Abstract
Intracellular pattern recognition receptors MDA5, RIG-I, and LGP2 are essential components of the cellular response to virus infection and are homologous to the DEXH box subfamily of RNA helicases. However, the relevance of helicase activity in the regulation of interferon production remains elusive. To examine the importance of the helicase domain function for these signaling proteins, a series of mutations targeting conserved helicase sequence motifs were analyzed for enzymatic activity, RNA binding, interferon induction, and antiviral signaling. Results indicate that all targeted motifs are required for ATP hydrolysis, but a subset is involved in RNA binding. The enzymatically inactive mutants differed in their signaling ability. Notably, mutations to MDA5 motifs I, III, and VI and RIG-I motif III produced helicase proteins with constitutive antiviral activity, whereas mutations in RIG-I motif V retained ATP hydrolysis but failed to mediate signal transduction. These findings demonstrate that type I interferon production mediated by full-length MDA5 and RIG-I is independent of the helicase domain catalytic activity. In addition, neither enzymatic activity nor RNA binding was required for negative regulation of antiviral signaling by LGP2, supporting an RNA-independent interference mechanism.
The first line of defense against virus infection is provided by the cellular antiviral response and innate immune system. The immediate response includes the induction of type I interferon (IFNα and IFNβ, referred to herein as IFN2) and other cytokines. Virus infections are sensed by cellular receptors that can recognize pathogen-associated molecular patterns such as viral nucleic acids (1). In addition to the transmembrane Toll-like receptors (2), a family of cytoplasmic RNA helicases has been identified that can detect cytosolic non-self RNA. Two members of this group are MDA5 (melanoma differentiation-associated gene 5) and RIG-I (retinoic acid-inducible gene I) (3). These proteins are characterized by the combination of two caspase activation and recruitment domain (CARD) motifs linked to an RNA helicase domain.
MDA5 and RIG-I can detect foreign RNAs and transmit a signal through the CARD-containing mitochondrial adaptor molecule IPS-1 (also named Cardif, MAVS, and VISA) (4–7). IPS-1 apparently serves as a scaffold for propagation of the signaling cascade that leads to the activation of transcription factors, including IRF-3 and NFκB (4, 7–9) which are responsible for transcriptional activation of a variety of antiviral effectors, including the IFNβ gene that is fundamental to the antiviral response.
Despite their similarities in domain structure and amino acid sequence, MDA5 and RIG-I are nonredundant and are involved in recognition of different types of non-self RNAs and therefore different viruses (10–12). RIG-I exhibits ligand specificity for short (<2 kbp) double-stranded RNA (dsRNA) and 5′-triphosphorylated single-stranded RNA (11, 13–17) and has been demonstrated to specifically recognize homopolymeric motifs, such as the polyuridine motif of the hepatitis C virus genomic 3′-nontranslated region (18). No specific natural ligand has been determined for MDA5, but it has preference for long (>2 kbp) dsRNA (17) and is a cytosolic receptor for the synthetic RNA, poly(I-C) (10, 11).
Recent data indicate that the two helicases also differ in regulation and signaling mechanisms. In RIG-I the C-terminal domain (CTD) contains a basic RNA binding pocket suitable for accommodation of 5′-triphosphate RNA (15, 16, 19). Sequence comparisons suggest the overall structure of the MDA5 CTD may be conserved but that MDA5 lacks key residues in the CTD RNA binding pocket (15, 16). In addition, the RIG-I CTD autoregulatory domain function was not observed for the MDA5 CTD (19).
A third related protein, LGP2, has high sequence similarity to MDA5 and RIG-I helicase domains but lacks the N-terminal CARD homology. Expression of LGP2 from a plasmid vector acts as a negative regulator of IFN production and antiviral signaling (19–22), but analysis of mice deficient for LGP2 suggests disparate functions for LGP2 in response to different viruses (23), acting as either a positive or negative regulator of antiviral signaling.
Great progress has been made in understanding the roles of MDA5, RIG-I, and LGP2 in RNA sensing and antiviral immune responses. Recent reports demonstrated in vitro unwinding activity of RIG-I for double-stranded oligonucleotides with 3′ overhangs. Interestingly, a reverse correlation between suitable unwinding substrates and immunogenic oligonucleotides was found (16). However, the significance of the DEXH box helicase domain for the role of proteins in innate immune responses still remains elusive.
MDA5, RIG-I, and LGP2 can be categorized as DEXH box RNA helicases belonging to the helicase superfamily 2 (24). This family is characterized by six highly conserved sequence motifs (I–VI) within the helicase domain (25). Structural studies of a variety of helicase family members reveal a general folding into two RecA-like globular domains, with domain 1 encompassing motifs I–III and domain 2 containing motifs IV–VI (26). Low resolution structural data of LGP2 (27) as well as the crystal structure of MDA5 domain 1 (PDB code 3b6e) suggest the general helicase fold to be conserved in these family members. Available evidence from structural and biochemical studies in other helicases has implicated specific motifs in different aspects of helicase functions. Motif I and motif II, also referred to as Walker A and B motif, respectively (28), are indispensable for ATP binding and hydrolysis. In most DEXH box helicases motif V and VI are also implicated in nucleotide binding (25, 29). In some studies, RNA binding has been assigned to motif IV and V (25, 29), whereas motif III has been implicated in forming intramolecular interactions between the domains. Indeed, interdomain interactions are also likely to occur between other motifs, to form a stabilized helicase catalytic core (30, 31).
To better understand the importance of helicase activity in cytoplasmic RNA sensing and antiviral signal transduction, we have systematically analyzed the catalytic core by site-directed mutagenesis. Results indicate that signaling by full-length RIG-I and MDA5 can occur independent of enzymatic activity. Mutations in the conserved helicase motifs produced a subset of RIG-I and MDA5 mutants that dissociate enzymatic activity from signal transduction activity, either producing enzymatically defective constitutive activators of IFNβ transcription or enzymatically active proteins defective in antiviral signaling. Furthermore, LGP2 mutants revealed that its ability to negatively regulate antiviral signaling is independent of both enzymatic activity and RNA binding activity.
EXPERIMENTAL PROCEDURES
Plasmids and Mutagenesis—FLAG-tagged MDA5 and RIG-I cDNA in expression vector pEFBos were provided by M. Gale, Jr. (University of Washington). LGP2 cDNA was obtained by PCR amplification of Human Universal Quick Clone II (Clontech) and cloned into p3xFLAG-CMV10 (Sigma) as described previously (20). Site-directed mutagenesis was carried out using QuickChangeXL mutagenesis kit (Stratagene). Introduced mutations were confirmed by DNA sequence analysis. For recombinant baculovirus production helicase cDNA sequences were amplified by PCR and cloned into pBac2cp (Novagen).
Cell Culture, Transfections, and Cytokine Treatment—Sf9 insect cells were maintained in Grace's insect cell medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). HEK293T, 2fTGH, U6A (a STAT2-deficient 2fTGH cell derivative), HeLa, Vero, and Huh 7.5 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% cosmic calf serum (HyClone) and 1% penicillin/streptomycin (Invitrogen). Transfections of HEK293T cells were carried out using the standard CaPO4 method (32) or FuGENE transfection reagent (Roche Applied Science). 2fTGH cells were transfected with Superfect (Qiagen). Transfection of HeLa, Vero, and Huh 7.5 cells and all poly(I-C) (Amersham Biosciences) transfections were performed with Lipofectamine transfection reagent (Invitrogen). All transfections were carried out according to manufacturers' protocol. Purified human IFNβ at 100,000 pg/ml was obtained from PBL InterferonSource and added to growth media at indicated final concentrations.
Protein Purification—HEK293T cells were transfected for expression of recombinant wild type or mutant helicase proteins with N-terminal FLAG tags. Cell lysates were precleared with Sepharose 6B beads (Sigma) and subsequently incubated with anti-FLAG M2 affinity gel (Sigma). Elution was achieved by addition of 3× FLAG peptide (Sigma). Proteins expressed by recombinant baculovirus infection of Sf9 insect cells were tagged with a His6 epitope, purified using Ni2+-nitrilotriacetic acid beads (Novagen), and eluted in buffer containing 250 mm imidazole.
ATP Hydrolysis Assay—500 ng of purified protein were incubated in 50 μl of ATPase reactions, containing 2 μg of poly(I-C), 0.5 mm ATP (Sigma), 0.66 nm [γ-32P]ATP (PerkinElmer Life Sciences) in 5 mm MOPS, pH 6.5, 0.3 mm MgCl2, 0.2 mm dithiothreitol, for 30 min at 37 °C. 10% of the reactions were loaded onto thin layer chromatography plates (Sigma) and analyzed by phosphorimaging with a Storm scanner (GE Healthcare) and subsequently quantified using ImageQuant™ software. The remaining reaction was analyzed by SDS-PAGE and subjected to silver staining.
Luciferase Reporter Gene Assays—Cells were cotransfected with reporter gene and expression vectors for Renilla luciferase as well as empty vector or the helicase of interest. The reporter gene contains the firefly luciferase open reading frame under the control of the interferon β promoter (–110). The next day cells were stimulated by infection with Sendai virus (Cantell strain, 3 × 106 PFU/well) infection or poly(I-C) (5 μg/well) transfection for 6 h before harvest. Relative luciferase activity was measured using Dual-Luciferase™ reporter assay (Promega). Data are plotted as average values (n = 3) with error bars representing standard deviation.
RT-PCR Analysis—HEK293T cells were transfected with helicase expression plasmids and 24 h later transfected with poly(I-C) or infected with Sendai virus (Cantell strain, 3 × 106 PFU/well). Total RNA was extracted using TRIzol reagent (Invitrogen). Samples were treated with DNase I (Invitrogen), and 1 μg of RNA was subjected to reverse transcriptase Superscript III (Invitrogen) for cDNA production. Quantitative real time PCR was carried out using the MX3000P real time PCR system (Stratagene) with SYBR green detection.
Antiviral Activity Assay—Antiviral responses were measured by cytopathic effect protection assay using VSV (Indiana strain) as a reporter virus. HEK293T cells were transfected for expression of helicase proteins. 24 h later supernatant was harvested, diluted, and added to 2fTGH cells. Following 8 h of incubation with the supernatant, 2fTGH cells were infected with VSV (Indiana strain) at 6 × 103 PFU/well in serum-free media for 1 h. Media were changed and cells incubated for 18 h before staining with 1% methylene blue in 50% ethanol.
RNA Binding Assay—For poly(I-C) interaction studies HEK293T cells were transfected for expression of helicase wild type and mutant proteins. Cell lysates were subjected to incubation with poly(I-C)-agarose beads (Sigma), washed in RNA binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl), and eluted by addition of SDS loading buffer. Bound protein was analyzed by SDS-PAGE and immunoblotting. Solution RNA binding assays were performed with immunopurified helicases proteins immobilized on anti-FLAG M2 affinity gel (Sigma). RNA substrates were prepared by in vitro transcription using Ambion Maxiscript T7 in the presence of [α-32P]UTP (PerkinElmer Life Sciences). dsRNA constructs were prepared by cotranscription and subsequent annealing of complementary strands. Approximately 500 ng of immobilized proteins and 1 μg of labeled RNA (at approximately 4 × 105 cpm/μg) were incubated for 2 h at 4 °C before washing in RNA binding buffer. Bound RNA was quantified by scintillation counting. Reactions were separated by SDS-PAGE and subjected to immunoblot analysis, and amounts of proteins were estimated using AutoChemi™ BioImaging Systems (UVP). Bound RNA was normalized to the relative amount of protein and presented as percent of wild type.
Limited Proteolysis—Proteins were expressed by infection of Sf9 insect cells growing in suspension culture with respective recombinant baculovirus and purified by Ni2+-nitrilotriacetic acid-agarose (Novagen). 10 μg of protein in 120-μl reactions containing 50 mm Tris-HCl, pH 7, 50 mm KCl, 4 mm dithiothreitol, 4 mm MgCl2, 6.5% glycerol were incubated with 10 ng of chymotrypsin. The samples were incubated at 37 °C, and aliquots were removed and stopped by addition of SDS-PAGE loading buffer at indicated time points. Samples were analyzed by SDS-PAGE and immunoblot using antiserum against MDA5 (antigen amino acids 587–600).
RESULTS
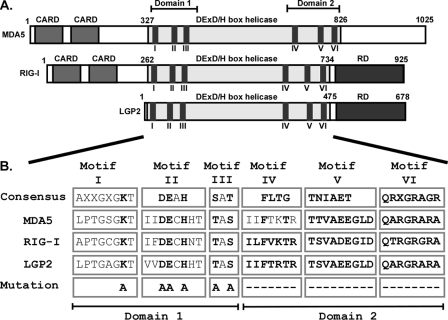
Identification and Mutation of Conserved Helicase Motifs—Comparison of the amino acid sequences of the MDA5, RIG-I, and LGP2 helicase domains with established consensus sequences of helicase families (33) clearly identifies characteristic helicase motifs clustered in two subdomains. Domain 1 contains motif I–III and domain 2 contains motif IV–VI (Fig. 1A). The sequences in individual motifs are highly conserved within MDA5, RIG-I, and LGP2 and despite slight variations from the consensus confidently characterize them as a related subset of DEXH box RNA helicases (Fig. 1B). Notably, the highest sequence identity is found between MDA5 and LGP2, which are 62% identical within helicase domain 2, encompassing motif IV–VI, whereas RIG-I and LGP2 or RIG-I and MDA5 are 43 and 44% identical in this region, respectively. The intervening sequences between motifs are also very similar among the three proteins but are poorly conserved with other DEXH box helicases. For example, the closest related human homologue is Dicer-1, sharing about 30% identity with LGP2, MDA5, and RIG-I within the helicase domain 1 and domain 2, but it lacks significant homology in surrounding regions.
FIGURE 1.
Conserved domains and helicase motifs of MDA5, RIG-I, and LGP2 proteins. A, diagrams illustrate the features of MDA5, RIG-I, and LGP2 proteins. In addition to the conserved DEX(D/H) box helicase regions, MDA5 and RIG-I contain two N-terminal CARD homologies that are important for interaction with downstream signaling molecules. RIG-I and LGP2 carry a regulatory domain (RD) at their C termini. The superfamily 2 RNA helicase domains are characterized by conserved amino acid sequence motifs, including I–VI. B, identification, sequence comparison, and mutations to helicase motifs I–VI. The DEXH box family motif consensus sequences (33) are aligned with corresponding sequences of MDA5, RIG-I, and LGP2. X indicates no amino acid preference at this position. Residues targeted for mutation are indicated in boldface type and were either substituted with alanine (A) or deleted (–). Introduced mutations are identical for all three helicases, with the exception of motif IV, which was subjected to two alanine substitution in MDA5 and whole motif deletions in RIG-I and LGP2.
To examine the role of individual conserved helicase motifs, a series of mutations were engineered in all three helicase proteins (Fig. 1B). Motif I, also known as the Walker A motif, is an NTP-binding motif with a critical lysine residue required for coordination of the NTP. This lysine was targeted by an alanine substitution. Motifs II and III were substituted entirely by alanine residues. Motifs IV, V, and VI are larger motifs and were deleted. An exception is motif IV of MDA5, which was subjected to two alanine substitutions to Phe-724 and Thr-727 instead of a motif deletion as in LGP2 and RIG-I. Expression of the mutant proteins in HEK293T cells resulted in similar levels of protein accumulation with the exception of LGP2 motif II. Independent preparations confirmed this mutant protein to be unstable, possibly the result of mutation-induced misfolding and subsequent proteasomal degradation.
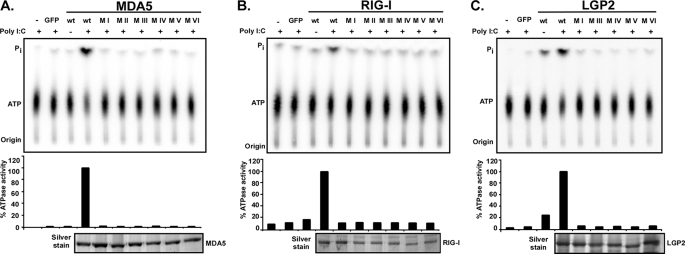
To test the effect of these mutations on enzymatic activity, ATP hydrolysis was tested (Fig. 2). Proteins were expressed in HEK293T cells, purified by immunoaffinity chromatography, and incubated with [γ-32P]ATP in the presence or absence of the synthetic dsRNA analogue poly(I-C). The ATP hydrolysis activity of wild type MDA5 and RIG-I proteins was strongly stimulated by poly(I-C) (Fig. 2, A and B), consistent with prior reports (3, 34–36). Wild type LGP2 exhibited significant ATP hydrolysis activity in the absence of poly(I-C), and the activity was enhanced by addition of RNA (Fig. 2C). For all three proteins, every mutation eliminated ATP hydrolysis activity. These results authenticate the identified helicase motifs as crucial for enzymatic activity.
FIGURE 2.
Mutations to RNA helicase motifs eliminate ATP hydrolysis activity. FLAG-tagged wild type and mutant MDA5 (A), RIG-I (B), and LGP2 (C) proteins were expressed in HEK293T cells and immunoaffinity-purified with FLAG M2 affinity gel. Eluted proteins were incubated with [γ-32P]ATP in the presence or absence of poly(I-C) and separated by thin layer chromatography (top). Origin and migration of free phosphate (Pi) and ATP are indicated. ATP hydrolysis activity was quantified by phosphorimage analysis and plotted as percentages of wild type protein activity in presence of poly(I-C) (center). Bottom panels show silver-stained SDS-PAGE demonstrating similar amounts of purified proteins in reactions. GFP, green fluorescent protein.
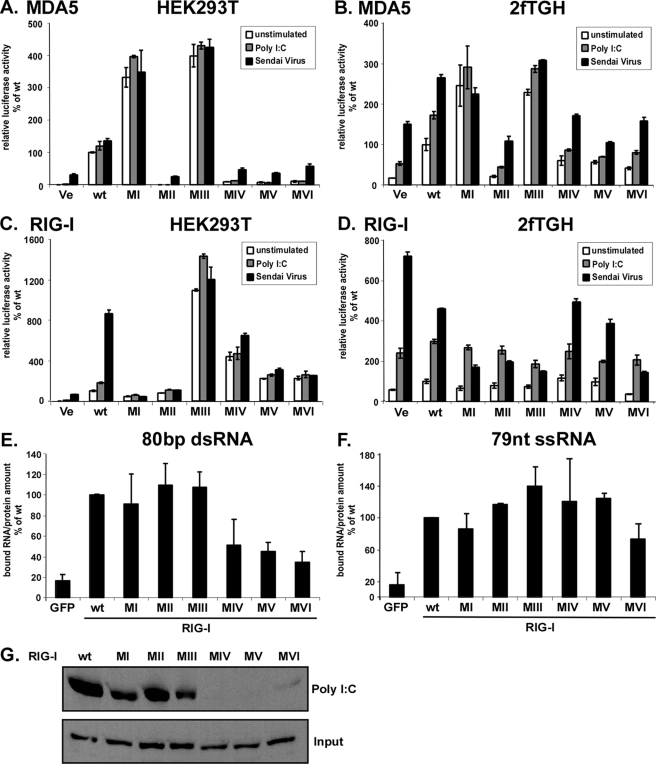
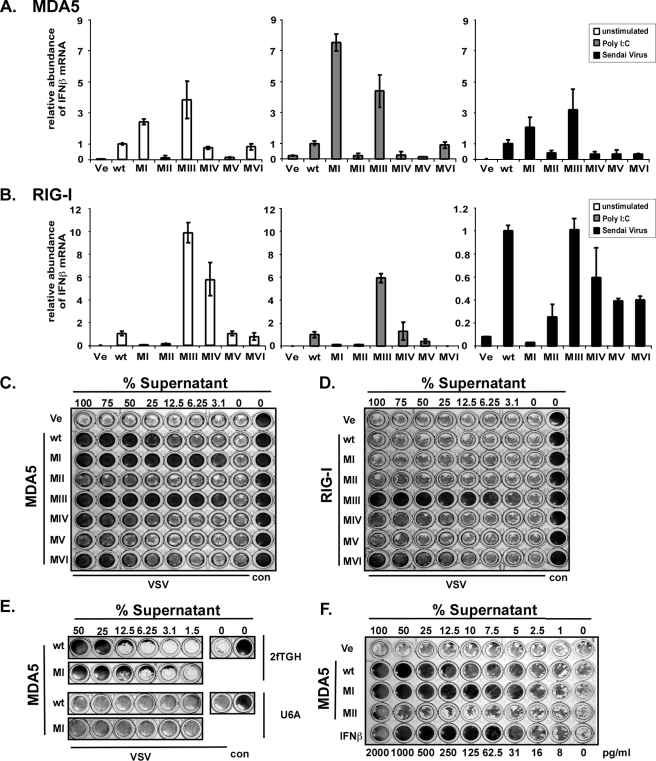
Signal Transduction by Mutant RIG-I and MDA5 Proteins—RNA sensing by MDA5 and RIG-I results in IFNβ gene expression. Targeted gene disruption in mice has indicated that poly(I-C) is primarily detected by MDA5, whereas RIG-I is required for detection of diverse viruses, including Sendai virus (11). It has been demonstrated previously that Sendai virus Cantell strain preparations are prone to defective interfering particles that can strongly activate the RIG-I system (37, 38). Irrespective of their in vivo specificity, however, it has been observed that in cell culture models, both helicases can be activated by poly(I-C) and Sendai virus (3, 21). Both of these stimuli were used to test the ability of the helicase wild type and mutants to induce the expression of an IFNβ promoter luciferase reporter gene (Fig. 3). We and others (15, 39) observe that HEK293T cells have a very low endogenous response to both poly(I-C) and Sendai virus in reporter gene assays. This response can be greatly elevated by ectopic expression of MDA5 (Fig. 3A) or RIG-I (Fig. 3C). As observed previously (19), expression of wild type MDA5 results in reporter gene activity, irrespective of stimulation, but the auto-regulated RIG-I does not activate the reporter gene in the absence of ligand stimulation. Our analysis of mutant proteins revealed that four MDA5 mutants, with mutations in motifs II or deletions in motifs IV, V and VI, were inactive for reporter gene transcription, resulting in similar profiles as the empty vector control. Two other mutants revealed an unexpected phenotype. Mutations to MDA5 motif I and motif III resulted in strongly induced reporter gene expression in the absence of stimulation, despite their inability to hydrolyze ATP. This strong constitutive activity was more than 3-fold compared with MDA5 wild type, even following poly(I-C) or Sendai virus stimulation (Fig. 3A). The observed differences were not because of differences in expression as evaluated by immunoblot analysis and titration experiments (data not shown).
FIGURE 3.
Signal transduction by MDA5 and RIG-I proteins and RIG-I RNA binding. Constitutive and inducible activation of the IFNβ promoter luciferase reporter gene by expression of the wild type and mutant MDA5 and RIG-I proteins. HEK293T (A and C) and 2fTGH (B and D) cells were transfected with luciferase reporter gene plasmids and expression vectors for helicase proteins MDA5 (A and B) or RIG-I (C and D). Parallel samples were left unstimulated (white) or were transfected with 5 μg/ml poly(I-C) (gray), or infected with 3 × 106 PFU Sendai virus (black), strain Cantell, for 6 h prior to lysis and luciferase assay. Values are normalized to the unstimulated wild type protein activity for each experiment. Representative experiments of four independent experiments are shown. Error bars depict standard deviation of triplicate samples. E–G, RNA binding by RIG-I wild type and mutants. E and F, RIG-I interaction with short RNA molecules. RIG-I wild type and mutant proteins were purified from HEK293T cells by immunoprecipitation with FLAG M2 affinity beads. Immobilized proteins were incubated with radioactively labeled 80-bp dsRNA (E) or 79-nt ssRNA (F) molecules and washed extensively. GFP, green fluorescent protein. Retained radioactivity was measured by scintillation counting, and counts/min are displayed as percent of wild type normalized to the total protein in each sample. Values are averaged from two independent experiments. G, HEK293T cell lysate expressing RIG-I wild type or mutant protein were incubated with poly(I-C)-coated agarose beads and analyzed by immunoblot with FLAG tag-specific antiserum. Ve, vector control; wt, wild type.
The expression of wild type RIG-I results in a small increase in reporter gene activation in the absence of stimulation, but both stimuli, in particular Sendai virus infection, resulted in enhanced luciferase activity (Fig. 3C). Motif I and II mutants were both inactive, in agreement with data reported previously (3, 21, 34), which was interpreted to suggest that the ATPase is required for RIG-I signaling. In contrast, mutation to RIG-I motif III produces a constitutively active protein. Stimulation with poly(I-C) slightly increased the activity of the motif III mutant. Deletions of RIG-I motifs IV, V, and VI produced intermediate phenotypes, with a basal activity higher than the unstimulated wild type protein. The activity of these mutants was not significantly enhanced by RNA or virus stimulation. Together, these data indicate that enzymatic activity of either MDA5 or RIG-I is not strictly required for antiviral signal transduction.
It has been demonstrated that expression of a RIG-I motif I mutant acts as a dominant negative inhibitor of endogenous RIG-I signaling (21). To verify the phenotypes observed for the motif mutants and test for dominant negative activities, similar luciferase reporter gene analysis was performed in the human fibrosarcoma cell line, 2fTGH. In contrast to HEK293T cells, 2fTGH exhibit a strong endogenous response to both poly(I-C) transfection and Sendai virus infection (Fig. 3, B and D). In these cells, the MDA5 mutants behaved similarly as in HEK293T cells; motifs I and III were found to be constitutively active, and other mutants were defective. None of the inactive MDA5 mutants behaved as dominant negative inhibitors of either poly(I-C) or Sendai virus-mediated signaling (Fig. 3B).
The RIG-I mutants exhibited a different phenotype in the signaling-competent 2fTGH cells. None of the mutants resulted in constitutive signaling activity, and although they did not have an appreciable effect on poly(I-C)-mediated reporter gene activation, Sendai virus stimulation was blocked in a dominant negative fashion by four of the mutants, including motifs I–III and VI. Proteins with deletions of motif IV or motif V did not substantially differ from wild type RIG-I in their response to Sendai virus (Fig. 3D).
Helicase Domain 2 Is Involved in RIG-I dsRNA Binding—RIG-I has been well described as an RNA binding molecule with stronger affinity to tested RNAs than MDA5 (11, 13–16, 19, 21). In addition to the helicase domain, the C-terminal regulatory domain of RIG-I was identified as an RNA-binding site accommodating 5′-triphosphate ends. The CTD alone can bind to 5′-triphosphorylated RNA molecules but with lower affinity than the full-length molecule (15, 16). The contributions of individual helicase domain motifs toward RIG-I RNA interaction were tested. RIG-I wild type and mutant proteins were evaluated for interaction with immobilized poly(I-C) or in vitro transcribed double-stranded (ds) or single-stranded (ss) RNA molecules in solution. Results indicate that deletions of motif IV, V, or VI impair binding to poly(I-C) or the 80-bp dsRNA, whereas binding of a 79-nt ssRNA molecule is largely unaffected by the mutations (Fig. 3, E–G).
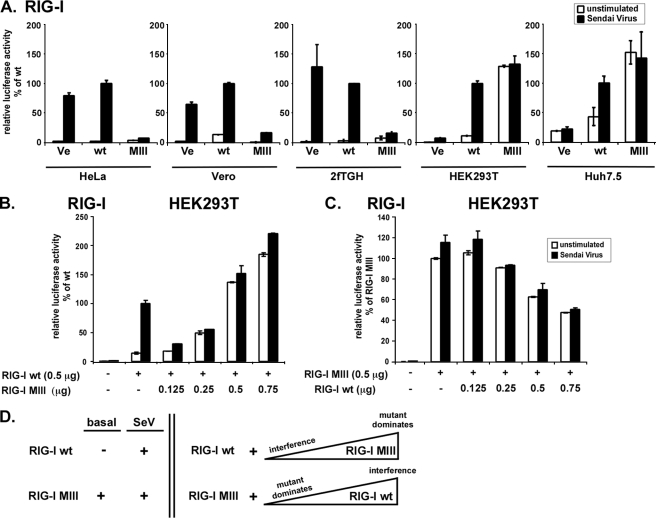
RIG-I Motif III Mutant Signaling Correlates with Endogenous Signaling Response and Interferes with RIG-I Wild Type Signaling—The differential activity of the RIG-I motif III mutant in the two different cell lines HEK293T and 2fTGH prompted further analysis of this mutant in several additional cell lines, including HeLa, Vero, and Huh 7.5. The RIG-I motif III mutant interfered with endogenous responses in 2fTGH, HeLa, and Vero lines but was constitutively active in HEK293T and Huh 7.5 (Fig. 4A). These two subsets of cells can also be characterized by their degree of endogenous RIG-I responses. 2fTGH, HeLa, and Vero all have strong endogenous RIG-I signaling, but HEK293T and Huh 7.5 lack endogenous RIG-I responses (15, 39, 40). Thus, the behavior of the expressed RIG-I motif III mutant reflects the endogenous signaling profile of the recipient cell line and exerts a dominant negative effect in RIG-I signaling-competent context but a constitutively active effect in signaling-deficient context.
FIGURE 4.
Differential activity of RIG-I motif III mutant reflects endogenous RIG-I signaling. A, luciferase reporter gene assay assessing RIG-I motif III mutant (MIII) reporter gene signaling activity in HeLa, Vero, 2fTGH, HEK293T, and Huh 7.5 cells. Cells were transfected with IFNβ promotor luciferase reporter gene plasmids and expression vectors for RIG-I (wt) or mutant. B and C, titration of RIG-I wild type and RIG-I motif III mutant. HEK293T cells were transfected with IFNβ reporter gene plasmids, 0.5 μg of RIG-I wild type expression vector, and increasing amounts of RIG-I motif III mutant expression vector (B). 0.5 μg of RIG-I motif III mutant expression vector and increasing amounts of RIG-I wild type expression vector were used in a similar assay (C). Each transfection reaction contained 1.25 μg of total DNA, using empty vector plasmid DNA for all assays. Parallel samples were left unstimulated (white) or infected with 3 × 106 PFU Sendai virus (black), strain Cantell, for 6 h prior to lysis and luciferase assay. Values are normalized to the stimulated wild type sample. Representative data for at least two independent experiments are shown. Error bars depict standard deviation of triplicate samples. Ve, vector control; wt, wild type. D, model scheme summarizing the apparent cross-talk between RIG-I (wt) and RIG-I motif III mutant (MIII) signaling activity when expressed alone (left) in absence (basal) or presence of Sendai virus (SeV), or when co-expressed in a titration experiment (right). Triangles indicate increasing abundance. For further explanation see text.
To test more directly the concept that the mutant protein influences wild type RIG-I signaling, a titration experiment was performed. HEK293T cells, which have minimal endogenous signaling responses, were transfected with a constant amount of RIG-I expression vector with or without increasing amounts of expression vector for RIG-I motif III mutant (Fig. 4B). Expression of the lowest amount of mutant protein interferes with RIG-I-mediated inducible signaling, but as the level of the mutant protein increases, its constitutive signaling property overwhelms the inducible activity, consistent with saturation of wild type RIG-I signaling. In a complementary experiment, titration of wild type RIG-I decreases the constitutive activity of the motif III mutant in a dose-dependent manner (Fig. 4C). These data demonstrate that the two RIG-I proteins influence each other's activity depending on their relative abundance (Fig. 4D).
Constitutive Antiviral Activity of MDA5 and RIG-I Mutants Results from IFN—To confirm the signaling ability of the mutant proteins, we measured the endogenous IFNβ response and antiviral strength of the helicase mutants. The level of IFNβ mRNA was measured in transfected HEK293T cells using real time RT-PCR (Fig. 5, A and B). Consistent with the luciferase assays, expression of MDA5 motif I or motif III mutant resulted in elevated IFNβ mRNA synthesis compared with MDA5 wild type in untreated, poly(I-C)-transfected, or Sendai virus-infected conditions (Fig. 5A). Analysis of the RIG-I mutants also confirmed the reporter gene data in HEK293T cells. Mutation to RIG-I motif III or deletion of motif IV activates the protein constitutively, and motif V and motif VI deletion mutants display activity similar to wild type RIG-I, and motif I and motif II are unable to promote transcription of IFNβ mRNA (Fig. 5B). A similar mutant RIG-I activity profile was observed after intracellular poly(I-C) stimulation. All RIG-I proteins except motif I mutant were stimulated by Sendai virus infection with the strongest activity for RIG-I wild type and motif III mutant.
FIGURE 5.
Constitutive IFNβ transcription and antiviral activity by mutant MDA5 and RIG-I. MDA5 (A) and RIG-I (B) wild type and mutant protein effect on IFNβ mRNA levels were measured by real time RT-PCR in HEK293T cells. Relative IFNβ mRNA abundance was measured in unstimulated cells (white) or in cells stimulated by 5 μg/ml poly(I-C) transfection (gray) or infection with 3 × 106 PFU Sendai virus (black) for 6 h prior to RNA extraction. IFNβ mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA, and data represent relative activity normalized to the wild type helicase protein under each condition. Representative data of three independent experiments are shown, and error bars represent standard deviation of duplicate samples. To measure antiviral activity, HEK293T cells expressing wild type or mutant MDA5 (C) or RIG-I (D) proteins were incubated for 36 h before the supernatant was diluted and transferred to freshly plated 2fTGH cells. After 8 h of incubation, the 2fTGH cells were infected with 6 × 103 PFU of VSV (Indiana strain). Cells were fixed 18 h post-infection and stained with methylene blue in 50% ethanol. E, IFN receptor signaling is required for antiviral responses. Data are similar to C but the diluted supernatants were added to 2fTGH and U6A (STAT2-deficient) cells for comparison. F, estimation of antiviral strength of expressed MDA5 wild type and motif I by comparison with purified IFNβ. In this example, equivalent protection end points were observed for 12.5% MDA5 wild type supernatant, 5% MDA5 motif I supernatant, and 31 pg/ml in IFNβ. Ve, vector control; wt, wild type; con, control.
To determine the antiviral activities of the constitutive helicase mutants in a biologically meaningful context, an antiviral cytopathic effect protection assay was carried out. HEK293T cells were transfected for expression of MDA5 or RIG-I constructs, and 24 h later supernatant was collected, diluted serially, and added to 2fTGH cells. After 8 h of incubation, these cells were washed and infected with VSV. Cells were fixed and stained with methylene blue to evaluate protection from virus-induced cytopathicity. The MDA5 motif I and motif III mutants produced higher constitutive protection than the wild type, consistent with their signaling profiles (Fig. 5C). Similarly, mutation to RIG-I motif III also resulted in constitutive antiviral activity leading to protection from virus infection (Fig. 5D). These results validate the IFNβ gene expression data as biologically relevant to antiviral protection.
To formally determine whether the antiviral activity of the mutant helicases is because of IFN production and signaling, a 2fTGH cell derivative lacking STAT2 expression, U6A, was used in an antiviral protection assay (Fig. 5E). 2fTGH and U6A cells were incubated with supernatants from HEK293T cells expressing either wild type MDA5 or motif I mutant and then infected with VSV. The 2fTGH cells were protected from VSV cytopathicity, but the U6A were not protected, implicating the involvement of a STAT2-dependent antiviral pathway such as type I IFN signaling. To estimate the relative antiviral strength of this mutant protein, purified IFNβ was titrated for comparison (Fig. 5F). Although absolute values varied slightly between experiments, the achieved antiviral strength in the supernatants of transfected cells typically compares with about 250 pg/ml IFNβ for MDA5 wild type. From the experiment shown in Fig. 4F, the supernatant of MDA5 motif I mutant can be estimated to be equivalent to 620 pg/ml IFNβ, ∼2.5 times greater than wild type.
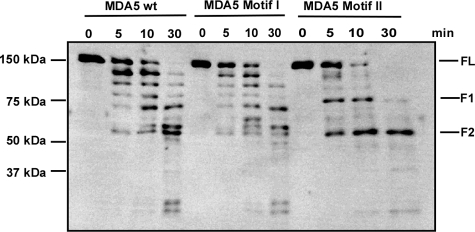
Distinct Conformation of Inactive MDA5 Protein—Recent models of RIG-I activation by RNA binding involve conformational changes that lead to exposure of the CARDs for interaction with downstream signaling components (15, 16, 19). We postulate that a conformational switch could also be the mechanism underlying the observed MDA5 activation or inactivation. To test this model, chymotrypsin was used to digest wild type MDA5, hyperactive mutant motif I, and inactive mutant motif II, which were designed into recombinant baculoviruses and purified from infected insect cells. Proteins were incubated with chymotrypsin and aliquots analyzed by immunoblot with an MDA5-specific antibody (Fig. 6). The fragmentation pattern of the two active proteins was nearly identical, suggesting similar conformations. In contrast, the inactive motif II mutant had a distinct digestion pattern yielding two unique sized fragments (F1 and F2), consistent with a difference in conformation from the active proteins.
FIGURE 6.
Differential protease sensitivity of active and inactive MDA5 variants. His6-tagged MDA5 wild type, motif I, and motif II mutant proteins were expressed in Sf9 insect cells by infection with recombinant baculovirus and affinity-purified. Proteins were subjected to limited digestion with chymotrypsin in a 1:1000 enzyme:substrate ratio for the indicated times. Samples were analyzed by immunoblot using MDA5-specific antiserum. FL, full-length MDA5; F1, fragment 1; F2, fragment 2.
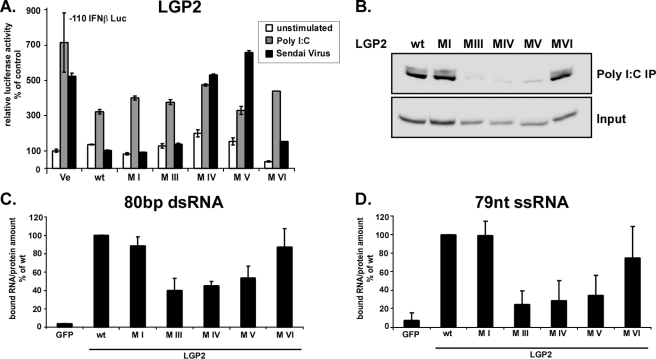
LGP2 Negative Regulation Is Independent of Enzymatic Activity and RNA Binding—Although the physiological role of LGP2 is not entirely understood, it is known that expression of LGP2 in cultured cells has the ability to negatively regulate antiviral signaling (19–22). The inhibition by LGP2 has been explained by RNA sequestration (21, 22), the action of a C-terminal repressor domain (19), or an inhibitory protein interaction with the signaling adaptor IPS-1 (20). We tested the ability of LGP2 helicase motif mutants to block IFN signaling. Wild type and mutant proteins were expressed in 2fTGH cells to determine their ability to repress the endogenous response to poly(I-C) or Sendai virus infection (Fig. 7A). Expression of LGP2 has only little effect on the response to poly(I-C) but strongly interferes with Sendai virus signaling. Mutations to LGP2 motifs I, III, and VI produced proteins that were still able to block antiviral responses similar to the wild type protein, but LGP2 with deletions to motif IV and V no longer suppressed signaling. The same observations were made when LGP2 mutants and wild type proteins were tested in HeLa cells (supplemental Fig. S1). Clearly the enzymatic activity of LGP2 is not required for its inhibitory action, but not all inactivating mutations retain negative regulatory activity.
FIGURE 7.
Negative regulation by LGP2 is independent of enzymatic activity and RNA binding. A, signaling interference reporter gene assay for LGP2. Human 2fTGH cells were transfected with plasmids coding for LGP2 or mutant proteins and luciferase reporters. After 24 h, cells were left unstimulated(white) or stimulated by transfection with 5 μg of poly(I-C) (gray) or infection with 6 × 103 PFU Sendai virus (black) for 6 h and assayed for IFNβ promoter luciferase reporter gene activity. B–D, RNA binding by LGP2 and mutants. B, HEK293T cell lysate expressing LGP2 wild type or mutant protein were incubated with poly(I-C)-coated agarose beads and analyzed by immunoblot with FLAG tag-specific antiserum. C and D, LGP2 interaction with short RNA molecules. LGP2 wild type and mutant proteins were purified from HEK293T cells by immunoprecipitation with FLAG M2 affinity beads. Immobilized proteins were incubated with radioactively labeled 80-bp dsRNA (C) or 79 nt of ssRNA (D) molecules and washed extensively. Retained radioactivity was measured by scintillation counting, and counts/min are displayed as percent of wild type normalized to the total protein in each sample. Values are averaged from two independent experiments.
We and others have observed that LGP2 is the strongest RNA binding molecule among these three helicases (19, 21). LGP2 can bind both single- and double-stranded RNA (27), but it is unclear whether the basic RNA binding pocket found in the RIG-I CTD is conserved and functional in LGP2 (15, 16). RNA binding activity of wild type and mutant LGP2 was tested for poly(I-C) bound to Sepharose beads and in vitro transcribed, 5′-triphosphorylated dsRNA or ssRNA in solution (Fig. 7, B–D). Results indicate that the mutations to motifs III and deletions of IV and V impair LGP2 RNA binding ability. One of these proteins, with mutation to motif III, retained the ability to act as a negative regulator. The phenotype of this mutant indicates that RNA sequestration is unlikely to be a significant mode of interference by LGP2, but it instead supports the two non-exclusive protein interaction mechanisms for LGP2-negative regulation (19, 20).
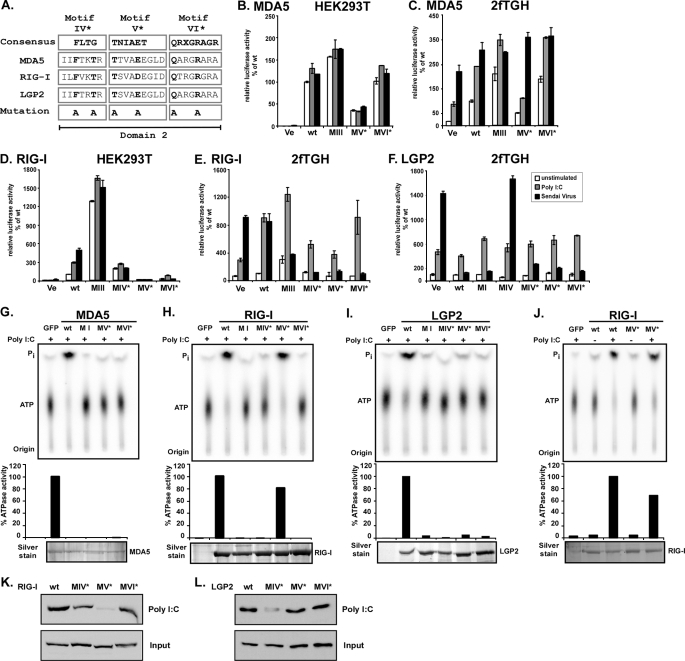
Point Mutations in Motif IV–VI Confirm Independence of Enzymatic Activity and Antiviral Signaling—The dramatic consequences of some of the deletion mutants may result from severe structural alterations. To more carefully examine these motifs, we designed additional point mutants to create more subtle deficiencies for MDA5 motif V and VI and RIG-I and LGP2 motif IV, V, and VI. Two conserved residues in each motif were targeted by alanine substitutions (Fig. 8A, indicated by *). All proteins were expressed and tested for ATP hydrolysis (Fig. 8, G–J). The point mutations in all three motifs of MDA5 and LGP2 extinguished enzymatic activity, as did the RIG-I mutants in motifs IV and VI. Point mutants to RIG-I motif V (motif V*) retained RNA-dependent ATP hydrolysis activity.
FIGURE 8.
ATP hydrolysis and antiviral signaling of point mutations in helicase domain 2. A, point mutations to helicase motifs IV–VI. The DEXH box family domain 2 consensus sequences (33) are aligned with corresponding sequences of MDA5, RIG-I, and LGP2. X indicates no amino acid preference at this position. Residues targeted for mutation are indicated in boldface type and were substituted with alanine (A). Introduced mutations are identical for all three helicases. B–F, constitutive and inducible activation of the IFNβ promoter luciferase reporter gene by expression of the wild type and mutant MDA5, RIG-I, and LGP2 proteins. HEK293T (B and D) and 2fTGH (C, E and F) cells were transfected with luciferase reporter gene plasmids and expression vectors for helicase proteins MDA5 (B and C) or RIG-I (D and E) or LGP2 (F). Parallel samples were left unstimulated (white) or were transfected with 5 μg/ml poly(I-C) (gray) or infected with 3 × 106 PFU Sendai virus (black), strain Cantell, for 6 h prior to lysis and luciferase assay. Values are normalized to the unstimulated wild type protein activity (B–E) or vector control (F) for each experiment. Representative experiments of two independent experiments are shown; error bars depict standard deviation of triplicate samples. Ve, vector control; wt, wild type. G–J, FLAG-tagged wild type and mutant MDA5 (G), RIG-I (H and J), and LGP2 (I) proteins were expressed in HEK293T cells and immunoaffinity-purified with FLAG M2 affinity gel. Eluted proteins were incubated with [γ-32P]ATP in the presence or absence (J) of poly(I-C) and separated by thin layer chromatography (top). Origin and migration of free phosphate (Pi) and ATP are indicated. ATP hydrolysis activity was quantified by phosphorimage analysis and plotted as percentages of wild type protein activity (center). Bottom panels show silver-stained SDS-PAGE demonstrating similar amounts of purified proteins in reactions. K and L, HEK293T cell lysate expressing FLAG-tagged RIG-I (K) or LGP2 (L) wild type or mutant protein were incubated with poly(I-C)-coated agarose beads and analyzed by immunoblot with FLAG tag-specific antiserum. GFP, green fluorescent protein.
In signal transduction assays, one of the MDA5 point mutants, MDA5 motif V*, was inactive, but the other MDA5 motif VI* was found to constitutively mediate signaling to the IFNβ promoter despite its lack of ATPase activity, producing a similar phenotype to the hyperactive MDA5 mutants motif I and motif III.
For RIG-I, both the enzymatically defective motif IV* and motif VI* mutants and the enzymatically intact mutant motif V* were unable to induce IFNβ promoter activity in HEK293T cells, but exhibited dominant negative interference with endogenous Sendai virus-induced antiviral signaling in 2fTGH cells (Fig. 8, D and E). This finding reinforces the independence of antiviral signaling and enzymatic activity. The point mutants in LGP2 domain 2 all retained the ability to efficiently block Sendai virus-induced IFNβ signaling (Fig. 8F), indicating that the observed lack of interference for deletion mutants in motif IV and V is the result of major structural changes that interfere with LGP2-inhibiting action.
When tested in poly(I-C) pulldown assays, RIG-I motif IV* and VI* mutants were slightly diminished in RNA binding compared with the wild type protein (Fig. 8K). Surprisingly, the RIG-I motif V* mutant that retained RNA-dependent ATP hydrolysis was not observed to form a stable RNA complex in this assay (Fig. 8K) or in solution RNA binding assays (data not shown). In contrast, LGP2 motif V* and motif VI* mutants bound poly(I-C) comparable with the wild type protein, but point mutations to LGP2 motif IV (motif IV*) were impaired for poly(I-C) binding (Fig. 8L).
DISCUSSION
The identification of cytoplasmic receptors for virus recognition that harbor DEXH box RNA helicase domains suggested a role for their intrinsic enzymatic activity in innate immune responses to infection. A systematic mutagenesis approach was used to investigate the importance of enzymatic activity of the helicase domain in RNA recognition, signal transduction, and antiviral functions of MDA5, RIG-I, and LGP2. Mutations targeting the six major RNA helicase motifs resulted in dramatic impairment of ATP hydrolysis activity for all three proteins, clearly implicating the defined motifs as part of the catalytic cores for these proteins. Despite their lack of enzymatic activities, several of the variant proteins retained or enhanced the ability to mediate signal transduction and antiviral responses.
For MDA5, mutation to either motif I or motif III resulted in proteins that retain their ability to induce the innate antiviral signaling response and were found to be hyperactive compared with the wild type protein. Expression of these mutants resulted in stimulation of IFNβ gene expression, and higher than normal basal antiviral responses, which are due to initiation of type I IFN signaling that is STAT2-dependent. Titration of the antiviral activity stimulated by the expressed MDA5 motif I mutant indicated an antiviral response comparable with that produced by about 600 pg/ml purified IFNβ. It is curious that an identical MDA5 motif I mutation was previously reported to have no influence on IFNβ reporter gene activation in L929 cells following infection with Newcastle disease virus (21). Despite this published finding, we have observed that our MDA5 motif I mutant is constitutively active for signaling in all the cell lines we tested, including L929 (supplemental Fig. S2) and HeLa (data not shown). Mutations to motif II or IV and deletions in motifs V and VI resulted in inactive MDA5 proteins that were inert for antiviral signaling. However, point mutations in MDA5 motif VI (motif VI*) also abrogated ATP hydrolysis but retained signaling capacity. None of the MDA5 mutants produced dominant negative effects, which is in contrast to what is observed for RIG-I mutants (3, 19, 21, 23) and suggests differences in interaction between the ectopically expressed mutants and endogenous signaling components. Differences in the RD region of the MDA5 C terminus might also explain the absence of auto- or trans-inhibition. We speculate that MDA5 has distinct conformations that are either active or inactive for antiviral signaling. In this scenario, the wild type protein could achieve equilibrium between the two conformations, resulting in basal signaling, but the engagement of ligand shifts the equilibrium toward the active conformation. The hyperactive mutants may represent stabilized active conformations, whereas inert mutants represent inactive forms. This model is supported by proteolysis experiments where it was found that the inactive MDA5 motif II mutant exhibits a distinct protease sensitivity compared with the two constitutively active proteins, MDA5 wild type and motif I mutant.
Constitutive hyperactivity was also observed with expression of the RIG-I motif III mutant, but the absolute effect of this protein differed among cell lines in a pattern that correlated with the endogenous RIG-I signaling capacity. In the case of HeLa, Vero, and 2fTGH cells, signaling by the endogenous RIG-I system is readily detected in a reporter gene assay, and the motif III mutant RIG-I protein acts as a dominant negative inhibitor of the endogenous signaling response. The same experiment carried out in HEK293T or Huh 7.5 cells, both of which lack robust endogenous RIG-I signaling, indicates that the motif III mutant RIG-I protein is constitutively active. Coexpression experiments confirmed the effects of RIG-I motif III mutant on wild type protein in a controlled experiment. When expressed alone, RIG-I exhibits inducible activity, and RIG-I motif III mutant is constitutively active. Coexpression results in dose-dependent interference with each other. Expression of RIG-I along with modest levels of RIG-I motif III mutant leads to interference with the inducible signaling, but higher concentration of the mutant protein results in dominant constitutive signaling. Conversely, constitutive signaling by RIG-I motif III mutant is observed with low expression of RIG-I, but high expression levels of RIG-I interfere with the constitutive activity of RIG-I motif III mutant. Thus, RIG-I diminishes the constitutive activity of the mutant in a dose-dependent manner, and the mutant interferes with inducible signaling by wild type RIG-I (Fig. 4D).
We speculate that this duality of function reflects the continued ability of motif III mutant to interact with and activate endogenous downstream signaling molecules, which results in the observed constitutive signaling in the absence of the intact endogenous RIG-I system. When expressed in the context of an intact endogenous RIG-I signaling system, the mutant interferes with normal signaling, possibly by hetero-oligomerization with the endogenous RIG-I or other downstream components of the signaling complex.
Another interesting RIG-I mutant is the point mutation to motif V, RIG-I motif V*, which retained ATP hydrolysis activity but is unable to mediate antiviral signaling and exhibited a dominant negative phenotype to Sendai virus-induced IFNβ transcription. This finding further supports a model in which enzymatic activity is independent of antiviral signaling.
In contrast to the positive-acting MDA5 and RIG-I, expression of LGP2 in cells results in inhibition of IFN antiviral responses (19–22). Several mechanisms have been proposed to account for the feedback inhibition functions of LGP2, including RNA sequestration (21, 22), heterodimer formation between LGP2 and RIG-I via the C-terminal regulatory domain (19), and protein-protein interaction between LGP2 and IPS-1 (20). Irrespective of the precise mechanism of LGP2 action, results demonstrate that the LGP2 inhibitory function is entirely independent of enzymatic activity, as demonstrated by mutant proteins that can still inhibit the virus-induced IFNβ transcriptional response despite loss of ATP hydrolysis activity. Mutations to LGP2 motifs I, III, deletion of motif VI, and point mutations to motif IV, V, and VI (IV*, V*, and VI*) result in proteins with negative regulatory capacity similar to the wild type LGP2. Mutation to motif III as well as deletion of motif IV and V resulted in proteins that are strongly impaired for binding to single- and double-stranded RNAs. In addition, LGP2 motif IV* mutant was also impaired in poly(I-C) binding. Taken together, mutation to LGP2 motif III and point mutations to motif IV (motif IV*) diminished RNA binding ability yet retained the ability to successfully inhibit IFNβ transcriptional responses induced by virus infection. This result demonstrates that RNA binding is dispensable for signaling inhibition. The other two RNA-binding defective mutants, deletions to LGP2 motif IV and V, are also defective in attenuation of antiviral signaling. In contrast, point mutants in these motifs (LGP2 motif IV* and motif V*) lack ATPase activity but retain signaling inhibition. This difference strongly supports that the deletions result in local protein misfolding creating a more severe defect.
Biochemical analysis of the three antiviral helicases revealed a number of differences concerning the involvement of conserved motifs in substrate interaction. Differences were observed in the RNA binding behaviors of RIG-I and LGP2 mutants. Deletions of motifs IV, V, and VI in RIG-I disrupted interactions with RNA, but the same deletion in LGP2 motif VI did not affect RNA binding. Instead, LGP2 motif III seemed to be required for the interaction with RNA, but RIG-I motif III mutant retained binding activity for short single- or double-stranded RNAs and was similar in the ability to bind to immobilized poly(I-C), although the activity was decreased compared with wild type RIG-I, in agreement with previous reports (16). More subtle point mutations revealed motif IV of LGP2 (motif IV*) to be important for RNA interaction. This is in agreement with previous findings that describe the phenylalanine residue of motif IV, one of the residues targeted in our mutant, as crucial for RNA binding and ATP hydrolysis in several yeast superfamily 2 helicases (41). For RIG-I, motif IV and motif VI mutants retain partial RNA interaction, but the motif V point mutant (motif V*) failed to bind poly(I-C). Curiously, this mutation of two highly conserved residues in motif V did not affect the RNA-induced ATPase function in RIG-I, but the exact same mutations in MDA5 and LGP2 disrupted ATP hydrolysis activity. Previous reports describing mutations in motif V of other DEXH box helicases, such as the yeast splicing factor Prp22 and the hepatitis C virus RNA helicase NS3, suggest the targeted threonine residue plays a role in linking ATP hydrolysis to the RNA cofactor (42, 43). It is possible that RIG-I motif V* has a more transient RNA interaction than the wild type protein, explaining the RNA-dependent ATPase, despite severely impaired RNA interaction in poly(I-C) pulldown assays. More detailed kinetic analysis of wild type and mutant protein will be required to identify the exact mechanism involved in RIG-I coupling of ATPase and RNA binding, but these observations highlighted several structure-function differences between RIG-I and LGP2.
In summary, a biochemical analysis of antiviral RNA helicase function in the innate immune response has identified conserved helicase domain sequence motifs that are essential to the enzymatic core and indispensable for ATP hydrolysis. Nonetheless, the proteins characterized in this study have dramatically diverse antiviral signaling properties, resulting in both constitutive or hyper-activated as well as dominant negative helicase protein variants. In addition, these proteins provide tools for further investigation of the mammalian cytoplasmic RNA recognition system.
Supplementary Material
Acknowledgments
We thank members of the Horvath Lab for critical evaluation and comments on the manuscript, Akihiko Komuro for generating LGP2 cDNA and the K30A mutant, and Michael Gale, Jr. (Washington University), and Charles Rice (Rockefeller University) for providing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant AI073919 (to C. M. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: IFN, type I interferon; IPS-1, interferon-β promoter stimulator-1; CARD caspase activation and recruitment domain; dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; nt, nucleotides; VSV, vesicular stomatitis virus; PFU, plaque-forming unit; MOPS, 4-morpholinepropanesulfonic acid; RT, reverse transcription; CTD, C-terminal domain.
References
- 1.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730–737 [DOI] [PubMed] [Google Scholar]
- 4.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981–988 [DOI] [PubMed] [Google Scholar]
- 5.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, J. (2005) Nature 437 1167–1172 [DOI] [PubMed] [Google Scholar]
- 6.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669–682 [DOI] [PubMed] [Google Scholar]
- 7.Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005) Mol. Cell 19 727–740 [DOI] [PubMed] [Google Scholar]
- 8.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2006) Nature 439 208–211 [DOI] [PubMed] [Google Scholar]
- 9.Saha, S. K., Pietras, E. M., He, J. Q., Kang, J. R., Liu, S. Y., Oganesyan, G., Shahangian, A., Zarnegar, B., Shiba, T. L., Wang, Y., and Cheng, G. (2006) EMBO J. 25 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gitlin, L., Barchet, W., Gilfillan, S., Cella, M., Beutler, B., Flavell, R. A., Diamond, M. S., and Colonna, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101–105 [DOI] [PubMed] [Google Scholar]
- 12.Loo, Y. M., Fornek, J., Crochet, N., Bajwa, G., Perwitasari, O., Martinez-Sobrido, L., Akira, S., Gill, M. A., Garcia-Sastre, A., Katze, M. G., and Gale, M., Jr. (2008) J. Virol. 82 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994–997 [DOI] [PubMed] [Google Scholar]
- 14.Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., and Reis e Sousa, C. (2006) Science 314 997–1001 [DOI] [PubMed] [Google Scholar]
- 15.Cui, S., Eisenacher, K., Kirchhofer, A., Brzozka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K. K., Krug, A., and Hopfner, K. P. (2008) Mol. Cell 29 169–179 [DOI] [PubMed] [Google Scholar]
- 16.Takahasi, K., Yoneyama, M., Nishihori, T., Hirai, R., Kumeta, H., Narita, R., Gale, M., Jr., Inagaki, F., and Fujita, T. (2008) Mol. Cell 29 428–440 [DOI] [PubMed] [Google Scholar]
- 17.Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., Hiiragi, A., Dermody, T. S., Fujita, T., and Akira, S. (2008) J. Exp. Med. 205 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito, T., Owen, D. M., Jiang, F., Marcotrigiano, J., and Gale, M., Jr. (2008) Nature 454 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito, T., Hirai, R., Loo, Y. M., Owen, D., Johnson, C. L., Sinha, S. C., Akira, S., Fujita, T., and Gale, M., Jr. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komuro, A., and Horvath, C. M. (2006) J. Virol. 80 12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y. M., Gale, M., Jr., Akira, S., Yonehara, S., Kato, A., and Fujita, T. (2005) J. Immunol. 175 2851–2858 [DOI] [PubMed] [Google Scholar]
- 22.Rothenfusser, S., Goutagny, N., DiPerna, G., Gong, M., Monks, B. G., Schoenemeyer, A., Yamamoto, M., Akira, S., and Fitzgerald, K. A. (2005) J. Immunol. 175 5260–5268 [DOI] [PubMed] [Google Scholar]
- 23.Venkataraman, T., Valdes, M., Elsby, R., Kakuta, S., Caceres, G., Saijo, S., Iwakura, Y., and Barber, G. N. (2007) J. Immunol. 178 6444–6455 [DOI] [PubMed] [Google Scholar]
- 24.Gorbalenya, A. E., and Koonin, E. V. (1993) Curr. Opin. Struct. Biol. 3 419–429 [Google Scholar]
- 25.Cordin, O., Banroques, J., Tanner, N. K., and Linder, P. (2006) Gene (Amst.) 367 17–37 [DOI] [PubMed] [Google Scholar]
- 26.Caruthers, J. M., and McKay, D. B. (2002) Curr. Opin. Struct. Biol. 12 123–133 [DOI] [PubMed] [Google Scholar]
- 27.Murali, A., Li, X., Ranjith-Kumar, C. T., Bhardwaj, K., Holzenburg, A., Li, P., and Kao, C. C. (2008) J. Biol. Chem. 283 15825–15833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J. (1982) EMBO J. 1 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner, N. K., and Linder, P. (2001) Mol. Cell 8 251–262 [DOI] [PubMed] [Google Scholar]
- 30.Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S., and Yokoyama, S. (2006) Cell 125 287–300 [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. L., Morgenstern, K. A., Griffith, J. P., Dwyer, M. D., Thomson, J. A., Murcko, M. A., Lin, C., and Caron, P. R. (1998) Structure (Lond.) 6 89–100 [DOI] [PubMed] [Google Scholar]
- 32.Ausubel, F. M., B. R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (eds) (1994) Current Protocols in Molecular Biology, pp. 9.1.4–9.1.11, John Wiley and Sons, Inc., New York
- 33.Abdelhaleem, M., Maltais, L., and Wain, H. (2003) Genomics 81 618–622 [DOI] [PubMed] [Google Scholar]
- 34.Gee, P., Chua, P. K., Gevorkyan, J., Klumpp, K., Najera, I., Swinney, D. C., and Deval, J. (2008) J. Biol. Chem. 283 9488–9496 [DOI] [PubMed] [Google Scholar]
- 35.Kang, D. C., Gopalkrishnan, R. V., Wu, Q., Jankowsky, E., Pyle, A. M., and Fisher, P. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacsovics, M., Martinon, F., Micheau, O., Bodmer, J. L., Hofmann, K., and Tschopp, J. (2002) Curr. Biol. 12 838–843 [DOI] [PubMed] [Google Scholar]
- 37.Strahle, L., Garcin, D., and Kolakofsky, D. (2006) Virology 351 101–111 [DOI] [PubMed] [Google Scholar]
- 38.Yount, J. S., Kraus, T. A., Horvath, C. M., Moran, T. M., and Lopez, C. B. (2006) J. Immunol. 177 4503–4513 [DOI] [PubMed] [Google Scholar]
- 39.Joo, C. H., Shin, Y. C., Gack, M., Wu, L., Levy, D., and Jung, J. U. (2007) J. Virol. 81 8282–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumpter, R., Jr., Loo, Y. M., Foy, E., Li, K., Yoneyama, M., Fujita, T., Lemon, S. M., and Gale, M., Jr. (2005) J. Virol. 79 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banroques, J., Cordin, O., Doere, M., Linder, P., and Tanner, N. K. (2008) Mol. Cell. Biol. 28 3359–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider, S., Campodonico, E., and Schwer, B. (2004) J. Biol. Chem. 279 8617–8626 [DOI] [PubMed] [Google Scholar]
- 43.Lin, C., and Kim, J. L. (1999) J. Virol. 73 8798–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.