Abstract
In bacteria, the majority of exported proteins are transported by the general Sec pathway from their site of synthesis in the cytoplasm across the cytoplasmic membrane. The essential SecA ATPase powers this Sec-mediated export. Mycobacteria possess two nonredundant SecA homologs: SecA1 and SecA2. In pathogenic Mycobacterium tuberculosis and the nonpathogenic model mycobacterium Mycobacterium smegmatis, SecA1 is essential for protein export and is the “housekeeping” SecA, whereas SecA2 is an accessory SecA that exports a specific subset of proteins. In M. tuberculosis the accessory SecA2 pathway plays a role in virulence. In this study, we uncovered basic properties of the mycobacterial SecA2 protein and its pathway for exporting select proteins. By constructing secA2 mutant alleles that encode proteins defective in ATP binding, we showed that ATP binding is required for SecA2 function. SecA2 mutant proteins unable to bind ATP were nonfunctional and dominant negative. By evaluating the subcellular distribution of each SecA, SecA1 was shown to be equally divided between cytosolic and cell envelope fractions, whereas SecA2 was predominantly localized to the cytosol. Finally, we showed that the canonical SecA1 has a role in the process of SecA2-dependent export. The accessory SecA2 export system is important to the physiology and virulence of mycobacteria. These studies help establish the mechanism of this new type of specialized protein export pathway.
In bacteria, proteins exported beyond the cytosol to the bacterial cell envelope or secreted into the extracellular environment are important for establishing cell wall structure, acquiring nutrients, and responding to changes in the environment. Surface and secreted proteins of bacterial pathogens are also ideally positioned to interact with host cells; consequently, these proteins and the systems that export them are important for virulence (1). Studies of bacterial protein export systems could yield new targets for antimicrobial therapies.
Many bacterial pathogens use specialized systems to export virulence factors (2, 3). In mycobacteria and a small subset of Gram-positive bacteria, there are accessory Sec systems that can be considered specialized protein export systems (4–11). The accessory Sec pathway is linked to virulence in several cases including Mycobacterium tuberculosis, Listeria monocytogenes, and Streptococcus gordonii (5, 12–14). The common constituent of the accessory Sec pathways studied so far is an accessory SecA homolog called SecA2. SecA2 is homologous to the essential SecA (SecA1) that is part of the general Sec pathway present in all bacteria (15).
Studies of the general Sec pathway show that the canonical SecA plays a central role in the process of guiding proteins that are synthesized as precursors with N-terminal signal sequences across the cytoplasmic membrane through a channel comprised of the SecYEG proteins (16, 17). Energy for protein translocation is provided by the ATPase activity of SecA. With repeated cycles of ATP binding and hydrolysis, SecA undergoes conformational changes that allow it to ratchet precursor proteins through the SecYEG translocase (18). ATPase activity is essential to SecA function (19). Mutants defective in SecA ATP binding/hydrolysis are unable to complement temperature-sensitive secA mutants (19–21).
In mycobacteria, there are two SecA proteins called SecA1 and SecA2. SecA1 is essential and is the housekeeping SecA of the mycobacterial general Sec pathway (4, 22, 23). In contrast, SecA2 is not essential in mycobacteria. Mutants with the secA2 gene deleted have been constructed in pathogenic M. tuberculosis and in nonpathogenic Mycobacterium smegmatis (4, 5). The ΔsecA2 mutant of M. tuberculosis is attenuated in mouse and macrophage models of infection, which demonstrates the importance of SecA2 to M. tuberculosis virulence (5, 13). The ΔsecA2 mutant of M. smegmatis exhibits a growth defect on rich agar and hypersensitivity to sodium azide (4). Two-dimensional gel analysis comparing exported proteins from wild type versus ΔsecA2 mutant M. tuberculosis and M. smegmatis identifies a small number of proteins that depend on SecA2 for export (5, 24). In M. tuberculosis, the SodA protein is one of the few proteins identified as requiring SecA2 for its secretion (5). In M. smegmatis, two lipoproteins with N-terminal signal sequences (Msmeg1712 and Msmeg1704) require SecA2 for their export to the cell wall (24).
Both SecA1 and SecA2 have ATP-binding Walker Box motifs, and the purified M. tuberculosis proteins bind and hydrolyze ATP in vitro (25). Previously, we showed that M. tuberculosis SecA2 with a mutated Walker Box is defective in ATP binding in vitro (25). To study the biological significance of ATPase activity in vivo, we tested the effect of amino acid substitutions in the Walker Box of SecA2 when expressed in M. smegmatis and M. tuberculosis. The bulk of our studies here focused on a particular Walker Box mutant of M. smegmatis SecA2, SecA2 K129R. We showed SecA2 K129R to be unable to fulfill its role in exporting SecA2-dependent proteins. We further discovered that in several assays SecA2 K129R not only failed to complement ΔsecA2 mutant phenotypes but behaved as a dominant negative. In the process of characterizing the dominant negative SecA2 protein, we showed that wild type SecA2 protein is predominantly localized to the cytosol, which is notably different from the subcellular distribution of SecA1 and SecA2 K129R.
The accessory SecA2 system of mycobacteria is unlike the accessory SecA2/Y2 systems of S. gordonii and Streptococcus parasanguinis in that it lacks an accessory SecY2 protein for export (6, 8, 26, 27). Without an obvious translocase, we considered the possibility that SecA2 uses the canonical Sec machinery (SecA1/SecYEG) to promote export of its select subset of proteins. Using M. smegmatis strains engineered to deplete SecA1, we showed that export of the SecA2-dependent substrate Msmeg1712 was severely impaired in the absence of SecA1. This result indicates SecA1 is required for SecA2-dependent export. It represents the first piece of evidence indicating a role for a canonical SecA1 in exporting substrates of an accessory SecA2 pathway.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Conditions—M. smegmatis was grown in Middlebrook 7H9 or on 7H10 (BD Biosciences) supplemented with 0.2% (w/v) glucose, 0.5% (v/v) glycerol, and 0.1% (v/v) Tween 80 (Fisher); or Mueller Hinton (BD Biosciences) supplemented with 0.1% (v/v) Tween 80. M. tuberculosis was grown in 7H9 or on 7H10 supplemented with 0.5% (v/v) glycerol and 1× ADS (0.5% (w/v) bovine serum albumin, 0.2% (w/v) dextrose, and 0.85% (w/v) NaCl). For cultures of mycobacteria, the antibiotics kanamycin (Acros Chemicals) and hygromycin (Roche Applied Science) were added as needed at 20 or 50 μg/ml, respectively. Luria Bertani was used to grow Escherichia coli cultures. When needed, kanamycin or hygromycin was added at 40 or 150 μg/ml, respectively. All of the plasmids used in this study are listed in supplemental Table S1, and cloned sequences were verified by DNA sequencing (University of North Carolina at Chapel Hill DNA sequencing facility and Eton Biosciences). Throughout this work, mc2155 and H37Rv were used as the wild type strains of M. smegmatis and M. tuberculosis, respectively. NR116 (described below) was used as the ΔsecA2 mutant of M. smegmatis, and mc23112 (5) was used as the ΔsecA2 mutant of M. tuberculosis.
Construction of ΔsecA2 Mutant M. smegmatis NR116—The previously published M. smegmatis ΔsecA2 mutant (mc22522) is an in-frame, unmarked deletion of approximately one-third of the secA2 gene (4). For this study a complete in-frame unmarked deletion of M. smegmatis secA2 (NR116) was used in which only three codons of the secA2 open reading frame remain. All of the phenotypes observed for mc22522 were also observed for NR116. NR116 was constructed in the following manner: (i) secA2 suicide plasmid pNR6. Sequences immediately upstream of secA2 were PCR-amplified from M. smegmatis genomic DNA using primers 5′-GTCGACCGACAGGTTCCAGCCGTAGAA-3′/5′-GTCGACCACGGCGTCAGTTGTGCCTCG-3′ and sequences immediately downstream of secA2 were PCR-amplified using the primers 5′-GTCGACTAGGCCCCAGCCATTAGGTTC-3′/5′-TGATATCGAGCACCTCCCAGCCCCATTC-3′. The upstream PCR product was cloned into the vector pCC1 using the Copy Control PCR cloning kit (Epicenter) to generate pNR3. The downstream PCR product was cloned into the vector pCR2.1 (Invitrogen) to generate pNR4. A 1087-bp SalI fragment of pNR3 was ligated into SalI-cut pNR4. The resulting plasmid, pNR5, contained a 2385-bp deletion of secA2, leaving only three codons. A 1737-bp MscI-EcoRV fragment containing the secA2 deletion was cut from pNR5 and cloned into the EcoRV site of the counterselectable suicide plasmid pYUB657, yielding vector pNR6. (ii) Two step allelic exchange. The ΔsecA2 mutant strain NR116 was constructed by two-step allelic exchange as described previously (4, 28, 29). Wild type M. smegmatis strain mc2155 was electroporated with pNR6, and hygromycin-resistant transformants were selected. Transformants were screened by Southern blot to identify a single-cross-over integration of pNR6 at the chromosomal secA2 locus yielding strain SCO5. To resolve the single cross-over strain, a saturated culture of SCO5 was diluted 1:100 into 7H9 without hygromycin and grown overnight at 37 °C. This culture was then plated onto 7H10 supplemented with 4.5% (w/v) sucrose to select against the sacB marker encoded on the backbone of the suicide vector. Sucrose-resistant clones were screened for hygromycin sensitivity. Sucrose-resistant, hygromycin-sensitive clones were screened by Southern blot analysis (data not shown) for evidence of a second recombination event leading to chromosomal deletion of secA2 (30). Briefly, genomic DNA was isolated from M. smegmatis strains and digested with BamHI. The probe used was the 1093-bp PCR product containing the sequence immediately upstream of secA2, used to generate pNR3. The resulting inframe, unmarked ΔsecA2 deletion mutant derived from SCO5 was named NR116.
Construction of secA2 Walker Box Mutant Plasmids pNR7 and pNR25—The QuikChange site-directed mutagenesis kit (Stratagene) was used to mutate the DNA encoding the Walker Boxes of secA2 using plasmid templates pMB162 for M. tuberculosis secA2 and pYA810 for M. smegmatis secA2 (4, 24). Both pMB162 and pYA810 vectors integrate at the chromosomal attB site of mycobacteria, and both plasmids express their respective secA2 genes from the hsp60 promoter. The Walker A motif of M. tuberculosis SecA2 was mutated in plasmid pMB162 using the following primers: 5′-CGGTGAGGGCAGAACCCTTGCC-3′ and 5′-CGGCAAGGGTTCTGCCCTCACC-3′. The resulting mutant plasmid contained secA2 K115R and was named pNR7. To create the comparable Walker A mutation in M. smegmatis secA2, which is K129R, primers 5′-CGGGTGAGGGCAGGACGCTGGC-3′ and 5′-GCCAGCGTCCTGCCCTCACCCG-3′ were used with pYA810 as the template to generate plasmid pNR25.
Construction of Inducible secA2 K129R Plasmid pNR54 (Tet ON)—M. smegmatis secA2 was PCR-amplified with primers 5′-AGGATCCATCCGGAGGAATCACTT-3′ and 5′-AGGATCCCTAGTGGTGGTGGTGGTGGTGGCGGAACACACCCGGCAGG-3′ and cloned into pCC1 to yield pNR52. Next, a 2.4-kb BamHI fragment was cloned from pNR52 into similarly cut pMP715 (kind gift from Dr. Martin Pavelka, University of Rochester). The resulting plasmid pNR53 has the secA2 gene cloned adjacent to the TetO operators. The vector pMP715 is an episomal plasmid that carries the Tet operator sequence from pSE100 (23), encodes the TetR repressor protein from pMC1s (31), and carries a kanamycin resistance gene. As with the precursor plasmids pSE100 and pMC1s, plasmid pMP715 can be used to achieve tetracycline regulation of gene expression (23) using anhydrotetracycline (Atc).6 To mutate Walker Box residue K129, pNR53 was subjected to site-directed mutagenesis as described above. The resulting plasmid pNR54 carries secA2 K129R under control of the regulatable promoter. Induction of SecA2 K129R in M. smegmatis transformed with pNR54 was achieved by treating cultures with 100 or 200 ng/ml Atc for 21 h at 37 °C.
Construction of Conditional secA1 Strains—As described previously, the SecA1 depletion strain MSE10 was used (23). For this study, we additionally introduced the conditional secA1 allele into the ΔsecA2 mutant background. NR116 was transformed with the suicide plasmid pKIsecA1 (kind gift from Dr. Sabine Ehrt, Weill Cornell Medical College), and hygromycin-resistant transformants were selected on 7H10 plates. Transformants were screened by Southern blot (data not shown) to detect integration of pKIsecA1 at the native secA1 locus in the chromosome. Genomic DNA was isolated from M. smegmatis strains and digested with BamHI and EcoRI. The probe used was a 410-bp SphI-MscI fragment from pKIsecA1. The resulting strain was named JM693. To achieve regulation of SecA1 in strains MSE10 and JM693, each strain was transformed with pNR55 (described below) and treated with or without 600 ng/ml Atc.
Construction of Msmeg1712-HA revTetR Plasmid pNR55 (Tet OFF)—To monitor the effect of SecA1 depletion on export of Msmeg1712-HA, we constructed a plasmid that constitutively expresses both Msmeg1712-HA and the reverse TetR repressor (revTetR). To construct pNR55, a 2392-bp NheI fragment containing revTetR was excised from pTEK-4S0X (23) and ligated into the NheI site of pHSG85 (24). pHSG85 was constructed by cloning a NotI-HindIII fragment containing the msmeg1712 gene and the promoter region from pHSG83 (24) into similarly cut pMV261.kan. Vector pNR55 produces Msmeg1712 tagged at the C terminus with the hemagglutinin (HA) epitope.
Macrophage Infections—Bone marrow macrophages were elicited from the femurs of C57BL/6 mice, as described previously (13, 25). 2.5 × 105 macrophages/well were seeded into 8-well chamber slides. M. tuberculosis strains were grown to mid-exponential phase, washed with phosphate-buffered saline containing 0.05% Tween 80, diluted in tissue culture medium (Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 2 mm glutamine, and 1× nonessential amino acids; Invitrogen), and added to the macrophages at a multiplicity of infection of 1.0. Macrophages were infected for 4 h at 37 °C in 5% CO2. On days 0, 3, and 5 post-infection, the contents of triplicate wells for each infection were lysed with 1× phosphate-buffered saline 0.05% Tween 80. The lysates were diluted and plated onto Middlebrook 7H10 agar to enumerate intracellular bacteria during infection.
Azide Sensitivity Assay—200 μl of saturated M. smegmatis culture was mixed with 3.5 ml of molten 7H9 top agar and then poured onto a 7H10 bottom agar plate. Sterile 6-mm filter discs were placed onto the surface of the cooled top agar. 10 μl of 0.15 m sodium azide was then added to the disc. The plates were inverted and incubated for 2 days at 37 °C, and the resulting zones of inhibition were measured. Each strain was tested in triplicate.
Epitope-tagged Expression Vectors for Msmeg1704 and Msmeg1712—To monitor the export of Msmeg1704 and Msmeg1712, both proteins were tagged with the HA epitope at the C terminus and expressed from hygromycin-resistant episomal plasmids pNR35 (Msmeg 1712-HA) and pNR36 (Msmeg1704-HA) (24). These vectors were constructed by replacing the kanamycin marker of pHSG51 and pHSG58 with the 1.5-kb SmaI-EcoRV hygromycin resistance fragment from pYUB412.7
Subcellular Fractionation—To determine the subcellular localization of proteins in M. smegmatis, we fractionated bacterial whole cell lysates as described previously (24). Whole cell lysates were generated by five passages through a French pressure cell. For Msmeg1704-HA and Msmeg1712-HA localization, the lysates were separated by differential ultracentrifugation into cell wall (27,000 × g pellet), membrane (100,000 × g pellet), and soluble (100,000 × g supernatant) fractions. For SecA1 and SecA2 localization experiments, the lysates were separated into cell envelope (100,000 × g pellet) and soluble (100,000 × g supernatant) fractions. In all cases, protein derived from the same amount of starting cells for each fraction was analyzed by SDS-PAGE and immunoblots.
Immunoblot Conditions—The anti-SecA2 (23), anti-GroEL (HAT5/IT-64, from the World Health Organization antibody collection), anti-MspA (gift from Dr. Michael Niederweis (32)), and anti-HA (Covance) antibodies were used at 1:20,000 dilutions. The anti-SecA1 antibody (23) was used at a 1:50,000 dilution. In some experiments, secondary antibodies conjugated to horseradish peroxidase (Bio-Rad) were used along with Western Lightening Chemiluminescent detection reagent (PerkinElmer Life Sciences). For quantitative westerns, secondary antibodies conjugated to alkaline phosphatase were used and detected using the ECF reagent (GE Healthcare). Fluorescence was quantified using a phosphorimager and ImageQuant 5.2 (Molecular Dynamics).
RESULTS
M. smegmatis Is a Valid Model for Studying the M. tuberculosis SecA2 System—Because of the challenges that accompany working with M. tuberculosis (slow growth and Biosafety Level 3 practices), the fast growing nonpathogen M. smegmatis is often used as a model mycobacterium (33). M. smegmatis has a functional SecA2 export system (4, 24). Previously, we showed that M. tuberculosis SecA2 can function in M. smegmatis as demonstrated by the ability of M. tuberculosis SecA2 to complement phenotypes of an M. smegmatis ΔsecA2 mutant (4). To further demonstrate functional conservation between the M. tuberculosis and M. smegmatis SecA2 systems, we tested the ability of M. smegmatis SecA2 to function in M. tuberculosis.
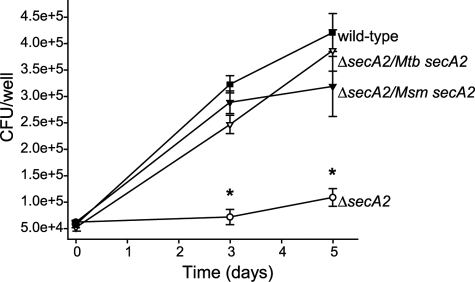
M. smegmatis secA2 Complements the Macrophage Growth Defect of a M. tuberculosis ΔsecA2 Mutant—M. tuberculosis strain H37Rv can grow inside macrophages over a period of several days, whereas the ΔsecA2 mutant is defective in intracellular growth (13). We tested the ability of M. smegmatis SecA2 to function in M. tuberculosis by infecting murine bone marrow-derived macrophages with wild type M. tuberculosis (H37Rv), the ΔsecA2 mutant mc23112, the ΔsecA2 mutant complemented with wild type M. tuberculosis secA2 (ΔsecA2/Mtb secA2), or the ΔsecA2 mutant expressing wild type M. smegmatis secA2 (ΔsecA2/Msm secA2) (Fig. 1). As shown previously (13), H37Rv and the ΔsecA2 mutant complemented with M. tuberculosis secA2 grew similarly in macrophages over a 5-day period of infection, whereas the ΔsecA2 mutant failed to grow in these macrophages. Introduction of M. smegmatis secA2 encoded on pYA810 in single copy at the chromosomal attB site also enabled the ΔsecA2 mutant to grow in macrophages.
FIGURE 1.
Both M. tuberculosis and M. smegmatis secA2 can complement the macrophage growth defect of a M. tuberculosis ΔsecA2 mutant. Murine bone marrow-derived macrophages were infected at a multiplicity of infection of 1.0 with the following strains: H37Rv (wild type), mc23112 (ΔsecA2 mutant), mc23112 complemented with wild type M. tuberculosis secA2 encoded on pMB162 (ΔsecA2/Mtb secA2), and mc23112 complemented with wild type M. smegmatis secA2 encoded on pYA810 (ΔsecA2/Msm secA2). Colony forming units (CFU) were determined by plating macrophage lysates. The infection was performed with triplicate wells for each strain. The error bars represent ± standard deviation of the mean. The data are representative of two independent experiments. *, p < 0.05.
The M. tuberculosis ΔsecA2 Mutant Has a Smooth Colony Phenotype That Is Complemented by M. smegmatis secA2—In contrast to the dull, rough, irregularly shaped colonies of wild type M. tuberculosis, colonies of the ΔsecA2 mutant are round and smooth with a glossy appearance. Notably, this difference is only detected when the bacteria are grown on media containing Tween 80 detergent. When grown on 7H10 plates without Tween 80, wild type and ΔsecA2 mutant colonies are indistinguishable (data not shown). To further test the ability of M. smegmatis secA2 to complement the M. tuberculosis ΔsecA2 mutant, we grew the same M. tuberculosis strains used in the macrophage experiments on 7H10 plates supplemented with 0.05% Tween 80. After 3 weeks of growth, the plates were photographed. As shown in Fig. 2A, the ΔsecA2 mutant was noticeably smoother than wild type H37Rv. Expression of M. tuberculosis secA2 or M. smegmatis secA2 in the ΔsecA2 mutant restored the rough colony morphology to the ΔsecA2 mutant.
FIGURE 2.
Complementation of a smooth colony phenotype of the M. tuberculosis ΔsecA2 mutant. A, wild type and ΔsecA2 mutant M. tuberculosis strains were transformed with pMV306.kan (empty vector), wild type M. tuberculosis secA2 encoded on pMB162 (Mtb secA2), or wild type M. smegmatis secA2 encoded on pYA810 (Msm secA2). B, wild type and ΔsecA2 mutant M. tuberculosis strains were transformed with Walker Box mutant secA2 alleles from M. tuberculosis encoded on pNR7 (secA2 K115R), or M. smegmatis secA2 K129R encoded on pNR25 (secA2 K129R). All transformants were grown on 7H10 plates containing 0.05% Tween 80 and photographed after 3 weeks of growth at 37 °C.
With the above assays, we showed that M. smegmatis SecA2 can substitute for M. tuberculosis SecA2 during growth in macrophages and in standard laboratory conditions. These experiments demonstrate functional conservation between M. tuberculosis SecA2 and M. smegmatis SecA2, and they argue that M. smegmatis is a valid model for studying SecA2 function in protein export.
ATP Binding Is Required for SecA2 Function—M. tuberculosis SecA2 is able to bind and hydrolyze ATP in vitro (25). A lysine to arginine substitution (K115R) in the Walker Box of M. tuberculosis SecA2 significantly decreases ATP binding in vitro (25). To assess the biological significance of ATP binding to SecA2, we tested the ability of SecA2 Walker Box mutants to function in M. smegmatis. Plasmid pNR25, expressing M. smegmatis secA2 K129R (the equivalent of the K115R mutation in M. tuberculosis secA2) was generated by site-directed mutagenesis of wild type secA2 carried on pYA810. Wild type and ΔsecA2 mutant M. smegmatis strains were transformed with pNR25 or pYA810, and the transformants were tested for ΔsecA2 mutant phenotypes, starting with the export of SecA2-dependent proteins.
M. smegmatis secA2 K129R Fails to Support Export of SecA2-dependent Proteins—Msmeg1704 and Msmeg1712 are lipoproteins that depend on SecA2 for export to the cell wall of M. smegmatis (24). To directly assess whether ATP binding is required for SecA2 function in promoting protein export, we tested whether the Walker Box mutant secA2 K129R can support export of these two SecA2-dependent proteins. To monitor export, each protein was tagged with the HA epitope at the C terminus. The tagged proteins were expressed in wild type (WT) M. smegmatis, ΔsecA2 mutant (Δ), ΔsecA2 mutant complemented with wild type secA2 (Δ/WT), or the ΔsecA2 mutant expressing the secA2 K129R allele (Δ/KR). Equivalent expression of SecA2 and SecA2 K129R in the complemented strains was confirmed by Western blot (data not shown). To assay export, the cells were lysed, and the whole cell lysate was then fractionated into cell wall, membrane, and soluble (SOL) fractions by differential ultracentrifugation. Proteins derived from an equivalent number of starting cells were analyzed for each fraction by SDS-PAGE and Western blot with anti-HA antibodies (Fig. 3A). As expected, the ΔsecA2 mutant failed to export Msmeg1704-HA and Msmeg1712-HA to the cell wall fraction (24). Moreover, both proteins accumulated in the soluble fraction of the ΔsecA2 mutant (Δ), which is consistent with an export defect and subsequent retention of unexported protein in the cytoplasm. As was also demonstrated previously, expression of wild type secA2 in the ΔsecA2 mutant (Δ/WT) complemented the export defect of Msmeg1704-HA and Msmeg1712-HA to the cell wall. However, when secA2 K129R was expressed in a ΔsecA2 mutant (Δ/KR), Msmeg1704-HA and Msmeg1712-HA failed to reach the cell wall and accumulated in the soluble fraction. These results indicated that SecA2 K129R is unable to function in exporting M. smegmatis SecA2-dependent proteins.
FIGURE 3.
M. smegmatis secA2 K129R fails to complement and exacerbates M. smegmatis ΔsecA2 mutant phenotypes. A, WT, ΔsecA2 mutant (Δ), ΔsecA2 mutant complemented with wild type secA2 (Δ/WT), and ΔsecA2 mutant expressing secA2 K129R (Δ/KR) M. smegmatis strains were transformed with vectors expressing HA-tagged Msmeg1704 (pNR36) or Msmeg1712 (pNR35). Whole cell lysates (WCL) were prepared from each strain and used to prepare cell wall (CW), membrane (MEM), and SOL fractions. Protein derived from an equal number of starting cells was analyzed by SDS-PAGE and immunoblot using anti-HA antibodies. B, M. smegmatis colonies grown on Mueller Hinton agar for 4 days at 37 °C. Shown from left to right are wild type M. smegmatis (WT) and ΔsecA2 mutant (Δ) strains carrying empty vectors, the ΔsecA2 mutant expressing wild type secA2 (Δ/WT), the ΔsecA2 mutant expressing secA2 K129R (Δ/KR), and wild type M. smegmatis expressing secA2 K129R (WT/KR). Colonies of the Δ/KR strain appeared upon extended incubation. C, the same strains in B were tested for sensitivity to 0.15 m sodium azide spotted on a filter disc. The size of the zone of clearing reports on azide sensitivity of each strain.
M. smegmatis secA2 K129R Fails to Complement and Exacerbates Phenotypes Associated with a M. smegmatis ΔsecA2 Mutant—The M. smegmatis ΔsecA2 mutant exhibits a growth defect on rich agar medium, such as Mueller Hinton, when compared with wild type bacteria (4). This growth phenotype is restricted to plate media; no growth defect is observed when the ΔsecA2 mutant is grown in liquid broth (data not shown). The ΔsecA2 mutant also exhibits increased sensitivity to azide, an inhibitor of ATPases (4). We further tested the functionality of SecA2 K129R by assessing its ability to complement the rich agar and azide phenotypes of the ΔsecA2 mutant.
M. smegmatis secA2 K129R failed to complement both the rich agar growth defect (Fig. 3B) and azide hypersensitivity (Fig. 3C) of the ΔsecA2 mutant. Moreover, expression of SecA2 K129R resulted in phenotypes more severe than that of the ΔsecA2 null mutant. M. smegmatis secA2 K129R Is a Dominant Negative Allele—When secA2 K129R was expressed in wild type M. smegmatis it led to the appearance of phenotypes like those exhibited by the ΔsecA2 mutant. Such a merodiploid strain (secA2+/secA2 K129R) exhibited a rich agar growth defect and azide sensitivity phenotype that is comparable with that seen with the ΔsecA2 mutant (Fig. 3, B and C). Because this strain carries wild type secA2 at the chromosomal locus and has secA2 K129R integrated in the chromosomal attB site, this result indicated that secA2 K129R is dominant negative.
Dominant negative mutations produce a gene product that interferes with the function of the wild type gene product in merodiploids to generate a null mutant phenotype (34). Dominant negative mutations usually occur when a mutant protein can still interact with the same proteins as the wild type protein, but the mutation blocks its function such that only nonproductive protein complexes form (35, 36).
Dominant negative alleles are typically dosage-dependent (34, 37, 38). To directly demonstrate a dosage-dependent dominant negative phenotype, we used an inducible promoter system to test the effects of high level expression of SecA2 K129R in a controlled fashion. The inducible system we used is based on the Tet operator (Tet ON) (23, 31). We cloned secA2 K129R downstream of two TetO sites on plasmid pMP715, which additionally carries the repressor protein TetR, to generate pNR54. TetR binds to the TetO sites and blocks expression of secA2 K129R. However, in the presence of the inducer molecule Atc, TetR will dissociate from TetO sites allowing expression of secA2 K129R. The level of secA2 K129R expression can be controlled by adjusting the concentration of Atc.
Wild type and ΔsecA2 mutant M. smegmatis strains were electroporated with pNR54, and the resulting transformants were grown in Mueller Hinton broth ± Atc induction. Growth was monitored by measuring optical density. After 21 h of induction, serial dilutions of each culture were plated onto Mueller Hinton plates ± Atc supplementation. As the concentration of inducer increased in the growth medium, a corresponding increase in SecA2 K129R expression was detected by SecA2 immunoblots (data not shown). High level expression of SecA2 K129R in both wild type and ΔsecA2 mutant strain backgrounds led to reduced growth in rich liquid medium as revealed by growth curves (Fig. 4, A and B). This was the first time we have observed any growth defect in liquid medium that is related to SecA2. As expression of SecA2 K129R increased, an increasingly severe rich agar growth defect was also observed in both wild type and ΔsecA2 M. smegmatis (Fig. 4, C and D). In fact, growth on agar was inhibited with the highest concentration of Atc tested. As a control, we confirmed that overexpression of wild type SecA2 from the same Tet ON system did not cause the same effects (data not shown). These results showed that the rich agar growth defect associated with SecA2 K129R is dosage-dependent, and they are consistent with secA2 K129R being a true dominant negative. They also showed that high level expression of SecA2 K129R inhibits growth of M. smegmatis.
FIGURE 4.
SecA2 K129R is dosage-dependent. Wild type and ΔsecA2 mutant M. smegmatis strains were transformed with pNR54 encoding inducible secA2 K129R under control of the Tet ON promoter. Strains were grown in Mueller Hinton broth with different concentrations of Atc (closed circle, 0 ng/ml; open circle, 100 ng/ml; closed triangle, 200 ng/ml). Growth of wild type (A) and ΔsecA2 mutant (B) was monitored by measuring A600 nm. At 21 h post-inoculation, serial dilutions of the wild type (C) and ΔsecA2 mutant (D) from each culture in the growth curve were plated onto Mueller Hinton plates, all of which contained 500 ng/ml Atc. Shown are colonies on plates incubated at 37 °C for 3–4 days. Even after extended incubation, colonies did not appear from cultures treated with 200 ng/ml Atc during liquid growth.
Walker Box Mutant secA2 Alleles Are Dominant Negative in M. tuberculosis—The Walker Box mutant alleles of secA2 were also tested in M. tuberculosis. Plasmids expressing M. tuberculosis SecA2 K115R (pNR7) and M. smegmatis SecA2 K129R (pNR25) were introduced into wild type and ΔsecA2 mutant M. tuberculosis, and the resulting strains were plated on 7H10 agar containing Tween 80 (Fig. 2B). In the ΔsecA2 mutant, secA2 K115R and secA2 K129R failed to complement the smooth colony phenotype of the ΔsecA2 mutant, as expected. When expressed in wild type M. tuberculosis, M. tuberculosis secA2 K115R and M. smegmatis secA2 K129R changed the rough colony appearance of wild type to a smooth phenotype similar to that of the ΔsecA2 mutant. These results are consistent with secA2 K115R and secA2 K129R also functioning as dominant negative alleles in pathogenic M. tuberculosis.
Differences in the Subcellular Localization of SecA1, SecA2, and SecA2 K129R—In E. coli, the single SecA is evenly distributed between cell envelope and soluble fractions (39). To establish the subcellular localization of SecA1, SecA2, and SecA2 K129R, we generated cell envelope (comprised of cell wall and cell membrane) and soluble (comprised of cytosol) fractions of M. smegmatis and performed quantitative immunoblot analysis on these fractions with anti-SecA1 and anti-SecA2 antibodies. Immunoblots for the cell wall protein MspA (M. smegmatis porin A), and the soluble cytoplasmic chaperone GroEL were used to establish the purity of the fractions. These experiments demonstrated SecA1 to be equally distributed between cell envelope and soluble fractions (Fig. 5, A and E). This result is like that reported for E. coli SecA (39) and is consistent with SecA1 being the “housekeeping” secretion factor of mycobacteria. The same subcellular fractionation was also performed on the ΔsecA2 mutant and showed that absence of SecA2 had no impact on expression or localization of SecA1 (Fig. 5B).
FIGURE 5.
Subcellular localization of SecA1, SecA2, and SecA2 K129R in M. smegmatis. Cultures of WT (A), ΔsecA2 mutant (Δ, B), ΔsecA2 mutant complemented with secA2 (Δ/WT, C), and ΔsecA2 mutant expressing secA2 K129R (Δ/KR, D) were used to prepare whole cell lysate (WCL), SOL, and cell envelope (ENV) fractions. Protein derived from an equal number of cells was analyzed using anti-SecA1 and anti-SecA2 antibodies. The cell wall porin MspA and the cytoplasmic chaperone GroEL are controls for the fractionation. Percent localization to a given fraction for SecA1 (E) and SecA2 (F) was determined by quantitative immunoblot analysis of the fractions and is reported as the percentage of the total (SOL fraction plus cell envelope). The error bars indicate the standard error of the mean of three independent replicates for each strain.
Subcellular localization of SecA2 revealed a distinguishable feature of the SecAs. Unlike SecA1, in wild type M. smegmatis SecA2 was predominantly localized to the soluble cytosolic fraction (Fig. 5, A and F). M. smegmatis SecA2 protein expressed in a complemented strain was also found to be predominantly cytosolic (Fig. 5C).
When we evaluated the subcellular distribution of SecA2 K129R, we found a striking difference from the cytosolic localization of wild type SecA2. SecA2 K129R was predominantly associated with the cell envelope fraction (Fig. 5, D and F). The shift in the localization pattern of SecA2 K129R suggests the dominant negative protein exerts its effects while trapped in a complex at the membrane. This is consistent with a model of SecA2 normally interacting in a transient manner with a membrane-embedded channel to deliver proteins for export and/or to energize the translocation process.
SecA1 Depletion Impacts Export of the SecA2-dependent Substrate Msmeg1712—If SecA2 uses the canonical SecA1/SecYEG machinery to export its select subset of proteins, then the export of SecA2-dependent proteins should also depend on these essential secretion factors. To study any contribution of essential Sec proteins to SecA2-dependent export, conditional sec strains are required. Recently, an M. smegmatis strain MSE10 was constructed where the chromosomal copy of secA1 can be conditionally regulated with the tetracycline operator/repressor system (23). Using the repressible (Tet OFF) system, the revTetR repressor binds TetO in the presence of Atc to block expression of SecA1 (31).
To test the contribution of SecA1 in SecA2-dependent export, conditional SecA1 strains were engineered to additionally express Msmeg1712-HA. This was achieved with plasmid pNR55 that constitutively expresses both the revTetR repressor protein and Msmeg1712-HA. This plasmid was introduced into MSE10, which contains secA1 cloned behind the TetO operator in the wild type M. smegmatis strain mc2155 (23). Plasmid pNR55 was also introduced into strain JM693, which carries the same secA1 conditional allele in a ΔsecA2 mutant background (see “Experimental Procedures”). The resulting strains were grown in Mueller Hinton with or without Atc for 21 h to deplete SecA1. The Atc treatment did not impair growth of the cultures. The cells were harvested by centrifugation, lysed, and fractionated by differential ultracentrifugation. Subcellular fractions were then analyzed by SDS-PAGE and immunoblot to assess the export of Msmeg1712-HA to the cell wall.
The addition of Atc resulted in greater than 95% reduction of SecA1, as determined by quantitative Western blots. Importantly, under the conditions used depletion of SecA1 did not have any global effect on protein expression as shown by immunoblot analysis of control proteins GroEL or MspA. Thus, our Atc treatment conditions specifically influenced SecA1. As expected, MspA, which is a previously reported SecA1-dependent exported protein, was not exported to the cell wall fraction of M. smegmatis when SecA1 was depleted (23).
In the presence of SecA1 (i.e. –Atc), Msmeg1712-HA was exported to the cell wall and membrane fractions in wild type M. smegmatis (MSE10), as expected. Strikingly, when SecA1 was depleted from MSE10 by Atc addition, Msmeg1712-HA no longer efficiently localized to the cell wall fraction (Fig. 6, A and B). Quantification of our blots showed a 90% reduction in Msmeg1712-HA in the cell wall fraction of the SecA1-depleted strain (+Atc) compared with the untreated strain (–Atc). Furthermore, Msmeg1712-HA specifically accumulated in the soluble fraction when SecA1 was depleted (+Atc), which is consistent with an export defect for Msmeg1712-HA.
FIGURE 6.
Expression of SecA1 and SecA2 is required for export of Msmeg1712-HA. (A) Strain MSE10 (WT/conditional secA1), was either left untreated or treated with 600 ng/ml Atc for 21 h to deplete SecA1. Cell wall (CW), membrane (MEM), and SOL subcellular fractions were prepared from whole cell lysate (WCL) of these cultures. For each fraction, protein derived from an equal number of starting cells was analyzed by SDS-PAGE and immunoblot. The percentage of total Msmeg1712-HA localized to a given fraction from ±Atc-treated MSE10 (B) or ±Atc-treated JM693 (C)(ΔsecA2/conditional secA1) cultures was quantified using phosphorimaging and is reported as the percentage of the total (cell wall plus membrane plus SOL). The results are the mean of three independent experiments ± standard error.*, p < 0.05.
A similar experiment was conducted with the JM693 strain that has the conditional secA1 in the ΔsecA2 mutant background (Fig. 6C). As expected because of the ΔsecA2 mutation, even in the absence of Atc this strain exhibited an export defect for Msmeg1712-HA. The export defect seen when only SecA2 was absent was comparable with that observed when only SecA1 was depleted (+Atc) in MSE10 (Fig. 6, B and C). When SecA1 was depleted (+Atc) in the ΔsecA2 mutant background of JM693, the defect in Msmeg1712-HA export was no greater than that seen when only secA2 was deleted/absent (–Atc) (Fig. 6, B and C).
These results indicated that SecA1 and SecA2 are both required for optimal export of Msmeg1712-HA. On their own, neither SecA1 nor SecA2 are sufficient to export Msmeg1712-HA. These data are the first to show that the canonical SecA1 has a role in exporting a substrate of the accessory SecA2 pathway.
DISCUSSION
The accessory SecA2 system of mycobacteria is a recent discovery. In this study we showed that ATP binding is important to the biological function of SecA2, we identified SecA2 as being predominantly localized to the cytosol, and we demonstrated a role for SecA1 in SecA2-dependent export. In addition, we showed that failure to express any SecA2 is better than expressing a Walker Box mutant secA2 allele, and we showed secA2 Walker Box mutants to be dosage-dependent dominant negative alleles. These results provide valuable information for understanding the mechanism of SecA2 export.
SecA2 Must Bind ATP to Function in Vivo—A previous study showed that SecA1 and SecA2 of M. tuberculosis are functional ATPases in vitro (25). Mutations in the Walker A (K108R) and Walker B (D209N) motifs of E. coli SecA destroy ATP binding and render SecA nonfunctional in a variety of in vitro and in vivo assays (19, 40). In E. coli SecA, these Walker Box mutations also alter the subcellular localization of SecA by preventing its dissociation from the membrane as a consequence of failing to cycle between ATP binding and hydrolysis. Here, we tested the corresponding Walker Box mutations in M. tuberculosis SecA2 (K115R) and M. smegmatis SecA2 (K129R).
In M. tuberculosis, we showed that the K115R substitution in the Walker A motif of SecA2 was unable to complement a smooth colony morphology phenotype of the M. tuberculosis ΔsecA2 mutant. Substitution of a Walker Box residue (D216N) in the Walker B motif also rendered M. tuberculosis SecA2 nonfunctional (data not shown). SecA2 K115R is also unable to carry out the SecA2 function of promoting M. tuberculosis growth in macrophages (25). We believe that both the intracellular growth defect and smooth colony morphology of the ΔsecA2 mutant are due to the inability of the ΔsecA2 mutant to export proteins that rely on the accessory SecA2 system. In regard to the altered colony morphology of the M. tuberculosis ΔsecA2 mutant, which we report here for the first time, we speculate that it is due to defects in the bacterial cell surface that arise from failure to export select cell wall biosynthetic enzymes. Using M. smegmatis and SecA2 K129R, we directly demonstrated the importance of SecA2 ATP binding to the process of exporting Msmeg1704 and Msmeg1712.
Walker Box Mutant secA2 Alleles Are Dominant Negative—The SecA2 Walker Box mutant alleles were not only nonfunctional, as shown by the failure to complement, but in several assays the Walker Box mutant alleles led to phenotypes worse than that of a ΔsecA2 null mutant. Dominant negative mutations often exhibit this type of behavior because many dominant negative proteins still bind to their interacting proteins and result in production of nonfunctional complexes (35–38). When expressed in wild type M. tuberculosis or M. smegmatis, the Walker Box mutant secA2 alleles were, in fact, dominant negative. Moreover, by using an inducible secA2 K129R allele expressed in M. smegmatis, we showed that the severity of phenotypes associated with SecA2 K129R was dosage-dependent.
There is a precedent for substitutions in the Walker Box lysine residue that we altered being dominant negative. As mentioned above, the E. coli SecA K108R mutant was instrumental in demonstrating the importance of ATPase activity for the canonical SecA (19). In an in vitro translocation assay, SecA K108R inhibits proOmpA export in the presence of wild type SecA. Although not specifically referred to as a dominant negative in this work, this result is consistent with SecA K108R being dominant negative. The DotB ATPase that is part of the Type IV secretion system of Legionella pneumophila and the PilQ ATPase that is required for R64 pilus biogenesis are other examples of ATPases that upon mutation of their Walker Box motifs behave as dominant negatives (35, 36).
The Subcellular Localization of SecA1 and SecA2 Differs—Previous studies in mycobacteria show that SecA1 and SecA2 are not functionally redundant; one SecA cannot compensate for the other (4). In this work, we demonstrated a distinct difference between SecA1 and SecA2 in their subcellular localization. SecA1 was equally distributed between cell envelope and cytoplasmic fractions of M. smegmatis, as is the case for E. coli SecA. In contrast, SecA2 was found predominantly in the cytoplasmic fraction. Fractions prepared from M. tuberculosis whole cell lysates showed a similar difference in subcellular localization of SecA1 and SecA2 (data not shown). This result is significant in suggesting that the roles played by SecA1 and SecA2 in protein export differ. Interestingly, the localization of the two SecA homologs of S. parasanguinis also differs (41). In S. parasanguinis, the canonical SecA is again found equally distributed in cell envelope and soluble fractions. However, opposite to our finding, in S. parasanguinis SecA2 is predominantly associated with the cell envelope.
A Model to Explain SecA2 Function and the Phenotypes of Walker Box Mutant secA2 Alleles—From the results of our study, we developed a model for SecA2 function in mycobacteria (Fig. 7). In a wild type cell, we propose that SecA2 predominantly resides in the cytosol, but it interacts in a transient fashion with a membrane-embedded translocase to promote export of a select subset of proteins. The Walker Box mutation (K129R) renders SecA2 catalytically “dead.” However, this dead protein appears able to still interact with its binding partners but because of the inability to hydrolyze ATP remains locked in a complex. In this way, the dead SecA2 protein could remain at the membrane titrating up interacting proteins, which is consistent with the altered subcellular distribution of SecA2 K129R. If the titrated SecA2-interacting proteins have additional functions in the cell, this would lead to phenotypes more severe than those exhibited by a ΔsecA2 null mutation. In fact, this is what we observed. With this model in mind, as the levels of SecA2 K129R increase we would also expect the observed phenotypes to become more severe. Because of the growth inhibitory phenotypes observed upon high level expression of SecA2 K129R, we believe the proteins tied up by SecA2 K129R are important to an essential process. Unlike the accessory SecA2/Y2 systems of S. gordonii and S. parasanguinis, the accessory SecA2 system in mycobacteria lacks an accessory SecY homolog or obvious translocase (6, 27, 42). Consequently, the essential SecA1/SecYEG translocase is an attractive candidate for being the interacting protein complex tied up by SecA2 K129R and for being the translocase used by SecA2. However, it is also possible that SecA2 interacts with an unidentified membrane-embedded translocase to export precursors out of the cytoplasm.
FIGURE 7.
Model to explain SecA2 function and the phenotypes of Walker Box secA2 alleles. SecA2 exports select proteins across the cytoplasmic membrane through a mechanism involving SecA1 and a membrane-embedded translocase. Amino acid substitution in the ATP-binding site of SecA2, as in SecA2 K129R, results in a dominant negative SecA2 protein trapped at the translocase and compromising an essential process.
If SecA2 uses the canonical SecA1/SecYEG translocase to export its subset of proteins, then proteins that depend on SecA2 for export should also depend on SecA1/SecYEG. Here we showed that when SecA1 was depleted in wild type M. smegmatis, Msmeg1712 was not efficiently exported to the cell wall. The export defect observed upon SecA1 depletion was comparable with that exhibited by a ΔsecA2 mutant. This result reveals a role for the essential SecA1 in the SecA2-dependent process of exporting Msmeg1712.
Although we showed a requirement for SecA1 in mycobacterial SecA2 export, the precise nature of the role of SecA1 is not yet clear. One possibility is that SecA1 and SecA2 directly interact to form a heterodimer capable of exporting a subset of proteins, like Msmeg1712, through the SecYEG translocase. In E. coli, SecA forms dimers, although the exact role of dimers in protein translocation remains in dispute (43–45). However, as of now, there is no evidence of a physical interaction between SecA1 and SecA2. An alternative is that the role of SecA2 is to recognize precursor substrates in the cytoplasm that are normally overlooked by SecA1. Upon recognition, SecA2 may then deliver these precursor proteins to the SecA1/SecYEG translocase complex for export. This type of function for SecA2 would be compatible with its primarily cytosolic location. Another possibility is that SecA2 works with SecA1 to promote export via a novel translocase.
A final possibility, which we consider less likely, is that the contribution of SecA1 is to assemble integral membrane proteins into a novel translocase for SecA2-dependent proteins. It appears that E. coli SecA can participate in assembling certain domains of some multi-spanning integral membrane proteins (46). However, the signal recognition particle GTPase, not SecA, is the requisite factor required for integral membrane insertion, and a role for SecA may be rare because there are integral membrane proteins clearly shown to not require SecA (47, 48). Furthermore, there is no evidence that SecA1 functions in assembling cytoplasmic membrane proteins of mycobacteria.
Our results from studying the Walker Box mutants are consistent in both M. tuberculosis and M. smegmatis, and we demonstrated the ability of each SecA2 to function in cross-species complementation experiments. However, there currently remains a discrepancy in the types of exported proteins identified as dependent on SecA2 for export in each of these mycobacteria. So far, SecA2-dependent proteins identified for M. tuberculosis lack obvious signal sequences (SodA) (5), whereas the two known SecA2-dependent proteins (Msmeg1704 and Msmeg1712) of M. smegmatis contain signal sequences (24). We believe this difference reflects our limited appreciation of the proteins exported by the SecA2 export system in each species. For this reason, we are continuing our search for additional SecA2-dependent proteins.
Clearly, identifying proteins that interact with SecA2 will be important for fully understanding the relationship between the essential SecA1/SecYEG pathway and the accessory SecA2 pathway. Dominant negative alleles, often in combination with suppressor mutations, can be extremely useful in identifying proteins that work together in a biological pathway. During the course of the work described here, we isolated spontaneous suppressors of the dominant negative secA2 K129R allele in M. smegmatis. Going forward, we believe these suppressors will be useful in identifying proteins that work with SecA2 in protein export. It is our hope that a thorough understanding of the accessory SecA2 system will aid the identification of new targets for anti-mycobacterial therapies.
Supplementary Material
Acknowledgments
We thank Martin Pavelka and Brendaliz Santiago for providing pMP715 and Sabine Ehrt for providing technical advice, strains, and plasmids. We also thank Michael Niederweis for anti-MspA antibodies. Finally, we thank Carolyn Teschke, Meghan Feltcher, and other members of the Braunstein lab for technical assistance and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI054540.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: Atc, anhydrotetracycline; WT, wild type; HA, hemagglutinin; SOL, soluble.
S. Bardarov and W. R. Jacobs, Jr., unpublished observation.
References
- 1.Finlay, B. B., and Falkow, S. (1997) Microbiol. Mol. Biol. Rev. 61 136–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdallah, A. M., Gey van Pittius, N. C., Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Grauls, C. M., Appelmelk, B. J., and Bitter, W. (2007) Nat. Rev. Microbiol. 5 883–891 [DOI] [PubMed] [Google Scholar]
- 3.Gerlach, R. G., and Hensel, M. (2007) Int. J. Med. Microbiol. 297 401–415 [DOI] [PubMed] [Google Scholar]
- 4.Braunstein, M., Brown, A. M., Kurtz, S., and Jacobs, W. R., Jr. (2001) J. Bacteriol. 183 6979–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein, M., Espinosa, B., Chan, J., Belisle, J. T., and Jacobs, W. R. J. (2003) Mol. Microbiol. 48 453–464 [DOI] [PubMed] [Google Scholar]
- 6.Rigel, N. W., and Braunstein, M. (2008) Mol. Microbiol. 69 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siboo, I. R., Chaffin, D. O., Rubens, C. E., and Sullam, P. M. (2008) J. Bacteriol. 190 6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensing, B. A., and Sullam, P. M. (2002) Mol. Microbiol. 44 1081–1094 [DOI] [PubMed] [Google Scholar]
- 9.Chen, Q., Wu, H., and Fives-Taylor, P. M. (2004) Mol. Microbiol. 53 843–856 [DOI] [PubMed] [Google Scholar]
- 10.Caspers, M., and Freudl, R. (2008) Arch. Microbiol. 189 605–610 [DOI] [PubMed] [Google Scholar]
- 11.Lenz, L. L., and Portnoy, D. A. (2002) Mol. Microbiol. 45 1043–1056 [DOI] [PubMed] [Google Scholar]
- 12.Lenz, L. L., Mohammadi, S., Geissler, A., and Portnoy, D. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtz, S., McKinnon, K. P., Runge, M. S., Ting, J. P., and Braunstein, M. (2006) Infect. Immun. 74 6855–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong, Y. Q., Bensing, B. A., Bayer, A. S., Chambers, H. F., and Sullam, P. M. (2008) Microb. Pathog. 45 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driessen, A. J., and Nouwen, N. (2008) Annu. Rev. Biochem. 77 643–667 [DOI] [PubMed] [Google Scholar]
- 16.Papanikou, E., Karamanou, S., Baud, C., Frank, M., Sianidis, G., Keramisanou, D., Kalodimos, C. G., Kuhn, A., and Economou, A. (2005) J. Biol. Chem. 280 43209–43217 [DOI] [PubMed] [Google Scholar]
- 17.Hendrick, J. P., and Wickner, W. (1991) J. Biol. Chem. 266 24596–24600 [PubMed] [Google Scholar]
- 18.Schiebel, E., Driessen, A. J., Hartl, F. U., and Wickner, W. (1991) Cell 64 927–939 [DOI] [PubMed] [Google Scholar]
- 19.Mitchell, C., and Oliver, D. (1993) Mol. Microbiol. 10 483–497 [DOI] [PubMed] [Google Scholar]
- 20.Klose, M., Schimz, K. L., van der Wolk, J., Driessen, A. J., and Freudl, R. (1993) J. Biol. Chem. 268 4504–4510 [PubMed] [Google Scholar]
- 21.van der Wolk, J., Klose, M., Breukink, E., Demel, R. A., de Kruijff, B., Freudl, R., and Driessen, A. J. (1993) Mol. Microbiol. 8 31–42 [DOI] [PubMed] [Google Scholar]
- 22.Sassetti, C. M., Boyd, D. H., and Rubin, E. J. (2003) Mol. Microbiol. 48 77–84 [DOI] [PubMed] [Google Scholar]
- 23.Guo, X. V., Monteleone, M., Klotzsche, M., Kamionka, A., Hillen, W., Braunstein, M., Ehrt, S., and Schnappinger, D. (2007) J. Bacteriol. 189 4614–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons, H. S., Wolschendorf, F., Abshire, M., Niederweis, M., and Braunstein, M. (2007) J. Bacteriol. 189 5090–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou, J. M., D'Lima, N. G., Rigel, N. W., Gibbons, H. S., McCann, J. R., Braunstein, M., and Teschke, C. M. (2008) J. Bacteriol. 190 4880–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, H., Bu, S., Newell, P., Chen, Q., and Fives-Taylor, P. (2007) J. Bacteriol. 189 1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., 3rd, Tekaia, F., Badcock, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R., Devlin, K., Feltwell, T., Gentles, S., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., and Barrell, B. G. (1998) Nature 393 537–544 [DOI] [PubMed] [Google Scholar]
- 28.Pavelka, M. S., Jr., and Jacobs, W. R., Jr. (1999) J. Bacteriol. 181 4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braunstein, M., Bardarov, S. S., and Jacobs, W. R. J. (2002) Methods Enzymol. 358 67–99 [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 31.Ehrt, S., Guo, X. V., Hickey, C. M., Ryou, M., Monteleone, M., Riley, L. W., and Schnappinger, D. (2005) Nucleic Acids Res. 33 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niederweis, M., Ehrt, S., Heinz, C., Klocker, U., Karosi, S., Swiderek, K. M., Riley, L. W., and Benz, R. (1999) Mol. Microbiol. 33 933–945 [DOI] [PubMed] [Google Scholar]
- 33.Reyrat, J. M., and Kahn, D. (2001) Trends Microbiol. 9 472–474 [DOI] [PubMed] [Google Scholar]
- 34.Appling, D. R. (1999) Methods 19 338–349 [DOI] [PubMed] [Google Scholar]
- 35.Sexton, J. A., Pinkner, J. S., Roth, R., Heuser, J. E., Hultgren, S. J., and Vogel, J. P. (2004) J. Bacteriol. 186 1658–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai, D., Horiuchi, T., and Komano, T. (2001) J. Biol. Chem. 276 17968–17975 [DOI] [PubMed] [Google Scholar]
- 37.Sun, Y., Dong, Z., Nakamura, K., and Colburn, N. H. (1993) FASEB J. 7 944–950 [DOI] [PubMed] [Google Scholar]
- 38.Budd, M. E., Choe, W., and Campbell, J. L. (2000) J. Biol. Chem. 275 16518–16529 [DOI] [PubMed] [Google Scholar]
- 39.Cabelli, R. J., Dolan, K. M., Qian, L. P., and Oliver, D. B. (1991) J. Biol. Chem. 266 24420–24427 [PubMed] [Google Scholar]
- 40.Jilaveanu, L. B., Zito, C. R., and Oliver, D. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7511–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, Q., Wu, H., Kumar, R., Peng, Z., and Fives-Taylor, P. M. (2006) FEMS Microbiol. Lett. 264 174–181 [DOI] [PubMed] [Google Scholar]
- 42.Cole, S. T., Eiglmeier, K., Parkhill, J., James, K. D., Thomson, N. R., Wheeler, P. R., Honore, N., Garnier, T., Churcher, C., Harris, D., Mungall, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R. M., Devlin, K., Duthoy, S., Feltwell, T., Fraser, A., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., Lacroix, C., Maclean, J., Moule, S., Murphy, L., Oliver, K., Quail, M. A., Rajandream, M. A., Rutherford, K. M., Rutter, S., Seeger, K., Simon, S., Simmonds, M., Skelton, J., Squares, R., Squares, S., Stevens, K., Taylor, K., Whitehead, S., Woodward, J. R., and Barrell, B. G. (2001) Nature 409 1007–1011 [DOI] [PubMed] [Google Scholar]
- 43.Das, S., Stivison, E., Folta-Stogniew, E., and Oliver, D. (2008) J. Bacteriol. 190 7302–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, H., Na, B., Yang, H., and Tai, P. C. (2008) J. Bacteriol. 190 1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Or, E., and Rapoport, T. (2007) FEBS Lett. 581 2616–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbanus, M. L., Scotti, P. A., Froderberg, L., Saaf, A., de Gier, J. W., Brunner, J., Samuelson, J. C., Dalbey, R. E., Oudega, B., and Luirink, J. (2001) EMBO Rep. 2 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serek, J., Bauer-Manz, G., Struhalla, G., van den Berg, L., Kiefer, D., Dalbey, R., and Kuhn, A. (2004) EMBO J. 23 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner, S., Pop, O., Haan, G. J., Baars, L., Koningstein, G., Klepsch, M. M., Genevaux, P., Luirink, J., and de Gier, J. W. (2008) J. Biol. Chem. 283 17881–17890 [DOI] [PubMed] [Google Scholar]
- 49.Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., Bansal, G. P., Young, J. F., Lee, M. H., and Hatfull, G. F. (1991) Nature 351 456–460 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.