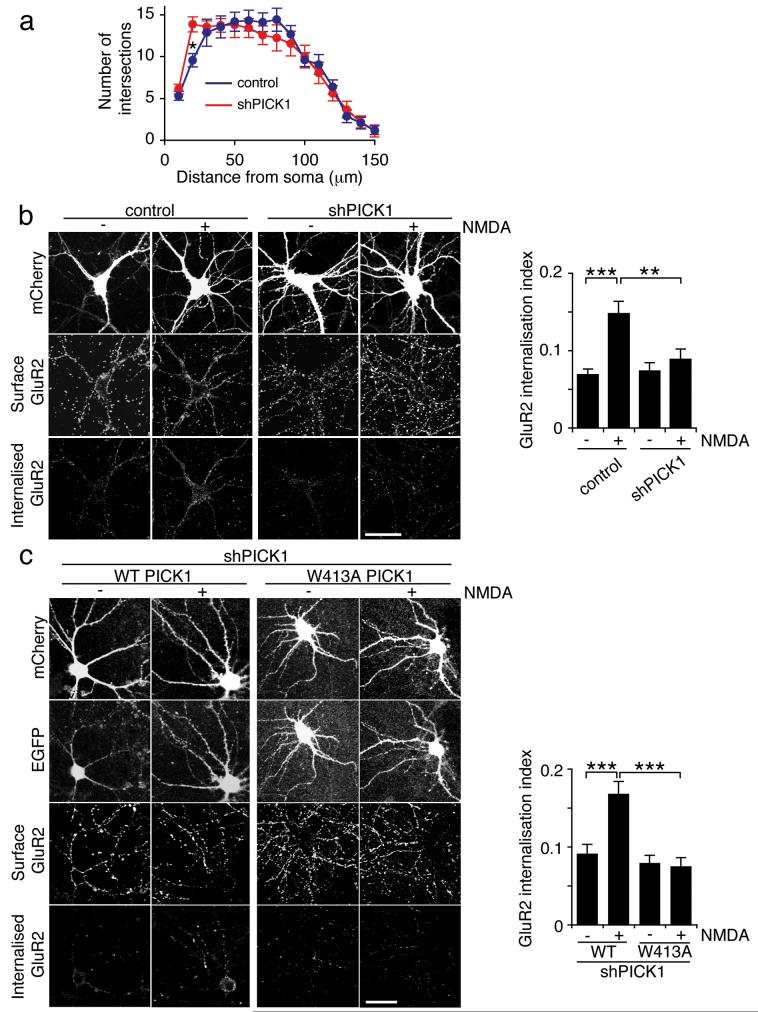
To analyse the role of PICK1 in regulating the neuronal actin cytoskeleton, we generated shRNA constructs to reduce PICK1 expression in hippocampal neurons. The constructs co-express mCherry fluorescent protein under the control of the neuronal CamKII promoter. PICK1 shRNA results in efficient PICK1 knockdown in transfected COS cells (see Supplementary Information, Fig. S2 online), and in hippocampal neurons (Fig 4a). In addition, neurons transfected with shPICK1 at 9-10 DIV show a dramatic change in morphology 5 days later (Fig. 4a). Neurons transfected at 13-14 DIV are more resistant to these morphological changes (see Fig. 5). Sholl analysis of neurons transfected at 10 DIV indicates a significant increase in the number of processes close to the cell body (at 10 and 20 μm radii) and significantly fewer distal processes (at 80-120 μm radii) compared to controls (Fig. 4b). CamKII staining confirms that all transfected cells are indeed neurons (see Supplementary Information, Fig. S2 online). The subcellular localisations of GluR2 and Arp2/3 are altered in shPICK1-transfected neurons (see Supplementary Information, Fig. S2 online). Cotransfection of a shRNA-resistant WT-PICK1 construct carrying silent mutations rescues neuronal morphology, confiming that the RNAi effect is indeed a direct result of PICK1 knockdown. In striking contrast, W413A-PICK1 does not rescue shRNA-induced changes (Fig. 4c and d). These experiments suggest that inhibition of Arp2/3 by PICK1 is required for correct neuronal morphology, and that the morphological changes that occur after PICK1 knockdown are due to over-activity of the Arp2/3 complex. To investigate this further, we used the CA domain of N-WASP to inhibit Arp2/331 in neurons with reduced PICK1 levels. Expression of N-WASP-CA reduced the aberrant proximal branching, but did not rescue the effects on distal processes (Fig. 4c and d). Interestingly, co-transfection of shPICK1 with shRNA-resistant KK251,252EE-PICK1, which does not bind F-actin, has an intermediate effect, and only partially rescues the aberrant proximal branching. Overexpressing each of these PICK1 constructs on a control mCherry background has no significant effect on gross neuronal morphology (see Supplementary Information, Fig. S3 online). To directly analyse the effect of PICK1 on the neuronal actin cytoskeleton, we used fluorophore-conjugated phalloidin to visualise F-actin in shRNA-transfected neurons. In neurons transfected with control plasmids, F-actin is found concentrated in dendritic spines and the cell periphery, as demonstrated previously11-13. 5 days following transfection with PICK1 shRNA, F-actin has an altered distribution, correlating with the aberrant morphology (Fig. 4e). Furthermore, the total phalloidin staining is significantly higher, providing additional evidence that PICK1 functions as an inhibitor of actin polymerisation in neurons. Cotransfection with shRNA-resistant WT-PICK1, but not W413A-PICK1, rescues the RNAi effects on the actin cytoskeleton (Fig. 4e).
Figure 4.
Inhibition of Arp2/3 activity by PICK1 is required for appropriate neuronal morphology in developing neurons.
(a) Knockdown of PICK1 expression disrupts neuronal morphology. Dissociated hippocampal neurons were transfected with mCherry-PICK1shRNA or mCherry control constructs at 10 DIV, and stained for PICK1 5 days later (middle panels). mCherry fluorescence is shown in the top panels, bottom panels are merged images. Note the untransfected cells adjacent to the transfected cells for comparison. Graph shows intensity of PICK1 staining for control and PICK1shRNA transfected cells. n=7 cells, **p<0.01. Data are means +/− SEM. Scale bar: 20 μm.
(b) Sholl analysis for control and shPICK1 transfected neurons. Concentric circles with increasing radii were superimposed on mCherry fluorescence images of neurons, and the number of processes crossing each radius was counted. n=10 cells, *p<0.05. Data are means +/− SEM.
(c) WT-PICK1 rescues neuronal morphology in shPICK1-treated neurons, PICK1 mutants do not. Dissociated hippocampal neurons were transfected with mCherry-PICK1shRNA and shRNA-resistant WT-PICK1-IRES-EGFP, W413A-PICK1-IRES-EGFP, KK251,252EE-PICK1-IRES-EGFP or N-WASP CA-IRES-EGFP as indicated. Top panels: mCherry fluorescence, bottom panels: EGFP fluorescence. Scale bar: 20 μm.
(d) Sholl analysis for rescue experiments. Dissociated hippocampal neurons were transfected as in (c). Concentric circles with increasing radii were superimposed on mCherry fluorescence images of neurons, and the number of processes crossing each radius was counted. n=15-20 cells. Data are means +/− SEM. Significant differences at 20 μm: p<0.05: sh/KK251,252EE, WT/N-WASP-CA, p<0.005: sh/WT, WT/W413A, WT/KK251,252EE, sh/N-WASP-CA. Significant differences at 100 μm: p<0.05: WT/KK251,252EE, p<0.01: WT/W413A, sh/WT, WT/N-WASP-CA.
(e) RNAi-knockdown of PICK1 increases total F-actin in hippocampal neurons. Dissociated hippocampal neurons were transfected with mCherry-PICK1shRNA or mCherry control, and WT-PICK1-IRES-EGFP or W413A-PICK1-IRES-EGFP as indicated, and stained for F-actin with Alexa350-phalloidin. Top panels: mCherry, middle panels: EGFP, bottom panels: Alexa350-phalloidin. Graph shows quantification of total phalloidin staining normalised to cell area. n=30 cells, *p<0.05, **p<0.01. Data are means +/− SEM. Scale bar: 20 μm.
Figure 5.
The PICK1-Arp2/3 complex interaction is required for NMDA-induced AMPAR endocytosis in neurons.
(a) PICK1 knockdown in more mature neurons has minimal effect on morphology. Dissociated hippocampal neurons were transfected with mCherry-PICK1shRNA or mCherry control constructs at 13 DIV and subjected to Sholl analysis five days later. Concentric circles with increasing radii were superimposed on neuron images (mCherry fluorescence), and the number of processes crossing each radius was counted. n=18 cells, *p<0.05. Data are means +/− SEM.
(b) PICK1 knockdown blocks NMDA-induced AMPAR endocytosis. Dissociated hippocampal cultures were transfected with either mCherry-PICK1shRNA or mCherry control constructs at 13 DIV. Internalised GluR2 in response to NMDA treatment was analysed at 18 DIV by antibody-feeding using anti-GluR2. Cells were fixed and subsequently stained for surface and internalised GluR2 using different fluorescent secondary antibodies. Representative images are shown for all conditions, and graph shows GluR2 internalisation index (internalised/surface+internalised). n=18 cells, **p<0.01; ***p<0.001. Data are means +/− SEM. Scale bar: 20 μm.
(c) WT-PICK1 rescues NMDA-induced AMPAR endocytosis in shPICK1-treated neurons, W413A-PICK1 does not. Hippocampal neurons were co-transfected with mCherry-PICK1shRNA and shRNA-resistant WT-PICK1-IRES-EGFP or W413A-PICK1-IRES-EGFP at 13 DIV. GluR2 internalisation was analysed at 18 DIV as in (a), above. n=18 cells, ***p<0.001. Data are means +/− SEM. Scale bar: 20 μm.
To investigate whether the increased branching close to the cell body represents bona fide dendritic branching, we carried out anti-MAP2 immunocytochemistry on control and shPICK1-transfected neurons. Sholl analysis of the MAP2 immunofluorescence demonstrates that there is an increase in proximal dendritic branching in shPICK1-transfected cells compared to controls (see Supplementary Information, Fig. S4 online). Qualitative analysis indicates that not all processes induced by PICK1 knockdown are MAP2-positive (see Supplementary Information, Fig. S4 online), suggesting that not all aberrant actin-based protrusions result in the formation of a dendrite.
Inhibition of Arp2/3-mediated actin polymerisation by PICK1 is required for NMDA-induced endocytosis of GluR2-containing AMPARs
Since PICK1 is crucial for regulated AMPAR endocytosis17-21,29,32, we asked whether the inhibition of actin polymerisation via PICK1-Arp2/3 interactions provides a driving force for receptor trafficking.
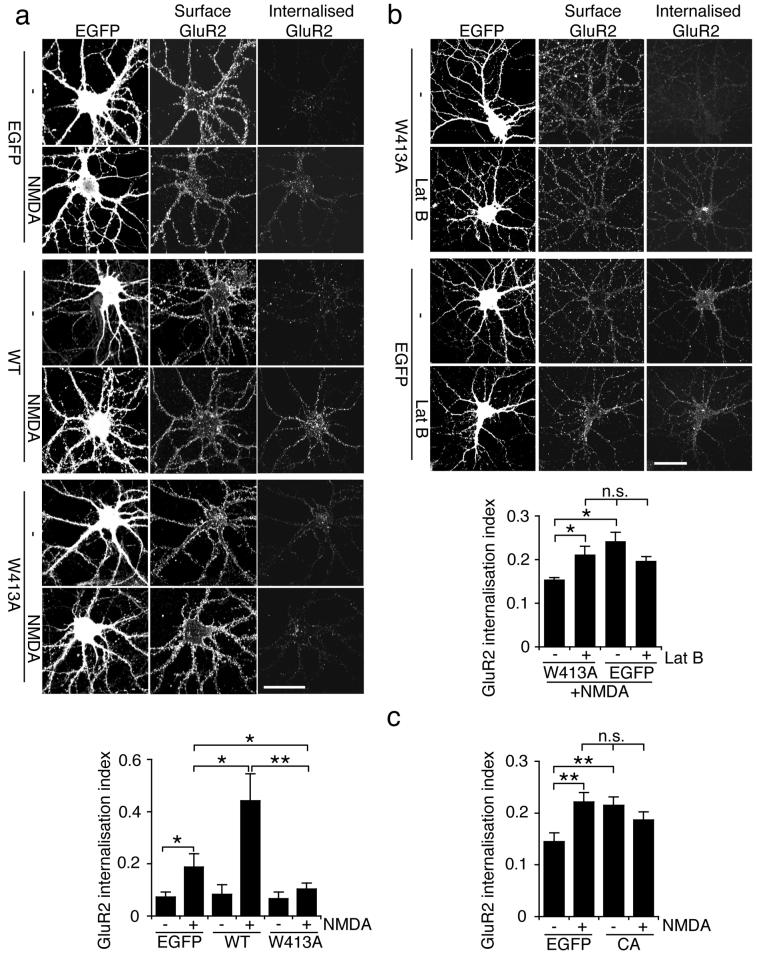
Since aberrant neuronal morphology might in itself affect trafficking events, we used an alternative approach to study AMPAR trafficking. We found that neurons transfected at 13-14 DIV were more resistant to morphological changes compared to those transfected at 9-10 DIV (Fig. 5a). We utilised a “chemical LTD” protocol, whereby NMDARs are activated by bath-application of NMDA to induce AMPAR internalisation19,29,33. Antibody-feeding immunocytochemistry was carried out at 18 DIV on cultured hippocampal neurons transfected at 13 DIV to quantify the degree of internalisation of GluR2 occurring in response to NMDA treatment. Control neurons show a two-fold increase in GluR2 endocytosis in response to NMDA, which is completely blocked in shPICK1-transfected neurons (Fig 5b). Neurons co-transfected with shPICK1 and shRNA-resistant WT-PICK1 exhibit restored NMDA-induced AMPAR endocytosis. However, in neurons co-transfected with shPICK1 and W413A-PICK1, NMDA-induced AMPAR endocytosis is completely abolished (Fig 5c).
To provide further evidence that PICK-Arp2/3 interactions are required for regulated AMPAR internalisation, we also employed a dominant negative approach. As demonstrated before, overexpression of WT-PICK1 enhances NMDA-induced AMPAR endocytosis compared to EGFP-expressing controls19. In stark contrast, W413A-PICK1 blocks NMDA-induced internalisation of GluR2-containing AMPARs (Fig. 6a). Overexpression of neither protein influences transferrin receptor internalisation (see Supplementary Information, Fig. S5 online). Since NMDA-induced AMPAR endocytosis is blocked by a PICK1 mutant that is unable to inhibit actin polymerisation, we asked whether this block could be reversed by pharmacological inhibition of actin polymerisation. Figure 6b shows that W413A-PICK1 expressing hippocampal neurons exposed to 20μM latrunculin B exhibit a comparable level of NMDA-induced AMPAR internalisation to EGFP-expressing control cells, supporting the hypothesis that W413A-PICK1 blocks trafficking because of an inability to inhibit actin polymerisation. Note that this level of latrunculin treatment does not alter neuronal morphology, consistent with a previous report34. To provide additional evidence that inhibition of Arp2/3 complex is required for AMPAR internalisation, we analysed AMPAR internalisation in neurons expressing the N-WASP CA domain, which inhibits endogenous Arp2/3 complex31. Inhibition of actin polymerisation by CA domain expression results in increased basal AMPAR internalisation that occludes the NMDA-induced increase in endocytosis (Fig. 6c).
Figure 6.
Inhibition of Arp2/3-mediated actin polymerisation by PICK1 is required for NMDA-induced AMPAR endocytosis in neurons.
(a) W413A-PICK1 overexpression blocks NMDA-induced internalisation of GluR2-containing AMPARs. Dissociated hippocampal neurons were transduced with either WT-PICK1-IRES-EGFP, W413A-PICK1-IRES-EGFP, or empty-IRES-EGFP Sindbis virus. Internalised GluR2 in response to NMDA treatment was analysed by antibody-feeding using anti-GluR2. Cells were fixed and subsequently stained for surface and internalised GluR2 using different fluorescent secondary antibodies. Representative images are shown for all conditions, and graph shows GluR2 internalisation index (internalised/surface+internalised). n=12 cells, *p<0.05; **p<0.01. Data are means +/− SEM. Scale bar: 20 μm.
(b) Latrunculin B reverses the blockade of NMDA-induced AMPAR endocytosis by W413A-PICK1. Dissociated hippocampal neurons transduced with W413A-PICK1-IRES-EGFP or empty-IRES-EGFP Sindbis virus were exposed to 20 μM Lat B or vehicle for 1h, and treated with NMDA. Note all cells are NMDA-treated in this experiment. Representative images are shown for all conditions, and graph shows GluR2 internalisation index (internalised/surface+internalised). n=25 cells, *p<0.05, n.s. p>0.05. Data are means +/− SEM. Scale bar: 20 μm.
(c) Arp2/3 inhibition by N-WASP CA domain enhances AMPAR endocytosis. Dissociated hippocampal neurons were transfected with N-WASP CA-IRES-EGFP or empty-IRES-EGFP. Internalised GluR2 in response to NMDA treatment was analysed by antibody-feeding using anti-GluR2. Cells were fixed and subsequently stained for surface and internalised GluR2 using different fluorescent secondary antibodies. Graph shows GluR2 internalisation index (internalised/surface+internalised). n=20 cells, **p<0.01. Data are means +/− SEM.
We also carried out internalisation assays in COS cells expressing GluR2myc and WT- or W413A-PICK1. WT-PICK1 mediates robust GluR2 internalisation19. In contrast, GluR2 endocytosis in W413A-PICK1 expressing cells is similar to levels in the absence of PICK1 (see Supplementary Information, Fig. S5 online), indicating that interactions with Arp2/3 are required for PICK1-mediated GluR2 endocytosis. Interestingly, latrunculin treatment has a different effect in COS cells compared to neurons, and results in reduced levels of GluR2 endocytosis in both WT- and W413A-PICK1 transfected cells. This suggests differences in actin-based endocytic mechanisms between neurons and COS cells.
Taken together, these data demonstrate that the inhibition of Arp2/3-mediated actin polymerisation by PICK1 is required for the NMDA-induced endocytosis of GluR2-containing AMPARs in neurons.
Interaction with GluR2 C-terminus enhances inhibition of Arp2/3 complex by PICK1
The dual role for PICK1-Arp2/3 interactions in AMPAR trafficking and cell morphology suggests that PICK1 may tonically inhibit the Arp2/3 complex, yet provide an enhanced, localised and transient inhibition to drive AMPAR endocytosis. Therefore, we investigated the possibility that the inhibition of actin polymerisation by PICK1 is regulated by interacting with GluR2. The PDZ domain forms an intramolecular interaction with the BAR domain, inhibiting non-PDZ interactions with additional proteins. This conformation is disrupted by binding of a PDZ ligand, exposing the BAR domain/C-terminal region to additional interactors29 (also see Supplementary Information, Fig. S6 online). Since actin and Arp2/3 complex bind non-PDZ regions of PICK1, we asked if GluR2 C-terminus (R2C) could modulate F-actin and Arp2/3 binding by altering the conformation of PICK1. Addition of R2C results in significant increases in PICK1-actin and PICK1-Arp2/3 interactions (Fig. 7a and b). These results suggest that GluR2 binding might enhance the inhibitory effect of PICK1 on Arp2/3-mediated actin assembly. As predicted, R2C dramatically enhances the inhibition of actin polymerisation by PICK1, resulting in nearly complete blockade of actin assembly (Fig. 7c).
Figure 7.
PICK1-mediated inhibition of actin polymerisation is enhanced by GluR2.
(a) GluR2 C-terminus (R2C) enhances the PICK1-actin interaction. Immobilised GST-PICK1 alone or complexed with his6mycR2C was incubated with 300 μg brain homogenate. Bound proteins were detected by immunoblotting. n=4, ***p<0.001. Data are means +/− SEM.
(b) R2C enhances the PICK1-Arp2/3 interaction. Immobilised GST-PICK1 alone or complexed with his6mycR2C was incubated with 10 nM of purified Arp2/3 complex. n=4, **p<0.01. Data are means +/− SEM.
(c) R2C enhances inhibition of actin polymerisation by PICK1. In vitro polymerisation of 2.5 μM pyrene-labelled actin in the presence of 25 nM Arp2/3, 100 nM GST-VCA, 800 nM his6PICK1 and 2 μM R2C as shown.
(d) Deleting the PICK1 PDZ domain increases Arp2/3 binding. Immobilised GST-WT-PICK1 or GST-Δ105-PICK1 was incubated with 10 nM purified Arp2/3. n=3, **p<0.01. Data are means +/− SEM.
(e) Deletion of the PICK1 PDZ domain enhances competition with VCA for binding to Arp2/3. Purified Arp2/3 was immunoprecipitated with anti-p34 antibody. Immunocomplexes were incubated with 100 nM GST-VCA with or without 500 nM his6WT-PICK1 or his6Δ105-PICK1. n=3, **p<0.01. Data are means +/− SEM.
(f)PICK1-actin binding is regulated by an intramolecular interaction in PICK1. Immobilised GST-Δ105-PICK1 alone or complexed with his6135Δ-PICK1 was incubated with 300 μg brain homogenate. n=3, ***p<0.001. Data are means +/− SEM.
(g) PICK1-Arp2/3 binding is regulated by an intramolecular interaction in PICK1. Immobilised GST-Δ105-PICK1 alone or complexed with his6135Δ-PICK1 was incubated with 10 nM purified Arp2/3. n=3, **p<0.01. Data are means +/− SEM.
(h) Deletion of the PICK1 PDZ domain enhances inhibition of actin polymerisation. In vitro polymerisation of 2.5 μM pyrene-labelled actin in the presence of 25 nM Arp2/3, 100 nM GST-VCA, 800 nM his6WT-PICK1 or his6Δ105-PICK1 and 1.5 μM his6135Δ-PICK1 as shown.
(i) Δ105-PICK1 over-rescues aberrant branching induced by PICK1 knockdown. Dissociated hippocampal neurons were co-transfected with mCherry control or mCherry-PICK1shRNA and empty-IRES-EGFP, WT-PICK1-IRES-EGFP, Δ105-PICK1-IRES-EGFP, or 135Δ-PICK1-IRES-EGFP at 10 DIV and subjected to Sholl analysis five days later. n=15-25 cells, *p<0.05 comparing WT-PICK1 to Δ105-PICK1. Data are means +/− SEM. Scale bar: 20 μm.
Since the proposed mechanism for this allosteric modulation is displacement of the PDZ domain from the BAR domain, we analysed the binding of F-actin and Arp2/3 to PICK1 with the PDZ domain deleted (Δ105-PICK1). Figure 7d shows that Arp2/3 binding to PICK1 is enhanced by PDZ deletion. Consistent with this, we demonstrate in Figure 1e that binding to F-actin is also enhanced by PDZ deletion. Furthermore, Δ105-PICK1 competes with VCA for Arp2/3 binding much more efficiently than WT-PICK1 (Fig. 7e). Adding purified PDZ domain (135Δ-PICK1) to Δ105-PICK1 inhibits binding to both actin and Arp2/3, confirming an allosterically-regulated interaction with PICK1 (Fig. 7f and g). Consistent with these observations, PDZ domain deletion potentiates the inhibition of actin polymerisation by PICK1 (Fig. 7h). Adding the free PDZ domain (135Δ-PICK1) to this reaction reduces the inhibition of actin assembly by Δ105-PICK1 (Fig. 7h), confirming that the intramolecular interaction between the PDZ domain and the BAR domain plays a crucial role in controlling the inhibition of actin polymerisation by PICK1. We investigated the effect of PDZ-deletion of PICK1 in hippocampal neurons. Δ105-PICK1 expression reduces branching in proximal dendrites in shPICK1 treated neurons to a greater extent than WT-PICK1 (Fig. 7i), suggesting that PDZ deletion enhances the influence of PICK1 on the actin cytoskeleton in neurons. 135Δ-PICK1 does not rescue neuronal morphology, presumably because this protein does not bind actin or Arp2/3.
These data show that the PICK1-R2C complex has a greater effect on reducing actin assembly compared to PICK1 alone, providing a mechanism for regulating PICK1 so that it is less active when the PDZ domains are unoccupied, and maximally active when bound to AMPARs.
DISCUSSION
Here, we describe a molecular mechanism for regulating neuronal actin dynamics by PICK1. PICK1 binds F-actin and the Arp2/3 complex, and inhibits Arp2/3-mediated actin assembly activated by the N-WASP VCA domain. This mechanism has two distinct roles in neuronal function; first, it is critical for development of neuronal morphology, and second, it is required for stimulated AMPAR endocytosis.
Our data demonstrate that constitutive inhibition of Arp2/3-mediated actin polymerisation by PICK1 is required to establish appropriate neuronal morphology. One of the characteristics of neurons with reduced PICK1 expression is an increase in projections from the cell body and proximal dendrites. This effect is reversed by inhibition of Arp2/3 activity, confirming that PICK1 prevents aberrant actin-based branching by suppressing Arp2/3 activity. A second morphological effect of PICK1 knockdown is a reduction in neurite length. This is consistent with previous reports demonstrating a requirement for Arp2/3 inhibition in neurite elongation31. However, inhibition of Arp2/3 activity by N-WASP-CA does not rescue neurite length, suggesting that alternative specific properties of PICK1 protein are involved in dendrite outgrowth. It has been shown that PICK1 is involved in regulation of growth cone function via a PDZ domain interaction with the netrin-1 receptor UNC535. This is consistent with our observation that PDZ-deleted PICK1 does not rescue the distal effects of PICK1 knockdown as efficiently as WT-PICK1. The morphological effects of PICK1 knockdown appear to be restricted to immature neurons, suggesting that following a certain developmental period, PICK1 is less critical for maintaining appropriate gross morphology of the dendritic arborisation. Further studies are needed to investigate the precise role of PICK1 in neuronal development.
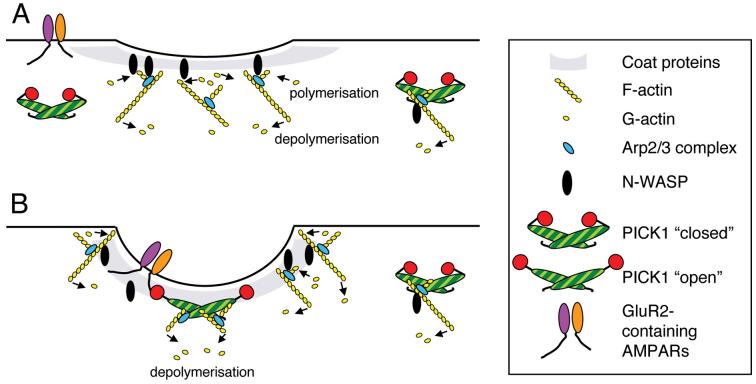
Recent studies have defined a role for actin dynamics in endocytosis in mammalian cell lines2,8-10. It has been proposed that actin polymerisation occurs around the nascent endocytic vesicle to provide mechanical forces for late stages of invagination and vesicle scission. Actin assembly occurring early and directly at the nascent coated pit may create forces to oppose membrane invagination2 (Fig. 8). Therefore, the initial invagination of the plasma membrane would require a phase of reduced F-actin, which may be followed by a highly localised, restricted burst of actin polymerisation around the neck of the coated pit to drive scission and subsequent propulsion away from the plasma membrane2,10. Indeed, actin assembly at clathrin-coated pits in fibroblasts has been observed as a transient late event, following clathrin and dynamin recruitment by several seconds, suggesting that the early induction of membrane curvature occurs under conditions of low F-actin36,37. We propose that PICK1 functions to inhibit actin assembly at endocytic sites until membrane invagination has been initiated (Fig. 8).
Figure 8.
A proposed model for the regulation of actin polymerisation by PICK1.
(a) In the absence of receptor cargo, PICK1 maintains a low-level tonic inhibition of actin polymerisation, sufficient to prevent inappropriate membrane protrusions that would occur as a result of high levels of F-actin assembly. Treadmilling cortical actin restricts invaginations by maintaining membrane tension.
(b) When GluR2 binds PICK1, PICK1-actin and PICK1-Arp2/3 interactions are enhanced and actin polymerisation is locally inhibited. Actin depolymerisation continues at these sites, resulting in a net loss of F-actin around the nascent coated pit. The resulting reduction in membrane tension allows the membrane to invaginate. Clathrin and associated coat proteins and the BAR domain of PICK1 may contribute to membrane curvature.
Although consistent with Zhou et al.34, who demonstrate that depolymerisation is the important shift in actin dynamics to trigger AMPAR endocytosis, two of our results were surprising to us. The reversal of the W413A-PICK1-mediated block of AMPAR endocytosis by the actin depolymerising drug latrunculin and the enhanced basal AMPAR endocytosis following Arp2/3 inhibition by N-WASP CA domain, both suggest that assembly of actin filaments may be less important for AMPAR internalisation than proposed for endocytic events in non-neuronal cells, and that the high density of F-actin in dendritic spines may be the dominant factor that constrains AMPAR trafficking. Whilst CA domain enhances basal trafficking that occludes NMDA-induced internalisation, overexpression of WT-PICK1 has no impact on basal trafficking, but instead enhances the NMDA effect. This difference between the two Arp2/3 inhibitors is likely to reflect the Ca2+-sensitivity of PICK1. In the absence of NMDAR activation, PICK1 binds weakly to GluR2 and therefore does not influence AMPAR trafficking19. Presumably PICK1 only inhibits Arp2/3 strongly enough to drive AMPAR internalisation when bound to GluR2. In contrast, CA domain inhibits Arp2/3 potently and ubiquitously enough to influence receptor trafficking, with no dependence on Ca2+ signals or additional protein interactions.
Since the BAR domain binds lipid membranes30 and may be involved in generating membrane curvature22,38,39, inhibition of Arp2/3-mediated actin polymerisation by PICK1 could contribute to membrane bending by reducing membrane tension and the rigidity of the cortical actin-plasma membrane module39. The KK251,252 motif that we identify as the actin binding site on PICK1 has previously been implicated in mediating the binding of the BAR domain to phospholipid membranes29,30 an observation that we confirm in this study. This strengthens the hypothesis that membrane bending is linked to actin-based processes via PICK1. Other proteins with BAR-like domains also have lysine-based motifs that mediate interactions with both phospholipids and F-actin40.
The dual role of the PICK1-Arp2/3 interaction in both trafficking and in the constitutive maintenance of the actin cytoskeleton may depend upon the PDZ-regulated allosteric switch. Under basal conditions, PICK1 PDZ domains may be unoccupied, resulting in a low, tonic level of Arp2/3 regulation that contributes to the development of neuronal morphology. NMDAR-mediated Ca2+ influx stimulates the interaction of GluR2 with the PICK1 PDZ domain19, resulting in enhanced interactions with F-actin and with Arp2/3 and hence enhanced inhibition of actin polymerisation specifically at the site of AMPAR endocytosis (see Fig. 8). This provides a mechanism for ensuring a tight temporal and spatial synchronisation of cargo recruitment and membrane invagination.
We have identified PICK1 as a novel regulator of the actin cytoskeleton in neurons. This work provides important mechanistic information about the modulation of the neuronal cytoskeleton and highlights PICK1 as a key molecule linking the fundamental cellular processes of cytoskeletal remodelling and protein traffic.
EXPERIMENTAL PROCEDURES
Plasmids and plasmid construction
His6 proteins were expressed from pET28 (Novagen), GST-fusions from pGEX4T-1 (Pharmacia). For COS cells, WT- and W413A-PICK1 were expressed from pcDNA3.1. For Sindbis viruses, WT- and W413A-PICK1 were ligated into pIRES-EGFP, then into pSinRep(nsP2S). shRNA and mCherry reporter were expressed from a modified pFIV (System Biosciences). mCherry was driven by the CamKII promoter, shRNA by the H1 RNA pol III promoter. The PICK1 shRNA sequence was as follows: 5′AAAGTACTATAATGACTGCTATTTCAAGAGAATAGCAGTCATTATAGTAC 3′. The control construct did not contain shRNA. WT, W413A, KK251,252EE, Δ105 and 135Δ-PICK1 and N-WASP CA domain expressed from pIRES-EGFP were used for neuronal transfections. For rescue experiments, WT-PICK1, W413A-PICK1 and KK251,252EE-PICK1 with silent mutations: TAc AAc GAt TGt TAc GCg (mutations in lower case) were expressed from pIRES-EGFP.
Preparation of recombinant proteins
His6- and GST-fusions were expressed and purified as described41, except that his6-fusions were eluted in 10 mM Tris pH 7.5, 60 mM NaCl, 2 mM DTT, 400 mM imidazole. PICK1flag was expressed in cortical neuronal cultures using Sindbis virus19. Protein was immunoprecipitated using anti-flag antibody-conjugated agarose beads. After washing, proteins were eluted from the beads with 1 mg/ml flag peptide.
Antibodies
Anti-p34 (Arp2/3 complex; BD biosciences); anti-β-actin (Sigma); anti-pan-actin (Sigma); anti-FLAG (M2, Sigma); anti-Myc (Santa Cruz); anti-PICK1 (rabbit, ABR; goat, Santa Cruz; mouse NeuroMab); anti-PSD95 (NeuroMab); anti-GluR2 (Chemicon, Zymed), anti-GST (Novagen); anti-MAP2 (Sigma); anti-N-WASP (Santa Cruz).
Buffers
Lysis Buffer: 25 mM HEPES pH 7.5, 125 mM NaCl, 0.5% TX-100, protease inhibitor cocktail (Roche).
Buffer A: 10 mM Tris pH 7.5, 0.2 mM ATP, 0.5 mM DTT, 0.2 mM CaCl2.
Polymerisation buffer: 50 mM KCl, 10 mM Tris pH 7.5, 2 mM MgCl2, 0.2 mM ATP, 0.2 mM DTT.
XB buffer: 10 mM HEPES pH 7.5, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 0.1% Tween-20. HBS: 20 mM HEPES pH 7.4 (Tris), 140 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2, 9% D-glucose.
Co-immunoprecipitation from neuronal cultures
Co-IPs were performed essentially as described19. Extracts of 12-15 DIV hippocampal cultures were prepared in lysis buffer. 3μg anti-PICK, anti-p34 or control IgG was used for IP.
GST pull-down assays
For Arp2/3 binding experiments, 5 μg of GST or GST-PICK1 was immobilised on glutathione-agarose beads in lysis buffer for 30 min at 4°C. In allostery assays, 500 nM his6R2C or his6135Δ were included at this stage. After washing twice in lysis buffer, beads were incubated with 10 nM purified Arp2/3 in XB buffer for 1.5 h at 4°C. Beads were washed three times in XB buffer, and bound proteins detected by immunoblotting.
Pull-downs from brain homogenate were the same except that rat brain homogenates prepared in lysis buffer (300 μg) were incubated with immobilised GST-fusions for 1 h at 4°C. Beads were washed three times in lysis buffer containing 0.1% TX-100.
Monomeric G-actin was purified as described below, diluted to 1 μM and centrifuged at 200,000 g for 1 h at 4°C to remove any residual F-actin. G-actin solution was then incubated with immobilised GST-PICK1 or GST-VCA in buffer A for 1 h at 4°C. Beads were washed three times in buffer A.
G-actin purification
Rabbit muscle acetone powder was mixed with fresh buffer A at a concentration of approximately 40 μM. Following centrifugation, supernatants were filtered through muslin cloth. Actin was polymerized by adding 50 mM KCl, 2 mM MgCl2 and 1 mM ATP and incubated at room temperature for 30 min. Samples were incubated for a further 90 min at 4°C and tropomyosin was subsequently removed by addition of 0.6 M KCl. Actin was centrifuged at 200,000 g for 3 h at 4°C and pellets resuspended in 5 ml buffer A. Samples were then dialysed for 48 h with 3 changes of buffer A to isolate monomeric actin. G-actin was clarified by centrifugation at 200,000 g for 1 h at 4°C to remove any residual F-actin and diluted to a final concentration of 1 μM.
Actin filament co-sedimentation assay
WT and mutant GST-PICK1 (100 nM) were added to G-actin (0-5 μM) in polymerisation buffer, incubated at room temperature for 1 h to allow F-actin assembly, and then centrifuged for 20 min at 250,000 g. Pellets and supernatants were analysed by SDS-PAGE and Coomassie staining.
Subcellular fractionation
This was carried out as described25.
Actin polymerisation assays
Lyophilised Pyrene-labelled rabbit muscle actin (Cytoskeleton) at 5 μM in buffer A was centrifuged at 70,000 rpm in a Ti70.1 rotor (Beckman) for one hour at 4°C to remove potential oligomers or actin filaments in the solution. Following centrifugation, supernatants were allowed to adjust to room temperature for one hour before commencing the assay. Calcium bound to pyrene-actin monomers was pre-exchanged with magnesium by incubating with 0.1 volumes of 10x ME buffer (500 μM MgCl2, 2 mM EGTA) for two minutes prior to the addition of polymerisation buffer. Unless otherwise indicated, polymerisation reactions contained 2.5 μM pyrene-actin, 25 nM purified Arp2/3, 100 nM GST-VCA and various concentrations of his6-PICK1 (wild type and mutants) or his6-R2C made up to a total volume of 300 μl using buffer A. Polymerisation was initiated by the addition of polymerisation buffer to a final concentration of 50 mM KCl, 10 mM Tris pH 7.5, 0.2 mM DTT, 2 mM MgCl2, and 1 mM ATP (along with the other constituents) to the Ca-Mg exchanged pyrene-actin followed by extensive trituration. Reactions were immediately transferred to a fine quartz cuvette and fluorescence data collected in a Spex Fluoromax fluorimeter at an excitation wavelength of 365 nm and emission wavelength of 386 nm at a constant 25°C. Fluorescence data was collected for 800-1500 seconds and all experiments were carried out at least three times with similar results. Note that the fluorimeter and the xenon-arc lamp were allowed to warm up one hour prior to performing the assays to ensure stable signal strength. To minimise photobeaching and to limit signal-noise ratio the excitation slits were set to a minimal 0.1 value. Moreover to avoid Schlieren lines, samples were extensively triturated in the quartz cuvette and all stock solutions filtered. Cuvettes were also thoroughly washed with ddH20 and dried between each run to remove actin oligomers. Pyrene-labeled actin, GST-WASP VCA and Arp2/3 complex were purchased from Cytoskeleton Inc.
Competition assays
Purified Arp2/3 complex was immunoprecipitated with 2 μg anti-p34 antibody immobilized on protein A beads (Sigma). After one wash with XB buffer, immunocomplexes were incubated with 100 nM GST-VCA with or without 500 nM his6WT-PICK1 or mutants for 1 h. Beads were then washed twice in XB and proteins detected by immunoblotting.
Quantification of SDS-PAGE and Immunoblots
Films of immunoblots from at least 3 independent experiments were scanned and analysed using Image J. Error bars are standard errors of the mean, and 2-tailed t-tests were carried out to determine significant differences.
Immunocytochemistry
For trafficking experiments live hippocampal neurons prepared from E18 wistar rats (DIV15-20), treated with fluorodeoxyuridine and uridine to inhibit glial growth, were surface labelled with anti-GluR2 (30 μg/ml) for 30 mins at room temperature in HBS plus 2 μM TTX. After two washes in HBS neurons were treated with 50 μM NMDA for 3 min at 37°C, followed by 9 min after drug washout. Neurons were fixed for 5 min with 4% paraformaldehyde (PFA) and stained with anti-mouse Cy5 secondary. Following 15 min fixation in PFA, cells were permeabilised and stained with anti-mouse Cy3 or Alexa350 secondary. For simple staining experiments, neurons were fixed in PFA, permeabilised and stained with appropriate antibodies or Alexa-conjugated phalloidin (Invitrogen). Transferrin receptor endocytosis assays were carried out as described41 using Alexa-conjugated transferrin. Images were acquired on a LSM510 confocal microscope (Zeiss) and analyzed using ImageJ software. Quantification of internalised GluR2 was carried out by thresholding the EGFP or mCherry fluorescence signal in ImageJ to define outlines of neurons, and the ratio of internalised over total (surface + internalised) mean GluR2 fluorescence within this area was calculated. Quantification of PICK1 and phalloidin staining was carried out by thresholding the mCherry fluorescence signal in ImageJ to define outlines of neurons, and the mean intensity fluorescence signal for PICK1 or phalloidin quantified within this area.
Sholl analysis of hippocampal neurons
Neurons were transfected at 9-10 DIV using Lipofectamine 2000, and analysed 5 days later. Intensity thresholds of mCherry or EGFP fluorescence or MAP2 immunofluorescence signal were set in Image J, concentric circles with 10 μm differences in radius were drawn around the cell body, and the number of processes crossing each circle was counted.
Lipid binding assays
These were carried out as described30,38. Brain lipid extracts (Folch extract I, Sigma) were resuspended in 20 mM HEPES, pH 7.4, 150 mM NaCl and 1 mM DTT. GST-fusion proteins (2.5 μM) were then incubated with 1 mg/ml lipid extract for 15 min at 37°C before high-speed centrifugation at 35,000 rpm for 15 min in a Beckman Ti70.1 rotor. Supernatant and pellet protein samples were then resolved by SDS-PAGE and visualised by Coomassie staining.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Giles Cory for invaluable discussions and advice on actin biology. We also thank Prof. A. Clarke and Dr. E. Compton-Daw for the use of and assistance with the fluorimeter, Prof E. Ziff for the KK251,252EE construct, Dr. L.J. King, Dr. J. Mellor, Prof. J. Henley for critical reading of the manuscript. J.G.H is a fellow of the Wellcome Trust, D.R. is funded by an MRC studentship. This work was supported by ENI-NET.
Footnotes
AUTHOR CONTRIBUTIONS
D.L.R. planned and performed the biochemistry and some imaging experiments.
S.M. supervised generation of shRNA, planned and performed some imaging experiments.
E.L.J. generated shRNA constructs.
J.G.H planned and performed imaging experiments, mutagenesis/cloning, supervised the project and wrote the paper.
REFERENCES
- 1.da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nature Rev. Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 2.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nature Rev. Mol. Cell. Biol. 2006;7:404–14. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 3.Stradal TE, Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr. Opin. Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Meyer G, Feldman EL. Signaling mechanisms that regulate actin-based motility processes in the nervous system. J. Neurochem. 2002;83:490–503. doi: 10.1046/j.1471-4159.2002.01185.x. [DOI] [PubMed] [Google Scholar]
- 5.Luo L. Actin cytoskeleton regulation in Neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 2002;18:601–35. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- 6.Kakimoto T, Katoh H, Negishi M. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J. Biol. Chem. 2006;281:29042–53. doi: 10.1074/jbc.M604025200. [DOI] [PubMed] [Google Scholar]
- 7.Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. Regulation of N-wasp and the arp2/3 complex by abp1 controls neuronal morphology. PLoS ONE. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qualmann B, Kessels MM, Kelly RB. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 2000;150:F111–6. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeng RL, Welch MD. Cytoskeleton: actin and endocytosis--no longer the weakest link. Curr. Biol. 2001;11:R691–4. doi: 10.1016/s0960-9822(01)00410-9. [DOI] [PubMed] [Google Scholar]
- 10.Merrifield CJ. Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–8. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Matus A, Brinkhaus H, Wagner U. Actin dynamics in dendritic spines: a form of regulated plasticity at excitatory synapses. Hippocampus. 2000;10:555–60. doi: 10.1002/1098-1063(2000)10:5<555::AID-HIPO5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 12.Rao A, Craig AM. Signaling between the actin cytoskeleton and the postsynaptic density of dendritic spines. Hippocampus. 2000;10:527–41. doi: 10.1002/1098-1063(2000)10:5<527::AID-HIPO3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 13.Dillon C, Goda Y. The actin cytoskeleton: integrating form and function at the synapse. Annu. Rev. Neurosci. 2005;28:25–55. doi: 10.1146/annurev.neuro.28.061604.135757. [DOI] [PubMed] [Google Scholar]
- 14.Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nature Neurosci. 2002;5:239–46. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- 15.Blanpied TA, Scott DB, Ehlers MD. Dynamics and regulation of clathrin coats at specialised endocytic zones of dendrites and spines. Neuron. 2002;36:435–49. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, et al. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron. 2007;55:874–89. doi: 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim CH, Chung HJ, Lee HK, Huganir RL. Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11725–30. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwakura Y, et al. N-methyl-D-aspartate-induced alpha-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid (AMPA) receptor down-regulation involves interaction of the carboxyl terminus of GluR2/3 with Pick1. Ligand-binding studies using Sindbis vectors carrying AMPA receptor decoys. J. Biol. Chem. 2001;276:40025–32. doi: 10.1074/jbc.M103125200. [DOI] [PubMed] [Google Scholar]
- 19.Hanley JG, Henley JM. PICK1 is a calcium-sensor for NMDA-induced AMPA receptor trafficking. EMBO J. 2005;24:3266–78. doi: 10.1038/sj.emboj.7600801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci. 2002;25:103–26. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 21.Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 22.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–8. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Itoh T, et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Tsujita K, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 2006;172:269–79. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–87. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 26.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 27.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci U. S. A. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nature Cell Biol. 2005;7:246–54. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- 29.Lu W, Ziff EB. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–21. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Jin W, et al. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J. Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strasser GA, Rahim NA, VanderWaal K,E, Gertler FB, Lanier LM. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 2004;43:81–94. doi: 10.1016/j.neuron.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Steinberg JP, et al. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron. 2006;49:845–60. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–25. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, Xiao M, Nicoll RA. Contribution of cytoskeleton to the internalization of AMPA receptors. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1261–6. doi: 10.1073/pnas.031573798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartoe JL, et al. Protein interacting with C-kinase 1/protein kinase Calpha-mediated endocytosis converts netrin-1-mediated repulsion to attraction. J. Neurosci. 2006;26:3192–205. doi: 10.1523/JNEUROSCI.3469-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nature Cell Biol. 2002;4:691–8. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 37.Merrifield CJ, Qualmann B, Kessels MM, Almers W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur. J. Cell Biol. 2004;83:13–8. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- 38.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 39.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–6. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 40.Mattila PK, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 2007;176:953–64. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanley JG, Khatri L, Hanson PI, Ziff EB. NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.