Abstract
The shikimate pathway enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) is the target of the broad spectrum herbicide glyphosate. The genetic engineering of EPSPS led to the introduction of glyphosate-resistant crops worldwide. The genetically engineered corn lines NK603 and GA21 carry distinct EPSPS enzymes. CP4 EPSPS, expressed in NK603 corn and transgenic soybean, cotton, and canola, belongs to class II EPSPS, glyphosate-insensitive variants of this enzyme isolated from certain Gram-positive bacteria. GA21 corn, on the other hand, was created by point mutations of class I EPSPS, such as the enzymes from Zea mays or Escherichia coli, which are sensitive to low glyphosate concentrations. The structural basis of the glyphosate resistance resulting from these point mutations has remained obscure. We studied the kinetic and structural effects of the T97I/P101S double mutation, the molecular basis for GA21 corn, using EPSPS from E. coli. The T97I/P101S enzyme is essentially insensitive to glyphosate (Ki = 2.4 mm) but maintains high affinity for the substrate phosphoenolpyruvate (PEP) (Km = 0.1 mm). The crystal structure at 1.7-Å resolution revealed that the dual mutation causes a shift of residue Gly96 toward the glyphosate binding site, impairing efficient binding of glyphosate, while the side chain of Ile97 points away from the substrate binding site, facilitating PEP utilization. The single site T97I mutation renders the enzyme sensitive to glyphosate and causes a substantial decrease in the affinity for PEP. Thus, only the concomitant mutations of Thr97 and Pro101 induce the conformational changes necessary to produce catalytically efficient, glyphosate-resistant class I EPSPS.
Glyphosate (N-phosphonomethylglycine) is a potent inhibitor of the shikimate pathway in plants, specifically targeting the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS,3 EC 2.5.1.19) (1). Glyphosate-based formulations exhibit broad spectrum herbicidal activity with minimal human and environmental toxicity (2, 3). The safety and efficacy of glyphosate, together with the existence of genetically modified, glyphosate-resistant crop varieties (4, 5), have combined to make glyphosate the most used herbicide in the world. Enzymes of the shikimate pathway are also regarded as attractive antimicrobial targets (6–9).
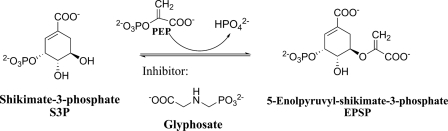
EPSPS catalyzes the transfer of the enolpyruvyl moiety of phosphoenolpyruvate (PEP) to the 5-hydroxy position of shikimate-3-phosphate (S3P) (Fig. 1). Binding of the first substrate, S3P, to the enzyme triggers a global conformational change from an “open” to a “closed” conformation. PEP and glyphosate bind in the active site, formed at the interface between the N- and C-terminal globular domains. Glyphosate inhibition is competitive with respect to PEP (10, 11), and structural studies confirmed that glyphosate occupies the PEP-binding site (12–15).
FIGURE 1.
Reaction catalyzed by EPSPS.
EPSPS from different organisms have been divided into two classes according to intrinsic glyphosate sensitivity: in Class I enzymes, found in all plants and in bacteria such as Escherichia coli and Salmonella typhimurium, catalytic activity is inhibited at low micromolar concentrations of glyphosate (16). Class II enzymes, found in bacterial species, including Staphylococcus aureus, Streptococcus pneumoniae, and Agrobacterium sp. strain CP4, are distinguished by their ability to sustain efficient catalysis in the presence of high glyphosate concentrations (5, 15–18).
Glyphosate insensitivity has been achieved in Class I EPSPS enzymes through natural selection, directed evolution, and site-directed mutagenesis. As suggested by the fact that glyphosate and PEP bind to the same site, EPSPS mutants with substantially decreased affinity for the inhibitor glyphosate typically also exhibited decreased affinity for the substrate PEP (16, 19). Single-site mutations such as T42M (20), G96A (21–23), and P101S (24–26) were found to be advantageous but insufficient for commercial glyphosate-resistant crops. Multisite mutations with more favorable properties were sought and discovered, including Petunia hybrida EPSPS mutants G101A/G137D and G101A/P158S (19), the E. coli EPSPS mutant G96A/A183T (27, 28), and the Zea mays EPSPS mutant TI02I/P106S (29–31). The T102I/P106S double mutant EPSPS (corresponding to T97I/P101S in E. coli; abbreviated as TIPS EPSPS) had particularly favorable characteristics. The TIPS mutations were introduced into the endogenous EPSPS enzyme of Z. mays (field corn, GA21 event) to produce the first commercial varieties of glyphosate-resistant maize. The Class II EPSPS from Agrobacterium sp. strain CP4 was eventually utilized to create transgenic glyphosate-resistant crops (NK603 corn event).
The distinct properties of the CP4 EPSPS have been recently elucidated (15). For the TIPS enzyme, however, the structural basis of glyphosate resistance was unknown. Here, we utilized EPSPS from E. coli as a model of the plant enzyme to investigate the kinetic and structural properties of the single-site T97I and the double-site TIPS mutant enzymes. The implications for glyphosate resistance with respect to genetic engineering and the likelihood of spontaneous mutations are discussed.
EXPERIMENTAL PROCEDURES
Chemicals and reagents were purchased from Sigma unless otherwise noted. S3P was synthesized and purified as described previously (18). The pET-24d vector (Novagen) containing the open reading frame of EPSPS from E. coli was used as a template for the mutations. Single-site T97I mutations were introduced in the wild-type EPSPS and in the P101S mutant EPSPS using the QuikChange II mutagenesis kit (Stratagene) and appropriate primers (MWG Biotech). The single mutant T97I and double mutant TIPS EPSPS enzymes were overexpressed in BL21(DE3)-competent cells (Novagen) and purified as previously described (22). After the final purification step, the enzymes were concentrated in 50 mm Tris, 1 mm dithiothreitol, and 1 mm EDTA using Centricon-30 devices (Millipore Corp., Billerica, MA) and stored at –80 °C. Protein concentration was determined using Coomassie reagent (Pierce) with bovine serum albumin as a standard.
Enzyme Kinetics—The enzymatic activities of WT, P101S, T97I, and TIPS EPSPS were measured by determining the amount of inorganic phosphate produced in the forward reaction with S3P and PEP as substrates in 96-well plates on a Spectra-Max 340PC plate reader (Molecular Devices, Sunnyvale, CA). Each 60-μl reaction mixture contained 50 mm HEPES (pH 7.5), 100 mm KCl, 2 mm dithiothreitol, and S3P, PEP, and glyphosate concentrations as indicated. The reactions were initiated by addition of enzyme and were stopped by addition of the ammonium molybdate-malachite green reagent (32). Change in absorbance at 650 nm was measured, and product formation was determined by comparison to inorganic phosphate standards. Enzymatic activity is expressed as micromoles of phosphate produced per minute of reaction per milligram of enzyme (units/mg).
The Km and Vmax values were determined by fitting data to the Michaelis-Menten equation. The Ki values were determined by fitting data to Equation 1,
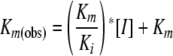 |
(Eq. 1) |
where Km(obs) is the Michaelis constant for PEP in the presence of glyphosate, [I] is the glyphosate concentration, and Km is the Michaelis constant for PEP in the absence of glyphosate. The IC50 values for EPSPS inhibition were determined by fitting data to Equation 2,
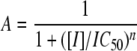 |
(Eq. 2) |
where A is the relative activity remaining in the presence of glyphosate, [I] is the concentration of glyphosate, and n is the Hill slope.
Crystallography—The T97I and TIPS mutant EPSPS were crystallized at 19 °C by the hanging drop, vapor-diffusion method in the presence of 5 mm S3P, with or without 5 mm glyphosate, using the sodium formate crystallization conditions described previously (22). The protein concentration in each case was 37.5 mg/ml, or 810 μm, maintaining a ligand-to-receptor molar ratio of ∼6:1. X-ray diffraction data were recorded at –180 °C using the rotation method on single flash-frozen crystals (detector: Rigaku HTC image plate; X-rays: CuKα, focused by mirror optics; Generator: Rigaku Micro-Max 007-HF (MSC, The Woodlands, TX)). The data were reduced with XDS (33). The program package CNS (34) was employed for phasing and refinement. Model building was performed with O (35). The structures were solved by molecular replacement using E. coli WT EPSPS (Protein Data Bank code 1G6S) (12) stripped of solvent molecules, ions, and ligands as the starting model. Refinement cycles were performed using data to the highest resolution with no sigma cut-off applied. Several rounds of minimization, simulated annealing (2500 K starting temperature), and restrained individual B-factor refinement were carried out. Data collection and refinement statistics are summarized in Table 1. Figs. were drawn with PyMOL,4 MolScript (37), Bob-Script (38), and Raster3D (39).
TABLE 1.
Summary of data collection and structure refinement
Values in parentheses refer to the highest resolution shell.
| T97I·S3P | T97I·S3P·glyphosate | TIPS·S3P | TIPS·S3P·glyphosate | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell dimensions | a = 57.7 Å, b = 85.1 Å, c = 87.5 Å; α = β = γ = 90° | a = 57.6 Å, b = 85.4 Å, c = 87.8 Å; α = β = γ = 90° | a = 57.5 Å, b = 84.7 Å, c = 87.1 Å; α = β = γ = 90° | a = 57.9 Å, b = 85.0 Å, c = 87.7 Å; α = β = γ = 90° |
| Resolution range | 20-1.75 (1.8-1.75) | 20-1.7 (1.75-1.7) | 20-1.7 (1.8-1.7) | 20-1.7 (1.8-1.7) |
| Unique reflections | 44,045 (3515) | 47,887 (3678) | 47,294 (7239) | 46,727 (6865) |
| Completeness (%) | 99.8 (100) | 99.1 (92.2) | 99.6 (98.2) | 96.7 (91.4) |
| I/σI | 19.2 (7.3) | 24.4 (11.3) | 30.4 (12.5) | 19.3 (10.0) |
| Rmerge (%)a | 5.4 (15.0) | 3.7 (9.7) | 2.9 (7.7) | 4.2 (9.4) |
| Structure refinement | ||||
| Protein atoms | 3,233 | 3,233 | 3,232 | 3,232 |
| Average B-factor (Å2) | 14.4 | 13.4 | 14.3 | 13.2 |
| Ligand atoms | 16 | 26 | 16 | 26 |
| Average B-factor (Å2) | 16.3 | 13.2 (S3P) 13.4 (glyphosate) | 17.5 | 12.7 (S3P) 21.4 (glyphosate) |
| Solvent molecules | 383 | 506 | 524 | 500 |
| Average B-factor (Å2) | 23.1 | 25.6 | 26.5 | 25.8 |
| r.m.s.d.b bonds (Å) | 0.01 | 0.01 | 0.011 | 0.011 |
| r.m.s.d. angles (°) | 1.6 | 1.6 | 1.6 | 1.6 |
| Rcryst (%)c | 16.4 | 15.6 | 16.3 | 15.9 |
| Rfree (%)d | 18.4 | 18.1 | 20.3 | 19.0 |
Rmerge = 100 × ΣhΣi|Ihi – Ih|/ΣhiIhi, where h is unique reflection indices.
r.m.s.d. = root mean square deviation from ideal values.
Rcryst = 100 × Σ|Fobs – Fmodel|/ΣFobs, where Fobs and Fmodel are observed and calculated structure factor amplitudes, respectively.
Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement (1190 for T97I·S3P, 1198 for T97I·S3P·glyphosate, 1183 for TIPS·S3P, and 1169 for TIPS·S3P·glyphosate).
RESULTS AND DISCUSSION
Amino acids 90–104 are strictly conserved in class I EPSPS from bacteria and plants (supplemental Table S1). We introduced the double mutation T97I/P101S (TIPS), the basis for glyphosate-resistant GA21 corn, and the single mutation T97I into E. coli EPSPS and studied the mutant enzymes by steady-state kinetics and crystallography.
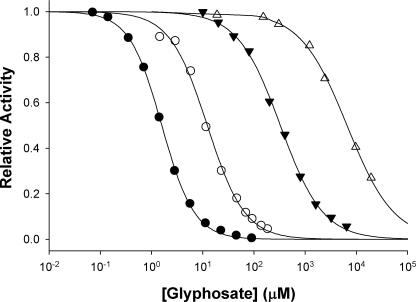
Enzyme Kinetics—The TIPS and T97I mutant enzymes were found to be catalytically active, and the enzymatic reactions displayed normal saturation kinetics. The kinetic constants of the wild-type (WT) and the P101S enzymes (12, 26) were re-determined in parallel to facilitate accurate comparisons. The WT EPSPS was the most active enzyme, with Vmax values of ∼57 units/mg and kcat/Km values of close to 106 m–1 s–1 for both substrates (Table 2 and supplemental Figs. S1–S6). The activity of the single-site mutant enzymes T97I and P101S were decreased 5- and 2.5-fold, respectively, and the activity of TIPS EPSPS was decreased nearly 9-fold. The substrate binding affinities, as reflected by the respective Km values, was only slightly decreased for the P101S and TIPS enzymes, but the T97I mutant showed nearly 9-fold increase in Km for PEP. Compared with the WT enzyme, the catalytic efficiencies with respect to PEP utilization were decreased 40- and 16-fold for the T97I and TIPS enzymes, respectively. Unlike WT EPSPS, which is very sensitive to glyphosate (Ki = 0.3 μm), the TIPS enzyme tolerates high glyphosate concentrations (Ki = 2.4 mm) (Fig. 2). By contrast, the P101S and T97I enzymes are still inhibited by moderate concentrations of glyphosate, with Ki values of 3 and 90 μm, respectively. Apparently, neither single-site mutation, T97I or P101S, is sufficient to enable glyphosate resistance and maintain high affinity for the substrate PEP. Only the simultaneous mutation of both residues renders the enzyme both insensitive to glyphosate and catalytically efficient.
TABLE 2.
Kinetic characteristics of mutant and wild-type EPSPS
| EPSPS | Km (PEP) | Km (S3P) | Vmax | kcat/Km (PEP) | kcat/Km (S3P) | IC50 | Ki |
|---|---|---|---|---|---|---|---|
| μm | μm | units/mg | m-1s-1 | m-1s-1 | μm | μm | |
| WT | 45 ± 5 | 48 ± 5 | 57 ± 1 | 9.3 × 105 | 9.1 × 105 | 1.6 ± 0.03 | 0.3 ± 0.07 |
| P101S | 71 ± 7 | 71 ± 6 | 23 ± 0.5 | 2.5 × 105 | 2.4 × 105 | 12 ± 0.5 | 3.0 ± 0.3 |
| T97I | 380 ± 21 | 77 ± 3 | 12 ± 0.1 | 2.3 × 104 | 1.2 × 105 | 330 ± 10 | 90 ± 6 |
| TIPS | 100 ± 6 | 71 ± 6 | 7 ± 0.2 | 5.7 × 104 | 7.4 × 104 | 6600 ± 190 | 2420 ± 87 |
FIGURE 2.
Inhibition of mutant and wild-type EPSPS by glyphosate. IC50 values of glyphosate inhibition were determined for EPSPS WT (•), P101S (○), T97I (▴), and TIPS (▵) at saturating concentrations of S3P (1 mm) and PEP concentrations equal to the respective Km values. Data were fit to Equation 2 yielding the IC50 values shown in Table 2. The data for the determination of the respective Ki values is shown in the supplemental material (Figs. S1–S6).
We next asked if analogs of the tetrahedral reaction intermediates, the most potent inhibitors of WT EPSPS described to date (40–42), would still act as potent inhibitors of the TIPS enzyme. The TIPS and WT enzymes showed nearly equal sensitivity to the inhibitors tested (supplemental Fig. S7). These findings support the hypothesis that inhibitors with more global coverage of the active site are not as affected by mutations that cause resistance to glyphosate (42).
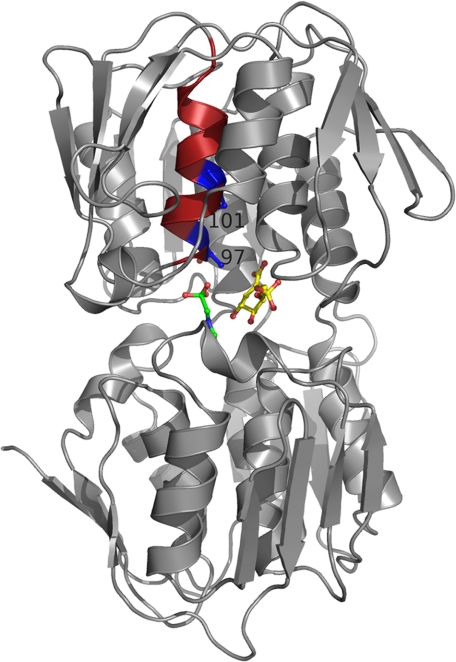
Protein Crystallography—To better understand the effect of the mutations on the catalytic efficiency and inhibition by glyphosate, the T97I and TIPS enzymes were co-crystallized with S3P alone and in complex with S3P and glyphosate; the resulting 1.7-Å resolution structures (Table 1) were compared with those of the WT (12) and P101S (26) enzymes. Thr97 and Pro101 are constituents of an α-helix in the N-terminal globular domain of EPSPS (Fig. 3). Neither of these two residues is directly involved in glyphosate binding. Pro101 is ∼9Å distant from glyphosate and the hydroxyl group of Thr97 interacts with the phosphonate moiety of glyphosate only through a bridging water molecule.
FIGURE 3.
Location of Thr97 and Pro101 in the structure of wild-type EPSPS from E. coli. EPSPS is composed of two globular domains that close upon binding of S3P and glyphosate. The two ligands (shown in yellow and green, respectively) are located in the interdomain cleft of the closed enzyme state. Displayed in maroon is the α-helix in the N-terminal domain containing residues Thr97 and Pro101 (shown in “blue”).
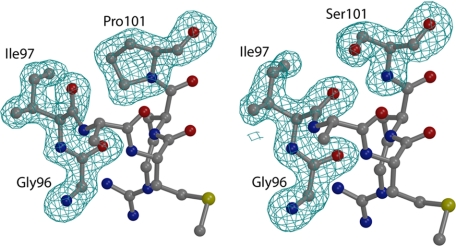
The altered amino acids are well defined in the respective electron density maps (Fig. 4). Glyphosate is bound in the same site and conformation as observed in the WT enzyme; however, potential steric clashes with the Cα atom of Gly96 occur in both mutant enzymes (Fig. 5). This leads to additional unfavorable interactions between the glyphosate molecule and other active site residues, particularly in the TIPS enzyme. The reduced binding potential of glyphosate in the respective mutant enzymes is reflected by an increase in the crystallographic B-factors (higher B-factor values reflect higher flexibility) (Fig. 6). Notably, the B-factors of S3P are only slightly increased for both mutant enzymes (supplemental Fig. S8). Thus, the crystallographic data seem to reveal the structural basis for the kinetic properties of these enzymes.
FIGURE 4.
Structure determination of the TIPS and T97I mutant EPSPS. Displayed is the electron density at 1.7-Å resolution, contoured at 3σ, derived from 1Fo – 1Fc Fourier syntheses omitting residues 96–97 and 101 during simulated annealing refinement of the S3P·glyphosate-bound structures of the T97I (left) and TIPS (right) enzymes.
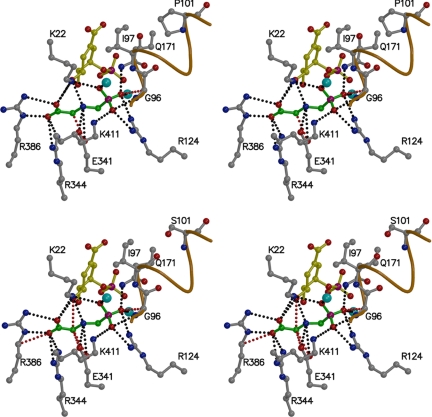
FIGURE 5.
The glyphosate binding sites of T97I and TIPS EPSPS (stereo views). Top: glyphosate (shown in green) bound to T97I EPSPS reveals potential steric clashes with the Cα atom of Gly96 (d = 3.13 Å) and with the side chain of Glu341 (d = 2.93 Å) (red dotted lines). Bottom: in TIPS EPSPS these clashes are more pronounced (Gly96: d = 3.02 Å; Glu341: d = 2.82 Å) and additional unfavorable interactions occur between glyphosate and S3P (d = 3.02 Å) and the Cδ atom of Arg386 (d = 3.11 Å). S3P is shown in yellow. The helix harboring residues 97–101 is indicated in orange. Black dotted lines indicate polar interactions. The cyan spheres denote water molecules.
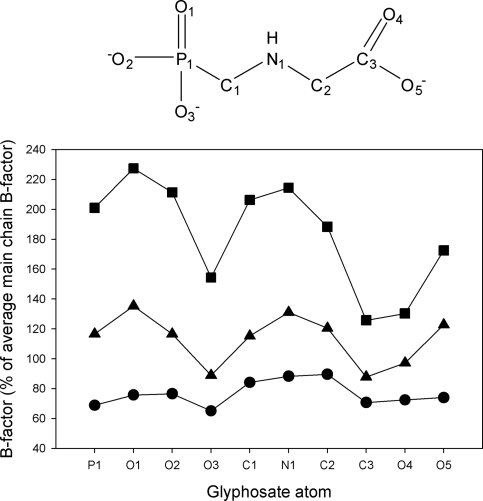
FIGURE 6.
Impact of the mutations on the glyphosate molecule. For WT EPSPS, the average B-factor of glyphosate (•; 7.7 Å2) is smaller than the average main-chain B-factor (10.0 Å2), reflecting the tight binding of the inhibitor. When bound to the T97I (▴) or the TIPS (▪) enzyme the B-factors are substantially increased, indicating the weaker binding potential as a result of these mutations.
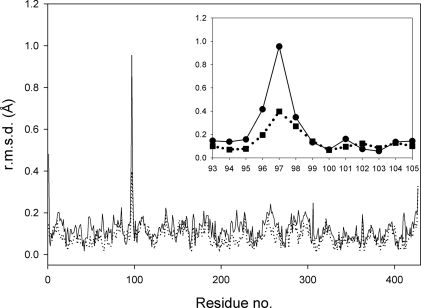
The overall structures of the T97I and TIPS enzymes are nearly identical to that of WT EPSPS; however, large root mean square deviation (r.m.s.d.) differences, up to 1 Å for the TIPS enzyme, occur in the main chain around residues 96–98 (Fig. 7). In the WT and P101S enzymes, the hydroxyl group of Thr97 is hydrogen-bonded with the side chain of Asn26 (Fig. 8). Upon mutation to isoleucine, the disruption of this polar interaction and the resulting repulsive forces between the Ile97 and Asn26 side chains impose changes in the backbone torsion angles of residues 96–98 (supplemental Table S2). In the T97I enzyme, the presence of the Pro101 ring holds the carbonyl oxygen of Ile97 in place, thereby constraining its rotational freedom. In the TIPS enzyme, in contrast, the additional Ser101 substitution allows the main chain of Ile97 to relax and the Ile97 side chain to move away from the glyphosate/PEP site. As a result, Gly96 is rearranged such that its Cα atom shifts toward the glyphosate binding site, thereby shortening the distance to the inhibitor's phosphonate moiety and effectively narrowing the binding site.
FIGURE 7.
Structural differences of the mutant enzymes. Plotted are the r.m.s.d. values of all 427 Cα atoms of the TIPS enzyme (solid line) and the T97I enzyme (dotted line) after superposition with WT EPSPS (PDB code 1G6S). The inset shows the r.m.s.d. values for the Cα atoms of residues 93–105. Superposition was performed with LSQkab of the CCP4 program suite (48, 36); the average r.m.s.d. value for TIPS and WT is 0.138 Å; for T97I and WT the average r.m.s.d value is 0.097 Å.
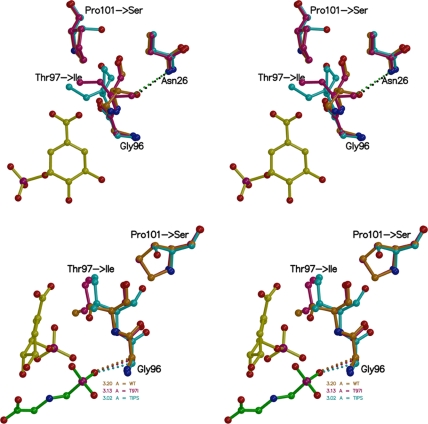
FIGURE 8.
Mutation-induced structural changes in EPSPS (stereo views). Top: EPSPS in complex with S3P. In the WT enzyme binary complex (shown in orange) Thr97 is in hydrogen bonding distance (2.8 Å) to the amide side chain of Asn96. In the T97I enzyme binary complex (maroon) the presence of the Pro101 ring holds Ile97 in place, and its side chain moves only slightly. In the TIPS enzyme binary complex (cyan) Ser101 allows larger conformational freedom for Ile97, and the isoleucine side chain swings away from Asn26. Bottom: EPSPS in complex with S3P and glyphosate bound. In the ternary complex, the mutations cause a shift of the Cα atom of Gly96 toward the phosphonate moiety of glyphosate, seen most drastically in the TIPS enzyme (cyan), thereby narrowing the inhibitor binding site. The view is ∼90° clockwise from the top.
Because glyphosate and PEP share the same binding site, one would expect intuitively that the TIPS enzyme to be less efficient in the utilization of PEP, yet only the single mutation drastically reduces the affinity for PEP. Because we cannot obtain the true ternary complexes with S3P and PEP bound to the EPSPS active site, we determined the respective EPSPS·S3P binary complexes to which PEP would bind during catalysis (Fig. 8). Even in the absence of glyphosate, the geometric constraints in the T97I enzyme lock the Ile97 side chain in close distance to the putative PEP site, likely to exert repulsive forces on the charged, polar PEP molecule during binding and/or catalysis. By contrast, Ile97 in the TIPS binary complex rotates even further away from the PEP site than in the ternary complex with glyphosate bound. Overall, it appears that the mutation-induced shifts of Gly96 predominantly impact the inhibitory potential of glyphosate, whereas the conformation of Ile97 influences the catalytic efficiency of the EPSPS enzyme.
CONCLUSIONS
The TIPS EPSPS enzyme shows high levels of glyphosate resistance while maintaining high affinity for its substrates, PEP and S3P. The single-site T97I enzyme is less sensitive to inhibition by glyphosate; moreover, in the absence of the compensating P101S mutation, it exhibits drastically decreased affinity for PEP. These phenomena can be attributed to specific structural changes in the active site. The glyphosate resistance exhibited by the TIPS enzyme is on the same order of magnitude as that observed for the class II EPSPS enzymes from S. aureus and CP4 EPSPS (15, 18). To date, TIPS EPSPS represents the only Class I enzyme with both high glyphosate resistance and unaltered affinity for PEP. The only other highly glyphosate-resistant class I mutant enzyme is G96A EPSPS from E. coli and K. pneumoniae, but this mutation causes a very large decrease in affinity for PEP (Km = 2.8 mm (22)). Mutations of the residue corresponding to Pro101 of E. coli EPSPS have been reported in a number of field-evolved glyphosate-resistant weeds (43–46). By contrast, mutations of Thr97 have never been observed. In fact, BLAST (47) analysis revealed that this residue is strictly conserved in aroA genes; notably, only EPSPS from Chlamydia species contain an isoleucine at the equivalent position. The decreased catalytic efficiency of the T97I mutant EPSPS with respect to utilization of PEP may explain why it has not been observed in glyphosate-resistant weeds and probably why it has not been exploited for the genetic engineering of crops. Although the TIPS enzyme is significantly less catalytically efficient than the wild-type enzyme, its high affinity for PEP apparently enables crops with this gene to have sufficient EPSPS activity to produce crop yields that are commercially competitive. Spontaneous double mutations of Thr97 and Pro101 in wild-type EPSPS are unlikely to occur. However, under the selective pressure caused by the presence of high glyphosate concentrations, it may be favorable for plants or bacteria with already established Pro101 mutations to acquire the additional mutation of Thr97, which would confer a very high level of resistance.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant 5R01GM070633.
The atomic coordinates and structure factors (codes 3FJX, 3FJZ, 3FK0, and 3FK1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Tables S1 and S2.
Footnotes
The abbreviations used are: EPSPS, 5-enolpyruvylshikimate-3-phosphate synthase; S3P, shikimate 3-phosphate; PEP, phosphoenolpyruvate; WT, wild type; TIPS, T97I/P101S; r.m.s.d., root mean square deviation.
W. L. DeLano (2003) PyMOL, DeLano Scientific, San Carlos, CA.
References
- 1.Steinrucken, H. C., and Amrhein, N. (1980) Biochem. Biophys. Res. Commun. 94 1207–1212 [DOI] [PubMed] [Google Scholar]
- 2.Williams, G. M., Kroes, R., and Munro, I. C. (2000) Regul. Toxicol. Pharmacol. 31 117–165 [DOI] [PubMed] [Google Scholar]
- 3.Smith, E. A., and Oehme, F. W. (1992) Vet. Hum. Toxicol. 34 531–543 [PubMed] [Google Scholar]
- 4.Padgette, S. R., Kolacz, K. H., Delannay, X., Re, D. B., LaVallee, B. J., CTinius, C. N., Rhodes, W. K., Otero, Y. I., Barry, G. F., Eichholz, D. A., Peshke, V. M., Nida, D. L., Taylor, N. B., and Kishore, G. M. (1995) Crop Sci. 35 1451–1461 [Google Scholar]
- 5.Dill, G. M., Jacob, C. A., and Padgette, S. R. (2008) Pest. Manag. Sci. 64 326–331 [DOI] [PubMed] [Google Scholar]
- 6.Bentley, R. (1990) Crit. Rev. Biochem. Mol. Biol. 25 307–384 [DOI] [PubMed] [Google Scholar]
- 7.Haslam, E. (1993) Shikimic Acid: Metabolism and Metabolites, pp. 1–6, John Wiley & Sons, Chichester, UK
- 8.Kishore, G. M., and Shah, D. M. (1988) Annu. Rev. Biochem. 57 627–663 [DOI] [PubMed] [Google Scholar]
- 9.Roberts, F., Roberts, C. W., Johnson, J. J., Kyle, D. E., Krell, T., Coggins, J. R., Coombs, G. H., Milhous, W. K., Tzipori, S., Ferguson, D. J., Chakrabarti, D., and McLeod, R. (1998) Nature 393 801–805 [DOI] [PubMed] [Google Scholar]
- 10.Boocock, M. R., and Coggins, J. R. (1983) FEBS Lett. 154 127–133 [DOI] [PubMed] [Google Scholar]
- 11.Steinrucken, H. C., and Amrhein, N. (1984) Eur. J. Biochem. 143 351–357 [DOI] [PubMed] [Google Scholar]
- 12.Schonbrunn, E., Eschenburg, S., Shuttleworth, W. A., Schloss, J. V., Amrhein, N., Evans, J. N., and Kabsch, W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1376–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschenburg, S., Kabsch, W., Healy, M. L., and Schonbrunn, E. (2003) J. Biol. Chem. 278 49215–49222 [DOI] [PubMed] [Google Scholar]
- 14.Park, H., Hilsenbeck, J. L., Kim, H. J., Shuttleworth, W. A., Park, Y. H., Evans, J. N., and Kang, C. (2004) Mol. Microbiol. 51 963–971 [DOI] [PubMed] [Google Scholar]
- 15.Funke, T., Han, H., Healy-Fried, M. L., Fischer, M., and Schonbrunn, E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franz, J. E., Mao, M. K., and Sikorski, J. A. (1997) Glyphosate: a Unique Global Herbicide, pp. 596–602 and 629–634, American Chemical Society, Washington, D. C.
- 17.Du, W., Liu, W. S., Payne, D. J., and Doyle, M. L. (2000) Biochemistry 39 10140–10146 [DOI] [PubMed] [Google Scholar]
- 18.Priestman, M. A., Funke, T., Singh, I. M., Crupper, S. S., and Schonbrunn, E. (2005) FEBS Lett. 579 728–732 [DOI] [PubMed] [Google Scholar]
- 19.Padgette, S. R., Re, D. B., Barry, G. F., Eichholtz, D. E., Delannay, X., Fuchs, R. L., Kishore, G. M., and Fraley, R. T. (1995) in Herbicide-resistant Crops: Agricultural, Environmental, Economic, Regulatory, and Technical Aspects (Duke, S. O., ed) Chap. 4, pp. 53–84, CRC Press, New York
- 20.He, M., Nie, Y. F., and Xu, P. (2003) Biosci. Biotechnol. Biochem. 67 1405–1409 [DOI] [PubMed] [Google Scholar]
- 21.Padgette, S. R., Re, D. B., Gasser, C. S., Eichholtz, D. A., Frazier, R. B., Hironaka, C. M., Levine, E. B., Shah, D. M., Fraley, R. T., and Kishore, G. M. (1991) J. Biol. Chem. 266 22364–22369 [PubMed] [Google Scholar]
- 22.Eschenburg, S., Healy, M. L., Priestman, M. A., Lushington, G. H., and Schonbrunn, E. (2002) Planta 216 129–135 [DOI] [PubMed] [Google Scholar]
- 23.Sost, D., and Amrhein, N. (1990) Arch. Biochem. Biophys. 282 433–436 [DOI] [PubMed] [Google Scholar]
- 24.Stalker, D. M., Hiatt, W. R., and Comai, L. (1985) J. Biol. Chem. 260 4724–4728 [PubMed] [Google Scholar]
- 25.Comai, L., Sen, L. C., and Stalker, D. M. (1983) Science 221 370–371 [DOI] [PubMed] [Google Scholar]
- 26.Healy-Fried, M. L., Funke, T., Priestman, M. A., Han, H., and Schonbrunn, E. (2007) J. Biol. Chem. 282 32949–32955 [DOI] [PubMed] [Google Scholar]
- 27.Eichholtz, D. A., Scott, G. C., and Murthy, K. G. (2001) U.S. Patent 6,225,114
- 28.Kahrizi, D., Salmanian, A. H., Afshari, A., Moieni, A., and Mousavi, A. (2007) Plant Cell Rep. 26 95–104 [DOI] [PubMed] [Google Scholar]
- 29.Spencer, M., Mumm, R., and Gwyn, J. (2000) U.S. Patent 6,040,497
- 30.Eichholtz, D. A., Gasser, C. S., and Kishore, G. M. (May 1, 2001) U. S. Patent 6,225,114
- 31.Lebrun, M., Sailland, A., Freyssinet, G., and Degryse, E. (2003) U.S. Patent 6,566,587
- 32.Lanzetta, P. A., Alvarez, L. J., Reinach, P. S., and Candia, O. A. (1979) Analyt. Biochem. 100 95–97 [DOI] [PubMed] [Google Scholar]
- 33.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795–800 [Google Scholar]
- 34.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and GL, W. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905–921 [DOI] [PubMed] [Google Scholar]
- 35.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard (1991) Acta Crystallogr. Sect. B Struct. Sci. 47 110–119 [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760–76315299374 [Google Scholar]
- 37.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24 946–950 [Google Scholar]
- 38.Esnouf, R. M. (1997) J. Mol. Graphics Model. 15 132–134 [DOI] [PubMed] [Google Scholar]
- 39.Merrit, E. A., and Bacon, D. J. (1997) Methods Enzymol. 277 505–524 [DOI] [PubMed] [Google Scholar]
- 40.Alberg, D. G., Lauhon, C. T., Nyfeler, R., Fassler, A., and Bartlett, P. A. (1992) J. Am. Chem. Soc. 114 3535–3546 [Google Scholar]
- 41.Priestman, M. A., Healy, M. L., Becker, A., Alberg, D. G., Bartlett, P. A., Lushington, G. H., and Schonbrunn, E. (2005) Biochemistry 44 3241–3248 [DOI] [PubMed] [Google Scholar]
- 42.Funke, T., Healy-Fried, M. L., Han, H., Alberg, D. G., Bartlett, P. A., and Schonbrunn, E. (2007) Biochemistry 46 13344–13351 [DOI] [PubMed] [Google Scholar]
- 43.Yu, Q., Cairns, A., and Powles, S. (2007) Planta 225 499–513 [DOI] [PubMed] [Google Scholar]
- 44.Baerson, S. R., Rodriguez, D. J., Tran, M., Feng, Y., Biest, N. A., and Dill, G. M. (2002) Plant Physiol. 129 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powles, S. B., and Preston, C. (2006) Weed Technol. 20 282–289 [Google Scholar]
- 46.Perez-Jones, A., Park, K.-W., Polge, N., Colquhoun, J., Mallory-Smith, C. A. (2007) Planta 226 395–404 [DOI] [PubMed] [Google Scholar]
- 47.Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990) J. Mol. Biol. 215 403–410 [DOI] [PubMed] [Google Scholar]
- 48.Kabsch, W. (1976) Acta Crystallogr. Sect. A 32 922–923 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.