Abstract
Proline dehydrogenase (PRODH) catalyzes the oxidation of L-proline to Delta-1-pyrroline-5-carboxylate. PRODHs exhibit a pronounced preference for proline over hydroxyproline (trans-4-hydroxy-L-proline) as the substrate, but the basis for specificity is unknown. The goal of this study, therefore, is to gain insights into the structural determinants of substrate specificity of this class of enzyme, with a focus on understanding how PRODHs discriminate between the two closely related molecules, proline and hydroxyproline. Two site-directed mutants of the PRODH domain of Escherichia coli PutA were created: Y540A and Y540S. Kinetics measurements were performed with both mutants. Crystal structures of Y540S complexed with hydroxyproline, proline, and the proline analog L-tetrahydro-2-furoic acid were determined at resolutions of 1.75 Å, 1.90 Å and 1.85 Å. Mutation of Tyr540 increases the catalytic efficiency for hydroxyproline three-fold and decreases the specificity for proline by factors of twenty (Y540S) and fifty (Y540A). The structures show that removal of the large phenol side chain increases the volume of the substrate-binding pocket, allowing sufficient room for the 4-hydroxyl of hydroxyproline. Furthermore, the introduced serine residue participates in recognition of hydroxyproline by forming a hydrogen bond with the 4-hydroxyl. This result has implications for understanding substrate specificity of the related enzyme human hydroxyproline dehydrogenase, which has serine in place of tyrosine at this key active site position. The kinetic and structural results suggest that Tyr540 is an important determinant of specificity. Structurally, it serves as a negative filter for hydroxyproline by clashing with the 4-hydroxyl group of this potential substrate.
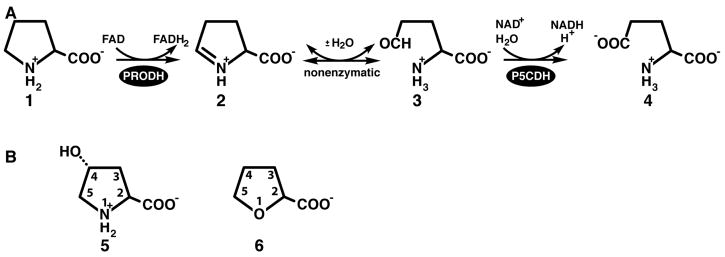
Amino acid oxidation is a central part of energy metabolism. The oxidation of proline, in particular, is increasingly being recognized as critical for cellular redox control, apoptosis and suppression of cancer (1–3). Proline dehydrogenase (PRODH) is the first of two enzymes of the proline catabolic pathway (Scheme 1A). It catalyzes the FAD-dependent oxidation of proline (1) to Δ1-pyrroline-5-carboxylate (P5C, 2). The cyclic imine P5C is hydrolyzed nonenzymatically to glutamate semialdehyde (3), which is subsequently oxidized to glutamate (4) by P5C dehydrogenase (P5CDH). PRODH and P5CDH are separate monofunctional enzymes encoded by distinct genes in eukaryotes and some bacteria, whereas they are combined into a single bifunctional enzyme called proline utilization A (PutA) in other bacteria. Inborn errors in proline oxidation enzymes lead to hyperprolinemia disorders (4), and deficiencies in PRODH have been linked to increased susceptibility to schizophrenia (5–9).
Scheme 1.
Bacterial monofunctional PRODHs and PutAs studied to date exhibit a pronounced preference for proline over hydroxyproline (trans-4-hydroxy-L-proline, 5 in Scheme 1B) as the substrate. For example, the PRODH domain of Escherichia coli PutA is over 100 times more efficient at oxidizing proline than hydroxyproline (vide infra). Also, it has been reported that hydroxyproline is not a substrate for the monofunctional PRODH from Thermus thermophilus (10). Given the high degree of amino acid sequence identity within PRODH active sites (10, 11), the marked preference for proline over hydroxyproline likely extends to the entire family of bacterial PRODHs/PutAs.
In humans, two forms of PRODH have evolved, one that prefers proline as the substrate and another specific for hydroxyproline (12–15). The proline-specific enzyme is encoded on chromosome 22 (NCBI RefSeq number NM_016335) and is known variously as POX (proline oxidase), PRODH2 and PIG6 (p53-induced gene 6). We will refer to it as POX here. The hydroxyproline-specific form is encoded on chromosome 19 (RefSeq number NM_021232) and is known as OH-POX. POX and OH-POX share 45 % amino acid sequence identity and have highly conserved active site sequences. Despite these apparent similarities, substrate crossover appears to be minimal, consistent with the observations that patients with genetic defects in the POX gene have normal plasma levels of hydroxyproline and those with defects in the OH-POX gene exhibit normal levels of proline (4, 13–15). (Normal plasma levels are 51 – 271 μM for proline and 1 – 46 μM for hydroxyproline (4).) The molecular-level basis for the difference in substrate specificity of POX and OH-POX is not known.
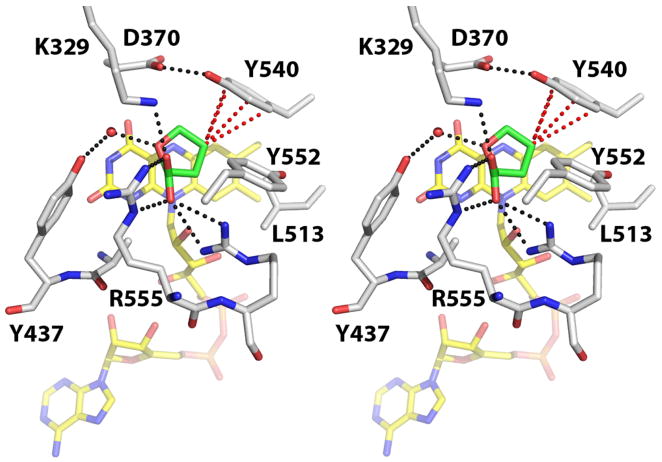
The goal of this study is to gain insights into how PRODHs distinguish between proline and hydroxyproline, two chemically and structurally similar molecules. Our focus here on an active site tyrosine residue was motivated by structural and sequence considerations. The 2.0 Å crystal structure of an E. coli PutA PRODH construct, PutA86-669 (residues 86-669 of PutA), complexed with the proline analog L-tetrahydro-2-furoic acid (THFA, 6) provided a model of enzyme – substrate interactions (16). In that structure, nine highly conserved residues contact the inhibitor (Figure 1). Of these residues, Tyr540 appeared important for enforcing the preference for proline over hydroxyproline because of its proximity to the C4 atom of the proline analog. Note that Tyr540 packs tightly against the C4-C5 locus of the inhibitor (Figure 1). This arrangement allows the phenol ring of Tyr540 to make four contacts shorter than 4.0 Å with the C4 atom of THFA (red dotted lines in Figure 1), with the closest contact being 3.3 Å. This observation suggests that Tyr540 might clash with the 4-hydroxyl group of hydroxyproline, which provides a way for the enzyme to discriminate hydroxyproline from the true substrate proline.
Figure 1.
Stereographic view of the active site of PutA86-669 complexed with THFA (PDB entry 1TIW). The FAD cofactor and THFA are colored yellow and green, respectively. Red dotted lines indicate close contacts (within 4.0 Å) between the C4 atom of THFA and the side chain of Tyr540. Black dotted lines indicate favorable electrostatic interactions. This figure and others were created with PyMOL (38).
Amino acid sequence data also points to a role for Tyr540 in specificity. Analysis of PRODH sequences shows that this tyrosine residue is found in all of the proline-specific enzymes, i.e., bacterial monofunctional PRODHs, PutAs (10) and human POX (Tyr548). In sharp contrast, the analogous residue in OH-POX - the only known hydroxyproline-specific member of the PRODH family - is Ser485. Furthermore, the Tyr548/Ser485 variation is the only obvious difference in predicted active site residues of POX and OH-POX. These observations further suggest that this position in the polypeptide chain is important for discriminating between proline and hydroxyproline.
Based on these observations, we hypothesized that Tyr540 is an important structural determinant of substrate specificity and that it functions as a negative filter for hydroxyproline by clashing with the 4-hydroxyl group of this potential substrate. This idea was explored by creating two mutants of the PutA PRODH construct PutA86-630 (residues 86-630 of E. coli PutA) in which Tyr540 is replaced by serine (Y540S) or alanine (Y540A). Herein we report the results of kinetic and structural studies of these mutants.
EXPERIMENTAL PROCEDURES
Mutagenesis, Expression and Purification
A PRODH domain construct consisting of residues 86-630 of E. coli PutA (PutA86-630) was used for this study. This enzyme was used rather than the PutA86-669 construct that was used previously for structural work because it yields higher resolution crystals with better reproducibility. We note that the additional C-terminal residues of PutA86-669 (i.e., 631-669) are disordered in the crystal, and their absence in PutA86-630 probably accounts for the improved crystallization characteristics.
Two mutants of PutA86-630, Y540S and Y540A, were created from a pET-23b (Novagen) construct encoding residues 86-630 of E. coli PutA with a C-terminal His tag. The mutations were introduced using the Quikchange kit (Stratagene) and verified at the DNA level by sequencing.
PutA86-630 and the two mutants were expressed using E. coli strain BL21 (DE3) pLysS. Cultures were grown in LB broth at 37 °C to OD600 ~ 0.6 and then induced with a final concentration of 0.3 – 0.5 mM isopropyl β-D-thiogalactoside. Induction occurred for 3 – 4 hours at 25 °C after which the cultures were harvested by centrifugation. The pelleted cells were resuspended in binding buffer (50 mM NaH2PO4 pH 8.0, 10 mM imidazole and 0.3 M NaCl) and frozen.
The proteins were purified using standard procedures as follows. Frozen cells were thawed at 4 °C and supplemented with five protease inhibitors (0.1 mM TPCK, 0.5mM AEBSF, 0.001 mM pepstatin, 0.01 mM leupeptin, 0.005 mM E-64). Cells were lysed by a French Press at 110 MPa and centrifuged at 15,000 rev min−1 for one hour at 4 °C. The supernatant was passed through a 0.45 μm filter (Millipore) and applied to 10 mL of Ni-NTA Superflow resin (Qiagen). The column was first washed with the binding buffer followed by a second wash of binding buffer containing 10 mM imidazole. The enzyme was eluted with 250 mM imidazole in binding buffer. Based on SDS-PAGE, fractions containing the protein of interest were pooled, supplemented with FAD to a concentration of 0.1 mM and dialyzed overnight in the dark at 4 °C into 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.5 mM EDTA and 5% glycerol. A second purification step was performed with a 5 mL HP HiTrap Q anion exchange column (GE Healthcare). The desired protein flowed through the column while the contaminating proteins bound to the resin. The sample was dialyzed overnight in the dark at 4 °C into 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.5 mM EDTA and 5% glycerol, and concentrated to 17 mg/mL using a Millipore Ultrafree-15 centrifugal filter. Protein concentration was determined using the BCA method (Pierce kit).
Crystallization of Y540S and Crystal Soaking
Crystals of PutA86-630 mutant Y540S were grown using the protocol reported for PutA86-669 (16). All crystallization experiments were performed at 22 °C using hanging drop vapor diffusion with drops formed by mixing equal volumes of the reservoir (2 – 5 μl) and protein (2 – 5 μl) solutions. The best crystals were grown over reservoir solutions containing 15 – 18 % (w/v) PEG3350 and 0.1 M sodium citrate buffer at pH 5.8 – 6.0. The protein stock solution consisted of 15 mg/mL Y540S and 10 mM of THFA (6 in Scheme 1B). Attempts to grow Y540S crystals in the absence of THFA were unsuccessful. Crystallization trials with Y540A were also unsuccessful.
Crystals of the Y540S/THFA complex were prepared for low temperature data collection by replacing the mother liquor with cryobuffer (18 % (w/v) PEG 3350, 15% PEG 200, 0.1 M citrate buffer pH 5.7) supplemented with 10 mM THFA. The cryoprotected crystals were picked up with Hampton loops and plunged into liquid nitrogen.
Crystals of Y540S complexed with hydroxyproline were prepared by soaking crystals of the Y540S/THFA complex in cryobuffer supplemented with hydroxyproline (trans-4-hydroxy-L-proline) present at saturating concentration (less than 1 M) and 30 mM sodium dithionite. The intense yellow color of the oxidized crystals was bleached over a period of several minutes, indicating that the FAD cofactor was reduced. When the bleaching was complete, the crystals were plunged into liquid nitrogen. Crystals of Y540S complexed with proline were prepared similarly, by soaking crystals of the Y540S/THFA complex in cryobuffer supplemented with 1 M L-proline (not saturating) and 30 mM sodium dithionite.
X-ray Diffraction Data Collection, Processing and Refinement
Crystals were analyzed at the Advanced Photon Source beamline 24-ID-C and the Advanced Light Source beamline 4.2.2. The space group is I222 with unit cell dimensions of a = 73 Å, b = 140 Å and c = 147 Å. There is one molecule per asymmetric unit, which corresponds to 61 % solvent and Vm of 3.2 Å3/Da. We note that this crystal form is the same one that we reported for PutA86-669 (16). We verified that the crystal structures of PutA86-669 and native PutA86-630 are identical, within experimental error (data not shown).
Data sets for the Y540S/THFA (1.85 Å resolution) and Y540S/hydroxyproline (1.75 Å resolution) complexes were collected at beamline 24-ID-C using an ADSC Q315 detector. Each consisted of 150 frames with oscillation width of 1.0° and detector distance of 250 mm. A 1.90 Å resolution data set for the Y540S/proline complex was obtained at beamline 4.2.2 using a NOIR-1 detector. This data set consisted of 220 frames collected with oscillation width of 0.5° and detector distance of 140 mm. The 24-ID-C data sets were processed with HKL2000 (17). The beamline 4.2.2 data set was integrated with MOSFLM (18) through the iMosflm graphical interface and scaled with SCALA (19) using the CCP4i interface (20).
Refinement calculations were performed with PHENIX (21) and model building was done with COOT (22). The starting model for rigid body and simulated annealing positional refinement was derived from the structure of PutA86-669 complexed with THFA (PDB code 1TIW). Active site side chains, the THFA ligand and solvent were removed prior to refinement. A common set of test reflections (5%) was used for all three refinements, and this set corresponded to the one previously used for refinement of PutA86-669/THFA structure. Data collection and refinement statistics are listed in Table 1.
Table 1.
Data Collection and Refinement Statistics for Y540S Structuresa
| Ligand | THFA | hydroxyproline | proline |
|---|---|---|---|
| Space group | I222 | I222 | I222 |
| Unit cell lengths (Å) | a = 72.6 | a = 73.4 | a = 73.1 |
| b = 140.1 | b = 142.6 | b = 140.9 | |
| c = 146.7 | c = 145.7 | c = 145.3 | |
| Wavelength (Å) | 0.97922 | 0.97949 | 1.00000 |
| Diffraction resolution (Å) | 46.1 – 1.85 (1.92 – 1.85) | 50.0 – 1.75 (1.78 – 1.75) | 27.7 – 1.90 (2.00 – 1.90) |
| No. of observations | 386345 | 453150 | 249908 |
| No. of unique reflections | 63740 | 76563 | 58713 |
| Redundancy | 6.1 (5.6) | 5.9 (5.5) | 4.3 (3.3) |
| Completeness (%) | 99.5 (95.7) | 99.3 (98.5) | 99.2 (95.1) |
| Rmerge(I) | 0.059 (0.481) | 0.047 (0.418) | 0.079 (0.417) |
| Average I/⌠ | 26.7 (2.1) | 37.8 (2.5) | 13.1 (3.3) |
| Wilson B-factor (Å2) | 24.2 | 27.0 | 21.4 |
| No. of protein chains | 1 | 1 | 1 |
| No. of atoms | 3937 | 3930 | 3808 |
| No. of protein residues | 468 | 468 | 471 |
| No. of water molecules | 266 | 252 | 143 |
| Rcryst | 0.198 (0.231) | 0.201 (0.231) | 0.212 (0.327) |
| Rfreeb | 0.227 (0.283) | 0.228 (0.260) | 0.242 (0.326) |
| RMSDc | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.008 |
| Bond angles (deg.) | 0.96 | 1.06 | 1.11 |
| Ramachandran plotd | |||
| Favored (%) | 98.5 | 98.9 | 98.9 |
| Allowed (%) | 1.5 | 1.1 | 1.1 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
| Average B-factors (Å2) | |||
| Protein | 33.3 | 39.2 | 39.8 |
| FAD | 19.0 | 24.0 | 22.4 |
| ligand | 23.2 | 37.9 | 26.5 |
| water | 34.3 | 38.4 | 31.8 |
| Coordinate error (Å)e | 0.21 | 0.21 | 0.26 |
| PDB accession code | 3E2R | 3E2Q | 3E2S |
Values for the outer resolution shell of data are given in parenthesis.
5% test set. A common set of test reflections was used for refinement of all structures.
Compared to the Engh and Huber parameters (36).
The Ramachandran plot was generated with RAMPAGE (37).
Maximum likelihood-based coordinate error reported by PHENIX.
Kinetic Characterization
The catalytic activities of PutA86-630, Y540S and Y540A were measured with proline and hydroxyproline serving as substrates. The assay used is based on reduction of dichlorophenolindophenol (DCPIP) as described previously for PutA proteins (23–26). One unit of activity is the quantity of enzyme that transfers electrons from 1 μmol of substrate to DCPIP per minute at 25 °C. All assays were performed in triplicate. The enzyme concentration was 0.12 μM in all assays except for those of the native enzyme with hydroxyproline. In this case, the enzyme concentration was increased to 3.0 μM because of the inherently slow kinetics of the native enzyme with this substrate.
Inhibition by the proline analog THFA was examined for all three enzymes using proline as the substrate and the enzyme at 0.12 μM. The kinetic parameters for proline (Vmax, Km) and Ki for THFA were determined using simultaneous nonlinear regression as described by Kakkar (27). In this approach, the rate data for a given enzyme at all inhibitor concentrations, including zero inhibitor concentration, are fit globally to Equation 1:
| (1) |
In Equation 1, v is the measured initial rate, Vmax is the maximal rate, [S] is the substrate concentration, Km is the Michaelis constant, [I] is the inhibitor concentration and Ki is the inhibition constant. Values of Vmax and Km for hydroxyproline were determined by fitting rate data to the Michaelis – Menten equation. Fitting calculations were performed with Origin 8 software.
RESULTS
Kinetic Characterization
Kinetic data for native PutA86-630, Y540S and Y540A using proline and hydroxyproline as substrates are shown in Figure 2. The corresponding steady-state kinetic parameters are listed in Table 2. Using the native enzyme with proline as the substrate, the kinetic parameters are Km = 0.059 M and kcat = 29 s−1, which corresponds to catalytic efficiency (kcat/Km) of 492 s−1M−1. As shown in Figure 2A, hydroxyproline is a relatively poor substrate. The kinetic parameters for the native enzyme with hydroxyproline as the substrate are Km = 0.75 M, kcat = 2.5 s−1 and catalytic efficiency of only 3 s−1M−1. The ratio of the two catalytic efficiencies provides a measure of the preference for proline over hydroxyproline as the substrate. This value is 164, indicating high specificity for proline. Note that the large substrate preference ratio reflects factors of ten in both Km and kcat favoring proline. To our knowledge, this result represents the first quantitative assessment of the substrate preference of any PRODH.
Figure 2.
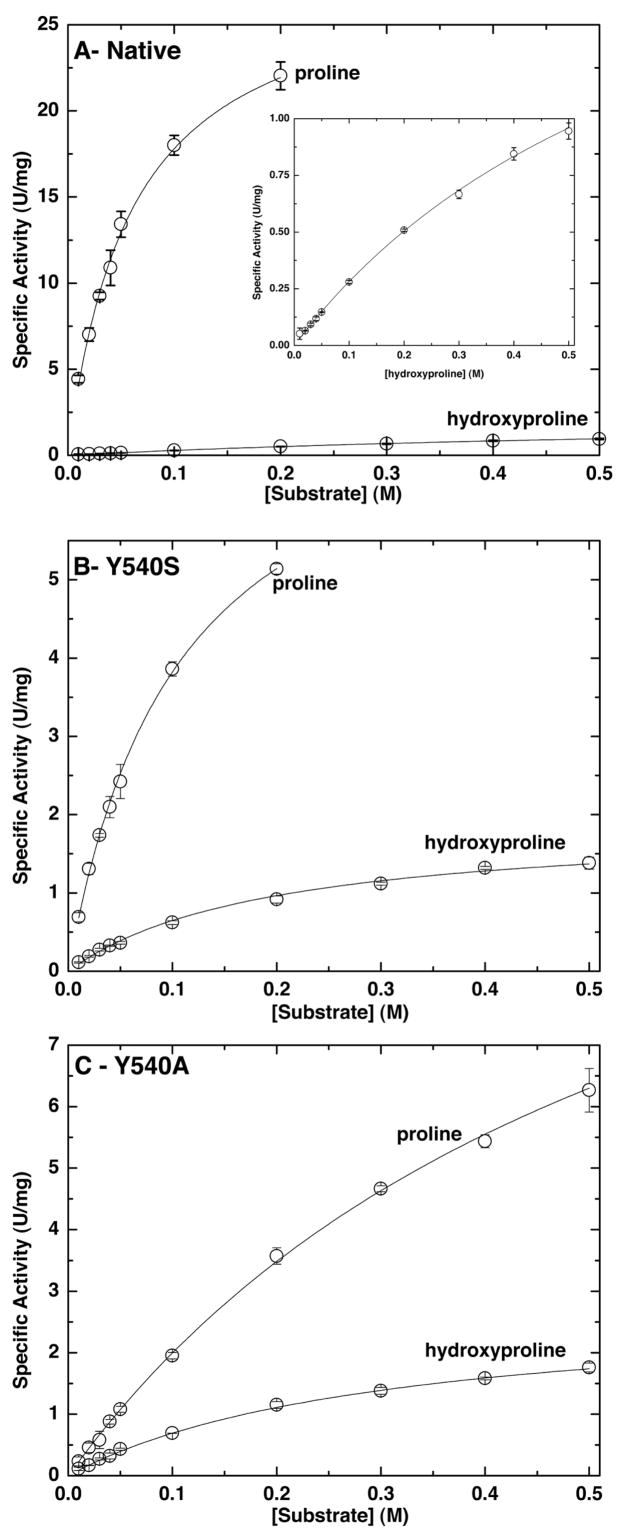
Steady-state kinetic data for (A) native PutA86-360, (B) Y540S and (C) Y540A. In each panel, two sets of data are shown corresponding to the specific activity using proline and hydroxyproline as the substrate. The curves represent the best fit of the data to the Michaelis – Menten equation. The inset in panel A shows an expansion of the hydroxyproline data.
Table 2.
Kinetic Parameters for PutA86-630, Y540S and Y540Aa
| Enzyme | Substrate | Km (M) | kcat (s−1) | kcat/Km (s−1M−1) | Preference for prolineb | Ki for THFA (mM) |
|---|---|---|---|---|---|---|
| PutA86-630 | proline | 0.059 ± 0.003 | 29. ± 1. | 492 | 164 | 0.084 ± 0.003 |
| PutA86-630 | hydroxyproline | 0.75 ± 0.05 | 2.5 ± 0.1 | 3 | - | - |
| Y540S | proline | 0.10 ± 0.01 | 7.8 ± 0.4 | 78 | 7 | 1.5 ± 0.1 |
| Y540S | hydroxyproline | 0.19 ± 0.01 | 2.0 ± 0.1 | 11 | - | - |
| Y540A | proline | 0.52 ± 0.03 | 13. ± 1. | 25 | 3 | 7.9 ± 0.5 |
| Y540A | hydroxyproline | 0.31 ± 0.01 | 2.9 ± 0.1 | 9 | - | - |
Enzyme assays were performed at 25°C.
Defined as (kcat/Km) for proline divided by (kcat/Km) for hydroxyproline.
Mutation of Tyr540 to either Ser or Ala substantially alters substrate specificity by increasing the catalytic efficiency for hydroxyproline and decreasing the efficiency for proline. As a result of these two effects, the proline/hydroxyproline preference ratios decrease from a value of 164 for the native enzyme to only seven and three, respectively, for Y540S and Y540A (Table 2). Thus, Tyr540 seems to be important for discriminating between these two substrates, as predicted from the structural considerations shown in Figure 1.
The mutants are more 3–4 times more efficient than the native enzyme at oxidizing hydroxyproline. The catalytic efficiencies of Y540S and Y540A for hydroxyproline are 11 and 9, respectively, compared to only 3 for the native enzyme. The increases are due almost entirely to reductions of Km. Note that the native and mutant enzymes have kcat values for hydroxyproline in the range 2.0 – 2.9 s−1. In contrast, Km for hydroxyproline decreases from 0.75 M for the native enzyme to 0.19 M for Y540S and 0.31 for Y540A. These results are consistent with the idea that Y540 of the native enzyme is a structural constraint that impedes the binding of hydroxyproline.
The mutations also have a large effect on the kinetics of the natural substrate, proline. The catalytic efficiency of Y540S for proline is 6-fold lower than that of the native enzyme. The efficiency of Y540A is 20-fold lower than native. The lower efficiencies reflect both decreases in kcat and increases in Km for the mutants. This result shows that Tyr540 is necessary for achieving maximal catalytic activity for proline in addition to interfering with the binding of hydroxyproline.
Inhibition by the proline analog THFA was also investigated as a means of understanding how Tyr540 shapes substrate recognition (Table 2). The values of Ki estimated for Y540S and Y540A are 1.5 mM and 7.9 mM, respectively. These values are substantially higher than the Ki of 0.084 mM for native PutA86-630. These results suggest that Tyr540 makes a significant contribution to the binding of the proline isostere THFA, and by inference, proline. The higher Ki of Y540A compared to Y540S suggests perhaps that the active site structure of Y540A is less native-like than that of Y540S. This conclusion is also supported by the higher Km values of Y540A compared to Y540S for both substrates.
Overall Structure of Y540S
Crystal structures of Y540S were determined to elucidate the structural perturbations caused by removal of the phenol group of Tyr540 and to gain insights into how Ser540 participates in the recognition of hydroxyproline (Table 1). The structure of the oxidized enzyme complexed with THFA was solved at 1.85 Å resolution. Structures of the reduced enzyme complexed with the substrates hydroxyproline and proline were determined at resolutions of 1.75 Å and 1.90 Å, respectively. The two substrate complexes are notable in that they are the first structures of any PRODH with a true substrate bound in the active site. Previous structures featured either competitive inhibitors (16, 28), mechanism-based inactivators (29), or no ligand (10).
Mutation of Tyr540 to Ser does not perturb the overall protein structure. As with the native enzyme, Y540S has a distorted (βα)8 barrel fold, with the FAD cofactor bound at the C-terminal ends of the strands of the barrel (Figure 3A). Residues 88-139 and 240-610 are highly ordered and form the core of enzyme, including the catalytic elements. Electron density is weak for the other residues, implying disorder in these regions, which is also the case for PutA86-669 (16).
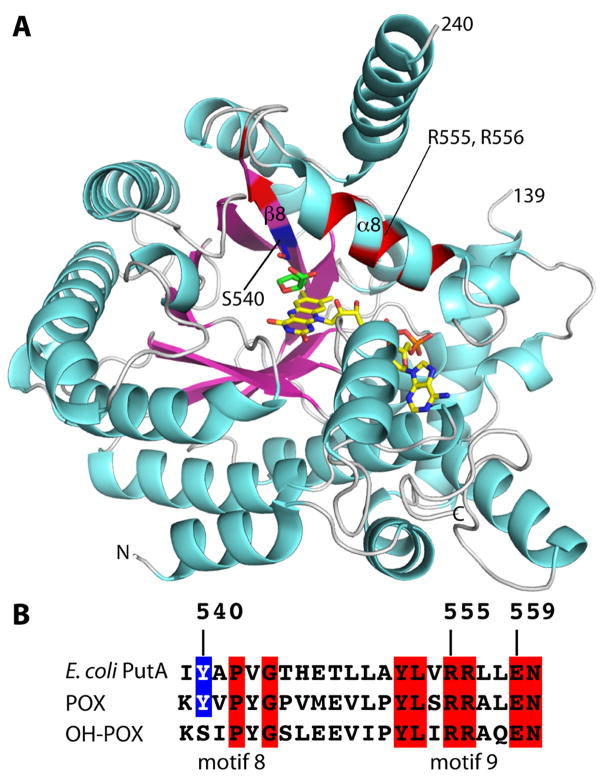
Figure 3.
Overall structure of the PutA PRODH domain. (A) Ribbon drawing of PutA86-630 mutant Y540S complexed with THFA. FAD and THFA are drawn as sticks and colored yellow and green, respectively. The side chain of Ser540, which is located on β8, is shown in blue sticks. The locations of Arg555 and Arg556 of α8 are indicated. Residues highlighted in blue and red correspond to the sequence alignment shown in panel B. (B) A section of an amino acid sequence alignment of E. coli PutA, POX and OH-POX in the region of conserved motifs 8 and 9, which corresponds to secondary structural elements β8 and α8 as shown in panel A.
The PRODH (βα)8 barrel differs from the classic one typified by triosephosphate isomerase in that the last helix of the fold, α8, sits atop the barrel and perpendicular to the strands, rather than alongside β8 and parallel to the strands (Figure 3A). This distortion is important because α8 contributes the active site residues Tyr552, Arg555 and Arg556 (Figure 1). Residue 540 is located in the middle of β8, which is the strand preceding the critical α8 helix (Figure 3A, blue patch).
The root mean square deviations for Cα atoms between the Y540S structures and that of PutA86-669 complexed with THFA (PDB code 1TIW) are 0.27 – 0.33 Å, which indicates that the mutant and native enzymes are identical at the fold level, within experimental error. Among the three Y540S structures, the root mean square deviations are even lower: 0.16 – 0.21 Å. Thus, the altered substrate specificity and kinetic parameters of the mutant enzymes are not due to global changes in protein conformation.
Substrate Recognition
For all three Y540S structures, electron density maps clearly indicated the presence of the bound ligand as well as the conformations of surrounding side chains and the locations of active site water molecules (Figure 4). In particular, the maps calculated for the hydroxyproline complex displayed a strong feature connected to the C4 atom of the ligand, which was not observed in maps calculated for the proline and THFA complexes (Figure 4). This feature was interpreted as representing the 4-hydroxyl group of hydroxyproline, indicating that hydroxyproline was bound in the active site rather than the THFA inhibitor that was used in co-crystallization. Apparently, the THFA ligand, which was presumably bound in the active site of the crystallized enzyme, was ejected during soaking and replaced with hydroxyproline, thus enabling us to capture the enzyme/substrate complex.
Figure 4.
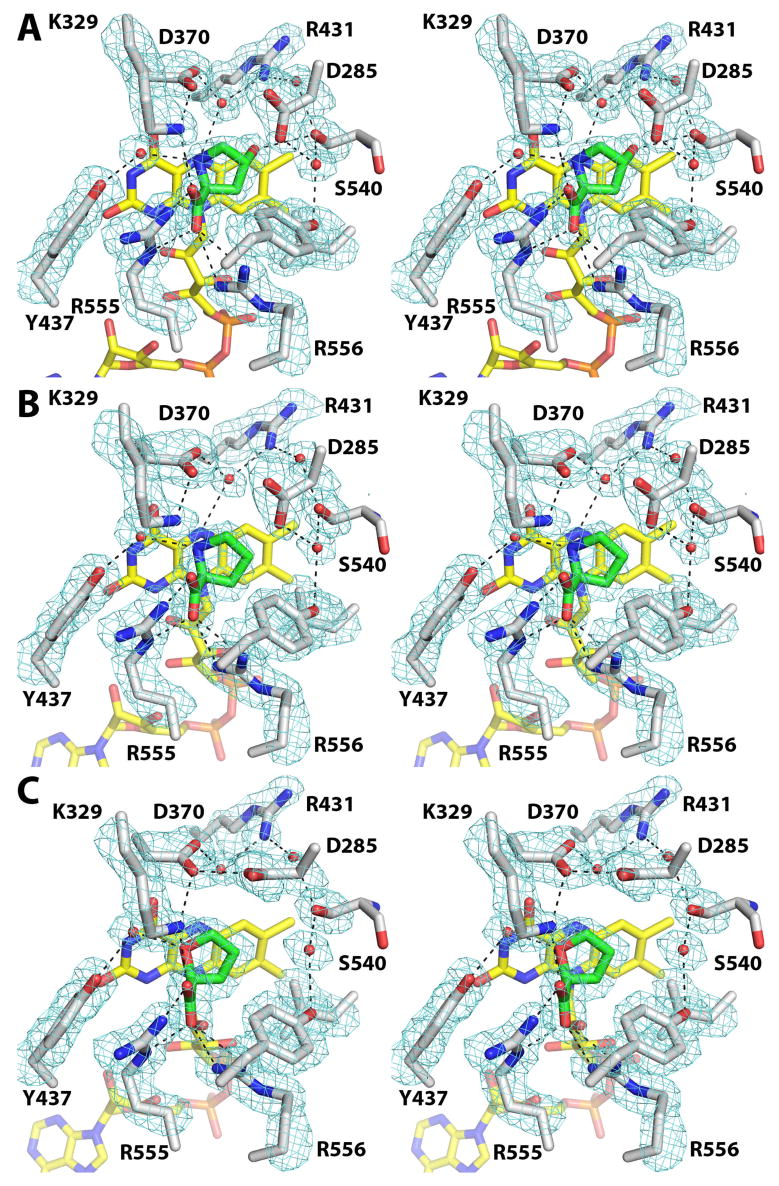
Stereographic views of the active sites of (A) Y540S/hydroxyproline, (B) Y540S/proline and (C) Y540S/THFA. The cages represent simulated annealing σA-weighted Fo - Fc maps contoured at 3 σ. Prior to calculation of each map, the ligand, surrounding side chains and water molecules were omitted, and simulated annealing refinement was performed with PHENIX.
Hydroxyproline, proline and THFA bind at the si face of the FAD isoalloxazine, with the ring of the substrate/inhibitor approximately parallel to and centered over the middle ring of the isoalloxazine (Figure 4). Thus, the active site ligand in all three Y540S structures occupies essentially the same space as THFA in the PutA86-669/THFA complex (compare Figures 1 and 4). In fact, most of the protein-ligand interactions observed in the PutA86-669/THFA complex are also found in the Y540S structures. These preserved features include the ionic interactions of Arg555, Arg556 and Lys329 with the ligand carboxylate, the water-mediated hydrogen bond with Tyr437 and the packing of Asp370, Ala436, Leu513 and Tyr552 against C atoms of the ligand/substrate.
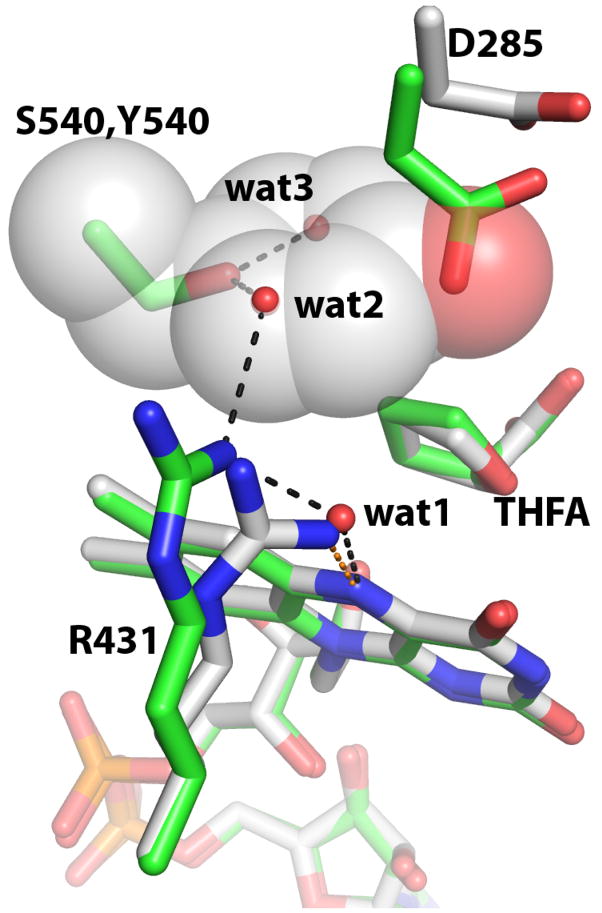
Electron density maps clearly indicated the conformation of Ser540 (Figure 4). In all three structures, the introduced side chain has the same χ1 angle as Tyr540 in the native enzyme, as shown by the comparison of the THFA complexes of Y540S and PutA86-669 in Figure 5. Note that the hydroxyl of Ser540 occupies the location of the Cγ atom of Tyr540 in the native enzyme.
Figure 5.
Conformational changes caused by mutation of Tyr540 to serine. The structures of Y540S/THFA (green) and PutA86-669/THFA (white) are shown. Tyr540 of PutA86-669 is represented as semitransparent spheres. Hydrogen bonds for Y540S and PutA86-669 are shown in black and orange, respectively. The three water molecules belong to the Y540S/THFA complex and are present in all three Y540S structures.
Ser540 does not directly contact THFA or proline, however, it makes an important interaction with hydroxyproline. The hydroxyl of Ser540 forms a hydrogen bond (3.1 Å) with the 4-hydroxyl of hydroxyproline (Figure 4A). Thus, Ser540 plays a direct role in recognition of this substrate. This result has implications for understanding substrate specificity of the human enzyme OH-POX (see Discussion).
Conformational Changes
Although the overall structure and most of the protein-ligand interactions are unchanged, the mutation of Tyr540 to Ser did cause conformational changes in two residues (Asp285, Arg431), as well as introduce three new solvent sites. These changes are observed in all three Y540S structures and are shown in a superposition of the THFA complexes of Y540S and PutA86-669 and (Figure 5).
Asp285 of the mutant enzyme is shifted by 2 Å from its position in the native enzyme (Figure 5). This movement brings the carboxylate group into the space corresponding to Tyr540 in the native enzyme. Presumably, this conformational change is a response to the cavity created by removal of the large tyrosine side chain. The exact position of Asp285 depends on the flavin redox state. (Compare, for example, the positions of Asp285 in Figures 4A and 4C.) Nevertheless, in all three structures, the carboxylate group of Asp285 penetrates the molecular volume corresponding to Tyr540 of the native enzyme.
Arg431 is the other side chain that moves in response to the mutation. In the native enzyme, Arg431 forms a hydrogen bond with the flavin N(5) (orange dots in Figure 5). Most flavin-dependent dehydrogenases, in fact, have such a hydrogen bond donor at this position, and this interaction is thought to be important for establishing the reduction potential of the flavin (30). Arg431 of the mutant enzyme is shifted 2.4 Å away from the N(5), which has allowed a water molecule (wat1 in Figure 5) to enter the active site and bind between the FAD N(5) and Arg431. Thus, the direct hydrogen bond of the native enzyme has been converted into a water-mediated one.
Changes in the active site solvent structure are also observed. There are three solvent sites in the mutant enzyme that are not present in the native enzyme (Figure 5). Wat1, as describe above, bridges Arg431 and FAD N(5). The other two water molecules (wat2, wat3) form hydrogen bonds with Ser540, and they occupy some of the space corresponding to Tyr540 of the native enzyme (Figure 5). As with the movement of Asp285, creation of the wat2 and wat3 sites helps compensate for the loss of molecular volume at residue 540. Moreover, the three new water molecules participate in an extensive hydrogen bond network linking Asp285, Ser327, Asp370, Arg431, Ser540 and Tyr552 (Figure 4). Thus, a major effect of the tyrosine-to-serine mutation is to increase the solvent content of the active site. The interactions provided by these new water molecules presumably help maintain the active site in the native-like conformation shown in Figure 4. Since Ser540 forms key hydrogen bonds with two of the three new water molecules, this solvent network is presumably destabilized and/or significantly altered in Y540A, which might explain why Y540A has higher Km and Ki values than Y540S.
DISCUSSION
Replacement of Tyr540 with the smaller side chains of Ala or Ser profoundly altered the kinetics of PutA86-630. The catalytic efficiency for the natural substrate, proline, substantially decreased, while the efficiency for hydroxyproline increased. The net effect is a shift from the overwhelming preference of the native enzyme for proline to modest preferences of the mutants for proline. These results suggest that Tyr540 is important for discriminating proline from hydroxyproline. Our structural analysis suggests that tyrosine at this location disfavors binding of hydroxyproline because of steric clash with the 4-hydroxyl group. Its removal, by mutation to Ala or Ser, releases this steric constraint and opens up enough room in the active site for the 4-hydroxyl group, as shown by the structure of Y540S complexed with hydroxyproline.
As described by White et al. (10), Tyr540 is part of a highly conserved sequence pattern known as motif 8, which is located on strand 8 of the PRODH barrel (Figure 3B). This motif precedes the conserved region located on helix 8 (denoted motif 9), which includes key residues that interact with the substrate: Arg555 and Arg556. The tyrosine residue of strand 8 (motif 8) is highly conserved. Tyrosine is found at this location in all bacterial monofunctional PRODHs and PutAs, as well as POX (Figure 3B). We thus predict that any PRODH having tyrosine in motif 8 will exhibit a pronounced preference for proline over hydroxyproline.
Interestingly, in OH-POX – the only member of the PRODH family that is specific for hydroxyproline – tyrosine is replaced by the smaller amino acid, serine (Ser485, see Figure 3B). In fact, the Tyr/Ser variation in motif 8 is the only significant difference between POX and OH-POX in residues predicted to directly contact the substrate (11). Indeed, these observations inspired creation of the Y540S mutant of PutA86-630.
There is renewed biomedical interest in OH-POX, which motivates further study of the structural and mechanistic attributes of this enzyme, including the determinants of substrate specificity such as those described here. Recent work from Phang’s group has shown that OH-POX, like its proline-specific counterpart POX, is induced by the tumor supressor p53 and contributes to apoptosis by serving as a superoxide - generating enzyme (31). Furthermore, recent studies suggest that hydroxyproline catabolism may play an important role in kidney stone (calcium oxalate) formation because hydroxyproline is a precursor of glyoxylate, and ultimately, oxalate (32–34). Therefore, it has been suggested that inhibition of hydroxyproline catabolic enzymes may be a possible new therapeutic strategy for treating individuals with a genetic predisposition to stone formation (35).
The present study sheds some light on the structural basis of substrate recognition by OH-POX. The structure of Y540S complexed with hydroxyproline showed that Ser540, which is analogous to Ser485 of OH-POX, forms a hydrogen bond with the 4-hydroxyl group of the substrate. Given the high degree of sequence conservation of active site residues other than the Tyr/Ser of motif 8 within the PRODH family, we suggest that an analogous hydrogen bond interaction involving Ser485 contributes to the specificity of OH-POX for hydroxyproline.
Although our data clearly show the importance of the tyrosine of motif 8 for enforcing substrate preference for proline, its mutation to either Ala or Ser did not reverse substrate preference. Thus, it remains unclear how OH-POX achieves high specificity for hydroxyproline. Structural and kinetic studies of OH-POX and POX will be needed to understand this unique aspect of substrate specificity within the PRODH family.
Acknowledgments
We thank Dr. David Valle for insightful discussions about substrate specificities of human PRODHs and Dr. Jay Nix of Advanced Light Source beamline 4.2.2 for help with data collection. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Part of this work is based upon research conducted at the Northeastern Collaborative Access Team beam lines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38.
Footnotes
This research was supported by the National Institutes of Health Grant GM-65546 (to JJT).
Coordinates and structure factors have been deposited in the Protein Data Bank under Accession Numbers 3E2Q, 3E2R and 3E2S.
Abbreviations: PRODH, proline dehydrogenase; P5C, Δ1-pyrroline-5-carboxylate; P5CDH, Δ1-pyrroline-5-carboxylate dehydrogenase; PutA, Proline utilization A; POX, proline-specific human proline oxidase/dehydrogenase; OH-POX, hydroxyproline-specific human proline oxidase/dehydrogenase; THFA, L-tetrahydro-2-furoic acid; PutA86-669, protein corresponding to residues 86 – 669 of E. coli PutA; PutA86-630, protein corresponding to residues 86 – 630 of E. coli PutA; Y540S, PutA86-630 site-directed mutant Tyr540Ser; Y540A, PutA86-630 site-directed mutant Tyr540Ala; DCPIP, dichlorophenolindophenol; PDB, Protein Data Bank.
References
- 1.Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810–1815. [PubMed] [Google Scholar]
- 2.Rivera A, Maxwell SA. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J Biol Chem. 2005;280:29346–29354. doi: 10.1074/jbc.M504852200. [DOI] [PubMed] [Google Scholar]
- 3.Pandhare J, Cooper SK, Phang JM. Proline oxidase, a proapoptotic gene, is induced by troglitazone: evidence for both peroxisome proliferator-activated receptor gamma-dependent and -independent mechanisms. J Biol Chem. 2006;281:2044–2052. doi: 10.1074/jbc.M507867200. [DOI] [PubMed] [Google Scholar]
- 4.Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Metabolic and molecular basis of inherited disease. McGraw Hill; New York: 2001. pp. 1821–1838. [Google Scholar]
- 5.Willis A, Bender HU, Steel G, Valle D. PRODH variants and risk for schizophrenia. Amino Acids. 2008 doi: 10.1007/s00726-008-0111-0. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, Lenane M, Robertson B, Wijsman EM, Rapoport JL, Gogos JA, Karayiorgou M. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti A. A compelling genetic hypothesis for a complex disease: PRODH2/DGCR6 variation leads to schizophrenia susceptibility. Proc Natl Acad Sci USA. 2002;99:4755–4756. doi: 10.1073/pnas.092158299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Abecasis GR, Heath SC, Knowles A, Demars S, Chen YJ, Roos JL, Rapoport JL, Gogos JA, Karayiorgou M. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc Natl Acad Sci U S A. 2002;99:16859–16864. doi: 10.1073/pnas.232186099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquet H, Raux G, Thibaut F, Hecketsweiler B, Houy E, Demilly C, Haouzir S, Allio G, Fouldrin G, Drouin V, Bou J, Petit M, Campion D, Frebourg T. PRODH mutations and hyperprolinemia in a subset of schizophrenic patients. Hum Mol Genet. 2002;11:2243–2249. doi: 10.1093/hmg/11.19.2243. [DOI] [PubMed] [Google Scholar]
- 10.White TA, Krishnan N, Becker DF, Tanner JJ. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J Biol Chem. 2007;282:14316–14327. doi: 10.1074/jbc.M700912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner JJ. Structural biology of proline catabolism. Amino Acids. 2008 doi: 10.1007/s00726-008-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramar R, Fitscha P. Studies on the dehydrogenation of proline and hydroxyproline in animal tissue. Enzymologia. 1970;39:101–108. [PubMed] [Google Scholar]
- 13.Efron ML. Familial Hyperprolinemia. Report Of A Second Case, Associated With Congenital Renal Malformations, Hereditary Hematuria And Mild Mental Retardation, With Demonstration Of An Enzyme Defect. N Engl J Med. 1965;272:1243–1254. doi: 10.1056/NEJM196506172722401. [DOI] [PubMed] [Google Scholar]
- 14.Efron ML, Bixby EM, Pryles CV. Hydrosyprolinemia. Ii A Rare Metabolic Disease Due To A Deficiency Of The Enzyme “Hydroxyproline Oxidase”. N Engl J Med. 1965;272:1299–1309. doi: 10.1056/NEJM196506242722501. [DOI] [PubMed] [Google Scholar]
- 15.Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, White TA, Schuermann JP, Baban BA, Becker DF, Tanner JJ. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43:12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. 2006;D62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 19.Evans P. Scaling and assessment of data quality. Acta Crystallogr. 2006;D62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 20.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. 2003;D59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 21.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Rad. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Abrahamson JL, Baker LG, Stephenson JT, Wood JM. Proline dehydrogenase from Escherichia coli K12. Properties of the membrane-associated enzyme. Eur J Biochem. 1983;134:77–82. doi: 10.1111/j.1432-1033.1983.tb07533.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown ED, Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J Biol Chem. 1992;267:13086–13092. [PubMed] [Google Scholar]
- 25.Brown ED, Wood JM. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J Biol Chem. 1993;268:8972–8979. [PubMed] [Google Scholar]
- 26.Vinod MP, Bellur P, Becker DF. Electrochemical and functional characterization of the proline dehydrogenase domain of the PutA flavoprotein from Escherichia coli. Biochemistry. 2002;41:6525–6532. doi: 10.1021/bi025706f. [DOI] [PubMed] [Google Scholar]
- 27.Kakkar T, Boxenbaum H, Mayersohn M. Estimation of Ki in a competitive enzyme-inhibition model: comparisons among three methods of data analysis. Drug Metab Dispos. 1999;27:756–762. [PubMed] [Google Scholar]
- 28.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10:109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White TA, Johnson WH, Jr, Whitman CP, Tanner JJ. Structural Basis for the Inactivation of Thermus thermophilus Proline Dehydrogenase by N-Propargylglycine(,) Biochemistry. 2008;47:5573–5580. doi: 10.1021/bi800055w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 31.Cooper SK, Pandhare J, Donald SP, Phang JM. A novel function for hydroxyproline oxidase in apoptosis through generation of reactive oxygen species. J Biol Chem. 2008;283:10485–10492. doi: 10.1074/jbc.M702181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight J, Jiang J, Assimos DG, Holmes RP. Hydroxyproline ingestion and urinary oxalate and glycolate excretion. Kidney Int. 2006;70:1929–1934. doi: 10.1038/sj.ki.5001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa Y, Hossain RZ, Ogawa T, Yamakawa K, Yonou H, Oshiro Y, Hokama S, Morozumi M, Uchida A, Sugaya K. Vitamin B6 deficiency augments endogenous oxalogenesis after intravenous L-hydroxyproline loading in rats. Urol Res. 2007;35:15–21. doi: 10.1007/s00240-006-0076-y. [DOI] [PubMed] [Google Scholar]
- 34.Marengo SR, Romani AM. Oxalate in renal stone disease: the terminal metabolite that just won’t go away. Nat Clin Pract Nephrol. 2008;4:368–377. doi: 10.1038/ncpneph0845. [DOI] [PubMed] [Google Scholar]
- 35.Coulter-Mackie MB. 4-Hydroxyproline metabolism and glyoxylate production: A target for substrate depletion in primary hyperoxaluria? Kidney Int. 2006;70:1891–1893. doi: 10.1038/sj.ki.5001987. [DOI] [PubMed] [Google Scholar]
- 36.Engh RA, Huber R. Accurate bond and angle parameters for x-ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- 37.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]