Abstract
Mitochondrial pre-messenger RNAs in kinetoplastid protozoa are substrates of uridylate-specific RNA editing. RNA editing converts non-functional pre-mRNAs into translatable molecules and can generate protein diversity by alternative editing. Although several editing complexes have been described, their structure and relationship is unknown. Here, we report the isolation of functionally active RNA editing complexes by a multistep purification procedure. We show that the endogenous isolates contain two subpopulations of ∼20S and ∼35–40S and present the three-dimensional structures of both complexes by electron microscopy. The ∼35–40S complexes consist of a platform density packed against a semispherical element. The ∼20S complexes are composed of two subdomains connected by an interface. The two particles are structurally related, and we show that RNA binding is a main determinant for the interconversion of the two complexes. The ∼20S editosomes contain an RNA-binding site, which binds gRNA, pre-mRNA and gRNA/pre-mRNA hybrid molecules with nanomolar affinity. Variability analysis indicates that subsets of complexes lack or possess additional domains, suggesting binding sites for components. Together, a picture of the RNA editing machinery is provided.
Keywords: African trypanosomes, electron microscopy, mass spectrometry, RNA editing, three-dimensional structure
Introduction
The RNA editing reaction in kinetoplastid protozoa such as Trypanosoma and Leishmania is characterised by a reaction cycle that inserts and deletes uridylates (U) into otherwise incomplete mitochondrial transcripts. Biochemically, the reaction can be divided into three steps, which are catalysed by proteins only. Proteins involved in RNA editing are assembled in large macromolecular machines known as editosomes (Madison-Antenucci et al, 2002; Stuart et al, 2005). The reaction is initiated by the formation of a guide (g)RNA/pre-edited mRNA duplex (Blum et al, 1990). This duplex functions as substrate for an endonuclease that cleaves the pre-mRNA at the first mismatch 5′ of the duplex. During insertion editing, a 3′-terminal uridylyl-transferase (TUTase) adds a number of U-nucleotides to the 3′-end of the 5′ pre-mRNA fragment, whereas for deletion editing, a 3′ → 5′ exoribonuclease removes terminal uridylates from the 5′ pre-mRNA fragment. Finally, an RNA ligase joins the two mRNA fragments. However, editosomes do not only contain endoribonuclease (Brecht et al, 2005; Carnes et al, 2005; Trotter et al, 2005), TUTase (Ernst et al, 2003), exoribonuclease (Brecht et al, 2005; Mian et al, 2006; Rogers et al, 2007) and RNA ligase activities (McManus et al, 2001), but also various additional polypeptides whose functions are poorly defined (Aphasizhev et al, 2003; Panigrahi et al, 2003a, 2003b, 2006).
Although significant progress has been made to unravel the protein inventory of editing complexes (Stuart et al, 2005), little is known as to the structural organisation and dynamics of this molecular machine. Especially, even though several editing complexes have been described, their relationship in the context of the catalytic cycle is not understood. Editing complexes include a 5–10S insertion subcomplex and a 5–10S deletion subcomplex each of which comprising three different proteins, an ∼20S complex that contains up to 20 proteins including those of both the 5–10S insertion and deletion subcomplexes as well as a 35–40S complex with biochemically poorly defined composition (Pollard et al, 1992; Schnaufer et al, 2003; Panigrahi et al, 2006). The presence of specific RNA classes has been shown for the 35–40S complex, which harbours both, gRNA and pre-mRNA (Pollard et al, 1992), whereas gRNAs are found only in a subset of ∼20S editing complexes (Rusché et al, 1997; Madison-Antenucci et al, 1998). In addition, it has been suggested that an 11S gRNA complex may be generated by the interaction of a 5S complex with gRNA serving as gRNA storage (Goulah and Read, 2007).
The understanding of the assembly of the RNA editing machinery is far from complete. A current model involves a stepwise pathway (Madison-Antenucci et al, 2002). An ∼20S gRNA maturation complex is formed by the interaction of polycistronic gRNAs with a pre-assembled core complex. This pre-assembled complex may be derived by the interaction of the 5–10S insertion and deletion subcomplexes with other proteins (Schnaufer et al, 2003). Polycistronic gRNAs are suggested to be cleaved in the ∼20S gRNA maturation complex (Grams et al, 2000). Ultimately, this ∼20S complex may interact with pre-edited/partially edited pre-mRNAs, resulting in the formation of an ∼35–40S complex, which was suggested to perform the catalytic reactions (Pollard et al, 1992). Together, this assembly model suggests that the 20S complex may be converted into the ∼35–40S particle and thus, both complexes may share a common core structure. However, the described data would also be consistent with an alternative model in which both complexes may represent functionally separate particles with individual assembly pathways in vivo. Due to their different functional roles, some proteins of the ∼20S complex may be exchanged for other components during the transition to the ∼35–40S particle resulting in different architectures of both complexes.
In any of these models, various intermediates can be discerned, each of which represents independent challenges for biochemical studies and structural biology. The structural analysis of such diverse assemblies is currently a major task for electron microscopy (EM), especially as the existing purification protocols can only incompletely separate mixtures of different functional states. Here, we have purified functional RNA editing complexes by a multistep purification approach. Endogenous complexes were analysed by biochemical, functional and structural means. We describe the structural relationship between the ∼20S and the ∼35–40S complexes and suggest how the ∼20S complex may be integrated into the larger complex, thereby providing a picture of the architecture of the RNA editing machinery. By variability analysis, we present a comprehensive view of the endogenous RNA editing machinery in trypanosomes at different assembly stages.
Results
Purification of native RNA editing complexes
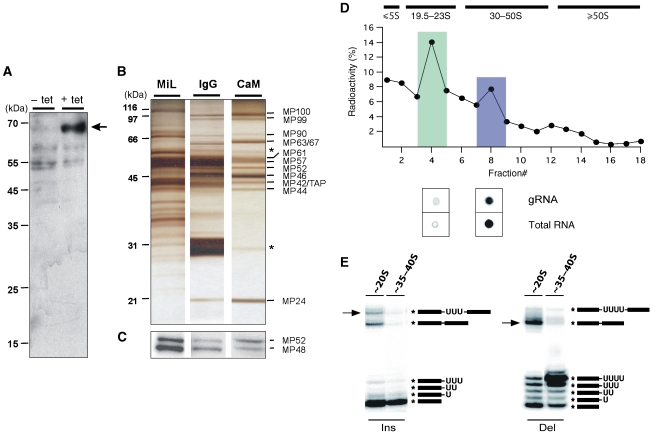
To isolate RNA editing complexes at native conditions, we used the tandem-affinity purification (TAP) protocol (Rigaut et al, 1999). To this end, we generated a Trypanosoma brucei cell line (29-13-MP42/TAP) that expresses a C-terminally TAP-tagged version of TbMP42. TbMP42 is a 42-kDa protein that was identified as an integral component of editosomes (Panigrahi et al, 2001b; Brecht et al, 2005; Niemann et al, 2008). Expression of TbMP42/TAP in the 29-13-MP42/TAP strain relies on the tetracycline (tet) repressor/operator system and, thus, can be conditionally regulated (Wirtz et al, 1999). Tet-induced insect stage 29-13-MP42/TAP trypanosomes showed a cell-doubling time identical to the parental 29-13 strain and were morphologically indistinguishable from wild-type cells (data not shown). Expression of TbMP42/TAP was verified by Western blotting (Figure 1A) and RNA editing complexes of tet-induced 29-13-MP42/TAP trypanosomes were enriched from non-ionic detergent lysates of mitochondrial vesicles isolated at isotonic conditions (Göringer et al, 1994; Hauser et al, 1996).
Figure 1.
Biochemical characterisation of endogenous TAP-tagged RNA editing complexes. (A) Western blot verification of the expression of the 64.3 kDa TAP-tagged version of TbMP42 (arrow). The whole cell protein extract was prepared from 29-13-MP42/TAP insect stage trypanosomes cultivated in the absence (−) or presence (+) of tet for 74 h. (B) Silver-stained protein pattern (2 μg each) of a mitochondrial lysate (MiL), an IgG column eluate (IgG) and a calmodulin column(CaM) eluate. Proteins were identified by mass spectrometry. Contaminants (β-tubulin, TEV protease) are marked by *. (C) Identification of the RNA editing ligases TbMP52 and TbMP48 through auto-adenylation in the presence of α-[32P]-ATP. (D) CaM eluates were radioactively labelled by tyrosin iodination (125I) and fractionated in linear 10–40% (v/v) glycerol gradients. The ∼20S (green) and ∼35–40S (blue) peak fractions were analysed for the presence of RNA by radioactive postlabelling methods and spotted onto PEI cellulose plates. The presence of gRNAs was identified by labelling with guanylyl transferase and α-[32P]-GTP. (E) RNA editing in vitro activity (Ins, insertion; Del, deletion) of TAP-tagged and glycerol gradient-purified ∼20S and ∼35–40S editing complexes. The electrophoretic mobilities of editing products (arrows), ligation products and of various reaction intermediates are given on the right. ‘*' indicates the position of the radioactive label.
Protein composition of native RNA editing complexes
The electrophoretic analysis showed a decrease in the complexity of the protein pattern from the mitochondrial lysate (MiL) to the calmodulin column (CaM) eluate, ultimately identifying 14 bands ranging in size from 18 to 100 kDa (Figure 1B). Mass spectrometry identified 12 of them as previously characterised editing proteins (TbMP100, TbMP99, TbMP90, TbMP67, TbMP63, TbMP61, TbMP57, TbMP52, TbMP46, TbMP44, TbMP42/TAP, TbMP24; nomenclature of Panigrahi et al, 2001a; Supplementary Tables 1 and 2). Two proteins, identified as β-tubulin and TEV protease, were considered contaminants. Of the two RNA editing ligases (TbMP48, TbMP52), only TbMP52 was identified by mass spectrometry. However, the presence of TbMP48 was verified by self-adenylation (Sabatini and Hajduk, 1995) in the presence of α-[32P]-ATP (Figure 1C).
Purified RNA editing complexes sediment as ∼20S and ∼35–40S particles
CaM eluates were further analysed by isokinetic ultracentrifugation in glycerol gradients using non-radioactive material and 125I labelled eluates to increase the detection limit (Figure 1D). In line with previously published studies (Pollard et al, 1992; Corell et al, 1996), the data show the presence of two complexes with apparent Svedberg (S) values of ∼20S and ∼35–40S. Using radioactive 5′ and 3′ post-labelling methods, we confirmed the presence of endogenous RNA, including gRNA, for the ∼35–40S complexes (Pollard et al, 1992; Corell et al, 1996), whereas ∼20S complexes did not contain detectable amounts of pre-bound RNA (Rusché et al, 1997). To test whether the two complexes were functionally active, we performed U-deletion and U-insertion RNA editing in vitro assays (Igo et al, 2000, 2002). The ∼20S complexes were fully competent to correctly edit synthetic pre-edited mRNAs in a gRNA-dependent fashion (Figure 1E). By contrast, ∼35–40S complexes only showed background activity (⩽2%) likely because the RNA-binding site is occupied with endogenous RNA.
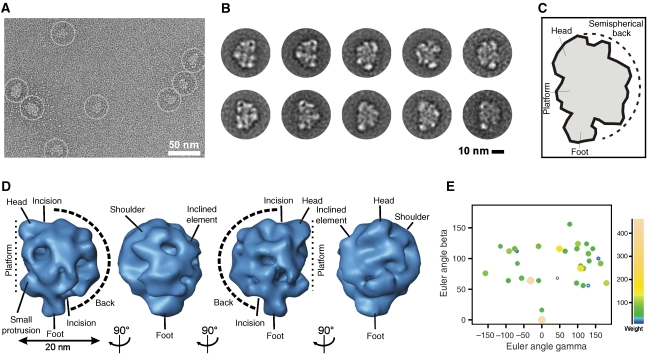
Electron microscopy of the ∼35–40S RNA editing complex
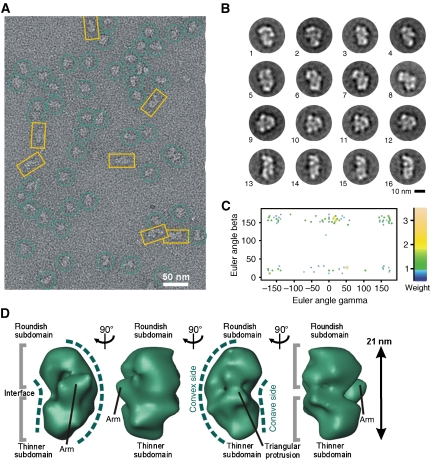
Using the TAP protocol (Rigaut et al, 1999) in combination with the GraFix method (Kastner et al, 2008), we purified ∼35–40S complexes for EM. The raw EM images of the ∼35–40S complexes displayed a monodisperse population of an asymmetric particle of up to ∼26 nm in size (Figure 2A). Imaged complexes revealed a compact globular shape with distinct structural features, suggesting that intact complexes had been purified.
Figure 2.
Electron microscopy of the ∼35–40S RNA editing complex. (A) Raw RT EM image of the ∼35–40S complex. (B) 2D class averages displaying a particle comprising an elongated platform density (oriented to the right, upper panel and to the left, lower panel) packed against a semispherical element (oriented to the left, upper panel and to the right, lower panel). (C) Structural landmarks of the complex. (D) Consensus structure of the ∼35–40S complex. Characteristic structural features are labelled. (E) Euler angle plot of the 3D RCTs included in the consensus map coloured according to weight as shown on the right.
Two-dimensional (2D) class averages revealed a characteristic distribution of densities (Figure 2B and C). Typical class averages displayed a particle composed of an elongated, straight to slightly convex platform density (oriented to the right in Figure 2B, upper row and to the left in the lower row) packed against a semispherical element (oriented to the left and right side in the upper and lower row). On both ends, the platform extends into a small head-like element oriented to the top and a larger foot-like density extending to the bottom of the structure. The majority of visualised particles adopted this morphology in several independent purifications. A small number of particles, however, differed in being composed of the platform density only suggesting that these particles may differ in composition.
Consensus structure of the ∼35–40S RNA editing complex
On the basis of 2D class averages, it is often difficult to distinguish whether given views belong to a common three-dimensional (3D) structure or represent broken or contaminating particles. We therefore applied a 3D averaging approach that combines the random conical tilt (RCT) (Radermacher, 1988) method with weighted averaging (Sigworth, 1998). This approach can be performed free of user-bias with heterogeneous data sets and can be expanded to a detailed variability analysis.
The obtained consensus structure displayed all structural characteristics described in the 2D class averages and visualised in the raw images (Figure 2D). Specifically, an elongated platform is seen to which small head-like and foot-like elements are attached on opposite sides. A small protrusion is visible in the lower part of the complex. A semispherical back is packed against the platform and forms a tight density network to it. The interface between both elements is marked by incisions in the upper and lower part. Notably, the semispherical back is asymmetric in its appearance: on one side a large protruding ‘shoulder' density is seen, whereas on the opposite side a smaller inclined element is visible. A plot shows the presence of a wide range of Euler angles (Figure 2E).
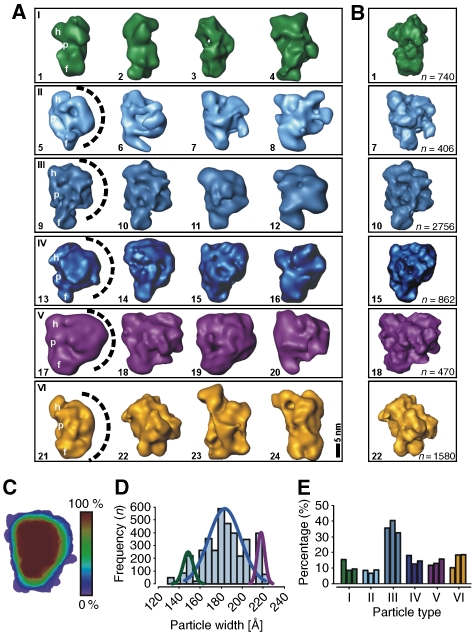
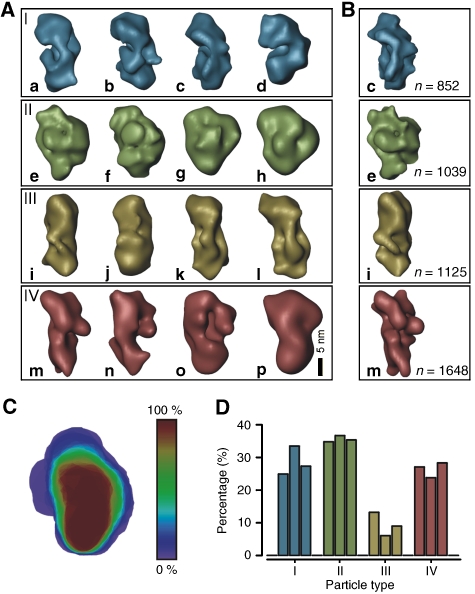
Variability analysis of the ∼35–40S RNA editing complex
The EM analysis suggested structural heterogeneity of the ∼35–40S complex, as some particles appeared to be composed of the platform density only. As the purified particles represent endogenous RNA editing complexes enriched at steady state conditions, these complexes may differ not only in the assembly stage, but also in their RNA and protein content. To address this aspect, we performed a 3D multivariate statistical analysis (MSA) (Liu et al, 2004) on the aligned 3D RCTs (Sander et al, 2006). Variability analysis revealed at least six morphologically different 3D subtypes (Figure 3A). Four of these subtypes (rows II–V) showed all the structural characteristics seen in the consensus structure (compare Figures 3A and 2D, first view). However, these subtypes differed considerably in width of the semispherical back (indicated as dashed line), ranging from small (row II) to extensive (row V). Notably, the first two structures shown in row III are very similar to the consensus structure. In contrast, structures shown in the row I were only composed of an elongated density that adopts different curvatures, while lacking the semispherical back. This confirms that the elongated 2D class averages described above miss this density. Furthermore, we also found a small subset of structures that did not show the typical structural features (row VI). We suggest that this population may either represent contaminating complexes or different assembly stages.
Figure 3.
Variability analysis of the ∼35–40S RNA editing complex. (A) 3D-MSA of individual aligned 3D RCT volumes indicated six subtypes of particle populations within the ∼35–40S sample. Landmarks are labelled in representative examples (h, head-like protuberance; p, platform; f, foot-like protuberance; the semispherical back is indicated as a dashed line). (B) Refined maps of representative 3D class averages subsequent to cMRA. The numbers of particles used to calculate the refined 3D structures are listed on the right. (C) Shape profiling of the ∼35–40S complex by colour-scale encoded overlays showing that the majority of complexes possesses a platform density packed against a semispherical back (red). (D) Histogram of the particle width suggesting at least three differently sized subpopulations. (E) cMRA of three independently purified samples revealing that the majority of particles belong to the subset shown in (A), row III (colour coding as in (A)) consistent with the consensus structure (Figure 2D).
Quantification of ∼35–40S editosomal subpopulations
To quantify the subpopulations, three different types of analysis were performed, (1) shape profiling (Figure 3C), (2) a width distribution analysis (Figure 3D) and (3) a competitive multireference alignment (cMRA) (Figure 3B and E). For a description of the overall architecture, all 3D class averages were colour-scale encoded according to their abundance and an overlay was created (Figure 3C). The majority of complexes (>80%) is composed of the described densities (red) in line with the consensus structure, whereas variable regions (blue) are found in the periphery of the semispherical back and in the size and position of the foot-like domain.
Quantification of the particles' width—a prominent individual feature of the 3D class averages—suggested an overlay of 3–5 Gaussian distributions (Figure 3D). First, a small subpopulation of complexes possesses a width of ∼15±1 nm (green curve) and represents the elongated particles as seen in Figure 3A, row I. The largest subpopulation (rows II–IV) has a width of ∼18.5±2 nm (blue curve). The third subpopulation (purple curve) comprises particles similar to the consensus structure except for the larger width of ∼21.5±1 nm (row V).
Finally, cMRA using the 3D class averages to create reference projections was performed. Thereby, each single-particle image is competitively aligned to reference projections of the 3D class averages so that the number of particles that fit best to each of the six subtypes can be depicted (Figure 3E) and refined 3D maps are obtained (Figure 3B). Significantly, we found no major differences between the three samples derived from independent editosome preparations, suggesting that the distribution of particle subtypes was preparation independent. In line with the shape profile (Figure 3C) and the distribution of the particle width (Figure 3D), we found that ∼40% of the particles adopted a conformation similar to the consensus structure (Figure 3E), whereas all other subpopulations occurred roughly equally in frequency (∼10–20%, each). Thus, the major particle subpopulation is composed of an elongated platform packed against a semispherical density, whereas the minor subpopulations lack the semispherical back or possess additional densities.
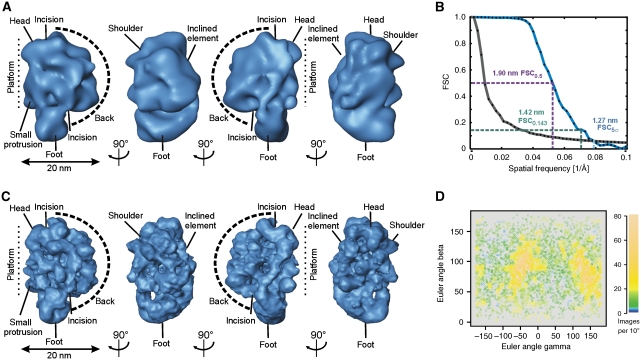
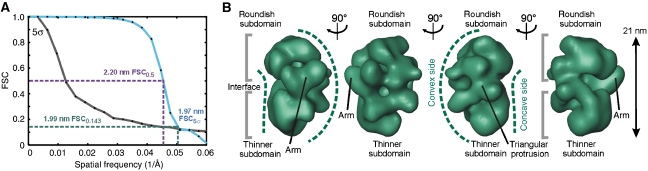
Structure refinement of the ∼35–40S RNA editing complex by cryo negative staining EM
The 3D map of the best-defined 3D class average (Figure 4A) was used as start-up reference to refine another data set of the ∼35–40S complex taken at liquid nitrogen temperature to a resolution of ∼12.7–19.0 nm using cMRA (Figure 4B and C). The Euler angle plot of all included particles indicated the presence of a wide variety of angular views with two more frequently found angular regions (Figure 4D). The initial consensus and the refined maps are very similar except for the higher resolution of the refined structure, resulting in more fine structural details in the refined map (Figure 4C). All described elements (compare landmarks in Figures 2D and 4C) were also visible in the refined reconstruction. The surface representation of the ∼35–40S complex is estimated to enclose a molecular mass of ∼1.45±0.15 MDa. By using the 3D map, the sedimentation coefficient can be predicted (Garcia de la Torre et al, 2001). Our calculation predicts a value of 35–41S, which is in agreement with the apparent sedimentation behaviour observed in the glycerol gradients.
Figure 4.
Refinement of the 3D map of the ∼35–40S RNA editing complex by cryo negative staining EM. (A) Surface representation of the best-defined 3D class average showing similar structural elements as seen in Figure 2D. (B) FSC suggested a resolution of 1.27–1.90 nm for the refined map. (C) Surface representation of the ∼35–40S RNA editing complex showing a wealth of fine structural details in the refined 3D map. The labelling of landmarks is similar to Figure 2D. (D) Euler angle plot of the refined data set. Relative particle numbers per unit area are colour coded as shown on the right.
Electron microscopy of the ∼20S RNA editing complex
The concentration of ∼20S particles seen in the EM was markedly higher by a factor of 5–10 as compared with the ∼35–40S complex. The raw EM images showed a monodisperse population of an elongated particle with dimensions of up to ∼21–26 nm (Figure 5A). Some of the particles appeared to adopt a straight conformation: these particles were particularly slim and elongated (yellow box). Other particles were broader and appeared to be in a more bent conformation (green circles).
Figure 5.
Electron microscopy of the ∼20S RNA editing complex. (A) Raw RT EM image of the ∼20–24S fractions showing an elongated particle of variable width including globular (green) and elongated (yellow) complexes. (B) 2D class averages of the ∼20S complex. (C) Euler angle plot of the ∼20S complex RCTs used to calculate the consensus map coloured according to weight as shown on the right. (D) Consensus model of the ∼20S complex displaying a bipartite shape. Landmarks are labelled.
These shapes were also reflected by the 2D class averages showing a variety of different views (Figure 5B). We found amongst others triangular (views 1–4), bent (views 5–8), semicircular (views 9–12) and elongated views (views 13–16), suggesting the sample to be heterogeneous. The majority of particles showed a bipartite appearance with two approximately equally sized subdomains connected by an interface. Both subdomains, however, differed in their structural details, suggesting that the particle is not a homodimer.
Consensus structure of the ∼20S RNA editing complex
Similar to the procedure used for the ∼35–40S complex, a consensus structure of the ∼20S complex was determined. The Euler angle plot disclosed preferential binding to the carbon with two preferred angular regions (Figure 5C). Overall, the complex has an elongated, slightly bent appearance (Figure 5D). This results in a concave–convex shape, displaying one concave and one convex contour on opposite sides. The particle is composed of two globular domains roughly equal in size, the upper domain of which being more roundish, and the lower one being somewhat thinner. Both subdomains interact extensively in an interface where a protruding arm is seen on one side (view 1) and a triangular protrusion emerges from the opposite side (view 3).
Variability analysis of the ∼20S RNA editing complex
To describe the structural variability suggested by the 2D class averages, 3D-MSA was performed using aligned RCTs. We identified four structural subgroups showing distinct structural features: representative 3D class averages are shown in Figure 6A and refined 3D maps subsequent to cMRA are shown in Figure 6B. The first subgroup comprises particles that show a clear separation into two approximately equally sized subdomains connected by an interface: the two subdomains adopt variable relative positions resulting in different curvatures of the particle (row I). The second subgroup (row II) also shows a slightly bipartite shape but displays, in addition, a small semispherical back similar to the ∼35–40S particles. The third and fourth subgroups (rows III and IV) differ from the former two in being more elongated but still exhibiting a bipartite shape. The fourth subtype possesses an additional domain attached to the body in the upper right region.
Figure 6.
Variability analysis of the ∼20S RNA editing complex. (A) Representative 3D class averages demonstrating the overall variability of ∼20S complexes. (B) Refined models of representative 3D class averages subsequent to cMRA. The numbers of particles used to calculate the refined 3D structures are listed on the right. (C) Shape profiling of the ∼20S complex by colour-scale encoded overlays revealing that the majority of complexes are composed of two subdomains connected by a broad interface (red). (D) Histogram of the overall abundance of structural subtypes within three data sets as determined by cMRA showing that about one third of the particles were assigned to the subgroups I and II (colour coding as in (A)).
Quantification of ∼20S editosomal subpopulations
An overlay of 3D shapes according to their proportion in the data set provides a graphical representation of particle shapes (Figure 6C) and also visualises variable regions. Notably, the majority of complexes has an elongated, slightly curved shape, whereas in some particles, additional densities are attached to the upper region.
Using cMRA, we quantified the abundance of the 3D subtypes for three data sets. Indeed, we found that the relative proportions of the 3D subtypes were similar in these data sets, suggesting that the distribution of particles belonging to the different subtypes is representative (Figure 6D). Consistently, ∼25–35% were assigned to the 3D subgroups I and II (Figure 6A), which exhibit the highest visual similarity to the consensus structure.
Refinement of the ∼20S RNA editing complex structure by cryo negative staining EM
The consensus structure of the ∼20S complex was refined by cMRA to ∼1.97–2.20 nm resolution using a data set imaged under cryogenic conditions (Figure 7A). The refined 3D reconstruction displays all structural features of the consensus structure; however, more fine structural characteristics are visible in the refined map consistent with the increase in resolution (Figure 7B). Similar to the consensus structure, the refined map revealed an asymmetric particle composed of two approximately equally sized subdomains connected by a density network at the interface. All densities described above were also visible (compare labelled elements in Figures 5D and 7B). The surface representation of the refined structure of the ∼20S editing complex was estimated to enclose a molecular mass of 800±80 kDa. The predicted sedimentation coefficient is ∼21–26S consistent with the experimentally determined apparent value of ∼20–24S.
Figure 7.
Refinement of the 3D map of the ∼20S RNA editing complex by cryo negative staining EM. (A) FSC suggests a resolution of 1.97–2.20 nm for the final 3D structure of the ∼20S complex. (B) Surface representation of the refined ∼20S complex demonstrating a tight network of structural elements. Landmarks are labelled as in Figure 5D.
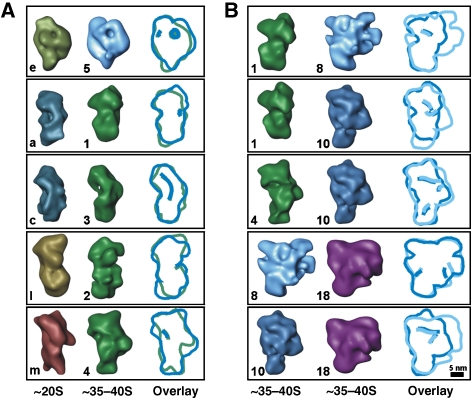
3D structural comparison of the RNA editing complexes
Recently, crystal structures of editing components have been described (Deng et al, 2004, 2005; Schumacher et al, 2006). However, these structures represent only small complexes or protein fragments and, thus, a direct identification of these components is usually not unambiguous at the resolution of the RNA editing complexes. We therefore applied the following strategy to determine the structural relationship between the analysed particles. By visual comparison, we identified pairs of ∼20S and ∼35–40S 3D class averages showing similar morphological features. The similar landmarks were used as anchor points to manually dock the ∼20S into the ∼35–40S complexes. Subsequently, this fit was refined by 3D alignment. Overall, the fitting experiments indicate that the ∼20S complexes may comprise a major portion of the platform of the ∼35–40S complexes and in some cases also parts of the semispherical back (Figure 8A for representative examples). In contrast, the majority of the semispherical back appears to be composed of other components (e.g., gRNA, pre-mRNA, additional proteins) that have no structural correlate in the ∼20S particles. Minor differences in the orientation of distinct domains are also indicated (e.g., the orientation of the head-like and foot-like protuberance in Figure 8A, first row). These minor structural differences suggest conformational and/or compositional changes of RNA editing complexes captured at the various stages.
Figure 8.
Structural relationship of the independently determined de novo structures of RNA editing complexes. For comparison, contour overlays are provided on the right and structures are labelled as in Figures 3A and 6A. The bandpass-filtered 3D structures may be rotated with respect to Figures 2, 3, 4, 5, 6 and 7 to maximise overlap. (A) 3D class averages of the ∼20S complex (left) were fitted into 3D class averages of the ∼35–40S complex (right). (B) Pairs of 3D class averages of the ∼35–40S complex suggest a common structural core to which additional domains are attached at the periphery.
This positioning is also supported by fitting experiments using 3D class averages from the 35–40S factions only. The elongated subpopulations may represent the major portion of the platform density of the larger ∼35–40S subtypes (Figure 8B, rows 1 and 2), whereas they may also contain parts of the semispherical back in other particles (row 3). Likewise, the largest ∼35–40S complexes show similar morphological features to smaller complexes (rows 4 and 5). Overall, we suggest that the various particle populations found in both gradient fractions are thus structurally related and share a common 3D structural core.
Interconversion of the two complexes
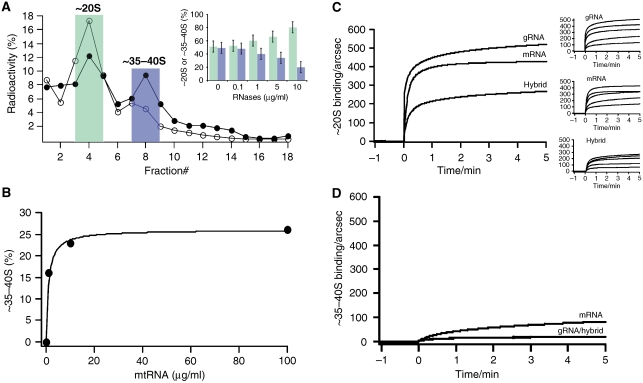
On the basis of the structural data, the molecular mass of the two complexes is estimated to 800±80 kDa (∼20S complex) and 1.45±0.15 MDa (∼35–40S complex) in line with the experimentally derived apparent sedimentation coefficients (Supplementary Table 3). The molecular mass of the ∼20S complex is further consistent with the sum of all proteins found in our analysis (∼790 kDa), assuming that every protein is present in one copy (Supplementary Table 2). Additional components such as gRNA, pre-/partially edited mRNA and/or additional proteins may thus be present in the larger complex (Supplementary Tables 2–4). However, if one assumes that at least one gRNA/pre-mRNA hybrid molecule interacts with the editosome, the RNA contribution can add up to ∼450 kDa. On the basis of this value, it is tempting to speculate that the presence of RNA represents the main compositional difference of the two complexes. As ∼35–40S complexes contain endogenous RNA (Figure 1D), we addressed this question by RNase digestion. The treatment of CaM eluates with a cocktail of single- and double-strand-specific RNases resulted in a concentration-dependent decrease in the amount of ∼35–40S complexes and at the same time in an increase of ∼20S editosomes (Figure 9A). This suggests an interconversion of the two complexes, which was further confirmed by incubating isolated ∼20S editosomes with T. brucei mitochondrial (mt) RNA. mtRNA contains gRNAs as well as pre-/partially edited mRNAs. Adding increasing amounts to ∼20S editosomes generated ∼35–40S complexes in a concentration-dependent fashion (Figure 9B).
Figure 9.
RNA is the major determinant for the interconversion of the two RNA editing complexes. (A) Representative glycerol gradient separation of 125I-labelled RNA editing complexes pre-treated with RNases A, T1 and V1 (filled circles: − RNases; open circles: + RNases). ∼20S complexes (green); ∼35–40S complexes (blue). Insert: RNase treatment results in a concentration-dependent decrease of the ∼35–40S complexes and an increase of the ∼20S complexes. Error bars represent relative errors. (B) Concentration-dependent formation of ∼35–40S complexes through binding of T. brucei mtRNA to 125I-labelled ∼20S editosomes. (C) Real-time surface plasmon resonance monitoring of the binding of ∼20S editing complexes to gRNA, pre-mRNA and gRNA/pre-edited mRNA hybrid molecules. Left: Sensograms of the binding reaction to the three RNAs at an ∼20S concentration of 40 nM. Right: Verification of the concentration-dependent binding reaction for all three RNAs (40–2 nM top to bottom). kass values varied between 0.1–0.25 × 104 M−1 s−1. Dissociation rate constants (kdiss) are in the range of 6–7 × 10−5 s−1. The calculated Kd values are 30 nM for the pre-edited mRNA, 65 nM for the gRNA and 54 nM for the gRNA/pre-mRNA hybrid. (D) Real-time monitoring of the RNA binding capacity of ∼35–40S editing complexes.
To derive quantitative data for the editosome/RNA interaction, we performed real-time RNA-binding experiments using a surface plasmon resonance (SPR)-based detection system. Figure 9C shows the binding curves of purified ∼20S complexes. The particles interact with synthetic gRNAs, pre-edited mRNAs and gRNA/pre-mRNA hybrid molecules in a concentration-dependent fashion. RNA binding is complete within ⩽1 min and is characterised by association rate constants (kass) in the range of 0.2 × 104 M−1 s−1 and dissociation rate constants (kdiss) of 0.7 × 10−6 s−1. The corresponding equilibrium dissociation constants (Kd) are in a nanomolar concentration range (gRNA: 65 nM, pre-mRNA: 30 nM, gRNA/pre-mRNA hybrid: 54 nM), indicating high affinity interactions between ∼20S editosomes and the three different RNA ligands. Importantly, none of the three RNAs is able to interact with ∼35–40S editosomes (Figure 9D).
Discussion
Here, we report the first 3D structural characterisation of the RNA editing machinery in T. brucei. Particles were isolated from the mitochondrial pool and, thus, represent the endogenous population of RNA editing complexes. The enriched complexes were functionally active in both, insertion and deletion RNA editing, and based on their sedimentation behaviour contained two major subpopulations: an ∼20S and an ∼35–40S complex. A variability analysis, however, identified various structural subtypes that differ in conformation and composition and also showed the most abundant and, therefore, representative structures.
Composition of RNA editing complexes
Our analysis identified 13 polypeptides in the TAP-tag enriched fractions. All of these proteins have documented links to RNA editing (Rusché et al, 1997; Aphasizhev et al, 2003; Panigrahi et al, 2003a, 2003b, 2006), and for some, functional roles have been reported (Stuart et al, 2005). These previous studies of ∼20S editing complexes have identified up to 20 proteins and estimated a molecular mass of up to 1.6 MDa (Panigrahi et al, 2001a). However, more recent studies suggested the existence of various subtypes of ∼20S complexes: while there appears to be a common set of proteins (TbMP99, TbMP81, TbMP63, TbMP57, TbMP52, TbMP48, TbMP46, TbMP44, TbMP42, TbMP24, TbMP18), other proteins appear to be present only in subsets of ∼20S complexes (Panigrahi et al, 2006). The ‘core' complex is remarkably similar to the protein composition of the editing complexes analysed herein (Supplementary Tables 1 and 2). Aside from TbMP81 and TbMP18, all ‘core' proteins were identified in our complexes. Furthermore, we also detected four additional proteins (TbMP100, TbMP90, TbMP67, TbMP61) that were found only in subsets of ∼20S complexes.
In contrast, the protein inventory of the ∼35–40S complexes is currently unknown, as comprehensive studies are lacking. Nonetheless, initial studies have identified some of the components and associated activities including RNA ligase (Pollard et al, 1992) and TUTase activity resulting in formation of a poly(U) tail in gRNA (McManus et al, 2000). The gRNA-binding protein gBP21 that accelerates gRNA/mRNA annealing (Müller et al, 2001) was found in the ∼35–40S fractions (Lambert et al, 1999). Also, TbRGG1 that preferentially binds to oligo(U) (Vanhamme et al, 1998) and REAP-1 that has increased affinity for pre-edited mRNA (Madison-Antenucci and Hajduk, 2001; Hans et al, 2007) were shown to be present in ∼35–40S fractions. In addition, gRNA and pre-mRNA were detected in ∼35–40S complexes (Pollard et al, 1992) in line with our results.
3D structure of the RNA editing complexes
By EM we draw the first picture of the endogenous RNA editing machinery on the 3D level. The ∼20S fractions showed a wide range of different shapes, the majority of which displayed a bipartite appearance. This bipartite shape is in line with biochemical data, suggesting that editing proteins are present as pairs or sets within the ∼20S complex (Panigrahi et al, 2006). In particular, insertion and deletion editing activities may reside in separate subcomplexes that are linked by a common set of shared proteins (Schnaufer et al, 2003). We therefore suggest that this separation of functional activities may also be reflected by the structural subdivision into two domains, whereas shared proteins might be located at the interface. For the ∼35–40S editing complexes, EM analysis showed two main structural features, an elongated platform and a semispherical domain, consistently reflecting the particles visible in the raw EM images and 2D class averages.
Structural diversity of RNA editing complexes
By 3D variability analysis, we revealed a significant structural diversity of both the ∼20S and the ∼35–40S complexes, which may be attributed to conformational and compositional differences. Heterogeneity of ∼20S complexes has been suggested recently on the basis of biochemical data (Panigrahi et al, 2006), but the extend was unexpected and exceeds previous experiences with many other particles (Leschziner and Nogales, 2007). In particular, the two subdomains of the ∼20S complexes adopt different relative positions, resulting in structures with straight to bent conformations. As a second source of heterogeneity, we identified an additional element attached to the upper domain in a subset of ∼20S particles. The situation becomes even more complex for the ∼35–40S particles for which we detected at least six subpopulations. As a main source of variability, we found a semispherical domain that differs significantly in size among the subpopulations.
Model for the assembly of the RNA editing machinery
The understanding of the molecular architecture of the RNA editing machinery is currently limited and a global view of this machine has not yet emerged. In particular, the unknown compositional and functional relationship of the purified editing complexes thus far leaves many open questions. Currently, several models for the relationship between the RNA editing complexes can be envisioned. In a first model, the ∼20S particle may represent an assembly/disassembly intermediate of the larger ∼35–40S complex, which has been suggested to be the catalytically active RNA editing machine in vivo (Madison-Antenucci et al, 2002). Both complexes may thus share a set of identical proteins forming a common core. In an alternative model, the ∼20S and ∼35–40S editing complexes may represent functionally separate complexes in vivo: the ∼20S particle functioning as a gRNA maturation complex (Grams et al, 2000) and the ∼35–40S particle representing the catalytically active RNA editing machine (Pollard et al, 1992). Due to the different functions, both complexes may differ in the overall architecture.
Here, our data are in line with the former model, as the various subpopulations of the ∼20S and ∼35–40S share a similar structural organisation (Figure 10). These structural elements may represent core components of the editing machinery found in the majority of the complexes. A comparison with the ∼35–40S fractions suggests that additional components are packed against the platform within a variable semispherical back. This domain thus represents a candidate to harbour most of the additional components. In line with previous studies (Pollard et al, 1992; Corell et al, 1996; Rusché et al, 1997), we showed that ∼35–40S complexes contain endogenous RNA, whereas ∼20S editosomes are essentially free of RNA. Therefore, we suggest that bound RNA represents the main compositional difference between the two complexes. The ∼20S editosomes associate with pre-edited mRNA and gRNAs thereby converting these to ∼35–40S complexes. The ∼20S complexes have an RNA-binding site, which binds gRNA, pre-mRNA and gRNA/pre-mRNA molecules with equal affinity and association rate constants. These data are further supported by the molecular mass calculations for the two particles and the experimentally determined apparent S values. In addition, the molecular mass of the ∼20S complex is consistent with the sum of all proteins found in our analysis. Depending on the degree of editing, the RNA components can add up to ∼450 kDa assuming that the editosome interacts with at least a single gRNA/pre-mRNA hybrid molecule. Note that glutaraldehyde crosslinks primary amines of proteins but does not react with RNA at the conditions used (Hopwood, 1975). Thus, the presence of ∼20S types of particles within the 35–40S region of the gradient is consistent with this characteristic. In addition, the association of a few low molecular mass polypeptides may contribute to the interconversion as well (Fisk et al, 2008). This emphasises the need for a detailed analysis of the protein composition and stoichiometry of the two complexes.
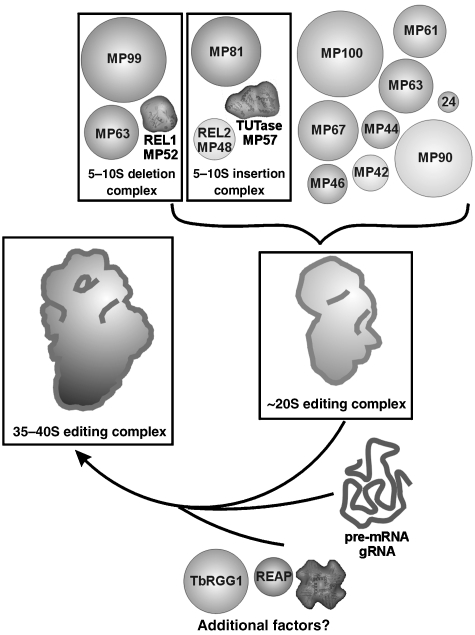
Figure 10.
Model for the assembly pathway of the RNA editing machine. The ∼20S complexes represent common core modules providing the majority of editing proteins. Through the interaction with gRNA, its target pre-mRNA and possibly additional proteins, the assembly is converted to the ∼35–40S complex, which is suggested to perform the RNA editing reaction.
In summary, we have characterised the two major RNA editing complexes of trypanosomes on a biochemical, functional and structural level and provide a model on how these complexes are related to each other. Our data suggest that the ∼20S and ∼35–40S editing complexes are not only biochemically related, but also share a common 3D structural core consistent with an assembly/disassembly model of the RNA editing machine. Together, we provide a basis for advancing our understanding of the architecture of the RNA editing machinery in African trypanosomes.
Materials and methods
Generation of the TbMP42/TAP cell line
The coding region of TbMP42 was amplified from genomic DNA of T. brucei strain 427. The resulting PCR fragment was cloned into pLEW100/TAP, a derivative of pLEW100 (Wirtz et al, 1999), containing the TAP-tag cassette derived from pBS1479 (Rigaut et al, 1999). The generated plasmid was named pLEW-TbMP42/TAP and was used for transfection.
Biochemical and functional analysis
Expression of TbMP42/TAP was induced by the addition of 1 μg/ml tet to the culture medium. Parasites from 20 l of cell culture were harvested at cell densities between 1–2 × 107 cells/ml. Editing complexes were isolated from mitochondrial preparations. The eluates were separated in 10% (w/v) SDS-containing polyacrylamide gels and visualised by silver staining. Proteins were identified by MALDI-ToF-analysis. RNA ligases were detected by self-adenylation (Sabatini and Hajduk, 1995). CaM eluates were analysed for their RNA content by phenol extraction followed by ethanol precipitation. RNA precipitates were 3′ and 5′ radiolabelled with [32P] following standard procedures. Guide RNAs were identified by incubation with guanylyl transferase (Blum and Simpson, 1990; Göringer et al, 1994) in the presence of α-[32P]-GTP. Radiolabelled RNA samples were spotted onto polyethyleneimine (PEI) cellulose plates before autoradiography. Purified complexes were tested for their activity using pre-cleaved RNA editing assays (Igo et al, 2000, 2002).
Protein iodination and RNA/editosome interaction measurements
Editing complexes were radioactively labelled by tyrosin iodination using Na125I and N-chloro tosylamide. Radioactive complexes were fractionated in isokinetic 10–40% (v/v) glycerol gradients at 4°C for 2 h at 100 000 g (Beckman TLS55 rotor). Eighteen 0.2 ml fractions were collected from the top of the gradients and quantified by scintillation counting. RNA digests were performed with increasing concentrations of RNases (A, T1, V1) for 1 h at 27°C. mtRNA was isolated from insect stage trypanosomes (Göringer et al, 1994). Radioactively labelled ∼20S complexes were incubated with increasing amounts of mtRNA for 30 min at 27°C and fractionated by density gradient centrifugation as above. Gradient fractions were TCA precipitated and quantified by scintillation counting. Editosome/RNA-binding affinities were measured by SPR (IAsys, Affinity Sensors).
Electron microscopy
A measure of 0.75–1.5 ml of the CaM eluate (7–10 μg) were loaded onto a 500μl cushion of 10% (v/v) glycerol in editing buffer followed by a linear 10–40% (v/v) glycerol and 0–0.1% glutaraldehyde gradient (Kastner et al, 2008). Samples were spun in a TH-641 rotor (Kendro, Hanau, Germany) for 14 h at 38 000 r.p.m. and 4°C. Gradient fractions were collected at 4°C from the bottom. EM grids from the ∼20S and the ∼35–40S fractions were prepared according to the sandwich carbon procedure using uranyl formate and either stored at room temperature (RT) or frozen in liquid nitrogen for cryo negative staining EM (Golas et al, 2003). EM images were taken on a CM200FEG (Philips/FEI, Eindhoven, The Netherlands) electron microscope on a TemCam-F415 (TVIPS, Gauting, Germany) (Sander et al, 2005) in tile mode or on Kodak SO-163 film scanned with a rotating drum scanner (Tango, Heidelberger Druckmaschinen, Germany). Single-particle images were selected manually and corrected for the contrast transfer function (Sander et al, 2003a).
Image processing
A detailed description of the data sets used in this study can be found in Supplementary Table 5. A total of 4288 tilt pairs (45°/0°) of the ∼35–40S complex were collected. Using iteratively refined 2D class averages untilted single-particle images were aligned using exhaustive MRA based on polar coordinates (Sander et al, 2003b) and classified by MSA-based classification (Frank and van Heel, 1982) in the context of IMAGIC-5 (van Heel et al, 1996). In the final iteration, the aligned images were divided into 214 classes. Of these, 175 classes were selected for further image processing based on the visual quality of the 2D class averages. Projection angles of the corresponding tilted images of selected zero-tilt classes were determined by the RCT method (Radermacher, 1988). Individual RCTs were combined into a consensus structure by an iterative approach using Gaussian noise as initial reference. A total of 12 188 tilt pairs of the ∼20S complex were treated likewise leading to a final classification into 609 classes. Individual 3D RCTs that were aligned according to the consensus structure were subjected to MSA-based classification, generating up to 58 3D maps. Representative 3D maps were selected for presentation as surface view (AmiraDev 2.3, TGS Europe, Merignac Cedex, France). Shape profiling was performed using colour-scale encoded overlays according to abundance. Three data sets each (8576, 2500 and 6786 images of the ∼35–40S complex; 24 866, 2613 and 3989 images of the ∼20S complex) were subjected to cMRA using 4° spacing of references generated from the 3D class averages. Altogether, 25 747 and 31 641 defocus-corrected particles of the ∼35–40S and ∼20S editing complexes, respectively, imaged at liquid nitrogen temperature using a Gatan cryoholder were selected for refinement by projection matching. Initial references were created from the best-defined 3D class average (∼35–40S complex) or the consensus structure (∼20S complex) and were iteratively refined to a fine angular separation of 2–4°. The resolution of the maps was estimated by Fourier-Shell-Correlation (FSC) (Harauz and van Heel, 1986). The molecular mass of the 3D reconstructions was estimated on the basis of its enclosed volume. The sedimentation coefficient was predicted using HYDROMIC (Garcia de la Torre et al, 2001). For fitting, 3Ds were first manually docked and the fit was subsequently refined using an exhaustive cross-correlation based real-space 3D alignment (Sander et al, 2006) with a final rotational/translational accuracy of 0.25°/0.5 nm.
Accession numbers
The EM maps have been deposited at the EMDB database under the accession numbers EMD-1594 and EMD-1595.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Berthold Kastner for discussion and suggestions, and Elke Schlüter and Nadja Pasqual for preparative work. The work was supported by a grant of the Bundesministerium für Bildung und Forschung (BMBF) and a European ‘3D Repertoire' grant to HS. HUG is supported as an International Research Scholar of the Howard Hughes Medical Institute (HHMI) and by the German Research Council (DFG-SFB579).
References
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L (2003) Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J 22: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L (1990) A model for RNA editing in kinetoplastid mitochondria: ‘guide' RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198 [DOI] [PubMed] [Google Scholar]
- Blum B, Simpson L (1990) Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62: 391–397 [DOI] [PubMed] [Google Scholar]
- Brecht M, Niemann M, Schlüter E, Müller UF, Stuart K, Göringer HU (2005) TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol Cell 17: 621–630 [DOI] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K (2005) An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci USA 102: 16614–16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corell RA, Read LK, Riley GR, Nellissery JK, Allen TE, Kable ML, Wachal MD, Seiwert SD, Myler PJ, Stuart KD (1996) Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol Cell Biol 16: 1410–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Ernst NL, Turley S, Stuart KD, Hol WG (2005) Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J 24: 4007–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG (2004) High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J Mol Biol 343: 601–613 [DOI] [PubMed] [Google Scholar]
- Ernst NL, Panicucci B, Igo RP Jr, Panigrahi AK, Salavati R, Stuart K (2003) TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell 11: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Fisk JC, Ammerman ML, Presnyak V, Read LK (2008) TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem 283: 23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, van Heel M (1982) Correspondence analysis of aligned images of biological particles. J Mol Biol 161: 134–137 [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J, Llorca O, Carrascosa JL, Valpuesta JM (2001) HYDROMIC: prediction of hydrodynamic properties of rigid macromolecular structures obtained from electron microscopy images. Eur Biophys J 30: 457–462 [DOI] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Lührmann R, Stark H (2003) Molecular architecture of the multiprotein splicing factor SF3b. Science 300: 980–984 [DOI] [PubMed] [Google Scholar]
- Göringer HU, Koslowsky DJ, Morales TH, Stuart K (1994) The formation of mitochondrial ribonucleoprotein complexes involving guide RNA molecules in Trypanosoma brucei. Proc Natl Acad Sci USA 91: 1776–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulah CC, Read LK (2007) Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J Biol Chem 282: 7181–7190 [DOI] [PubMed] [Google Scholar]
- Grams J, McManus MT, Hajduk SL (2000) Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J 19: 5525–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans J, Hajduk SL, Madison-Antenucci S (2007) RNA-editing-associated protein 1 null mutant reveals link to mitochondrial RNA stability. RNA 13: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, van Heel M (1986) Similarity measures between images. Exact filters for general geometry three-dimensional reconstruction. Optik 73: 146–156 [Google Scholar]
- Hauser R, Pypaert M, Hausler T, Horn EK, Schneider A (1996) In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Sci 109 (Part 2): 517–523 [DOI] [PubMed] [Google Scholar]
- Hopwood D (1975) The reactions of glutaraldehyde with nucleic acids. Histochem J 7: 267–276 [DOI] [PubMed] [Google Scholar]
- Igo RP Jr, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K (2000) Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol 20: 8447–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo RP Jr, Weston DS, Ernst NL, Panigrahi AK, Salavati R, Stuart K (2002) Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot Cell 1: 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, Uchtenhagen H, Urlaub H, Herzog F, Peters JM, Poerschke D, Lührmann R, Stark H (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods 5: 53–55 [DOI] [PubMed] [Google Scholar]
- Lambert L, Müller UF, Souza AE, Göringer HU (1999) The involvement of gRNA-binding protein gBP21 in RNA editing-an in vitro and in vivo analysis. Nucleic Acids Res 27: 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschziner AE, Nogales E (2007) Visualizing flexibility at molecular resolution: analysis of heterogeneity in single-particle electron microscopy reconstructions. Annu Rev Biophys Biomol Struct 36: 43–62 [DOI] [PubMed] [Google Scholar]
- Liu J, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Lucaveche C, Winkler H, Baumann BA, Squire JM, Irving TC, Reedy MK, Taylor KA (2004) Electron tomography of fast frozen, stretched rigor fibers reveals elastic distortions in the myosin crossbridges. J Struct Biol 147: 268–282 [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S, Grams J, Hajduk SL (2002) Editing machines: the complexities of trypanosome RNA editing. Cell 108: 435–438 [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S, Hajduk SL (2001) RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol Cell 7: 879–886 [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S, Sabatini RS, Pollard VW, Hajduk SL (1998) Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J 17: 6368–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Adler BK, Pollard VW, Hajduk SL (2000) Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol Cell Biol 20: 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Shimamura M, Grams J, Hajduk SL (2001) Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian IS, Worthey EA, Salavati R (2006) Taking U out, with two nucleases? BMC Bioinformatics 7: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller UF, Lambert L, Göringer HU (2001) Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J 20: 1394–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann M, Brecht M, Schlüter E, Weitzel K, Zacharias M, Göringer HU (2008) TbMP42 is a structure-sensitive ribonuclease that likely follows a metal ion catalysis mechanism. Nucleic Acids Res 36: 4465–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP (2003a) Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectrom 14: 728–735 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD (2006) Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 12: 1038–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Gygi SP, Ernst NL, Igo RP Jr, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, Stuart KD (2001a) Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol 21: 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Carmean N, Igo RP Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, Stuart KD (2001b) Four related proteins of the Trypanosoma brucei RNA editing complex. Mol Cell Biol 21: 6833–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K (2003b) Identification of novel components of Trypanosoma brucei editosomes. RNA 9: 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Harris ME, Hajduk SL (1992) Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J 11: 4429–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermacher M (1988) Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech 9: 359–394 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rogers K, Gao G, Simpson L (2007) Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J Biol Chem 282: 29073–29080 [DOI] [PubMed] [Google Scholar]
- Rusché LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B (1997) Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16: 4069–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini R, Hajduk SL (1995) RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J Biol Chem 270: 7233–7240 [DOI] [PubMed] [Google Scholar]
- Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Lührmann R, Stark H (2006) Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell 24: 267–278 [DOI] [PubMed] [Google Scholar]
- Sander B, Golas MM, Stark H (2003a) Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. J Struct Biol 142: 392–401 [DOI] [PubMed] [Google Scholar]
- Sander B, Golas MM, Stark H (2003b) Corrim-based alignment for improved speed in single-particle image processing. J Struct Biol 143: 219–228 [DOI] [PubMed] [Google Scholar]
- Sander B, Golas MM, Stark H (2005) Advantages of CCD detectors for de novo three-dimensional structure determination in single-particle electron microscopy. J Struct Biol 151: 92–105 [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K (2003) Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell 12: 307–319 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J (2006) Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism. Cell 126: 701–711 [DOI] [PubMed] [Google Scholar]
- Sigworth FJ (1998) A maximum-likelihood approach to single-particle image refinement. J Struct Biol 122: 328–339 [DOI] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK (2005) Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30: 97–105 [DOI] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K (2005) A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell 20: 403–412 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaili N, Göringer U, Benne R, Pays E (1998) Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J Biol Chem 273: 21825–21833 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information