In the history of biology, the early 1950s to the mid-1960s represent a period of landmark discoveries that led to the establishment of a new science named Molecular Biology. The discovery of the double helical structure of DNA by Watson and Crick in 1953 facilitated the formulation of specific questions related to the hottest questions of the time, gene replication and expression. Many ambitious and talented scientists with varied backgrounds (geneticists, biochemists, microbiologists, physicists, and others) initiated work toward similar goals. Thus, starting with the confirmation in Escherichia coli of the hypothesis that DNA replication was semiconservative and, shortly thereafter, the discovery of mRNA, the formulation of the operon model for regulation of gene expression, and finally the elucidation of the genetic code, a flood of important discoveries arrived during this period. The favorite experimental system during this time was E. coli and its bacteriophages, i.e. prokaryotic systems. But, as exemplified by the demonstration of the universality (with minor exceptions) of the genetic code as well as the earlier demonstration of the similarity in major metabolic pathways such as glycolysis and the tricarboxylic acid cycle from bacteria to humans, biologists assumed that the fundamental biological principles discovered in E. coli must be largely correct for other organisms. In fact, there was a famous saying (generally attributed to Jacques Monod), “What is true for E. coli is also true for elephants.” Thus, there was a kind of feeling among ambitious people who participated in this early development of molecular biology that most of the important questions in biology, specifically questions asked using E. coli as a model organism, had been answered. Many people thought that cellular differentiation and morphogenesis could be the next major question in molecular biology and started to switch from E. coli to other experimental organisms, mostly eukaryotes, to study the questions related to differentiation or other phenomena uniquely observed in eukaryotes.
The most extreme expression of this view came from Gunther Stent. Soon after the formal event marking the final decipherment of the genetic code, the Cold Harbor Symposium in 1966, he published an article in Science entitled “That Was the Molecular Biology That Was” (1), followed in 1969 by the publication of an expanded version as a book entitled “The Coming of the Golden Age: A View of the End of Progress” (2). After reviewing not only the history of progress in science, biology in particular, but also progress in other human activities, such as music and painting, Stent concluded that there may be no major conceptual breakthroughs remaining to be made in science. The exception perhaps would be in studying functions of the human brain, and this notion, the end of progress, might be equally true for art and other human activities. Such opinions were a strong influence on some early-day molecular biologists who had made significant contributions to the development of “classical molecular biology” using E. coli. This was exemplified by the exodus of many people from E. coli to other experimental systems at a time when many important problems in E. coli still remained unsolved. Like Seymour Benzer, who switched from E. coli molecular biology to Drosophila neurobiology in the late 1960s, Stent himself left E. coli molecular biology and switched to neurobiology using the leech as an experimental system. Among other early E. coli molecular biologists who switched their fields/systems, notably successful persons were, for example, George Streisinger, who initiated zebrafish research, and Sydney Brenner, who initiated research using the nematode worm Caenorhabditis elegans.
From Fermentation Biochemistry to Prokaryotic Molecular Biology
I started scientific research related to fermentation biochemistry as a student of the University of Tokyo in 1950. At that time, Japan was still struggling to recover from the destruction and social turmoil caused by its defeat in the Second World War. The country was isolated from the rest of the world, and we started to learn only very slowly about the progress in science that took place abroad during the war and immediately thereafter. By carrying out research related to microbial metabolism of organic acids such as tartaric acid (3), I learned some old biochemical and microbiological techniques used in the 1930s. Although I enjoyed doing the research and found some satisfaction in my modest achievements, I became keenly aware that my research would make no significant contribution to progress in the mainstream of basic science. Fortunately, I was able to come to the United States as a postdoctoral fellow in late 1957 and to work with three people prominent in the then emerging field of molecular biology. I stayed in the United States for three years, the first two of which I spent in Sol Spiegelman's laboratory at the University of Illinois, with summer intermissions spent with Jim Watson at Harvard. The third year I spent with Seymour Benzer at Purdue University in Indiana (Fig. 1). In addition, through these three people and through attending several meetings, I became acquainted with several noted molecular biologists, such as Gunther Stent, who played significant roles in the early days of molecular biology. In the summer of 1959, I took the Phage Course at Cold Spring Harbor, taught by George Streisinger, Frank Stahl, and Salvador Luria. Because I had significant experience in doing research (albeit in old-fashioned ways, as mentioned above), I had few problems in carrying out biochemical experiments aimed at some specific goals. In fact, the experiments I did with Benjamin Hall during the last three months in Spiegelman's laboratory yielded exciting results. We investigated whether small amounts of RNA synthesized in E. coli after phage T2 infection (discovered a few years earlier by Volkin and Astrachan (4)) were identical to rRNAs as predicted by the central dogma (as it was conceived then, i.e. before the discovery of mRNA). We discovered that the T2-specific RNAs (the name we gave to mRNA encoded by T2 genome) were clearly different from the E. coli rRNAs in their sizes and could be physically separated and concluded that that they must represent intermediate information carriers between phage DNA and phage-specific proteins (5). Because I had arranged to move to Benzer's laboratory, there was not enough time to do the follow-up experiments that I wished to do, and the work was published in an incomplete form. The real proof for the concept of mRNA was achieved only subsequently using the same phage-infected E. coli system by Brenner, Jacob, and Meselson (6) as well as Jim Watson's group (7).
FIGURE 1.
The mentors during my postdoctoral time. a, Sol Spiegelman with his wife, Helen Spiegelman (1958). b, J. D. Watson, S. Benzer, and myself (from left to right) at a meeting in 1990.
Although I was very uneasy in leaving the above-mentioned “important” experiments without completion, I was also eager to learn genetics in Benzer's laboratory. Because of the war and post-war turmoil, my basic science education in Japan was very fragmentary and inadequate, and none of the courses I took included genetics, even an elementary description of Mendelian genetics. So, as I learned biochemistry mostly through my actual participation in various research projects, mainly in fermentation biochemistry, I thought that I would be able to learn genetics through my actual participation in suitable research projects. Thus, I made an arrangement to work with Benzer, who was doing beautiful research on T4 phage genetics. I remember Jim Watson's comments on my plan; he said something like “you are going backward by starting `classical' phage genetics; you should come to Harvard to go forward.” I replied that I would go backward to learn genetics and continued in my decision to join Benzer's laboratory. I carried out mainly two projects. One was to identify the protein encoded by rII genes of phage T4. Benzer requested that I do it, saying that “you are a biochemist, and therefore, you should be able to identify the protein.” The second was to use genetic approaches to examine whether rII deletions make the distance between two outside markers shorter as one would expect from real physical deletions of DNA. Although the rII genetics worked beautifully, and I even made an unexpected new discovery, the presence of two kinds of heterozygotes in crosses of T4 (8), I failed to identify rII gene products. (The presence of two kinds of heterozygotes in T4 was later explained by the discovery of terminal redundancy of the phage T4 genome by George Streisinger (9).)
After three years of experience as a postdoctoral fellow working in these prominent laboratories in the mainstream of molecular biology, I was ready to start my own independent research as a (prokaryotic) molecular biologist. I returned to Japan in 1960 and took a position as an assistant professor in the Institute of Protein Research at Osaka University. Knowing the system in Japan at that time, I had applied to the National Institutes of Health for a research grant and received everything I asked for. Thanks to this financial support, I was able to organize my own laboratory and start research with complete independence. To avoid competition in the field of mRNA and to accomplish something unique, I decided to work on the mode of action of colicins. My long-term goal was to study membrane structure and function by analyzing biochemical changes that follow binding of colicin proteins to their specific receptors. We soon started to obtain various interesting results (10) and to have stimulating interactions with Salvador Luria, who also had a similar goal, switching from his celebrated classical work on phage genetics to colicin research around the same time (11). However, I also kept my interests in mRNA and ribosomes, and ribosome research eventually became the main research project in my laboratory during the time from the mid-1960s to 1980s. In the meantime, I moved back to the United States in 1963 and took a faculty position in the Department of Genetics at the University of Wisconsin, making the rather difficult personal decision of leaving my own country and committing myself to doing research in the United States.
Sabbatical Time at the University of Aarhus
In the mid-to-late 1960s, I had my own interesting unsolved questions related to ribosomes and colicins and was too busy to think seriously about other research subjects/organisms. However, a serendipitous event was to change that. In early 1970, I received an invitation from Niels Ole Kjeldgaard and Kjeld Marcker to spend a year as a visiting professor at the newly created Department of Molecular Biology at the University of Aarhus in Denmark, where they had recently moved. I knew both of them professionally. Marcker, working with Fred Sanger at the Medical Research Council (MRC) in Cambridge, England, discovered formylmethionyl-tRNA as an initiator tRNA in E. coli (12) and subsequently demonstrated the presence of a unique (unformylated) methionyl-tRNA functioning as an initiator tRNA in a eukaryotic system, too (13). We had recently demonstrated that the 30 S small ribosomal subunit binds formylmethionyl-tRNA (14) and that dissociation of the 70 S ribosome into 30 S and 50 S subunits is obligatory for initiation of protein synthesis in E. coli (15). Our discovery provided a functional explanation for the bipartite structure of the ribosome. At that time, many people were studying the mechanism of protein synthesis using in vitro systems containing poly(U) as mRNA and ribosomes consisting mostly of 70 S ribosomes. The synthesis of polyphenylalanine was measured using radioactive phenylalanine. Thus, it was generally assumed that the initiation of protein synthesis takes place on 70 S ribosomes. I presented our work at the Nucleic Acid Gordon Conference in 1967, which generated heated discussions with other workers, including collaborators of Marcker from the Cambridge MRC Laboratory.
As for Kjeldgaard, I knew his work with Ole Maaløe in Copenhagen on ribosome biosynthesis, in particular the discovery of growth rate-dependent control of ribosome synthesis (16). I also had an occasion to meet him in the ribosome meeting in Berlin in 1967. However, when I received the invitation, we had just recently demonstrated that the 50 S ribosomal subunit from Bacillus stearothermophilus could be reconstituted from its molecular components (17), as had been done for the E. coli 30 S subunit several years earlier (18). Although the concept that the tertiary structure of proteins is determined by the primary amino acid sequence had been established at that time, how more complex biological structures are generated was not necessarily clear. Our work demonstrated that the information for correct ribosome structures is present in their molecular components. In addition, ribosome reconstitution systems provided new ways to study ribosomes. Thus, to extend this initial work, my laboratory was busy working on various aspects of ribosome structures, functions, and in vitro assembly reactions. For this reason, I was initially hesitant to take a sabbatical leave, but I also thought that it might be a good time to pause and think about the future direction of my research and especially about the possibility of switching to eukaryotic molecular biology. Hearing that Marcker was starting several new projects related to gene expression in eukaryotic systems made this invitation attractive. Eventually, I accepted the invitation, or rather half of it, and spent about six months in Aarhus.
Marcker had brought some excellent young researchers from Cambridge. One of these young researchers, Alan Smith, had developed a mammalian in vitro protein-synthesizing system and identified the initiator tRNA. There had been several published observations suggesting that interferons inhibit mammalian protein synthesis in virus-infected cells but not in uninfected cells. In discussion of these observations with Marcker, we thought that the in vitro protein-synthesizing system developed by Alan could be useful to study the mechanism of the apparent specific inhibition of protein synthesis in virus-infected cells. Marcker was very interested in interferons and had obtained a “purified” preparation of an interferon. Because I had learned several techniques from Alan, e.g. how to make Ehrlich ascites tumor cells in mice and active extracts for protein synthesis, Marcker suggested that I pursue this project. After growing the mouse L cells and infecting them with a virus, I was unable to confirm the specific inhibition of virus growth by interferon, possibly because the activity of the interferon preparation was too weak. In retrospect, the laboratory was too new, and several projects that were going on were still in preliminary stages with the exception of the mammalian in vitro protein-synthesizing system. More fundamentally, my interest in mammalian molecular biology was rather opportunistic and not based upon my own specific questions. Still, I thought that my experience would be useful to my understanding and possible transition to eukaryotic molecular biology in the future.
My visit to Aarhus did not inspire me to switch to eukaryotic molecular biology, but my daily contact and discussion with Kjeldgaard's group made me realize that there were several unsolved and challenging questions related to ribosome biosynthesis remaining to be studied in E. coli. For example, to explain the growth rate-dependent control of ribosome synthesis, Maaløe had just published a theoretical paper proposing a “passive control” model of ribosomal protein (r-protein) synthesis (19). In addition, he suggested that some of the r-proteins might act as an inducer of rRNA synthesis, explaining how equimolar synthesis of rRNA and r-proteins might be achieved (19). To me, his model was too speculative, but it could be tested experimentally. Watching the Danish groups (people in both Maaløe's lab in Copenhagen and Kjeldgaard's lab in Aarhus) studying the questions using growing E. coli cells and repeating many measurements under many different conditions, I thought that such questions could be studied biochemically once the genes for these ribosomal components were isolated. Cloning technology had not yet been developed, but some E. coli genes had been isolated as transducing phages, such as λgal-transducing phages. In addition, many r-protein genes were thought to be clustered near the str gene, the first gene identified to encode a r-protein. In fact, this identification had been recently achieved in my laboratory using the ribosome reconstitution technique (20). Thus, by the end of my sabbatical, I decided not to switch from E. coli to other systems but to initiate research in a new direction, regulation of synthesis of ribosomes and ribosomal components in E. coli.
It took a few years, but we were successful in integrating λ phages near the str gene (and at some other loci) on the chromosome and in isolating many r-protein genes as transducing phages (e.g. Refs. 21 and 22). Furthermore, the breakthrough discovery of gene cloning technology together with other technological progress around the same time in the mid-1970s allowed isolation of all the genes for rRNA in E. coli (23, 24) and helped to expedite progress of our regulation studies. In this way, we discovered that co-regulation of the synthesis of most, if not all, r-proteins is achieved by a feedback repression using some r-proteins as repressors at the level of translation rather than at the level of transcription. We demonstrated that as long as rRNA synthesis continues, r-proteins synthesized are assembled into ribosomes and that when r-protein synthesis exceeds rRNA production, repressor r-proteins accumulated in the pool prevent further translation of r-protein mRNA. This model, coupling of r-protein mRNA translation with rRNA synthesis, explains near-stoichiometric production of all r-proteins relative to rRNA (25–27). At that time, because of the strong influence of the original operon model proposed by Monod and Jacob, virtually everyone who worked on gene regulation was focused on processes that operated at the level of transcription. Co-regulation of many unlinked genes by coupling of r-protein production from separate mRNAs with a single event, ribosome assembly, was also novel and broader than co-regulation of physically linked genes by a repressor in the original operon model. Thus, our model broke through a conceptual barrier. Soon molecular biologists were finding numerous regulatory mechanisms that involved the control of translation. I remember that some years later in 1986, when a symposium was organized by the British Microbiological Society to commemorate the twenty-fifth anniversary of the operon model, I was very happy to be able to discuss our model by contrasting it with the original operon model (27).
Switching to Yeast as a Model Eukaryote to Study Regulation of rRNA Synthesis
My serious interest in doing research using yeast started after we elucidated an outline of the mechanisms involved in the regulation of ribosome synthesis in E. coli in the early 1980s. After the discovery of the feedback regulation of translation of r-protein mRNA mentioned above, I thought that such a simple and beautiful principle that ensures near-stoichiometric production of all r-proteins relative to rRNA synthesized might be used in other organisms, including eukaryotes. Thus, I started to think about testing such a possibility experimentally. At that time, we were still busy in studying details of the mechanisms of translational regulation by repressor r-proteins we had identified for several E. coli r-protein operons. In addition, we had just discovered that growth rate-dependent regulation of rRNA synthesis involves a feedback mechanism (28) and were carrying out various experiments to establish and extend this conclusion. Nevertheless, I thought that testing the possible universality of the regulation models obtained in the E. coli system should be feasible if we selected a suitable eukaryote(s) and that this might be an occasion to switch to eukaryotic molecular biology.
In the mid-1970s, a series of technological developments (gene cloning and DNA sequencing in particular) revolutionized biology, enabling research scientists to isolate genes from almost any organism. Watching the remarkable progress in eukaryotic molecular biology, it was natural for me to think about testing the ideas on regulation using a yeast system. When I was a student working on projects related to fermentation biochemistry, I had occasion to grow some yeast and fungal strains. Thus, although I did not learn the physiology or genetics of these organisms, yeast was certainly not an unfamiliar organism. In addition, some of my former professional friends and former graduate/postdoctoral students had already switched to yeast from prokaryotic molecular biology, and I was witnessing their activities in the new fields. More directly, I was also following the research on yeast ribosomes, mostly using Saccharomyces cerevisiae, by people such as Jon Warner and Rudy Planta.
Initially, I was not certain whether it would be better to work on S. cerevisiae or Schizosaccharomyces pombe. S. cerevisiae was already being proven as an attractive model eukaryote, as exemplified by Lee Hartwell's work on the cell cycle. However, several young people trained in the S. cerevisiae system as well as established people who had switched from E. coli had already started to work on S. cerevisiae ribosomes, especially the obvious experiments such as cloning and characterizing r-protein genes. The field of yeast ribosomes appeared to be becoming crowded. I considered working on S. pombe mainly because relatively few people, mostly in Europe and Japan, were doing research using S. pombe, and they were primarily focused on the cell cycle. Thus, in the fall of 1983, I spent a week or so in the laboratory of Masayuki Yamamoto, learning about this organism and the necessary experimental techniques for yeast genetics. Yamamoto worked with me on E. coli r-protein gene expression as a postdoctoral student and then switched to S. pombe, focusing first on mitosis in Mitsuhiro Yanagida's lab at Kyoto University and then initiating research on meiosis in his own lab at the University of Tokyo. I also had discussions with Yanagida, who had recently isolated mutants of S. pombe defective in topoisomerases I and II (top1 and top2) and found that total RNA accumulation stopped under conditions in which both top1 and top2 activities were conditionally inactivated by temperature-sensitive (ts) mutations. I decided to use this double mutant to test the question of coupling of r-protein gene expression with rRNA accumulation. Separately, however, I asked one of the graduate students, who had just finished a small project related to E. coli ribosome research in his first year, whether he was willing to start to work on the yeast S. cerevisiae. The student, Mike Wittekind, was courageous enough to agree with this proposition. He spent a week or so in Tom Petes' lab, then at the University of Chicago, to learn necessary techniques and other basics and started our first S. cerevisiae project. My suggestion was to fuse the gene for the largest subunit of RNA polymerase I (Pol I) to the GAL1 promoter and construct a strain in which rRNA synthesis by Pol I could be diminished by shifting cultures from galactose to glucose medium. By growing such a strain in galactose and then shifting to glucose medium and thus repressing Pol I synthesis, one should be able to examine the question of coupling of r-protein gene expression with rRNA synthesis. In this way, we started to do yeast experiments using two different yeast systems but asking the same specific questions. However, progress of the yeast research was initially very slow, partly because our research was disrupted by the move of the laboratory from Madison to Irvine, CA, which took place from 1984 to 1985.
The experiments using S. pombe were carried out by a postdoctoral student, Masahiro Yamagishi. With the mutant strain obtained from Yanagida and the method of pulse-chase isotope labeling to measure synthesis rates of individual r-proteins, a method adapted from the one used routinely in our E. coli ribosome projects, we obtained clear results for many r-proteins analyzed; synthesis of r-proteins continued for a long time in the absence of rRNA synthesis, and overproduced r-proteins were slowly degraded (29). The simplest interpretation was the absence of feedback repression of r-protein synthesis, which was clearly different from the E. coli system. At that time, Jon Warner's group and others were carrying out experiments using cloned r-protein genes to see whether there was a feedback regulation as was observed for E. coli cells in similar gene dosage experiments done a few years earlier by our laboratory (25). Although the initial results obtained in the yeast system were interpreted to favor the model of translational feedback coupled with rRNA synthesis, it eventually became clear that the apparent absence of an increase in r-protein synthesis rate in proportion to gene dosage is due to a rapid degradation of overproduced r-proteins in the absence of rRNA to interact with and assemble into ribosomes (e.g. Refs. 30–33). The results of our experiments using S. pombe were certainly consistent with this conclusion obtained with the S. cerevisiae systems. Although it took time to construct strains, Mike Wittekind's experiments using S. cerevisiae also demonstrated unambiguous results, supporting the absence of coupling of r-protein synthesis with rRNA accumulation by translational feedback (34). In retrospect, it is not surprising that yeast and E. coli use different mechanisms to control the synthesis of ribosomal components. As I myself was well aware, it had long been realized that the specific mechanisms underlying certain biological functions, including mechanisms of gene regulation, are the consequences of evolutionary tinkering and may not necessarily be the same among diverse organisms, especially between prokaryotic and eukaryotic organisms. This concept, that evolutionary tinkering can lead to different organisms doing a given job in different ways, was discussed earlier by Francois Jacob (35). Nevertheless, I was hoping to find that the elegant mechanism to regulate synthesis of nearly all ribosomal components as found in E. coli was applicable to other organisms, including eukaryotes. In this sense, the results of our first venture using two different yeast systems were, in a way, disappointing. On the other hand, the realization of clear differences between eukaryotes and prokaryotes in such basic biological activities as ribosome synthesis encouraged me to explore eukaryotic molecular biology in hopes of finding other new and elegant products of evolution. Notably, designing experiments to test a specific model and obtaining the answer successfully gave me a confidence in switching from E. coli to yeast even at this late point in my career. Regarding the question of S. cerevisiae versus S. pombe as the experimental system for our research, I decided to use S. cerevisiae mainly because of the availability of more genetic information and reagents, including various mutant strains and plasmid vectors at that time. I thought that the presence of many research scientists using S. cerevisiae in related fields would be a great positive factor in providing stimulation for me and that I should simply avoid unnecessary competition with other workers.
Approaches to the Study of rRNA Transcription by Pol I in Budding Yeast
Regarding specific research subjects, I decided to focus on rRNA transcription and not on r-protein gene expression. At that time, many people were studying transcription of mRNA by Pol II using both yeast and other model eukaryotic organisms and in relation to regulation of gene expression, e.g. that observed during cellular differentiation. I thought that studying r-protein gene expression might become one of many similar studies related to transcription by Pol II. Transcription of genes for small RNAs, such as tRNAs or 5 S RNA, by Pol III was also being fairly well studied, perhaps because of the small gene size, and had already attracted some very able investigators. In contrast, studies of rRNA transcription by Pol I were not advanced compared with those of the other nuclear RNA polymerases. Specific initiation of transcription of rRNA genes had been demonstrated using extracts from various organisms such as mouse and human cultured cells. Biochemists such as Ingrid Grummt and Robert Tjian were busy working in fractionating extracts to identify factors required for correct transcription initiation. Others such as Ron Reeder and Tom Moss were using a frog oocyte injection system to study rRNA gene expression in connection with gene structures. However, there were no good genetic approaches to clarify biochemical mechanisms involved in rRNA transcription by Pol I. As for Pol I transcription in S. cerevisiae (referred to as “yeast” hereafter), extensive studies had been carried out by both Warner's lab and Planta's lab, but their studies were mostly using intact cells. There was neither an in vitro system to study rRNA transcription nor a useful genetic approach to identify factors involved in Pol I transcription. I thought that we would still be able to make some unique contributions in this area of eukaryotic molecular biology. Thus, whereas most of my lab members were still working on projects related to various aspects of ribosome synthesis in E. coli, I started to give more serious thoughts to yeast Pol I transcription.
Two obvious approaches I decided to take were (a) developing an in vitro system for specific Pol I transcription and (b) devising a method(s) to isolate mutants defective in rRNA synthesis. Regarding the first approach, Dan Riggs, a postdoctoral student, was successful in devising a method to make an in vitro system using yeast cell extracts (36). However, identifying factors by further fractionation of extracts appeared to be a daunting task. Thus, my hope was to use the second approach to isolate mutants and then use these to advance the biochemical identification of factors.
Genetic approaches to isolate mutants defective in rRNA synthesis were attempted previously. One notable approach was to screen ts yeast mutants for the one defective in RNA accumulation but not in protein accumulation. This was done by Hartwell, McLaughlin, and Warner (37) in conjunction with the celebrated screen for cdc mutants. Unfortunately, mutants obtained in this way turned out to be mostly defective in splicing of mRNA (38). Because most of the genes with introns in S. cerevisiae are r-protein genes, a mutant deficient in mRNA splicing will be able to produce essentially all of its proteins except for the r-proteins. Such mutants will synthesize rRNA, but the newly synthesized rRNA precursors will be degraded in the absence of simultaneous production of r-proteins so that the phenotype of these mutants will be continued accumulation of most (non-ribosomal) proteins by pre-existent stable ribosomes and a very large decrease in total RNA accumulation. Because accumulation of mature rRNAs is indirectly inhibited by mutations in many other genes, the question became how we could devise a method to select or screen for yeast cells with mutations that specifically and/or directly affect rRNA synthesis by Pol I. Fortunately, there were two events around that time that led to our success in devising a method of mutant isolation.
In the spring of 1989, Yasuhisa Nogi joined my laboratory as a postdoctoral student. He initially started yeast research as a graduate student in the laboratory of Yasuji Oshima, a noted yeast geneticist in Japan, and then continued the yeast work on galactose metabolism in Toshio Fukasawa's laboratory. Thus, he was familiar with yeast genetics and technology, and I started to discuss with him possible strategies for mutant isolation. After spending about one year or so without much success, in the fall of 1990, we noticed a paper just published by Janice Kranz and Connie Holm, who described a general method to identify a yeast gene(s) that is functionally homologous to an essential gene (X) isolated in other organisms (39). A multicopy (unstable) plasmid containing gene X together with ADE3 (and URA3, which allows the investigator to select either for or against cells containing the plasmid) was first constructed and was introduced into a yeast ade2 ade3 mutant strain. The ade2 mutation causes accumulation of an intermediate that is converted to a red pigment. Accumulation of this intermediate does not occur in the wild-type strain or in the ade2 ade3 double mutant. It was shown that if X functions in yeast and if a yeast gene(s) functionally homologous to X exists and is essential for growth, mutants defective in such genes would form red colonies because the growth of such mutants requires retention of gene X and the associated ADE3 on the plasmid (see the legend to Fig. 2 for the formation of red color in ade2). Thus, the yeast gene homologous to X could easily be isolated and identified by screening plasmid libraries for genes that complement growth of specific mutants (detected as red colonies) in the absence of the plasmid carrying gene X. This approach was called “cloning by function.” This color screening method described was very clever and motivated us to think about the use of such a color screening method for our mutant isolation. We realized that if we devised an artificial system on a plasmid that could functionally complement mutations in essential components in the Pol I machinery, the Kranz and Holm method could be used for our purpose. An obvious possible way to synthesize rRNA without functional Pol I or its essential transcription factor(s) is to use another polymerase, Pol II, and a fusion gene in which the DNA segment coding for the primary rRNA transcript, 35 S rRNA, is fused to a strong Pol II promoter. In the earlier studies on feedback regulation of rRNA synthesis in E. coli, we fused the E. coli precursor rRNA coding region to a strong λ phage promoter, PL, and successfully demonstrated the synthesis of rRNA from this fusion gene carried on a plasmid (40), so we had an optimistic view of the use of a fusion gene to synthesize functional rRNAs. Regarding a strong Pol II promoter for fusion, the GAL7 promoter was an obvious choice because Yasuhisa worked on yeast GAL7 before coming to my laboratory. Thus, the first experiment was to construct a plasmid carrying the 35 S rRNA coding region fused to the GAL7 promoter and to test whether functional rRNAs are synthesized by Pol II using such a fusion gene as template. By this time, in collaboration with Andre Sentenac's laboratory in Gif-sur-Yvette, France, Mike Wittekind in our laboratory had isolated several mutants, each carrying a ts mutation in the largest subunit, A190, of Pol I. By introducing the plasmid carrying the fusion gene into one of the ts mutants, we confirmed the expected results: the ts mutant carrying the plasmid formed colonies on galactose medium but not on glucose medium at a restrictive temperature of 37 °C (41). It was a moment of joy and great relief. With this result, we were convinced that the method we had conceived would work and immediately constructed the plasmid shown in Fig. 2. It should be noted that mutations inhibiting stable accumulation of rRNA indirectly at steps downstream from the rRNA transcription step, e.g. mutations inhibiting rRNA processing, r-protein production, or r-protein-rRNA interactions, are all expected to inhibit rRNA accumulation derived from transcription of the GAL7-35 S rRNA fusion gene on the plasmid and therefore would be excluded from this screen. Thus, using an ade2 ade3 mutant carrying this plasmid, we started to screen for red mutant colonies on galactose plates, followed by the test to confirm their dependence on galactose for growth. In this way, we were able to isolate many mutants that have mutations in genes required for the synthesis of rRNA and to define 12 genes, RRN1–RRN13 (RRN8 turned out not to represent a gene with the original criteria) (Ref. 42; reviewed in Refs. 43 and 44). They included the genes encoding five essential or nearly essential Pol I subunits not shared by Pol II or Pol III, as expected (see the legend to Fig. 3). The remaining seven encoded proteins had not been known previously. By characterizing these proteins, we were able to define two transcription factor complexes, UAF (upstream activation factor; six subunit proteins) and CF (core factor; three subunit proteins), and a single protein transcription factor, Rrn3p (Fig. 3). I should note that the cellular amounts of Pol I transcription factors, UAF, CF, and Rrn3p, are very small compared with Pol II general transcription factors. Therefore, the identification and purification of these factors would have been very difficult if we did not have genes for the protein or their subunit proteins. With the genes for at least some of their subunits available, we were able to use epitope tagging methods, which allowed detection as well as affinity purification of these transcription factors. In vitro systems using these purified components (45) were very useful to clarify mechanisms involved in the initiation of rRNA transcription by Pol I as well as the elongation of rRNA chains. For example, in a recent project to demonstrate the presence of coupling of Pol I elongation with co-transcriptional rRNA processing and ribosome assembly, we isolated Pol I rpa135 mutants (which carry mutations in the gene for Pol I subunit A135) that are hypersensitive to 6-azauracil as candidates defective in elongation. Using an in vitro system with purified components, we were able to compare the rate of transcription elongation of mutant Pol I with that of wild-type Pol I in vitro and to show a large (∼10-fold) reduction for mutant Pol I relative to wild-type Pol I (46).
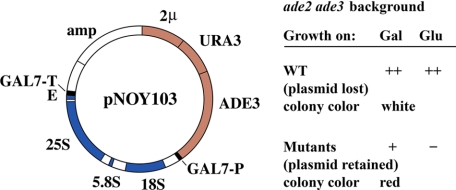
FIGURE 2.
Isolation of rrn mutants, which are defective in rRNA synthesis by Pol I. The yeast strain carried ade2 and ade3 mutations and the 2μ-based multicopy plasmid pNOY103. This plasmid carries ADE3 (and URA3 for plasmid selection and counterselection) and the hybrid gene in which the 35 S precursor rRNA coding region is fused to the GAL7 promoter (GAL7-P) and terminator (GAL7-T). It is known that yeast strains carrying ade2 accumulate an intermediate that is converted to a red pigment and that mutation ade3 blocks the pathway to this intermediate, thus preventing the formation of the red pigment. The starting (wild-type (WT)) strain does not form red colonies on rich galactose medium because the plasmid is unstable and not retained in most cells in colonies. Mutants (rrn mutants) that are specifically defective in the Pol I transcription machinery form red colonies on the same rich galactose medium because they can grow only by transcribing the fusion gene by Pol II and hence maintaining the plasmid (which carries ADE3) and accumulating red pigments. The rrn mutants are unable to grow on glucose because transcription from the GAL7 promoter is repressed by glucose.
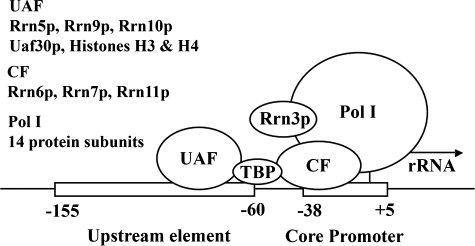
FIGURE 3.
Model for Pol I transcription initiation from the 35 S rRNA gene promoter. Pol I contains 14 protein subunits. Seven of them are unique to Pol I. Among the genes for these seven, deletion of the genes for two (A34.5 and A14) does not cause any significant growth defects. Mutants carrying alterations in these two genes were not found among rrn mutants, as expected, in the mutant screen shown in Fig. 2. The genes for the remaining five subunits, A190, A135, A12, A43, and A49, were identified as RRN1 (RPA190), RRN2 (RPA135), RRN4 (RPA12), RRN12 (RPA43), and RRN13 (RPA49), respectively, by the rrn mutant screening and were demonstrated to be essential or nearly essential for cell growth. Regarding Pol I transcription factors in mammalian Pol I systems, Rrn3p homologs of human (hRrn3) and mouse (TIFIA) have sequence homology to yeast Rrn3p. Human factor SL1 (and mouse TIFIB) consists of TATA-binding protein (TBP) and three protein subunits. SL1 is functionally homologous to the complex of TATA-binding protein and CF in yeast. However, there is no significant sequence similarity between the three subunits of SL1 and the three subunits of CF. No factor homologous to yeast UAF has been found in mammalian systems thus far.
Our helper plasmid system that uses Pol II for rRNA synthesis was also useful in studying roles of rDNA as well as rRNA sequence elements during rRNA processing and ribosome assembly, subjects studied by other investigators. The difficulty of mutating sequences of all tandemly repeated rRNA gene copies was overcome by the use of Pol I ts mutation combined with a Pol II-driven hybrid rDNA template carrying the specific sequence alterations to be studied. The effects of the mutational alterations on rRNA processing were analyzed first by growing a strain carrying the mutant hybrid rDNA on a helper plasmid at a permissive temperature and then shifting to a nonpermissive temperature and following the processing of rRNA transcribed from the mutant hybrid rDNA by Pol II in galactose medium (see e.g. Refs. 47 and 48).
When I decided to work on yeast in the mid-1980s, I continued several E. coli ribosome synthesis projects. When I found that we were able to isolate rrn mutants in early 1991, I decided to phase out the E. coli projects because of the confidence that the yeast research would be more exciting and challenging and that I should be able to succeed in discovering something unique to eukaryotes and interesting. Thus, except for graduate students who had already been working on E. coli as thesis projects, all the people in the laboratory started to participate in the yeast work. Our last E. coli research papers were published in 1994 (49, 50), formally ending our research using prokaryotic organisms.
In the early days of molecular biology, in the 1950s and 1960s, some of the people interested in solving genetic problems tried to use biochemical approaches without much training in biochemistry. There was a popular expression defining molecular biologists as “people practicing biochemistry without license.” In those early days, I had an inferiority complex with regard to my lack of a formal education and ignorance in classical (Mendelian) genetics, as mentioned earlier in this article. It was good for me to have decided to work in Seymour Benzer's laboratory from 1959 to 1960, doing research on T4 even though it was classical phage genetics. This experience helped me to use genetic approaches without a proper license in my subsequent research career, which covered both prokaryotic and eukaryotic molecular biology.
This use of genetic approaches in our yeast research was very satisfying to me. While working on E. coli ribosome projects and earlier on colicin projects, I tried to use various genetic approaches, but those were not very rewarding. For example, we isolated many mutants of E. coli defective in ribosome assembly as cold-sensitive mutants (51). However, except for mapping some of the mutations, the mutants did not give much insight regarding biochemical mechanisms involved in ribosome assembly, and those mutants had been lost when gene cloning methods became available. Perhaps one occasion when I felt happy with our genetic approaches related to E. coli projects was when I thought about the strategy for integration of a λ phage near the str-spc region of the E. coli chromosome (Ref. 21; details of the story described in Ref. 52). The strategy was successful and led to the isolation of many r-protein genes clustered in this region as transducing phages (e.g. Refs. 21 and 22). After switching to yeast, I certainly appreciated the “power of yeast genetics” and enjoyed research by combining genetic approaches with biochemistry. The remarkable progress in technology to manipulate DNA certainly helped genetic approaches to be more powerful compared with what was possible during the time of our E. coli ribosome research.
Legacy of the Early-day Molecular Biology
As often noted when one discusses the origin of molecular biology, there were two different schools that initiated new types of science in the early-to-mid twentieth century that eventually joined together to form the new field of molecular biology. One was the structural school developed in England that was practiced by x-ray crystallographers exemplified by people such as John Kendrew and Max Perutz. The other was the informational school initiated by people asking questions regarding the nature of genes as exemplified by the early phage group initiated by Max Delbrück. Gunter Stent's proclamation of the end of progress in Molecular Biology in 1968 (1, 2) was made as a molecular biologist of the informational school. It may be interesting to note that even regarding information transfer from genes to protein, several fundamental discoveries were made subsequently, e.g. discoveries of reverse transcriptase, introns and splicing, and small RNAs involved in regulation of gene expression, and that these discoveries were made mostly in research on eukaryotic systems. The general notion of the end of major progress in (prokaryotic) molecular biology, as publicized by people like Stent, stimulated the exodus of early molecular biologists from the prokaryotic systems in the late 1960s to 1970s and switching to new areas of research not explored in the earlier studies using prokaryotes. Features unique to multicellular organisms aside, it became obvious by the mid-1970s, especially after the successful demonstration of transformations of yeast cells with plasmid DNA carrying yeast genes (53, 54), that the yeast S. cerevisiae would be an excellent model organism for studies of unique features of eukaryotic cells. It is not surprising that quite a few early prokaryotic molecular biologists switched to yeast. One such person was David Botstein, who described the story of his switch to yeast in a highly interesting essay (55). In fact, I remember an occasion on which I talked with him at the Cold Spring Harbor Laboratory when I attended a meeting during a summer in the mid-1970s, almost certainly a meeting related to ribosome biosynthesis, and when I was still working on E. coli, including mapping of rRNA genes on the E. coli chromosome. David was telling me in an excited way about his recent discovery that even though >100 rRNA genes exist on the haploid yeast genome, these genes behave like a simple gene in genetic crosses, suggesting that they cluster at a single chromosome site, and that the frequency of recombination within the cluster during meiosis is greatly reduced (published in Ref. 56). As Botstein described in his article, quite a few former prokaryotic molecular biologists joined the “classical” yeast geneticist group and, together, made significant contributions to the rise of yeast to the position of the model organism for the study of eukaryotic cells that may correspond to the position of E. coli for prokaryotic cells. Some obvious research subjects for these former prokaryotic molecular biologists included, in addition to regulation of gene expression, functions and biosynthesis of cell organelles that are not present in prokaryotic cells, such as the nucleus (and subnuclear structures like chromosomes and nucleolus), mitochondria, endoplasmic reticulum, and Golgi apparatus as well as protein/RNA trafficking between these organelles and cytoplasm. When I decided to work on Pol I transcription of rRNA genes in yeast, I certainly thought about the nucleolus in yeast, and the molecular characterization of its structures and its functional significance were two of our research goals (for some of our later publications related to this subject, see Refs. 57 and 58).
It should be noted that contributions to the development of eukaryotic molecular biology made by the prokaryotic molecular biologists after switching to eukaryote research might have been through the style and rigor of reductionist approaches that were used in E. coli and phage research and maintained to study new questions in eukaryotic molecular biology. The beautiful work on yeast mating-type determination and interconversion done by Ira Herskowitz (e.g. Ref. 59), who started as a λ phage molecular biologist, may be the best example for this thesis. Although my switch from E. coli to yeast took place quite late compared with those who switched earlier in late 1960s to 1970s, I now feel that I have also maintained approaches and styles that resemble those I used while working on E. coli molecular biology.
Of course, the amazing technical progress in the past decade or so, which led to the successful complete sequencing of genomes of many organisms from E. coli and yeast to human, has drastically altered the nature of biological research. One can now analyze cellular activities as a whole using genomic, proteomic, and metabolomic approaches, i.e. approaches used by “systems biology.” Perhaps by combining the style and rigor used by the early-day molecular biologists with the new global approaches taken by systems biology, which include various modern new technologies that were unthinkable in those early days of molecular biology, one can hope that conceptual breakthroughs, which Stent concluded in 1968 would become rare in biology, may continue to be achieved.
Acknowledgments
I acknowledge continued grant support from the National Institutes of Health (from 1961 to the present; the current grant is GM35949) and the National Science Foundation (from 1964 to 1995). Quite a few of the early-day molecular biologists, whose names I have given in this article, are gone by now. They include my postdoctoral-time mentors, Sol Spiegelman and Seymour Benzer, and other teachers, colleagues, and friends: Francis Crick, Max Delbrück, Salvador Luria, George Streisinger, Ole Maaløe, Niels Ole Kjeldgaard, Rudy Planta, and Gunther Stent. Other notable early-day molecular biologists mentioned, Jacques Monod, John Kendrew, and Max Perutz, also have passed away. I miss them and cherish my memories of these early-day molecular biologists. Ira Herskowitz, who was much younger than I, also passed away prematurely. I dedicate this article to these people. I would also like to thank my former graduate students, postdoctoral fellows, and other staff scientists for their dedicated participation in research in my laboratory throughout my career. Finally, I thank James F. Crow, Melanie Oakes, Suzanne Sandmeyer, Robert Steele, Millard Susman, and Marian Waterman for their reading of the drafts of this article and for their useful suggestions and comments and Melanie Oakes and Catherine Cao for their help in the preparation of this article.
References
- 1.Stent, G. S. (1968) Science 160 390–395 [DOI] [PubMed] [Google Scholar]
- 2.Stent, G. S. (1969) The Coming of the Golden Age: A View of the End of Progress, Natural History Press, Garden City, NY
- 3.Nomura, M. (1953) J. Agric. Chem. Soc. Jpn. 27 46–50 [Google Scholar]
- 4.Volkin, E., and Astrachan, L. (1956) Virology 2 149–161 [DOI] [PubMed] [Google Scholar]
- 5.Nomura, M., Hall, B. D., and Spiegelman, S. (1960) J. Mol. Biol. 2 306–326 [Google Scholar]
- 6.Brenner, S., Jacob, F., and Meselson, M. (1961) Nature 190 576–581 [DOI] [PubMed] [Google Scholar]
- 7.Gros, F., Hiatt, H., Gilbert, W., Kurland, C. G., Risebrough, R. W., and Watson, J. D. (1961) Nature 190 581–585 [DOI] [PubMed] [Google Scholar]
- 8.Nomura, M., and Benzer, S. (1961) J. Mol. Biol. 3 684–692 [DOI] [PubMed] [Google Scholar]
- 9.Streisinger, G. (1992) in Phages and the Origins of Molecular Biology (Cairns, J., Stent, G. S., and Watson, J. D., eds) pp. 335–340, Cold Spring Harbor Laboratory Press, Woodbury, NY
- 10.Nomura, M. (1963) Cold Spring Harbor Symp. Quant. Biol. 28 315–324 [Google Scholar]
- 11.Luria, S. E. (1984) in A Slot Machine, A Broken Test Tube: An Autobiography, pp. 103–113, Harper & Row, New York
- 12.Marcker, K., and Sanger, F. (1964) J. Mol. Biol. 8 835–840 [DOI] [PubMed] [Google Scholar]
- 13.Smith, A. E., and Marcker, K. A. (1970) Nature 226 607–610 [DOI] [PubMed] [Google Scholar]
- 14.Nomura, M., and Lowry, C. V. (1967) Proc. Natl. Acad. Sci. U. S. A. 58 946–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guthrie, C., and Nomura, M. (1968) Nature 219 232–235 [DOI] [PubMed] [Google Scholar]
- 16.Maaløe, O., and Kjeldgaard, N. O. (1966) Control of Macromolecular Synthesis, W. A. Benjamin, Inc., New York
- 17.Nomura, M., and Erdmann, V. A. (1970) Nature 228 744–748 [DOI] [PubMed] [Google Scholar]
- 18.Traub, P., and Nomura, M. (1968) Proc. Natl. Acad. Sci. U. S. A. 59 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maaløe, O. (1969) Dev. Biol. 3 33–58 [Google Scholar]
- 20.Ozaki, M., Mizushima, S., and Nomura, M. (1969) Nature 222 333–339 [DOI] [PubMed] [Google Scholar]
- 21.Jaskunas, S. R., Lindahl, L., and Nomura, M. (1975) Proc. Natl. Acad. Sci. U. S. A. 72 6–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaskunas, S. R., Lindahl, L., and Nomura, M. (1975) Nature 256 183–187 [DOI] [PubMed] [Google Scholar]
- 23.Kenerley, M. E., Morgan, E. A., Post, L., Lindahl, L., and Nomura, M. (1977) J. Bacteriol. 132 931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellwood, M., and Nomura, M. (1980) J. Bacteriol. 143 1077–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fallon, A. M., Jinks, C. S., Strycharz, G. D., and Nomura, M. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 3411–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yates, J. L., Arfsten, A. E., and Nomura, M. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura, M. (1986) Symp. Soc. Gen. Microbiol. 39 199–220 [Google Scholar]
- 28.Jinks-Robertson, S., Gourse, R. L., and Nomura, M. (1983) Cell 33 865–876 [DOI] [PubMed] [Google Scholar]
- 29.Yamagishi, M., and Nomura, M. (1988) Curr. Genet. 13 305–314 [DOI] [PubMed] [Google Scholar]
- 30.Warner, J. R., Mitra, G., Schwindinger, W. F., Studney, M., and Fried, H. M. (1985) Mol. Cell. Biol. 5 1512–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ElBaradi, T. T. A. L., van der Sande, C. A. F. M., Mager, W. H., Raue, H. A., and Planta, R. J. (1986) Curr. Genet. 10 733–739 [DOI] [PubMed] [Google Scholar]
- 32.Maicas, E., Pluthero, F. G., and Friesen, J. D. (1988) Mol. Cell. Biol. 8 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsay, Y.-F., Thompson, J. R., Rotenberg, M. O., Larkin, J. C., and Woolford, J. L., Jr. (1988) Genes Dev. 2 664–676 [DOI] [PubMed] [Google Scholar]
- 34.Wittekind, M., Kolb, J. M., Dodd, J., Yamagishi, M., Memet, S., Buhler, J.-M., and Nomura, M. (1990) Mol. Cell. Biol. 10 2049–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob, F. (1977) Science 196 1161–1166 [DOI] [PubMed] [Google Scholar]
- 36.Riggs, D. L., and Nomura, M. (1990) J. Biol. Chem. 265 7596–7603 [PubMed] [Google Scholar]
- 37.Hartwell, L. H., McLaughlin, C., and Warner, J. (1970) Mol. Gen. Genet. 109 42–56 [DOI] [PubMed] [Google Scholar]
- 38.Rosbash, M., Harris, P. K. W., Woolford, J. L., and Teem, J. L. (1981) Cell 24 679–686 [DOI] [PubMed] [Google Scholar]
- 39.Kranz, J. E., and Holm, C. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6629–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gourse, R. L., Takabe, Y., Sharrock, R. A., and Nomura, M. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 1069–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nogi, Y., Yano, R., and Nomura, M. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 3962–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogi, Y., Vu, L., and Nomura, M. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 7026–7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nomura, M. (1988) in Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I (Paule, M., ed) pp. 155–172, R. G. Landes & Co., Austin, TX
- 44.Nomura, M., Nogi, Y., and Oakes, M. (2004) in The Nucleolus (Olson, M., ed) pp. 128–153, R. G. Landes & Co., Austin, TX
- 45.Keener, J., Josaitis, C. A., Dodd, J. A., and Nomura, M. (1998) J. Biol. Chem. 273 33795–33802 [DOI] [PubMed] [Google Scholar]
- 46.Schneider, D. A., Michel, A., Sikes, M. L., Vu, L., Dodd, J. A., Salgia, S., Osheim, Y. N., Beyer, A. L., and Nomura, M. (2007) Mol. Cell 26 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henry, Y., Wood, H., Morrissey, J. P., Petfalski, E., Kearsey, S., and Tollervey, D. (1994) EMBO J. 13 2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Côté, C. A., Greer, C. L., and Peculis, B. A. (2002) RNA (Cold Spring Harbor) 8 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito, K., Mattheakis, L. C., and Nomura, M. (1994) J. Mol. Biol. 235 111–124 [DOI] [PubMed] [Google Scholar]
- 50.Saito, K., and Nomura, M. (1994) J. Mol. Biol. 235 125–139 [DOI] [PubMed] [Google Scholar]
- 51.Guthrie, C., Nashimoto, H., and Nomura, M. (1969) Proc. Natl. Acad. Sci. U. S. A. 63 384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nomura, M. (1981) in Science and Scientists: Essays by Biochemists, Biologists and Chemists (Kageyama, M., Nakamura, K., Oshima, T., and Uchida, T., eds) pp. 133–145, Japan Scientific Societies Press, Tokyo
- 53.Hinnen, A., Hicks, J. B., and Fink, G. R. (1978) Proc. Natl. Acad. Sci. U. S. A. 75 1929–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beggs, J. D. (1978) Nature 275 104–109 [DOI] [PubMed] [Google Scholar]
- 55.Botstein, D. (1993) in The Early Days of Yeast Genetics (Hall, M. N., and Linder, P., eds) pp. 361–373, Cold Spring Harbor Laboratory Press, Woodbury, NY
- 56.Petes, T. D., and Botstein, D. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 5091–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oakes, M., Aris, J. P., Brockenbrough, J. S., Wai, H., Vu, L., and Nomura, M. (1998) J. Cell Biol. 143 23–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oakes, M. L., Siddiqi, I., French, S. L., Vu, L., Sato, M., Aris, J. P., Beyer, A. L., and Nomura, M. (2006) Mol. Cell. Biol. 26 3889–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herskowitz, I. (1989) Nature 342 749–757 [DOI] [PubMed] [Google Scholar]