Abstract
Mitochondrial pre-messenger RNAs (pre-mRNAs) in African trypanosomes require RNA editing in order to mature into functional transcripts. The process involves the addition and/or removal of U nucleotides and is mediated by a high-molecular-mass complex, the editosome. Editosomes catalyze the reaction through an enzyme-driven pathway that includes endo/exoribonuclease, terminal uridylate transferase and RNA ligase activities. Here we show that editing involves an additional reaction step, a 3′ nucleotidyl phosphatase activity. The activity is associated with the editing complex and we demonstrate that the editosomal proteins TbMP99 and TbMP100 contribute to the activity. Both polypeptides contain endo-exonuclease-phosphatase domains and we show that gene ablation of either one of the two polypeptides is compensated by the other protein. However, simultaneous knockdown of both genes results in trypanosome cells with reduced 3′ nucleotidyl phosphatase and reduced editing activity. The data provide a rationale for the exoUase activity of the editosomal protein TbMP42, which generates nonligatable 3′ phosphate termini. Opposing phosphates at the two pre-mRNA cleavage fragments likely function as a roadblock to prevent premature ligation.
INTRODUCTION
RNA editing in kinetoplast protozoa describes a post-transcriptional processing phenomenon that alters mitochondrial transcripts by the insertion and/or deletion of uridylyl residues. The reaction is mediated by a unique protein complex, which has been termed the editosome. Editosomes consist of about 20 polypeptides that process pre-edited mRNAs (pre-mRNAs) through a pathway of enzyme-driven reaction steps (1–3). The basal steps of the reaction cascade involve endo/exoribonuclease (4–8), terminal uridylyl transferase (9,10) and RNA ligase activities (11–13) in addition to accessory activities such as matchmaking-type RNA/RNA annealing factors (14–18) and RNA helicases (19,20). Although the structure of the editosome is unknown, the protein inventory of the complex is well studied. It contains polypeptides for every step of the above-described reaction cycle; however, many of the activities are present in pairs or even higher numbers. This redundancy is not understood but has been used to suggest that deletion and insertion-type RNA editing may be executed by separate subcomplexes (21). Within this context the 3′ to 5′ exoribonuclease (exoUase) activity of the editosome is of special interest. The activity is required for the release of U nucleotides from a deletional editing site as well as for removing excess Us from an insertion site (22). Three editosomal proteins have been shown to execute exoUase activity in vitro: TbMP100, TbMP99 and TbMP42 (23–25). TbMP100 and TbMP99 are highly similar proteins (29% identity and 46% similarity on the amino acid level) that contain 5′–3′ exonuclease as well as endo-exo-phosphatase (EEP) motifs (26,27). The two proteins are characterized by a single-strand (ss) and U-specific 3′ to 5′ exoUase in vitro activity, which is abolished by point mutations within the EEP domain (25). Gene ablation of TbMP100 through RNA interference (RNAi) results in a severe growth defect and the analysis of TbMP100-deficient mitochondrial extracts shows reduced in vitro insertion and deletion activities 200 h postinduction (24).
TbMP42, the third exoUase protein, has no canonical nuclease consensus motif (28). The polypeptide is characterized by two C2H2-type Zn-fingers at its N-terminus and a putative oligonucleotide/oligosaccharide-binding domain (OB-fold) at its C-terminus (28). Recombinant TbMP42 (r42) has been shown to bind nucleic acid substrates with nanomolar affinity and it executes both endo- and exoribonuclease activities in vitro (23). TbMP42-deficient mitochondrial extracts show reduced insertion-editing activity, as well as reduced endo- and exoribonuclease activity. All three activities can be restored through the addition of r42 to TbMP42-minus extracts (23). The in vitro observed endonucleolytic activity might be secondary in vivo since it was shown that TbMP90 and TbMP61 act as endoribonuclease in deletion- and insertion-type editing (29,30). The catalytic activity of TbMP42 resides within the OB-fold of the protein (23). It was further shown that r42 acts as a topology-dependent ribonuclease that is sensitive to base stacking (31). The enzyme relies on Zn2+-ions and likely follows a metal–ion catalysis reaction pathway (31). However, recent data show that upon knockdown of TbMP42 deletion-type editing is not affected (32,33). Hence, although all three proteins (TbMP100, 99 and 42) execute exoUase activity in vitro, their exact function in vivo remains controversial. Since functional redundancy is a noticeable feature of the editosomal protein inventory (3) it is not unexpected to find two or more proteins with exoUase activity. Whether these proteins execute their function at different editing sites, as proposed for the endonuclease TbMP67 (34), or belong to distinct editing subcomplexes remains elusive.
Here we present an analysis of the U removal reaction that is catalyzed by r42. We demonstrate that the enzyme generates 3′ nucleoside monophosphates, which results in nonligatable phosphate termini at the 3′ ends of the 5′ pre-mRNA cleavage fragments. This likely represents a roadblock to prevent premature ligation and we demonstrate that editosomes can resolve the situation by executing a 3′ nucleotidyl phosphatase activity. We further show that gene ablation of the two endo-exo-phosphatase motif-containing proteins TbMP100 and TbMP99 results in trypanosome cell lines with reduced nucleotidyl phosphatase activity and reduced RNA-editing activity. Thus, we suggest that RNA editing involves a 3′ nucleotidyl phosphatase activity.
MATERIALS AND METHODS
Trypanosome cell growth
Insect stage Trypanosoma brucei brucei strains 427 (35) and 29-13 (36) were grown in SDM-79 medium as described (37).
20S editosome preparation and RNA-editing assays
Mitochondrial vesicles were isolated, lyzed at isotonic conditions (38) and fractionated in linear 10–35% (v/v) glycerol gradients as described (39). The presence of the RNA-editing ligases TbMP52 and TbMP48 was monitored by autoadenylation (40). Samples were separated in SDS-containing 12% (w/v) polyacrylamide gels prior to phosphorimaging. RNA substrates for the precleaved insertion assay (PIA) and the precleaved deletion assay (PDA) were prepared, radioactively labeled and purified as described (23,41,42). Prior to assaying, 50 fmol radioactively labeled 5′ mRNA cleavage fragment (5′ CF) (specific activity ∼0.3 µCi/pmol) was annealed with 1 pmol gRNA and 1 pmol 5′ phosphorylated 3′ mRNA cleavage fragment (3′ CF) (5 min at 65°C and 10 min at RT). PIAs were carried out for 3 h at 27°C using 1–2.5 µg protein of fractionated mitochondrial lysate in editing buffer (EB) [20 mM HEPES/KOH pH 7.5, 30 mM KCl, 10 mM Mg(OAc)2] containing 0.2 mM DTT, 0.5 mM ATP and 0.04 µM UTP, whereas UTP was omitted in PDAs. Reaction products were resolved in denaturing polyacrylamide gels [18% (w/v), 8 M urea] and visualized by phosphorimaging. Product formation was quantified densitometrically using ImageGauge V4.23 (Fuji Film Science Lab). RNA sequences for PIA: 5′ CF (18 nt) – GGAAGUAUGAGACGUAGG, 3′ CF (13 nt) – AUUGGAGUUAUAG-NH2 and gRNA-PCA6 (34 nt) – CUAUAACUCCGAUAAACCUACGUCUCAUACUUCC. PDA: 5′ CF (22 nt) – GGAAAGGGAAAGUUGUGAUUUU, 3′ CF (15 nt) – GCGAGUUAUAGAAUA-NH2, gRNA-A6(14)PC_del (33 nt) – GGUUCUAUAACUCGCUCACAACUUUCCCUUUCC.
Exoribonuclease assay
Recombinant TbMP42 was prepared as described (23). Exoribonuclease assays with r42 were carried out in EB containing 5 mM CaCl2, 0.1 mM ZnSO4 and 1 M urea. Experimental procedures were as in (31) using the PDA RNA substrate.
Phosphatase assay
Fractionated mitochondrial lysate was incubated with 0.5 pmol of α-[32P]-mononucleoside 3′,5′ bisphosphate (5′-[32P]-pCp)-labeled phosphatase RNA substrates (spec. activity ∼0.04 µCi/pmol) in EB at 27°C for up to 3 h. Two RNA substrates were used: RNA31 (31 nt) – GGGAAAGUUGUGAUUUUUGCGAGUUAUAGCG and RNA8 (8nt) – GCGCUCCC. In pH optimization experiment, MES substituted for HEPES in a range of pH 3.8 to 8.0. Bivalent cation requirements were studied using EDTA, EGTA and 1,10 phenanthroline ranging from 0.1 to 50 mM. Reaction products were separated in 18% (w/v) polyacrylamide gels containing 8 M urea, analyzed by phosphorimaging and quantified using ImageGauge V4.23 (Fuji Film Science Lab). Dephosphorylation within the editing context used the setup as described for the PIA with the radioactive label introduced 3′ at the 5′ CF by ligation of 5′-[32P]-pCp resulting in 5′ hydroxyl- and 3′ phosphate containing 5′ CF. To obtain 5′/3′ hydroxyl-containing 5′ CF, the pCp-labeled RNA was treated with alkaline phosphatase.
Nucleotide analysis
A 55-nt-long RNA was produced by in vitro transcription as described (43) with the exception of using α-[32P]-CTP instead of α-[32P]-UTP to introduce the radioactive label. The RNA transcript was purified by size-exclusion chromatography, EtOH precipitation and subsequent gel purification. RNA (0.5 pmol) was incubated with 10 µg r42 for 2× 4 h at 37°C. Reaction products were resolved by two-dimensional (2D) thin-layer chromatography (TLC) on PEI-cellulose plates (44). First dimension: 50:28.9:1.1 iso-butyric acid:H2O:NH4OH [25% (v/v)] for 5 h; 2nd dimension: 100:60:2 (v/w/v) NaxHyPO4 (pH 6.8):(NH4)2SO4:n-propanol for 4 h. Plates were dried before running the second dimension and prior to phosphorimaging.
Gene silencing through RNAi and characterization of 99 single knockdown (skd), 100 skd and 99/100 double knockdown cell lines
Gene ablation of TbMP99 and TbMP100 was carried out based on the inducible RNAi system by Wang et al. (45) using the hairpin-vector. For the TbMP99 single knockdown (99 skd), a 374-bp DNA fragment (ORF positions 1150–1524) was used. For the TbMP100 single knockdown (100 skd), a 361-bp DNA fragment (ORF positions 1119–1480) was used and the bler-cassette of that vector was replaced with the blasticidinr (blast) cassette. Strain 29-13 (36) was transfected with either the 99-skd or the 100-skd RNAi construct as described (23). Clonal cell lines were established by limited dilution. To obtain the TbMP99/100 double knockdown (99/100 dkd) cell line, the clonal 99-skd cell line was transfected with the 100-skd RNAi construct and cloned out. Cells were grown and induced for 6 days as described (23). Total RNA extraction was carried out according to (46). Transcript abundance was determined by RT-PCR using the primer pair GGCATCAAGAATTTTCGGTGGAC and TTGTCACAACAACCTGTAGCACAC for the TbMP99 gene silencing and the primer pair TAAGAAGGCGAAGGAAGGGG and CGCATAATGCAGCAAGATGAC for the TbMP100 gene silencing. The transcript of the α-tub gene served as an internal control. PCR products were resolved in nondenaturing 5% (w/v) polyacrylamide gels and visualized by ethidiumbromide (EtBr)-staining.
Homology modeling
Sequences of TbMP99, TbMP100 (DB-source accession AY228172.1 and AY228171.1) and Synaptojanin (PDB-ID 1I9Y B) were aligned using T_coffee (47). Output files were transferred to the Swiss Model Alignment Interface (48) and homology models were calculated. Images were rendered using PyMOL (http://www.pymol.org).
RESULTS
The exoUase activity of TbMP42 generates 3′-phosphate termini
The editosomal protein TbMP42 is a single strand (ss)-specific endo/exoribonuclease that is sensitive to base stacking (31). The enzyme likely follows a Zn2+-dependent two-metal–ion reaction pathway and recombinant TbMP42 (r42) has been shown to cleave and process synthetic gRNA/pre-mRNA hybrid RNAs in a distributive fashion (23,31).
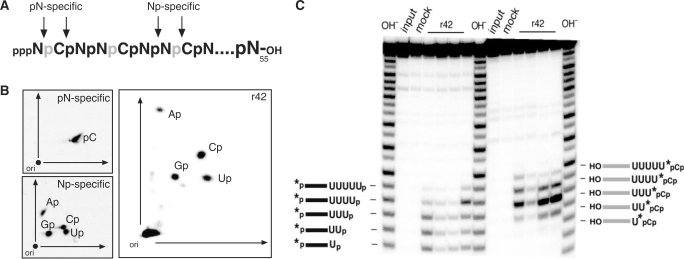
In order to analyze the biochemical consequences of the TbMP42-mediated trimming reaction we examined the reaction products of the exonucleolytic activity of r42. For that we used an RNA oligonucleotide with a length of 55 nt (RNA55) that was transcribed in vitro in the presence of α-[32P]-CTP (Figure 1A). Following transcription, RNA55 was digested with r42 and compared to control digests with nuclease P1 and a cocktail of RNase A, RNase T1 and RNase T2. RNase P1 has been shown to generate 5′ nucleoside monophosphates (5′ NMP; pN) (49), while RNase A, RNase T1 and RNase T2 all generate 3′ NMP's (Np) (50–52). Digestion products were separated by 2D TLC (44). Figure 1B shows a representative result. Due to its cleavage specificity nuclease P1 only generates radioactive 5′ CMP (pC). In the case of the 3′ NMP-generating RNases the radioactive label is ‘transferred’ to the 3′ neighboring nucleoside thereby generating all four radioactive 3′ NMPs: Ap, Cp, Gp and Up. Digestion of RNA55 with r42 shows the same specificity, which demonstrates that r42 proceeds through a reaction mechanism that generates 3′ nucleoside monophosphates (Np).
Figure 1.
Nucleotide analysis of the r42-mediated trimming reaction. (A) Schematic representation of the input RNA (RNA55). RNA55 was co-transciptionally labeled using α-[32P]-CTP (radioactive phosphates are depicted in gray) and arrows indicate potential cleavage sites of pN- and Np-specific ribonucleases. (B) 2D thin-layer chromatography of the hydrolysis products of RNA55 generated by digestion with r42 or with pN- and Np-specific ribonucleases. (C) Gelelectrophoretic analysis of r42-derived cleavage fragments in comparison to RNA fragments derived from alkaline hydrolysis (OH−). Panel to the left: The input RNA is 5′ [32P]-labeled (*) and was incubated with four different r42 preparations (left to right) at limiting protein concentrations. The generated fragments co-migrate with their corresponding OH ladder fragments and thus must carry 3′ phosphates. Panel to the right: Complementary analysis of the corresponding 3′ cleavage fragments (radiolabeled at their 3′ ends through the ligation of 5′-[32P] pCp). These fragments do not co-migrate with their OH-treated counterparts, thereby indicating the presence of 5′ OH groups. Schematic representations of the different RNAs are given on the left and right. Mock represents an RNA sample incubated in the absence of r42.
3′ phosphate ends are also generated at limiting r42 conditions using synthetic gRNA/pre-mRNA hybrids as substrate molecules. Figure 1C shows a representative analysis of four different r42 preparations. The electrophoretic mobility of r42-derived, 5′ [32P]-labeled RNA fragments was compared to fragments generated by alkaline hydrolysis. OH− treatment results in the formation of fragments with 3′ phosphate ends and as can be seen from Figure 1C these fragments co-migrate with all r42-derived fragments. Thus, r42 generates 3′ phosphate termini. Although the four protein preparations show different specific activities, their cleavage specificity is identical (Figure 1C). To further verify the presence of 3′ phosphate termini we analyzed the corresponding 3′ cleavage fragments through 3′ end-labeling with 5′-[32P] pCp. These fragments must carry a 5′ OH group and as a consequence should not co-migrate with 5′ [32P]-labeled fragments of a hydroxyl ladder. Figure 1C (panel to the right) clearly demonstrates this nonidentical electrophoretic behavior.
20S editosomes generate 3′-phosphate termini
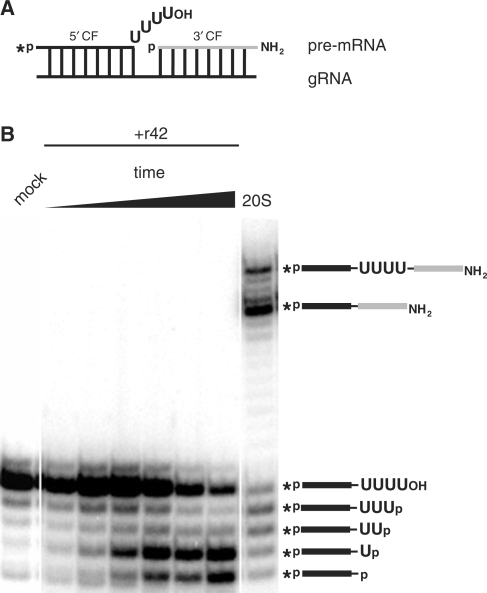
The described result raises the question whether the Np-generating specificity of r42 is relevant within the context of the RNA-editing reaction cycle. For that we compared the r42-derived exonucleolytic reaction products with RNA fragments generated by incubation with 20S editosomes. Using the precleaved U deletion in vitro editing system developed by Seiwert and Stuart (41), we monitored the gRNA-directed exonucleolytic degradation of 4 Us from a radiolabeled 5′ pre-mRNA fragment. The generated products were separated in denaturing, high-resolution polyacrylamide gels, in which the presence or absence of a terminal phosphate group can be visualized by an ∼0.5 nt offset (53). Figure 2 shows a typical experiment. r42 degrades the ss U-overhang of the input 5′ pre-mRNA fragment in a distributive fashion and the generated RNAs have an identical electrophoretic mobility as fragments generated by incubation with 20S editosomes. This indicates that the fragments not only have the same nt length but also the same phosphorylation status. Thus, the exoUase activities of 20S editosomes and of r42 generate 3′ phosphate termini.
Figure 2.
Comparison of exoUase reaction products derived from incubations with r42 and 20S editosomes. (A) Sketch of the input precleaved deletion mRNA/gRNA hybrid molecule (5′ cleavage fragment (CF), black, 3′ CF, grey). An asterisk designates the radioactive phosphate group. OH and NH2 indicate 3′ terminal hydroxyl or amino groups. (B) Electrophoretic analysis of the reaction products of the pre-mRNA/gRNA hybrid shown in (A) after incubation with r42 or 20S editosomes. A schematic representation of the processed RNAs is given on the right. Mock represents an RNA sample incubated in the absence of 20S editosomes or r42.
Editosomes contain 3′ nucleotidyl phosphatase activity
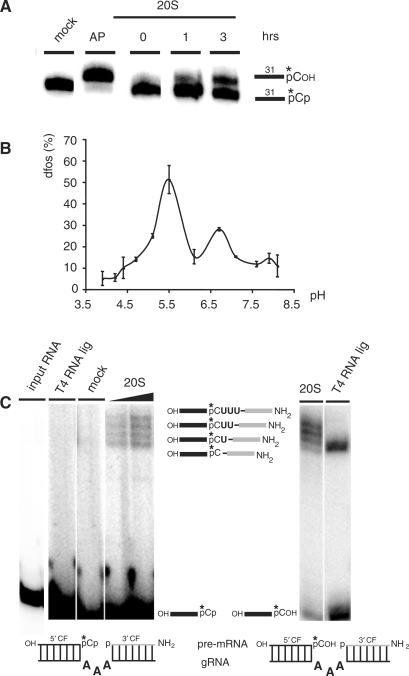
The result described above has two consequences for the RNA-editing reaction cycle. First, it implies that the 5′ pre-mRNA cleavage fragment after the exonucleolytic trimming reaction by TbMP42 must carry a 3′ phosphate. Second, since the 3′ pre-mRNA cleavage fragment has been shown to be 5′ phosphorylated (5), this creates a situation in which the two pre-mRNA fragments oppose each other with two nonligatable phosphate groups (54). As a consequence, we investigated whether enriched editosome preparations contain a 3′-specific nucleotidyl phosphatase activity to release the 3′-terminal phosphate from the 5′ pre-mRNA fragment. For our analysis we used a chemically synthesized RNA substrate with a length of 31 nt (RNA31). The RNA is characterized by a 5′ hydroxyl terminus and was 3′ phosphorylated through the ligation of radioactively labeled 5′-[32P]-pCp (RNA31-pCp). RNA31-pCp was incubated with 20S editosomes and the resulting reaction products were analyzed in denaturing polyacrylamide gels. Figure 3A shows a representative kinetic experiment. Over time, a radioactive RNA product with a decreased electrophoretic mobility is generated, which co-migrates with a control sample where RNA31-pCp was 3′ dephosphorylated using alkaline phosphatase (AP). This indicates that 20S editosomes contain 3′ nucleotidyl phosphatase activity. Nucleotidyl phosphatases frequently are acidic enzymes and require bivalent cations for their activity (55,56). Therefore, we analyzed the editosome-associated nucleotidyl dephosphorylation activity with respect to these two criteria. The activity has two pH-maxima at pH 5.5 and pH 6.7 (Figure 3B). Zn2+-cations are required for the dephosphorylation reaction and Ca2+-ions inhibit the reaction with an IC50 of ∼5 mM (data not shown). As described above, Mg2+-ion titration and EDTA chelation experiments identified two optima at 5 mM and 20 mM suggesting two separate activities (data not shown). Lastly, we tested whether the identified nucleotidyl phosphatase activity can be traced as an individual reaction step within the editing cycle. For that we generated a precleaved gRNA/pre-mRNA hybrid molecule in which the two pre-mRNA fragments face each other by two phosphate residues (Figure 3C). We confirmed that T4 RNA ligase cannot ligate the two pre-mRNA fragments; however, upon incubation with 20S editosomes the expected editing intermediates as well as the edited mRNA product are generated (Figure 3C). This indicates that 20S editosomes can dephosphorylate the 3′ end of a 5′ pre-mRNA cleavage fragment within the context of a gRNA/pre-mRNA hybrid RNA.
Figure 3.
Characterization of the 3′ nucleotidyl phosphatase activity of 20S editosomes. (A) A 31 nt ssRNA molecule (RNA31) was radioactively labeled (*) at its 3′ end using 5′-[32P]-pCp and incubated for up to 3 h with 20S editosomes or alkaline phosphatase (AP). Reaction products were resolved in urea containing polyacrylamide gels and are depicted in the margin to the right. (B) Plot of the dephosphorylation (dfos) activity of 20S editosomes at different pH values. (C) Dephosphorylation of RNA-editing substrate molecules as part of an RNA-editing reaction cycle. Left panel: Precleaved insertion editing of 20S editosomes using a 3′ phosphorylated 5′ pre-mRNA cleavage fragment (5′ CF). Mock, mock control; T4 RNA ligase, control ligation with T4 RNA ligase. Right panel: Precleaved insertion editing of 20S editosomes using a 3′ dephosphorylated 5′ CF. Reaction products and intermediates are sketched in between the two gels. The mock sample contains a single band and degradation was measured <1%.
TbMP99 and TbMP100 as candidate nucleotidyl phosphatases
Inspection of the protein inventory of the editosome revealed two candidate polypeptides for the above-described phosphatase activity. The proteins are known as TbMP99 and TbMP100 (26,27), which are characterized by the presence of a 5′ to 3′ exonuclease motif and a C-terminal endo-exo-phosphatase (EEP) domain. TbMP99 and TbMP100 have a high degree of identity/similarity (29%/46%) on the amino acid level and have been shown to possess exoUase activity in vitro (24,25).
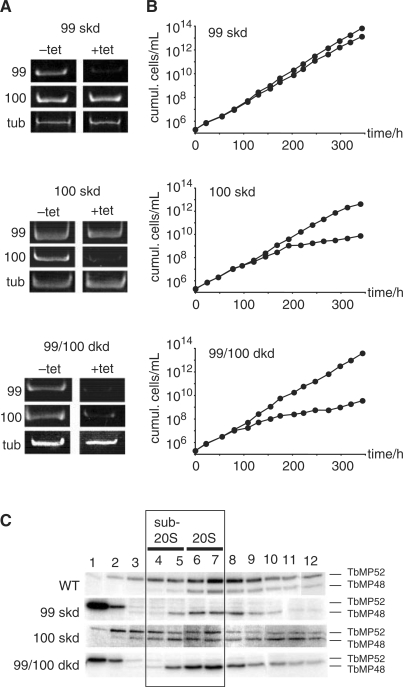
To investigate whether the two proteins contribute to the described 3′ nucleotidyl phosphatase activity we generated conditional kd trypanosome cell lines using RNAi (45). Three RNAi strains were constructed: skd cells for TbMP99 (99 skd) and TbMP100 (100 skd) and a dkd cell line (99/100 dkd) to downregulate both genes at the same time. All three strains were analyzed by RT-PCR to confirm gene ablation after induction with tetracycline (tet). Figure 4A shows that 6 days postinduction the transcript levels for TbMP99 and TbMP100 are at or below the level of detection in all three cell lines. Although TbMP99 and TbMP100 share a high degree of similarity, RNAi-mediated downregulation was specific for the individual mRNAs. Analysis of the growth behavior of the three parasite cell lines demonstrated no growth phenotype for the 99-skd cells (Figure 4B). However, 100-skd trypanosomes multiplied with a reduced growth rate starting at around 150 h postinduction (Figure 4B). The 99/100 dkd cells showed the strongest phenotype. A reduced growth rate was already apparent after 100 h of RNAi induction (Figure 4B).
Figure 4.
RNAi-mediated downregulation of TbMP99 and/or TbMP100. (A) RT-PCR analysis of TbMP99 and TbMP100 single knockdown (99 skd and 100 skd) and TbMP99/TbMP100 double knockdown cells (99/100 dkd). Upon induction (6 days) with tetracycline (+tet) the mRNA level of either TbMP99, TbMP100 or of both transcripts was ≤5%. tub, α-tubulin. (B) Growth behavior of 99 skd, 100 skd and 99/100 dkd trypanosomes. (C) Autoadenylation of the editing ligases TbMP52 and TbMP48 in gradient fractionated mitochondrial extracts from wild-type (WT), 99 skd, 100 skd and 99/100 dkd trypanosomes prepared 6 days postinduction. Sub-20S and 20S fractions are indicated.
As a follow-up we determined the integrity of editing complexes in mitochondrial extracts of all three cell lines. This was done by monitoring the distribution of the two core components of the 20S editosome, the RNA ligases TbMP52 and TbMP48 (57–59). Downregulation of TbMP99 resulted in a complete loss of TbMP52 from complexes ≥20S (Figure 4C). The majority of the signal (≥95%) was found at the top of the gradient, which indicates nonassembled protein. By contrast, TbMP48 was assembled in complexes ≥20S, although a shift towards sub-20S fractions (fractions 4/5) was observed. Knockdown of TbMP100 did not result in a disassembly of TbMP52. Both ligases were identified in a broad distribution ranging from sub-20S to 40S and higher. Lastly, mitochondrial extracts from 99/100 dkd cells showed a distribution identical to the 99-skd extracts (Figure 4C). Ligase TbMP52 was found at the top of the gradient while TbMP48 was shifted toward sub-20S.
Editosomes from 99/100 dkd cells have reduced RNA editing and reduced 3′ nucleotidyl phosphatase activity
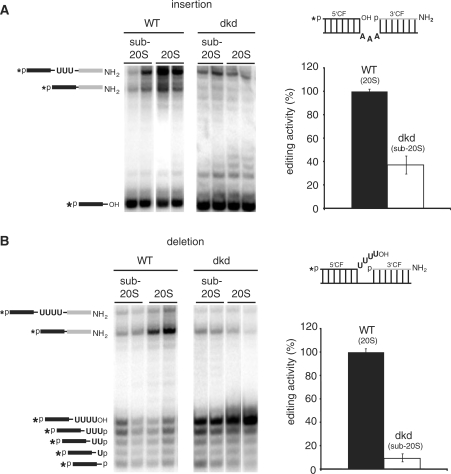
Enriched editosome preparations from all three RNAi cell lines were analyzed for their in vitro RNA-editing activity by monitoring both, U-deletion and U-insertion editing (41,42). Due to the reduced molecular mass and the altered hydrodynamic properties of the editing complexes in the kd-cell lines, we analyzed sub-20S as well as 20S editosome fractions. The two skd-cell lines (99 skd and 100 skd) showed activities identical to editosome preparations derived from uninduced cells (data not shown). By contrast, editosome preparations from 99/100 dkd cells were affected in their ability to process synthetic pre-edited mRNAs. Comparing 20S editosomes from uninduced cells with sub-20S fractions from the 99/100 dkd cell line (please note, the absence of TbMP99, TbMP100 and of ligase TbMP52 amounts to 250 kDa) revealed a drop in the insertion activity by ≥60% (Figure 5A). Similarly, deletion editing was reduced by ≥90% compared to the activity derived from uninduced cells (Figure 5B).
Figure 5.
RNA-editing activity of TbMP99/100-minus trypanosomes. (A) Precleaved insertion editing assay of mitochondrial 20S and sub-20S fractions of wild-type (WT) and TbMP99/100 double knockdown (99/100 dkd) cells. The assay monitors the gRNA-dependent insertion of 3U nucleotides. A representation of the pre-mRNA/gRNA hybrid is depicted on the right. 5′ pre-mRNA cleavage fragment (CF). black; 3′ CF, grey. The chemical identities of the 5′ and 3′ ends are as in Figure 2A. An asterisk indicates the radiolabel. Reaction products were separated in denaturing polyacrylamide gels and visualized by phosphorimaging. A schematic representation of the reaction products is given on the right. The graph shows a quantitative analysis of the editing activity. (B) Precleaved deletion editing assay of mitochondrial sub-20S and 20S fractions of WT and 99/100 dkd cells. The assay monitors the gRNA-dependent deletion of 4Us. All annotations are as in (A).
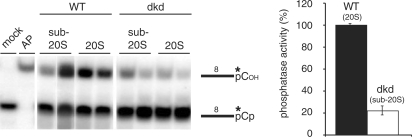
Editosome-containing fractions from the two skd-cell lines were fully competent to dephosphorylate a 3′-terminal phosphate from a ss substrate RNA (data not shown). However, sub-20S fractions from the 99/100 dkd cell line had only residual 3′ nucleotidyl phosphatase activity (reduction ≥80%) (Figure 6), which suggests that TbMP99 as well as TbMP100 can act as 3′ nucleotidyl phosphatases. Either one of the two proteins is sufficient to execute its function within the context of the editing complex.
Figure 6.
3′ Nucleotidyl phosphatase activity of wild-type (WT) and TbMP99/100-depleted (99/100 dkd) sub-20S and 20S editosomes. (A) Autoradiograph of a phosphatase assay using an 8-nt single-stranded RNA substrate molecule (RNA8). An asterisk represents the position of the radioactive label. Reaction products are given in the margin to the right. Mock: RNA8 incubated in the absence of 20S editosomes. AP, alkaline phosphatase (B) Quantitative analysis of the assay shown in (A).
DISCUSSION
The final reaction step in both, deletion- and insertion-type RNA editing is the re-ligation of the two processed pre-mRNA fragments. This requires the presence of a 3′ hydroxyl and a 5′ phosphate terminus at the editing site. The generation of 5′ phosphate ends during the initial endonucleolytic cleavage of the pre-mRNA has been demonstrated by Seiwert et al. (5) and this has led to the assumption that the editosome-associated exoribonucleolytic activity releases 5′ nucleoside monophosphates. A study by Rusché et al. (60) analyzed the reaction products of the exoUase only in one dimension and hence no distinction was made between a 5′ nucleoside monophosphate (pN) and a 3′ nucleoside monophosphate (Np). Here we show that the exoribonucleolytic activity of TbMP42 generates 3′ nucleoside monophosphates (Np). Moreover, the electrophoretic mobility of 3′ uridylyl overhangs processed by r42 compared to editing active fractions of mitochondrial extracts is identical. This suggests that either TbMP42 is the only active exoUase of the 20S editosome or that all other enzymes follow the same reaction mechanism. Otherwise, one should be able to detect mixed populations of 3′ uridylyl-overhangs terminating in a phosphate or a hydroxyl group in precleaved deletion assays, which is not the case.
In a full-round editing-reaction cycle a deletion-editing site is subject to an endoribonucleolytic cleavage reaction, which generates a 5′ phosphate at the 3′ cleavage fragment (5). The endoribonuclease activity of TbMP42 is not capable to generate these 5′P/3′OH ends. Recent studies suggest that this cleavage is executed by TbMP61 in insertion editing and by TbMP90 in the case of deletion-type editing (29,30). Thus, the in vitro endoribonuclease activity of TbMP42 is likely not executed in the initial endoribonucleolytic attack of the pre-mRNA. The exoUase activity of the editosome then creates 3′ phosphate termini thereby introducing a roadblock that prevents premature ligation. This roadblock is removed by a 3′-specific, acidic nucleotidyl phosphatase activity, which is present in mitochondrial extracts of African trypanosomes and is associated with a 20S high-molecular-mass complex. In that sense, the described reaction cascade serves as a protection against premature ligation events.
In insertion-type editing, the TUTase frequently adds more Us than specified by the gRNA (22). Thus, insertion editing requires an exoUase trimming reaction and as a consequence a phosphatase activity as well. This explains the >60% reduction of TbMP99- and TbMP100-depleted extracts in insertion editing. The concerted action of nuclease, phosphatase and ligase are also found in yeast tRNA splicing (61).
Further evidence for the described scenario can be derived from the analysis of the reaction products in the in vitro precleaved deletion-editing assay (PDA) (41). The dominant ligation product is the ‘faithfully’ edited mRNA with all four Us removed and the two mRNA fragments ligated. The second most abundant product is the unedited mRNA product. No U-residue is removed from the editing site but the two mRNA fragments are ligated, since initially the substrate has ligation compatible ends. All other ligation products (–3U, –2U and –1U) are at or below the level of detection. One possible explanation could be that the recruitment of the 3′-specific nucleotidyl phosphatase occurs in concert with the dissociation of the exoUase activity. Another explanation would be an exoUase with a processive mode of action. However, TbMP42 was shown to act in a distributive fashion (23).
These findings indicate a contribution of one previously not recognized enzymatic function to the editing-reaction cycle, complementing the current mechanism of the RNA-editing reaction cycle. A revised version of the RNA-editing reaction cycle including the 3′ specific nucleotidyl phosphatase is depicted in Figure 7. Together, based on our analysis we conclude a mode of action for TbMP42. The exoUase activity of the protein produces 3′ nucleoside monophosphates, which at a deletion-editing site results in two opposing phosphate residues. In order to resolve this situation for the subsequent ligation step, a 3′ specific nucleotidyl phosphatase is required. Such an activity is present in mitochondrial extracts of African trypanosomes and it co-sediments with the RNA-editing activity. Moreover, its pH optimum and its cofactor requirement suggest that the enzyme(s) belong to the group of 3′ acidic phosphatases. The characterization of the activity identified two potential candidates, TbMP99 and TbMP100. Both proteins carry an endo-exo-phosphatase motif (26,27) and have been shown to execute exoUase activity in vitro (24,25). However, whether this is of relevance in vivo remains controversial. Assigning nuclease or phosphatase activity to proteins containing an EEP domain is ambiguous (62). For example, both TbMP99 and 100 were homology-modeled to an EEP-domain containing exonuclease (63). However, both peptides can also be homology-modeled along the EEP-domain containing nucleotidyl phosphatase synaptojanin (Supplementary Figure 1).
Figure 7.
Reaction cycle of an RNA-editing deletion reaction including a nucleotidyl phosphatase activity (altered from ref. 64). Base pairing between the pre-edited mRNA and the gRNA are indicated by vertical lines. Colons represent G:U bp.
The 3′ specific nucleotidyl phosphatase is reduced upon knockdown of both TbMP99 and 100 in vitro. We propose that at least one peptide executes a nucleotidyl phosphatase activity and contributes this activity to the editing-reaction cycle. Recombinant protein should be able to complement for the RNAi-mediated loss of the nucleotidyl phosphatase. Likely, either one of the two proteins will be able to complement the activity. Depletion of TbMP100, but not of TbMP99 results in a growth phenotype. It is hence plausible to assume that complementation of TbMP100 with TbMP99 is suboptimal, whereas complementation of TbMP99 with TbMP100 is not. A similar phenomenon was observed for the RNA-editing ligases TbMP52 and TbMP48 (3). Since there is evidence for separate insertional and deletional editing (sub)complexes (21), it is tempting to speculate that the two enzymes contribute their function to different particles.
Taken together, the data provide a new facet of the biochemical consequences of TbMP42's function as an exonuclease in the RNA-editing reaction cycle. By carrying out its activity, premature ligation is prevented through the generation of a 3′ phosphate at the 5′ mRNA cleavage fragment. Thus, the protein provides a form of quality control after the ribonucleolytic trimming reaction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
International Research Scholar of the Howard Hughes Medical Institute (to H.U.G.); The German Research Council (DFG-SFB579 to H.U.G.). Funding for open access charge: DFG-SFB579.
Conflict of interest statement. None declared.
Supplementary Material
Acknowledgements
We thank Cordula Böhm for discussion and Verena Bihrer for her help in analyzing the 99/100 dkd trypanosome cell line.
REFERENCES
- 1.Madison-Antenucci S, Grams J, Hajduk SL. Editing machines: the complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 2.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carnes J, Stuart K. Working together: the RNA editing machinery in Trypanosoma brucei. In: Göringer HU, editor. RNA Editing. Heidelberg: Springer; 2008. pp. 143–164. [Google Scholar]
- 4.Cruz-Reyes J, Sollner-Webb B. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc. Natl Acad. Sci. USA. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 6.Piller KJ, Rusché LN, Cruz-Reyes J, Sollner-Webb B. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA. 1997;3:279–290. [PMC free article] [PubMed] [Google Scholar]
- 7.Aphasizhev R, Simpson L. Isolation and characterization of a U-specific 3′-5′-exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 2001;276:21280–21284. doi: 10.1074/jbc.M100297200. [DOI] [PubMed] [Google Scholar]
- 8.Igo RP, Jr, Weston DS, Ernst NL, Panigrahi AK, Salavati R, Stuart K. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot. Cell. 2002;1:112–118. doi: 10.1128/EC.1.1.112-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl Acad. Sci. USA. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst NL, Panicucci B, Igo RP, Jr, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 11.McManus MT, Shimamura M, Grams J, Hajduk SL. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusché LN, Huang CE, Piller KJ, Hemann M, Wirtz E, Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 14.Müller UF, Lambert L, Göringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blom D, Burg J, Breek CK, Speijer D, Muijsers AO, Benne R. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 2001;29:2950–2962. doi: 10.1093/nar/29.14.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller UF, Göringer HU. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–455. doi: 10.1093/nar/30.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher MA, Karamooz E, Zíková A, Trantírek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 19.Missel A, Göringer HU. Trypanosoma brucei mitochondria contain RNA helicase activity. Nucleic Acids Res. 1994;22:4050–4056. doi: 10.1093/nar/22.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Missel A, Souza AE, Nörskau G, Göringer HU. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 22.Byrne EM, Connell GJ, Simpson L. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 1996;15:6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 23.Brecht M, Niemann M, Schlüter E, Müller UF, Stuart K, Göringer HU. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl Acad. Sci. USA. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers K, Gao G, Simpson L. Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J. Biol. Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- 26.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panigrahi AK, Schnaufer A, Carmean N, Igo RP, Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Niemann M, Brecht M, Schlüter E, Weitzel K, Zacharias M, Göringer HU. TbMP42 is a structure-sensitive ribonuclease that likely follows a metal ion catalysis mechanism. Nucleic Acids Res. 2008;36:4465–4473. doi: 10.1093/nar/gkn410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Law JA, O'Hearn SF, Sollner-Webb B. Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion. RNA. 2008;14:1187–1200. doi: 10.1261/rna.899508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X, Ernst NL, Stuart KD. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol. Cell Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross GA. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 36.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasit. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 37.Brun R, Schönenberger M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 38.Hauser R, Pypaert M, Häusler T, Horn EK, Schneider A. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J. Cell Sci. 1996;109:517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- 39.Göringer HU, Koslowsky DJ, Morales TH, Stuart K. The formation of mitochondrial ribonucleoprotein complexes involving guide RNA molecules in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 1994;91:1776–1780. doi: 10.1073/pnas.91.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabatini R, Hajduk SL. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J. Biol. Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 41.Seiwert SD, Stuart K. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- 42.Kable ML, Seiwert SD, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 43.Homann M, Göringer HU. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 1999;27:2006–2014. doi: 10.1093/nar/27.9.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 45.Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 46.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 47.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 48.Kopp J, Schwede T. The SWISS-MODEL Repository: new features and functionalities. Nucleic Acids Res. 2006;34:D315–D318. doi: 10.1093/nar/gkj056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirsebom LA. RNase P RNA-mediated catalysis. Biochem. Soc. T. 2002;30:1153–1158. doi: 10.1042/bst0301153. [DOI] [PubMed] [Google Scholar]
- 50.Raines R. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K, Moore S. Ribonuclease T1. In: Boyer PD, editor. The Enzymes, V,. Vol. 15. New York: Academic Press; 1982. pp. 435–468. [Google Scholar]
- 52.Deshpande RA, Shankar V. Ribonucleases from T2 family. Crit. Rev. Microbiol. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- 53.Cruz-Reyes J, Piller KJ, Rusché LN, Mukherjee M, Sollner-Webb B. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1-generated sequencing ladders. Biochemistry. 1998;37:6059–6064. doi: 10.1021/bi972868g. [DOI] [PubMed] [Google Scholar]
- 54.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 55.Jilani A, Ramotar D. Purification and partial characterization of a DNA 3′-phosphatase from Schizosaccharomyces pombe. Biochemistry. 2002;41:7688–7694. doi: 10.1021/bi012213m. [DOI] [PubMed] [Google Scholar]
- 56.Deshpande RA, Wilson TE. Identification of DNA 3′-phosphatase active site residues and their differential role in DNA binding, Mg2+ coordination, and catalysis. Biochemistry. 2004;43:8579–8589. doi: 10.1021/bi049434n. [DOI] [PubMed] [Google Scholar]
- 57.Panigrahi AK, Gygi SP, Ernst NL, Igo RP, Jr, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao G, Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- 60.Rusché LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abelson J, Trotta CR, Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 62.Dlakic M. Is CdtB a nuclease or a phosphatase? Science. 2001;291:547. doi: 10.1126/science.291.5504.547a. [DOI] [PubMed] [Google Scholar]
- 63.Mian IS, Worthey EA, Salavati R. Taking U out, with two nucleases? BMC bioinformatics. 2006;7:305. doi: 10.1186/1471-2105-7-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ochsenreiter T, Hajduk S. The function of RNA editing in Trypanosomes. In: Göringer HU, editor. RNA Editing. Heidelberg: Springer; 2008. pp. 181–197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.