Abstract
Protein components of the Drosophila male ejaculate, several of which evolve rapidly, are critical modulators of reproductive success. Recent studies of female reproductive tract proteins indicate they also are extremely divergent between species, suggesting that reproductive molecules may coevolve between the sexes. Our current understanding of intersexual coevolution, however, is severely limited by the paucity of genetic and evolutionary studies on the female molecules involved. Physiological evidence of ejaculate–female coadaptation, paired with a promiscuous mating system, makes Drosophila mojavensis an exciting model system in which to study the evolution of reproductive proteins. Here we explore the evolutionary dynamics of a five-paralog gene family of female reproductive proteases within populations of D. mojavensis and throughout the repleta species group. We show that the proteins have experienced ongoing gene duplication and adaptive evolution and further exhibit dynamic patterns of pseudogenation, copy number variation, gene conversion, and selection within geographically isolated populations of D. mojavensis. The integration of these patterns in a single gene family has never before been documented in a reproductive protein.
IN internally fertilizing organisms, female reproductive tracts are the arena for a dynamic molecular interface between the sexes. Ejaculate–female interactions are essential to sperm fate and fertilization, guiding sperm through the female reproductive tract, preserving them in this environment, and ultimately mediating gamete fusion (reviewed in Neubaum and Wolfner 1999). Reproductive tract interactions also modulate critical postmating changes in female behavior and physiology, such as upregulating immune response, reformatting the female reproductive tract, and delaying female remating (reviewed in Robertson 2007; Wolfner 2007).
Despite the significance of ejaculate–female interactions for overall fitness, the male molecules involved in these processes exhibit dynamic evolutionary histories. Seminal proteins and sperm proteins have been observed to evolve rapidly in a broad range of taxa (reviewed in Swanson and Vacquier 2002; Clark et al. 2006; Panhuis et al. 2006). Similarly, lineage-specific gene duplications have been documented in Drosophila seminal fluid proteins (Cirera and Aguadé 1998; Wagstaff and Begun 2007; Almeida and Desalle 2008, 2009; Findlay et al. 2008), as well as fertilization proteins in both Drosophila and abalone (Loppin et al. 2005; Clark et al. 2007). Finally, Drosophila male ejaculates are known to undergo a high frequency of lineage-specific changes in seminal fluid content, by functionally coopting existing genes and acquiring novel genes from noncoding sequence (Begun and Lindfors 2005; Mueller et al. 2005; Begun et al. 2006; Findlay et al. 2008).
The rapid evolution of male ejaculates frequently is postulated to arise from molecular coevolution with interacting proteins in the female reproductive tract (Parker 1979; Eberhard 1996; Swanson and Vacquier 2002). If this is the case, female reproductive molecules are also expected to evolve rapidly. Recent evidence of adaptive evolution in Drosophila female reproductive tract proteins is consistent with this prediction (Swanson et al. 2004; Panhuis and Swanson 2006; Kelleher et al. 2007; Lawniczak and Begun 2007; Prokupek et al. 2008). Compared to the preponderance of studies of male ejaculates, however, the dynamics of female proteins remain largely unexplored.
Two, nonmutually exclusive mechanisms are hypothesized to result in reciprocal evolutionary change between male and female reproductive molecules. First, cryptic female choice could empower females to bias fertilization success toward certain males based on postcopulatory biochemical cues (Eberhard 1996). Cryptic female choice may lead to cyclical evolution of male trait and female preference, consistent with traditional models of runaway sexual selection (Fisher 1915; 1930). Alternatively, sexual conflict, or a difference in the reproductive interests of the two sexes (Parker 1979), is predicted to result in an evolutionary arms race between males and females (Rice 1996; Gavrilets 2000).
In this study, we explore the dynamics of a female reproductive tract protein gene family in the cactophilic fruit fly Drosophila mojavensis. A promiscuous mating system (reviewed in Markow 1996), as well as extensive evidence of ejaculate–female biochemical coadaptation (Knowles and Markow 2001; Pitnick et al. 2003; Knowles et al. 2005; Kelleher and Markow 2007) makes D. mojavensis an extraordinary system for the study of reproductive molecules. Specifically, interpopulation crosses exhibit significant differences from intrapopulation crosses in egg size (Pitnick et al. 2003), a mating-dependent increase in female desiccation resistance (Knowles et al. 2005), and the size and duration of the insemination reaction, an opaque mass that forms in the uterus after copulation (Knowles and Markow 2001). Similarly, interspecific crosses between D. mojavensis and its sister species D. arizonae [most recent common ancestor (MRCA) ∼0.7 MYA] (Reed et al. 2007; Matzkin 2008), exhibit considerable sperm mortality, failure in sperm storage, reduced oviposition, and aberrant insemination reactions, consistent with a breakdown in coadapted gene complexes (Kelleher and Markow 2007).
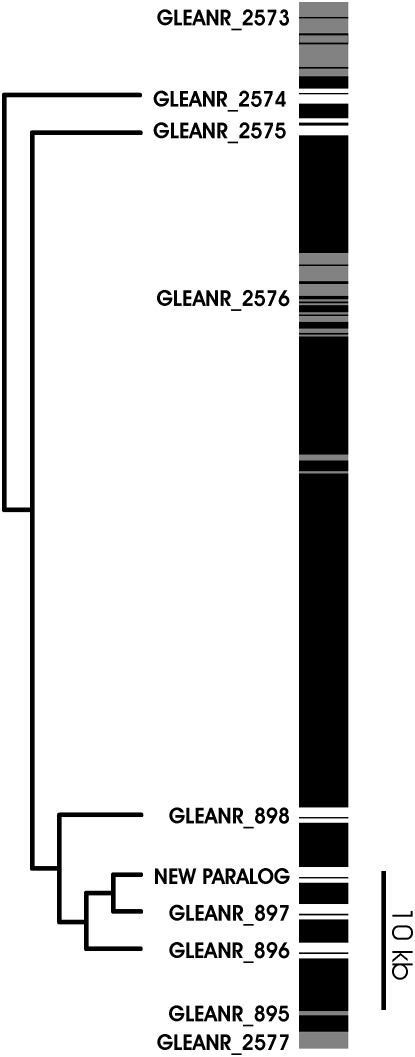
The gene family examined here is one of five lineage-specific protease gene families identified from D. arizonae female reproductive tracts and encodes five serine-endoprotease paralogs: Dmoj\GLEANR_2575 (GI17776), Dmoj\GLEANR_2574 (GI17775), Dmoj\GLEANR_896 (GI23802), Dmoj\GLEANR_897 (GI23804), and Dmoj\GLEANR_898 (GI23805) (Figure 1; Kelleher et al. 2007). Although the specific function of these enzymes remains unknown, they are predicted secreted proteins expressed only in the lower female reproductive tract, implying specialized interaction with the male ejaculate (Kelleher et al. 2007). Serine-endoprotease activity in D. arizonae female reproductive tracts, furthermore, is regulated by mating, pointing to a direct relationship between reproduction and proteolytic function (E. S. Kelleher and J. E. Pennington, unpublished results).
Figure 1.—
Genomic arrangement of the female reproductive tract protease gene family examined in this study. The exon structure of six paralogs (open) and neighboring coding sequences (shaded) are indicated along an ∼50-kb region of D. mojavensis chromosome 3. The position of novel paralog, not present in the sequenced strain of D. mojavensis, was determined by PCR. Scale is indicated by 10-kb size marker.
If female reproductive tract proteases are coevolving with the male ejaculate, two predictions follow about their evolutionary dynamics. First, the coevolutionary trajectory within each population should exert unique selective pressures on the proteins involved. To explore this hypothesis we compare patterns of variation and deviations from neutrality at these loci between the four geographically isolated populations of D. mojavensis: Baja Peninsula, Catalina Island, mainland Sonora, and Mojave Desert (Machado et al. 2007; Reed et al. 2007; Figure 2). Second, ongoing coevolution with interacting proteins predicts a history of adaptive evolution across the repleta species group. We therefore examine patterns of divergence at these loci from five repleta group species and two outgroups. We discuss our results in terms of our predictions, as well as the emerging role of gene duplication in reproductive protein evolution.
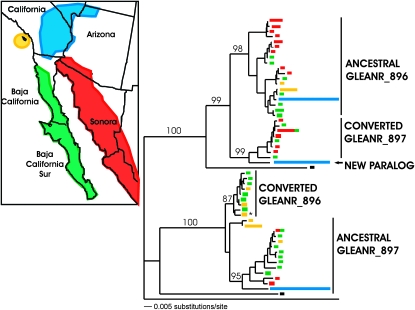
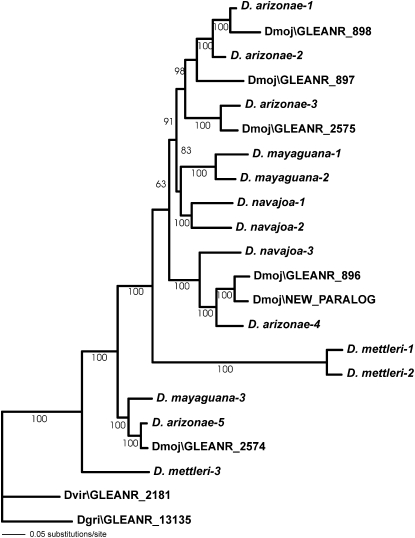
Figure 2.—
Neighbor-joining analysis sampled GLEANR_896, GLEANR_897, and new paralog haplotypes. Bar length indicates number of sampled individuals corresponding to each haplotype, and bar color is indicative of geographic locality. * denotes a GLEANR_897 ancestral allele that does not group with the remainder of its haplogroup. Neighbor-joining bootstrap values are indicated above the relevant branch.
MATERIALS AND METHODS
Flies:
D. mojavensis were collected from Catalina Island (2001), Mojave Desert (2002), Baja Peninsula (2002), and mainland Sonora (2007) by J. Bono, L. Reed, and L. Matzkin. D. arizonae were collected in Tucson, Arizona (2000), by L. Matzkin. D. navajoa, D. mettleri, and D. mayaguana were obtained from the Tucson Drosophila Stock Center, now located at the University of California at San Diego. All flies used in population analyses were maintained as isofemale lines. Between 7 and 14 isofemale lines were sampled for each population and locus (supplemental Table 1).
Sequencing:
Genomic DNA was isolated from whole flies using the DNeasy Kit (QIAGEN) according to manufacturer instructions. For D. mojavensis and D. arizonae, standard PCR was performed using internal, paralog-specific primers (Figure 1). In cases where gene conversion obscured paralog identity (GLEANR_896 and GLEANR_897), additional flanking primers were used to ensure gene-specific amplification. For D. navajoa, D. mettleri, and D. mayaguana universal primers for the entire gene family were used to amplify and clone PCR products. Cloned PCR products were sequenced using M13F and M13R primers. All sequencing was performed on an ABI 3700 DNA sequencer with Big Dye Terminator chemistry. D. grimshawi and D. virilis sequences were obtained from their sequenced genomes (http://rana.lbl.gov/drosophila/). Primers and PCR conditions are available from the authors upon request. Base calling and assembly were performed in Sequencher 4.8.
Inverse polymerase chain reaction:
Genomic DNA from a single Mojave Desert isofemale line was digested with each of four restriction enzymes according to manufacturer instructions (New England Biolabs): AciI, MboI, MseI, and TaqI. Digested fragments were then incubated with ∼20 units DNA ligase (Fermentas) at 17° overnight to generate circularized DNA. Circularized DNA was then used for standard PCR with inverted primers specific to the novel paralog. Primers and PCR conditions are available from the authors upon request.
Reverse transcriptase polymerase chain reaction:
Total RNA was extracted from 20 adult males, 20 adult female reproductive tracts (oviduct, spermathecae, seminal receptacle, parovaria, uterus), and 20 adult female carcasses (no female reproductive tract) from a Mojave Desert isofemale line using TRIZOL reagent (Invitrogen), according to manufacturer instructions. RNA was treated with Dnase I (NEB) and reverse transcribed with the iScript cDNA synthesis kit (Roche). Resultant cDNA was diluted to 5 ng/μl for all three samples and used as template for standard PCR with ribosomal protein 32 (control) and paralog-specific (experimental) primers. Quantity of resultant product was compared on a 1% agarose gel stained with ethidium bromide. Primers and PCR conditions are available from the authors upon request.
Polymorphism analyses:
Haplotypes were phased in Arlequin (http://lgb.unige.ch/arlequin/software/), and subsequent polymorphism analyses, estimation of population parameters, and tests of selection were performed in DNAsp (Rozas and Rozas 1995) and SITES (http://lifesci.rutgers.edu/∼heylab/ProgramsandData/Programs/SITES/SITES). Sample sizes, sequence lengths, estimates of polymorphism, site frequency spectra tests, and McDonald–Kreitman tests (McDonald and Kreitman 1991) for all loci are presented in supplemental Table 1. Significance of site frequency spectra statistics was assessed by coalescent simulations under the conservative assumption of no recombination. For tests requiring an outgroup, one or more D. arizonae orthologs were used.
Gene conversion was detected by GENECONV (http://www.math.wustl.edu/∼sawyer/geneconv/) within an alignment of all unique haplotypes for all paralogs using the method of Sawyer (1989). Briefly, gene conversion tracts between pairs of sequences are identified by stretches of complete identity interspersed between two regions of considerable mismatch or one region of mismatch and the end of the alignment. Statistical significance of these fragments is determined by permutation tests. Neighbor-joining gene trees (Saitou and Nei 1987) were constructed in Paup*4.0b10 (Swofford 2000).
HKA tests:
Polymorphism data from all 10 random loci in Machado et al. (2007) were partitioned into the four geographic populations of D. mojavensis and a single D. arizonae outgroup sequence. Polymorphism and divergence for these loci were measured in DNAsp (Rozas and Rozas 1995), and neutrality was assessed by the method of Hudson et al. (1987), implemented in HKA (http://lifesci.rutgers.edu/∼heylab/heylabsoftware.htm#HKA). For the complete set of 10 loci, significant deviations from neutrality were detected in all four populations of D. mojavensis. To identify a neutral sample, loci with large deviations from expected values were sequentially removed until the P-value of the HKA test was >0.1. The neutral sample was then compared against experimental loci using HKA.
Phylogenetic analyses:
Consensus sequences were used to eliminate mutations introduced by cloning or Taq DNA polymerase. Sequences were additionally screened by eye to identify PCR recombinants. No such chimeric sequences were found. Phylogenetic relationships were inferred with Mr. Bayes (http://mrbayes.csit.fsu.edu/authors.php).
Codon-based analyses of adaptive evolution:
Nested maximum-likelihood models of codon evolution were implemented in the codeml program of PAML (Yang 1997) and compared using likelihood ratio tests. Two tests of positive selection were performed. In the first test the neutral model (M1) is compared with the selection model, in which a class of sites is permitted to exhibit dN/dS (ω) > 1 (M2). In the second test, a β-distribution of site classes in which the most rapidly evolving is constrained to ω ≤ 1 (M7) was compared to a similar model in which the most rapidly evolving site class is permitted to exhibit ω > 1 (M8). Multiple initial values of ω were used to ensure convergence on the likelihood optima.
Two additional tests were implemented to determine if specific branches on the phylogeny had experienced adaptive evolution. First, a free-ratios model, in which each branch is allowed to have a different dN/dS, was compared to a model where the dN/dS of the branch of interest was fixed to 1 (Yang 1998). Second, a branch site model, in which the branch of interest is allowed a rapidly evolving class of sites, ω > 1, was compared to a similar model in which ω is fixed to 1 (Yang et al. 2005).
Three-dimensional modeling:
Bayes empirical Bayes positively selected sites predicted under M8 (Yang 1997; Yang et al. 2005), catalytic sites (reviewed in Polgar 2005), and protease inhibitor sites (reviewed in Srinivasan et al. 2006) were mapped to a predicted three-dimensional (3D) model for GLEANR_898 obtained from Swiss-Model (Schwede et al. 2003).
We tested for an association between positively selected sites and protease inhibitor sites using a permutation test previously implemented in Clark et al. (2007).
The test statistic was the mean distance from each selected site to the nearest inhibitor site. Each permutation identified a random set of selected sites, equal in number to those observed, and calculated the statistic for that set. Buried, core sites were not considered for random sets, because they evolve at a relatively slower rate than surface sites and are rarely inferred as positively selected. This exclusion makes the test more conservative. Buried sites were those with ≤10% surface exposure per residue as calculated by GETAREA (Fraczkiewicz and Braun 1998). A P-value was determined as the fraction of random permutations with a mean distance equal to or lower than the observed mean distance between selected and inhibitor sites. The test for clustering of positively selected sites was similar except that the test statistic was the mean pairwise distance between all selected sites as described in Clark and Swanson (2005).
RESULTS
A novel gene duplicate in the Mojave Desert population:
Consistent, reproducible heterozygosity in sequence data for GLEANR_896 in multiple individuals from seven isofemale lines derived from the Mojave Desert population suggested the acquisition of a novel paralog. Flanking sequence upstream of the novel paralog generated by inverse PCR identified a breakpoint with the repetitive element dmoj_2 (http://insects.eugenes.org/species/cgi-bin/gbrowse/dmoj/). Although this repetitive element made subsequent inverse PCR uninformative, test PCRs pairing a primer on the breakpoint with multiple primers in the coding sequences of GLEANR_896 and GLEANR_897 amplified an ∼2-kb fragment between the breakpoint and the 3′ end of GLEANR_897. The sequence of this fragment included an additional breakpoint between dmoj_2 and the 3′ flanking sequence of GLEANR_897. We thus hypothesize that the new paralog maps to the intergenic sequence between GLEANR_897 and GLEANR_898 (Figure 1).
Using the breakpoint between the new paralog and dmoj_2 we were able to design paralog-specific primers and obtain sequence for 13 of 14 sampled isofemale lines from the Mojave Desert. We were unable to amplify the new paralog from any isofemale lines from mainland Sonora, Catalina Island, or the Baja Peninsula. Southern blots further confirmed that this paralog is absent from all sampled isofemale lines from these three localities (not shown).
To determine if and where the new paralog is expressed we performed semi-quantitative RT–PCR on sexually mature adult males, sexually mature lower female reproductive tracts, and sexually mature female carcasses lacking their female reproductive tracts (supplemental Figure 1). Similar to the other five paralogs, the novel paralog was expressed exclusively in females, with enriched expression in lower female reproductive tracts (supplemental Figure 1). Resultant cDNA was sequenced to verify paralog identity. Collectively, these data indicate that the Mojave Desert population recently has acquired a novel paralog, whose expression pattern suggests female-specific reproductive function.
Ectopic recombination:
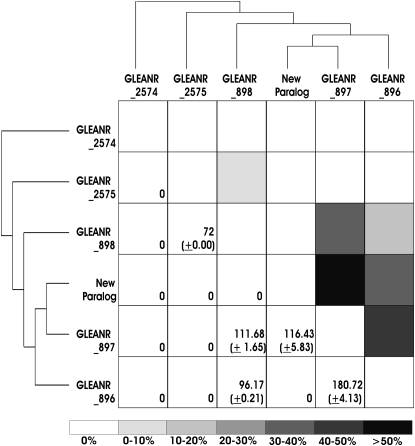
Ectopic recombination, through both nonallelic homologous recombination and gene conversion, facilitates exchange of genetic information between paralogous members of a multigene family. It is critical to describe ectopic recombination in population data, as this process can significantly alter patterns of polymorphism in duplicated genes (Innan 2003; Thornton 2007). We employed GENECONV (Sawyer 1989) to identify pairs of divergent paralogous haplotypes that share regions of complete identity, indicative of gene conversion (Figure 3). No gene conversion tracts were detected between the most basal duplicate, GLEANR_2574, and any other paralog, suggesting this paralog evolves independently (Figure 3). Significant fragments, however, were detected for at least one haplotype of all other paralogs in the gene family (Figure 3).
Figure 3.—
Ectopic recombination. An alignment of all unique haplotypes was used to detect significant fragments of complete identity in GENECONV, on the basis of the method of Sawyer (1989). Branching relationships are from Kelleher et al. (2007) and this publication. Note that there is some ambiguity concerning the placement of GLEANR_897. The percentage of pairwise comparisons between paralogs that show evidence of gene conversion is indicated by shading in the boxes above the diagonal. The average length of identified conversion tracts between paralogs, and the standard deviation of this estimate, are indicated in the boxes below the diagonal.
The highest frequency of significant converted fragments, as well as the longest average fragment length, were observed between the adjacent, closely related duplicates GLEANR_896, GLEANR_897, and the new paralog (Figure 3; also see table of polymorphism, supplemental Table 2). Gene genealogies of GLEANR_896 and GLEANR_897 haplotypes, furthermore, revealed that these loci are not reciprocally monophyletic, suggesting extensive ectopic recombination between paralogous lineages (Figure 2, supplemental Table 2). In contrast, no recombination is detected between genetically and physically distant paralogs GLEANR_896 and GLEANR_2575 (Figure 3). Ectopic recombination, therefore, is negatively associated with both phylogenetic and physical distance.
In many cases, it was impossible to infer the directionality of gene conversion, in terms of a donor and recipient paralog. For GLEANR_896 and GLEANR_897, however, putatively ancestral haplotypes group with the D. arizonae ortholog, while converted haplotypes group with the alternate paralog (Figure 2). Ancestral haplotypes, furthermore, are found in all four populations, while converted haplotypes are population specific. Thus, converted haplotypes of GLEANR_896 have been recipients of genetic variation from ancestral GLEANR_897 donors, and reciprocally, converted haplotypes of GLEANR_897 have been recipients of genetic variation from ancestral GLEANR_896 donors (Figure 2). The approximate gene conversion tract length was 518 bp for GLEANR_896 conversion haplotypes and 443 bp for GLEANR_897 conversion haplotypes (of ∼700 aligned bases), on the basis of visual examination of polymorphic sites (see supplemental Table 2).
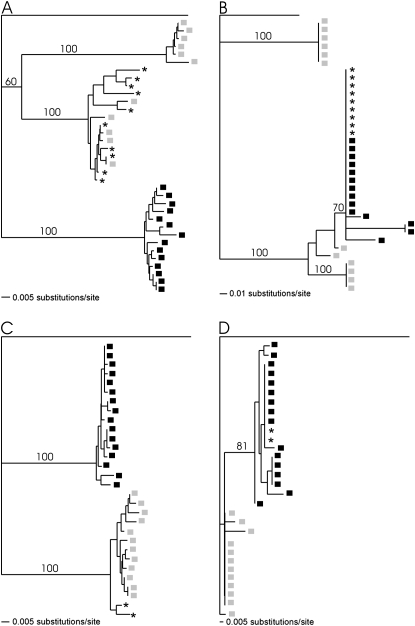
Ectopic recombination involving the genetically more distant paralogs, GLEANR_898 and GLEANR_2575, was not extensive enough to degrade allelic monophyly. Gene genealogies of converted and unconverted regions were therefore compared separately to determine if the evolutionary history of these two portions of the gene could be confidently inferred (Figure 4). In two cases, gene conversion tracts from a set of recipient haplotypes grouped with all haplotypes from a donor paralog with high bootstrap support (Figure 4), indicating the direction of gene conversion.
Figure 4.—
Directional gene conversion in GLEANR_2575, GLEANR_898, and GLEANR_897. Solid boxes denote individual haplotypes of donor paralogs. Shaded boxes indicate unconverted haplotypes of the recipient paralog. * denotes converted haplotypes of the recipient paralog. (A) Neighbor-joining analysis of GLEANR_897 (shaded) and GLEANR_898 (solid) haplotypes, excluding a 52-bp gene conversion tract. (B) Neighbor-joining analysis of GLEANR_897 (shaded) and GLEANR_898 (solid) haplotypes, including only a 52-bp gene conversion tract. (C) Neighbor-joining analysis of GLEANR_898 (shaded) and GLEANR_2575 (solid) haplotypes, excluding a 72-bp gene conversion tract. (D) Neighbor-joining analysis of GLEANR_898 (shaded) and GLEANR_2575 (solid) haplotypes, including only a 72-bp gene conversion tract.
To explore the contribution of genetic exchange between paralogs to genetic variation within populations, we estimated nucleotide diversity (π) for both the complete set of sampled alleles from a given population, as well as for the sample with all recipient alleles excluded. In all cases, our estimate of π was lower when recipient alleles were excluded (Table 1). In four cases, furthermore, the observed decrease was greater than two standard deviations, indicating that ectopic recombination contributes significantly to standing variation within populations (Table 1).
TABLE 1.
Ectopic recombination contributes to genetic variation
| Population | Donor | Recipient | Nc | Nnc | π | SD (π) | π (nc) |
|---|---|---|---|---|---|---|---|
| Baja Peninsula | GLEANR_898 | GLEANR_897 | 7 | 5 | 0.0689 | 0.0127 | 0.0553 |
| Baja Peninsula | GLEANR_896 | GLEANR_897 | 4 | 8 | 0.0689 | 0.0127 | 0.0110*** |
| Baja Peninsula | GLEANR_897 | GLEANR_896 | 7 | 6 | 0.0787 | 0.0082 | 0.01378*** |
| Mainland Sonora | GLEANR_2575 | GLEANR_898 | 1 | 6 | 0.0136 | 0.0026 | 0.0108 |
| Mainland Sonora | GLEANR_897 | GLEANR_898 | 1 | 11 | 0.0564 | 0.0180 | 0.0453 |
| Mainland Sonora | GLEANR_897 | GLEANR_896 | 9 | 12 | 0.0564 | 0.0180 | 0.0240 |
| Catalina Island | GLEANR_2575 | GLEANR_898 | 1 | 6 | 0.0085 | 0.0039 | 0.0000* |
| Catalina Island | GLEANR_897 | GLEANR_896 | 5 | 2 | 0.0725 | 0.0238 | 0.00673* |
For individual paralogs, nucleotide diversity was estimated for the complete data set (π), as well as for the data set with conversion alleles excluded [π (nc)]. Nc, the number of sampled recipient conversion alleles; Nnc, the number of sampled alleles that were not recipients of gene conversions. * denotes greater than two standard deviations below π. ** denotes greater than three standard deviations below π. *** denotes greater than four standard deviations below π.
Segregating pseudogenes:
Functional redundancy between recent duplicates is predicted to result in relaxed evolutionary constraint at individual paralogs, allowing for the acquisition of deleterious mutations or complete loss of function (Ohno 1970; Hughes 1994; Force et al. 1999). Consistent with this prediction, we found evidence of three distinct pseudogene haplotypes in two different paralogs, GLEANR_2575 and GLEANR_898. In the Baja Peninsula population, one premature stop codon and one frameshift deletion are found in GLEANR_2575. These mutations occur prior to the first of three amino acid residues that comprise the catalytic triad (reviewed in Polgar 2005), as well as residues that determine substrate binding affinity (Sprang et al. 1988), thus rendering the protease completely nonfunctional. Both alleles were resequenced to verify the mutations did not reflect amplification or sequencing errors. One converted allele of GLEANR_897 sampled from mainland Sonora also contained a frameshift deletion, although insufficient DNA remained for resequencing of this individual. This frameshift occurs between the second and third amino acids in the catalytic triad, but prior to all residues that determine substrate binding affinity, and likely also renders the protease nonfunctional.
Pseudogene haplotypes often reflect relaxed purifying selection, but can also be maintained as balanced polymorphisms (Hexter 1968; Wiesenfeld 1968), or sweep rapidly through populations in cases of adaptive gene loss (Stedman et al. 2004; Wang et al. 2006). GLEANR_2575 alleles sampled from the Baja Peninsula and GLEANR_897 alleles from mainland Sonora do not exhibit deviations from neutrality in McDonald–Kreitman tests (McDonald and Kreitman 1991), nor do they show a significant skew in the site frequency spectra (supplemental Table 1). There is no evidence, therefore, that pseudogene haplotypes observed here confer a selective advantage.
Deviations from neutrality at GLEANR_898:
Standard McDonald–Kreitman (McDonald and Kreitman 1991) tests for GLEANR_898 indicate an excess of nonsynonymous polymorphism, relative to divergence, in the Baja Peninsula, Catalina Island, and mainland Sonora populations (Table 2). Intriguingly, both mainland Sonora and Catalina Island exhibit segregating conversion alleles at this locus (Table 1). Although Catalina Island no longer exhibits a deviation from neutrality when segregating conversion alleles are excluded from the analysis, the G-test for mainland Sonora remains significant (Table 2).
TABLE 2.
Deviations from neutrality in GLEANR_898
| Standard MK test
|
Standard MK test (no conversion)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Polymorphic | Fixed | Test | Polymorphic | Fixed | Test | Tajima's D | ||
| Baja Peninsula | Syn. + nc | 9 | 35 | G-test | −0.69 | |||
| Nonsyn. | 13 | 12 | ** | NS | ||||
| Catalina Island | Syn. + nc | 14 | 31 | G-test | 0 | 15 | NA | −1.69 |
| Nonsyn. | 13 | 10 | * | 0 | 7 | ** | ||
| Mojave Desert | Syn. + nc | 6 | 35 | G-test | −1.43 | |||
| Nonsyn. | 4 | 12 | NS | * | ||||
| Mainland Sonora | Syn. + nc | 12 | 31 | G-test | 3 | 17 | G-test | −0.10 |
| Nonsyn. | 18 | 12 | ** | 13 | 7 | ** | NS | |
McDonald–Kreitman tests utilized D. arizonae as an outgroup. Lineage-specific McDonald–Kreitman tests were polarized with Dmoj\GLEANR_2575. Syn., synonymous; nonsyn., nonsynonymous. *P < 0.05; **P < 0.01.
Balancing or diversifying selection is one possible explanation for an excess of nonsynonymous polymorphism in a McDonald–Kreitman framework. In general, these selective regimes are accompanied by other patterns, such as an excess of intermediate frequency polymorphism, the appearance of two well-differentiated haplogroups that exhibit significant linkage disequilibrium, or an excess of polymorphism relative to divergence when compared to other loci (Hudson et al. 1987). No excess of intermediate frequency polymorphism is observed either in mainland Sonora or in the Baja Peninsula for GLEANR_898, as Tajima's D (Tajima 1989) is slightly negative in both cases (Table 2). Two well-differentiated haplotype groups, furthermore, are not apparent in gene genealogies of this locus (not shown). Zns, a measure of the correlation in allele frequencies across polymorphic sites (Kelly 1997), does not indicate significant linkage disequilibrium at this locus (not shown). Finally, an HKA test (Hudson et al. 1987) detects no excess of polymorphism, relative to neutral loci (not shown). The data, therefore, provide little evidence that balancing selection is operating on the GLEANR_898 locus in mainland Sonora or the Baja Peninsula.
An alternate explanation for the observed excess of replacement variation is that these sites represent weakly deleterious variants that contribute only to polymorphism, but not to divergence. If so, the majority of these variants should be segregating at low frequency (Kimura 1983; Nachman 1998). Site frequency spectra for silent and replacement sites in GLEANR_898 were therefore compared separately for the mainland Sonora and Baja California populations (Table 3). In both populations, Tajima's D (Tajima 1989) is slightly more positive for replacement sites than for silent sites, the opposite of what is expected for mildly deleterious variants (Table 3). The observed excess of nonsynonymous polymorphism, therefore, does not appear to arise from weakly deleterious mutations.
TABLE 3.
Estimates of Tajima's D for silent and replacement sites in GLEANR_898
| Tajima's D
|
||
|---|---|---|
| Baja Peninsula | Mainland Sonora | |
| All | −0.69 | −0.10 |
| Silent | −0.88 | 0.70 |
| Replacement | −0.48 | −0.35 |
A third explanation for the observed deviation from neutrality is that a recent relaxation in functional constraint may allow for the acquisition of replacement mutations that were not tolerated under the previous selective regime (Takahata 1993). Although this scenario is difficult to verify empirically, it is plausible for a multigene family that may be undergoing antagonistic molecular coevolution. The degree of ectopic recombination, as well as the frequency of segregating pseudogenes, suggests that the paralogs sampled here are at least partially functionally redundant. If coevolving interactors change their evolutionary “strategy,” paralogs with formerly critical function could experience relaxed selective constraint.
Evolutionary history of the novel paralog:
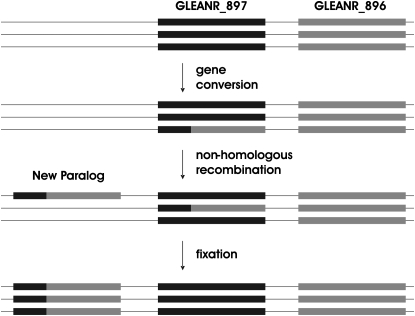
Neighbor-joining analysis indicates that the novel paralog found in the Mojave Desert is most similar to converted alleles of GLEANR_897 from mainland Sonora and the Baja Peninsula (Figure 2). Because the new duplicate is a chimera of GLEANR_896 and GLEANR_897, but is not nested between these two paralogs (Figure 1), the conversion haplotype and the gene duplication could not have resulted from a single event of nonallelic homologous recombination. The new paralog, therefore, likely has arisen via tandem duplication of a segregating conversion allele of GLEANR_897. Although it is impossible to determine the history of this gene with confidence, Figure 5 outlines a mechanism for the creation of the Mojave Desert chromosome with the fewest mutational steps. First, a gene conversion event from GLEANR_896 to GLEANR_897 creates a converted GLEANR_897 allele. Second, unequal crossing over, mediated by homologous or repetitive flanking sequence, results in a tandem gene duplication event. Third, this duplicated chromosome rises to high frequency in the Mojave Desert population.
Figure 5.—
Hypothesized mechanism for the origin of a new paralog in the Mojave Desert population. Two tandem duplicates, GLEANR_897 (solid) and GLEANR_896 (shaded), are indicated on a chromosome. A gene conversion event results in a novel allele GLEANR_897. Unequal crossing over between an ancestral and a converted chromosome then results in a novel tandem duplicate. The duplicated chromosome is then fixed in the Mojave Desert.
It is intriguing that the duplication event in the Mojave Desert population unites the converted and unconverted haplotypes of GLEANR_897 on a single chromosome. This result is reminiscent of models in which two alleles are maintained as a balanced polymorphism, and a subsequent gene duplication experiences immediate directional selection due to heterosis (Spofford 1969; Ohno 1970; Otto and Yong 2002; Walsh 2003; Proulx and Phillips 2006). If GLEANR_897 converted and unconverted haplotypes represent a balanced polymorphism, the GLEANR_897 converted haplotype should have arisen by a single ancestral gene conversion event, prior to the divergence of the mainland Sonora, Baja Peninsula, and Mojave Desert populations (0.45–0.68 MYA) (Reed et al. 2007; Matzkin 2008).
Although, all GLEANR_897 haplotypes group together with high bootstrap support (Figure 2), this is not necessarily indicative of a single mutational origin for the converted haplotype. If ectopic recombination between paralogs is more frequent, or more frequently tolerated, in certain genetic regions, similar chimeric haplotypes could be generated continuously by gene conversion. If so, a considerable number of shared polymorphisms are expected between GLEANR_897 converted alleles and GLEANR_896 ancestral alleles within the converted region. The number of private and shared polymorphisms in converted GLEANR_897 alleles and GLEANR_896 ancestral alleles within the converted region are presented in Table 4. In the mainland Sonora population only one polymorphism is shared between converted and ancestral alleles (Table 4), suggesting that converted alleles are not continuously sampling genetic variation from ancestral haplotypes. In Baja Peninsula, where eight shared polymorphisms are seen, the polymorphisms are associated with only two ectopic recombination events. Collectively, therefore, the data do not suggest a high frequency of gene conversion from GLEANR_896 ancestral alleles to GLEANR_897 converted alleles. This result is in stark contrast to GLEANR_896 converted and GLEANR_897 ancestral alleles, which exhibit a high frequency of shared polymorphisms indicative of ongoing gene conversion (Table 4).
TABLE 4.
Private and shared polymorphisms in ancestral and converted haplogroups of GLEANR_896 and GLEANR_897
| Haplogroup | Population | Length converted region | Ancestral S | Shared S | Converted S | % ancestral shared | % converted shared |
|---|---|---|---|---|---|---|---|
| GLEANR_897 converted | Baja Peninsula | 443 bp | 20 | 8 | 3 | 28.57 | 72.73 |
| GLEANR_897 converted | Mainland Sonora | 443 bp | 22 | 1 | 4 | 4.30 | 20.00 |
| GLEANR_896 converted | Baja Peninsula | 518 bp | 3 | 6 | 2 | 66.67 | 80.00 |
| GLEANR_896 converted | Catalina Island | 518 bp | 1 | 2 | 1 | 66.67 | 66.67 |
Ancestral S, private polymorphisms in the ancestral (or donor) haplogroup in the converted region. For GLEANR_897 converted, the ancestral haplogroup is GLEANR_896 ancestral, and for GLEANR_896 the ancestral haplogroup is GLEANR_897 ancestral. Shared S, number of shared polymorphisms between ancestral and converted haplogroups. Converted S, private polymorphisms in the converted (or recipient) haplogroup within the converted region. % ancestral shared, the percentage of polymorphisms in the ancestral haplogroup that are shared with the converted haplogroup. % converted shared, the percentage of polymorphisms in the converted haplogroup that are shared with the ancestral haplogroup.
If the GLEANR_897 converted haplotype is an old balanced polymorphism, it is predicted to have acquired and maintained its own set of genetic variation. Consistent with this hypothesis, this haplogroup exhibits one silent and two replacement polymorphisms, fixed or at high frequency (>60%) among these alleles, which are not present in any other haplotype of GLEANR_896 or GLEANR_897 in D. mojavensis. A third amino acid variant, fixed in the GLEANR_897 haplogroup, is present in only one sampled haplotype of GLEANR_896 and was entirely absent from unconverted haplotypes of GLEANR_897. Intriguingly, the three amino acid variants, also found in the new paralog from the Mojave Desert, are shared with D. arizonae GLEANR_896. Sites that are shared with an outgroup are inferred to represent the ancestral state. Thus, the conversion tract in GLEANR_897 converted haplotypes appears to be derived from an ancestral allele of GLEANR_896 that is no longer segregating in any D. mojavensis population. Ancestral variation is expected if the converted haplotype resulted from an ancient gene conversion event that occurred prior to the radiation of the mainland Sonora, Baja Peninsula, and Mojave Desert populations.
The confounding nature of gene conversion makes it problematic to present a compelling argument that the maintenance of GLEANR_897 converted and ancestral haplotypes is the result of balancing selection. Extensive gene conversion generates slightly positive values of Tajima's D (Tajima 1989), and furthermore, makes this statistic extremely conservative because the variance of the test statistic is over estimated (Innan 2003). Similarly, HKA tests are inappropriate assessments of balancing selection for duplicates undergoing gene conversion because recombining paralogs are on average more polymorphic than single-copy loci (Innan 2003; Thornton 2007). Nonetheless, our data do suggest that the two haplotypes are old, have been retained in two of four geographically isolated populations of D. mojavensis for at least 0.45 MY, and have duplicated in a third population. The degree of linkage disequilibrium in both mainland Sonora (Zns = 0.69, P = 0.01) and the Baja Peninsula (Zns = 0.82, P = 0.00), furthermore, indicates little recombination has occurred between haplogroups during this time. Determining the role of natural selection in maintaining the GLEANR_897 converted and ancestral polymorphism will present an important challenge for future studies.
Although our data suggest that gene duplication was preceded by allelic divergence between the GLEANR_897 ancestral and GLEANR_897 converted haplotypes, GLEANR_897 converted haplotypes are separated from the new paralog by an average of 20 nucleotide differences (∼3%). The majority of these differences are inside the gene conversion tract. To explore if gene duplication may have been followed by a period of adaptive evolution, we estimated the corrected ratio of nonsynonymous to synonymous divergence (dN/dS) for this branch in the portion of the alignment contiguous with the conversion tract (Yang 1998). Although the branch leading to the novel paralog does exhibit dN/dS of 1.25, consistent with adaptive evolution, this value does not provide a significantly better fit to the data than a model where the value is fixed to 1 (P = 1.00). A branch-site model, in which only a subset of sites on this branch were hypothesized to experience positive selection, similarly did not provide a significantly better fit to the data than a model that does not incorporate adaptive evolution (P = 1.00; Yang et al. 2005). Although these analyses provide little evidence for adaptive protein evolution following gene duplication, it is important to remember that their statistical power is extremely limited for branches where few changes have occurred.
Directional selection:
Although segregation of deleterious mutations clearly suggests relaxed purifying selection at some loci in this multigene family, we also find evidence for positive directional selection, a frequent observation among reproductive proteins (reviewed in Swanson and Vacquier 2002; Clark et al. 2006; Panhuis et al. 2006). Catalina Island flies show an excess of low-frequency polymorphism at GLEANR_898 and GLEANR_897, a possible indicator of recent directional selection (Table 5). Similarly, Mojave Desert flies exhibit an excess of low-frequency polymorphism at GLEANR_898, and no segregating sites at GLEANR_897, GLEANR_896, or the new paralog (Table 4; supplemental Table 1). A reanalysis of seven autosomal and three sex-linked random loci sampled in Machado et al. (2007) does not detect any significant skew toward positive or negative values in site frequency spectra tests for either of these populations (supplemental Table 3). The observed excess of rare polymorphism, therefore, does not appear to result from demographic processes such as a recent population expansion. Gene conversion, furthermore, is known to skew Tajima's D marginally positive (Innan 2003; Thornton 2007), making the observation of significantly negative values highly unexpected.
TABLE 5.
HKA and site-frequency spectra analysis of GLEANR_898, GLEANR_897, and GLEANR_896
| Population | Locus | Inheritance | Intraspecific length | S | Interspecific length | D | Tajima's D |
|---|---|---|---|---|---|---|---|
| Catalina Island | 996a | Autosomal | 856 | 2 | 827 | 38.50 | −0.71 |
| 5239a | Autosomal | 870 | 1 | 870 | 13.25 | −0.61 | |
| 5246a | Autosomal | 872 | 0 | 849 | 22.00 | NA | |
| A4125a | Autosomal | 880 | 4 | 871 | 49.00 | −0.78 | |
| X100a | Sex linked | 875 | 0 | 849 | 46.00 | NA | |
| GLEANR_898 | Autosomal | 710 | 21 | 710 | 54.29 | −1.69** | |
| GLEANR_897 | Autosomal | 682 | 34 | 682 | 88.28 | −1.45* | |
| Neutral χ2 = 2.77 | P = 0.54 | ||||||
| Neutral + 898 χ2 = 8.8493 | P = 0.12 | ||||||
| Neutral + 897 χ2 = 8.3066 | P = 0.14 | ||||||
| Neutral + 898 + 897 χ2 = 9.23 | P = 0.16 | ||||||
| Mojave Desert | 996a | Autosomal | 856 | 1 | 827 | 40.25 | −0.61 |
| 1343a | Autosomal | 886 | 1 | 869 | 9.25 | −0.61 | |
| 5239a | Autosomal | 870 | 3 | 870 | 14.75 | −0.75 | |
| 5246a | Autosomal | 870 | 1 | 850 | 16.25 | −0.61 | |
| A4115a | Autosomal | 824 | 2 | 824 | 16.50 | −0.71 | |
| A4125a | Autosomal | 917 | 4 | 908 | 48.00 | 0.65 | |
| X100a | Sex linked | 911 | 3 | 890 | 47.50 | 0.17 | |
| GLEANR_898 | Autosomal | 710 | 4 | 710 | 49.22 | −1.43* | |
| GLEANR_897 | Autosomal | 691 | 0 | 691 | 100.91 | NA | |
| GLEANR_896 | Autosomal | 697 | 0 | 697 | 75.00 | NA | |
| Neutral χ2 = 2.59 | P = 0.84 | ||||||
| Neutral + 898 χ2 = 3.10 | P = 0.87 | ||||||
| Neutral + 897 χ2 = 14.06 | P = 0.05 | ||||||
| Neutral + 896 χ2 = 11.21 | P = 0.13 | ||||||
| Neutral + 898 + 897 χ2 = 22.40 | P = 0.008 | ||||||
S, number of segregating sites; D, divergence from D. arizonae ortholog. χ2 and P-values for multiple HKA tests performed in HKA (http://lifesci.rutgers.edu/∼heylab/heylabsoftware.htm#HKA) are reported. *P < 0.05; **P < 0.01.
Sequences from Machado et al. (2007).
To further test the hypothesis of directional selection, polymorphism and divergence between our experimental loci and a group of loci that behave neutrally (Machado et al. 2007; see materials and methods) were compared by the HKA test (Hudson et al. 1987; Table 5). When including GLEANR_896 and GLEANR_897 in the data set, no deviations from neutrality were detected for the Catalina Island population (Table 5). It is important to note, however, that the HKA test is extremely conservative for duplicate genes experiencing ectopic recombination, as the expected level of polymorphism is higher than for single-copy loci (Innan 2003; Thornton 2007). For the Mojave Desert population, GLEANR_897, as well as a test that included GLEANR_898, GLEANR_897, and GLEANR_896, both showed an excess of divergence consistent with directional selection. Although we cannot infer the causative mutation responsible for these patterns, it is intriguing that the selective sweep is associated with a chromosome harboring a novel duplicate. The novel duplicate could be adaptive because of its specific sequence, or alternatively, simply because it represents an additional gene copy.
Duplication and adaptive evolution in the repleta species group:
To further elucidate the evolutionary history of this gene family, we sequenced paralogs across five repleta group species, D. mojavensis, D. arizonae, D. navajoa, D. mayaguana, and D. mettleri. Sequence data from the D. grimshawi and D. virilis genomes provided appropriate outgroups. Bayesian phylogenetic inference of 22 orthologs and paralogs indicates that the genes exist as a single copy in D. grimshawi and D. virilis, whereas three or more copies exist in all repleta group species (Figure 6). The radiation of the gene family, therefore, appears lineage specific to the repleta species group. D. mojavensis, D. navajoa, D. mayaguana, and D. mettleri, furthermore, all exhibit two paralogs that are more closely related to each other than to any other sequence in the alignment. This pattern, common to multigene families, suggests either ongoing gain and loss of individual paralogs or concerted evolution by extensive ectopic recombination (reviewed in Nei and Rooney 2005). GENECONV detected a significant fragment in at least one paralog from D. mojavensis, D. arizonae, D. mettleri, and D. mayaguana, indicating ectopic recombination contributes to divergence of this multigene family. No significant fragments are found between lineage-specific paralogs from D. navajoa or D. mayaguana however, suggesting these are authentic lineage-specific duplicates. Observation of a novel paralog and segregating pseudogenes in the polymorphism data further supports the assertion that lineage-specific gain and loss is an ongoing process in the evolution of this gene family.
Figure 6.—
Bayesian phylogeny of 22 orthologs and paralogs from seven Drosophila species. Posterior probabilities are indicated.
To determine if the gene family has experienced positive selection within the repleta species group, we implemented maximum-likelihood codon based models in PAML (Yang 1997). For this analysis, all nodes with a posterior probability of <90 (Figure 6) were collapsed to polytomies to prevent spurious results due to inaccuracy in the tree topology. For two different tests of positive selection, a model that allowed for a class of sites that evolves adaptively (dN/dS > 1) provided a significantly better fit to the data than a model that did not (Table 6). The detected signature of adaptive evolution is consistent with our previous analysis (Kelleher et al. 2007).
TABLE 6.
Maximum-likelihood codon-based analysis of positive selection in the repleta species group
| Data set | M1 | M2 | LRT | P | P(s) | ω | BEB selected sites |
|---|---|---|---|---|---|---|---|
| Full alignment | −7424.47 | −7388.35 | 72.24 | 1.91E-17 | 0.08 | 2.80 | 68, 132, 133, 135, 253 |
| Exclude D. mettleri-1, | −6642.72 | −6619.94 | 45.55 | 1.49E-11 | 0.07 | 2.63 | 68, 132, 133, 135, 253 |
| D. mettleri-2 | |||||||
| Exclude D. mettleri-1, D. mettleri-2, | −5726.11 | −5703.86 | 44.49 | 2.55E-11 | 0.08 | 2.79 | 68, 132, 133, 135, 253 |
| Dmoj\GLEANR_897, D. arizonae-4, | |||||||
| D. arizonae-5, Dvir\GLEANR_2181 | |||||||
| Exclude D. mettleri-1, D. mettleri-2, | −5265.49 | −5258.06 | 14.88 | 1.15E-04 | 0.03 | 3.02 | 253 |
| Dmoj\GLEANR_896, | |||||||
| Dmoj\GLEANR_898, | |||||||
| Dmoj\NEW_PARALOG, | |||||||
| D. arizonae-1, D. arizonae-2, | |||||||
| D. arizonae-5, D. mayaguana-1 | |||||||
| Data set | M7 | M8 | LRT | P | P(s) | ω | BEB selected sites |
| Full alignment | −7426.01 | −7376.89 | 98.23 | 3.72E-23 | 0.13 | 2.11 | 68, 73, 112, 113, 132, 133, 135, 179, 184, 187, 204, 208, 211, 209, 253 |
| Exclude D. mettleri-1, D. mettleri-2 | −6643.22 | −6608.53 | 69.37 | 8.16E-17 | 0.13 | 1.92 | 68, 73, 112, 132, 133, 135, 179, 187, 204, 253 |
| Exclude D. mettleri-1, D. mettleri-2, | −5734.58 | −5698.60 | 71.96 | 2.19E-17 | 0.14 | 2.08 | 68, 73, 112, 113, 132, 133, 135, 179, 187, 204, 208, 209, 253, 257 |
| Dmoj\GLEANR_897, D. arizonae-4, | |||||||
| D. arizonae-5, Dvir\GLEANR_2181 | |||||||
| Exclude D. mettleri-1, D. mettleri-2, | −5269.09 | −5253.85 | 30.48 | 3.38E-08 | 0.11 | 1.70 | 68, 112, 113, 133, 135, 179, 187, 253 |
| Dmoj\GLEANR_896, Dmoj\GLEANR_898, | |||||||
| Dmoj\NEW_PARALOG, D. arizonae-1, | |||||||
| D. arizonae-2, D. arizonae-5, D. mayaguana-1 |
M1, M2, M7, and M8 denote codon models implemented in PAML (Yang 1997). LRT, the value of the likelihood ratio test between nested models. P, the probability of the LRT under a χ2 distribution. P(s), proportion of sites in the positively selected site class. ω, estimated dN/dS of the positively selected site class. BEB selected sites, Bayes empirical Bayes predicted selected sites for the given selection model (Yang et al. 2005).
Two aspects of our data could lead to an incorrect inference of adaptive evolution in this type of analysis. First, sequences from D. navajoa, D. mayaguana, and D. mettleri were obtained from cloned PCR products, meaning there could be mutations in the aligment that have been introduced by Taq DNA polymerase. All cloned sequences in the alignment, however, are a consensus of three or more colonies except D. mettleri-1 and D. mettleri-2, and should therefore be free of PCR introduced mutations. A reanalysis of the data with D. mettleri-1 and D. mettleri-2 excluded still yields highly significant tests, indicating the inference of adaptive evolution is not the result of PCR error (Table 6).
The observed gene conversion in our alignment could also lead to spurious results in codon-based analysis of adaptive evolution, as recombination is known to cause false positives for this class of tests (Anisimova et al. 2003). To avoid this problem, two subsets of the alignment that included only one of a pair or group of sequences with evidence for gene conversion were created (Table 6). Analyses of these pruned alignments were still highly significant, indicating that the observed adaptive evolution is independent of gene conversion.
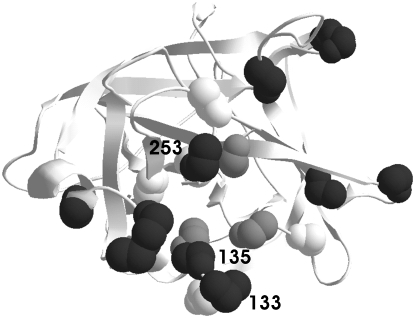
Depending on the data set, likelihood analysis suggests that between 3 and 13% of sites are experiencing positive selection, with an estimated dN/dS between 1.7 and 3.02 (Table 6). Bayes empirical Bayes selected sites (Yang et al. 2005), furthermore, are remarkably congruent between different data sets and different models (Table 6; Figure 7). Selected sites (solid), often are observed to be closely associated with sites important to protease inhibitor susceptibility and resistance (Figure 7; reviewed in Srinivasan et al. 2006). Indeed, three selected sites and protease inhibitor interaction sites occur at the same residue: a statistically significant excess (Fisher's exact test, P = 0.0085).
Figure 7.—
Predicted 3D structure of GLEANR_898. Bayes empirical Bayes selected sites identified under M8 (Yang 1997; Yang et al. 2005) identified with at least two data sets are solid. Sites that are determinants of protease inhibitor susceptibility are open (reviewed in Srinivasan et al. 2006). Shaded sites comprise the catalytic triad (reviewed in Polgar 2005). Selected sites 133, 135, and 253 also are determinants of inhibitor susceptibility (Srinivasan et al. 2006).
To further explore if selected sites and protease inhibitor interaction sites are associated in three-dimensional space, we compared the average pairwise distance between each selected site and the closest protease inhibitor interaction site to 10^6 sets of randomly sampled sites. Selected sites are significantly closer to protease inhibitor interaction sites than expected by chance (P = 0.02220), indicating that these two groups of sites are physically associated within the structure of the protein. This result does not reflect a spurious association of a cluster of selected sites with a single protease inhibitor site, as selected sites are not significantly clustered with each other (P = 0.31839).
DISCUSSION
Several aspects of our data suggest that the protease gene family examined here evolves nonindependently as a functionally redundant complex. First, we observed ectopic recombination between five of six paralogs within this gene family. Although our data do not indicate whether ectopic recombination is a source of adaptive genetic variation, in many cases conversion tracts were segregating at intermediate or high frequency, indicating that these mutations are not significantly deleterious. Considerable interchange of divergent sequence implies functional overlap between the encoded proteins.
Paralogs with partially or completely overlapping functions are expected to experience relaxed evolutionary constraint (Ohno 1970; Hughes 1994; Force et al. 1999). Consistent with this prediction, we find two indicators of relaxed constraint at three different loci in this multigene family. First, GLEANR_898 exhibits an excess of replacement polymorphism but no evidence for balancing selection or the segregation of weakly deleterious mutations. This deviation from neutrality, therefore, may indicate that a recent relaxation in functional constraint has allowed for the accumulation of mutations that were not tolerated in the previous selective regime (Takahata 1993; Nachman 1998). Second, we discovered three distinct pseudogene haplotypes in two different paralogs. In all three cases, the relevant mutations likely rendered the protein completely nonfunctional. The prevalence of such haplotypes in our sample would suggest that purifying selection is relatively weak.
Although relaxed constraint may imply these proteases have little or no important function, evidence for adaptive evolution within this gene family would suggest otherwise. Our analysis of divergence across the repleta species group asserts that these genes are evolving rapidly and adaptively, consistent with a critical role in organismal fitness. The Mojave Desert population, furthermore, exhibited an elevated ratio of divergence to polymorphism in GLEANR_897, as well as an excess of rare variants at the adjacent GLEANR_898, indicative of recent directional selection in this genomic region. Although we found no compelling evidence of adaptive evolution in the remaining three populations, this may reflect the limited framework for detecting deviations from neutrality in the complex scenario of multiple paralogs undergoing gene conversion (Innan 2003; Thornton 2007).
We propose that the observed pattern of relaxed constraint paired with positive directional selection reflects an intriguing evolutionary mechanism employed by repleta group females. By tolerating a larger array of genetic variation, generated by single base pair mutations, ectopic recombination, and gene duplication, females can more rapidly explore adaptive space to generate novel advantageous variants. This strategy has long been hypothesized to explain the complex evolutionary histories of vertebrate MHC alleles, and their role in immune response, although the empirical data remain controversial (reviewed in Martinsohn et al. 1999; Nei and Rooney 2005). Interestingly, several single-copy reproductive proteins exhibit a similar pattern of elevated polymorphism within populations, but exhibit rapid, adaptive evolution between species (Swanson et al. 2001; Galindo et al. 2003; Turner and Hoekstra 2006, 2008; Gasper and Swanson 2006; Hamm et al. 2007; Moy et al. 2008).
Mathematical models of sexual conflict predict that females can halt the evolutionary chase of a male interactor by splitting into two divergent haplogroups (Gavrilets and Waxman 2002; Hayashi et al. 2007). Although our data provide no compelling evidence of balancing selection, it is easy to envision how a complex of paralogs that duplicate and recombine could be adaptive in the context of sexually antagonistic coevolution. Determining the relative roles of sexual conflict and cryptic female choice in shaping the evolutionary history of the proteases examined here, however, will require a significantly more detailed understanding of their biochemical and physiological functions.
If the gene family examined here is engaged in an evolutionary dynamic with components of the male ejaculate, its history within populations is expected to be a unique reflection of this coevolutionary trajectory. Consistent with this prediction, the patterns of pseudogenation, duplication, gene conversion, and adaptive evolution exhibited by the female reproductive proteases examined in this study are largely population specific. Ectopic recombination between GLEANR_896 and GLEANR_897 is biased in opposite directions between the mainland Sonora and Catalina Island populations. Pseudogene haplotypes and acquisition of a novel paralog were also confined to a single population. Finally, all deviations from neutrality were population specific, as predicted if the selective pressure experienced by this gene family is determined by a distinct intersexual dynamic.
The identities of male interactors for the female proteases examined here remain obscure; however, it is intriguing that positively selected sites in this gene family are significantly associated with residues known to determine protease inhibitor susceptibility (reviewed in Srinivasan et al. 2006). Protease inhibitors are found in the male ejaculates of both D. mojavensis (Wagstaff and Begun 2005), and D. melanogaster (Swanson et al. 2001; Findlay et al. 2008). Consistent with the hypothesis that male protease inhibitors regulate female proteases, trypsin and elastase-like serine-endoprotease activity in D. arizonae female reproductive tracts is observed to decrease after mating (E. S. Kelleher and J. E. Pennington, unpublished results). Adaptive evolution of female proteases, therefore, may reflect molecular coevolution with protease inhibitors in the male ejaculate, as previously suggested for D. melanogaster reproductive proteases and inhibitors (Wong et al. 2008).
We previously have hypothesized that the proteases examined here may play a role in the degradation of the insemination reaction in mated females (Kelleher et al. 2007). This opaque mass that fills the uterus after mating (Patterson 1946) differs in severity between the four populations of D. mojavensis (Knowles and Markow 2001). Male and female contributions to this process, furthermore, are thought to coevolve antagonistically between the sexes (Knowles and Markow 2001). It is exciting, therefore, that the evolutionary history of the novel paralog is correlated with insemination reaction mass size differences between populations. Specifically, the Mojave Desert population exhibits the largest reaction mass in intrapopulation crosses (Knowles and Markow 2001), as well as a gene duplication event that engendered permanent heterozygosity for the converted and unconverted alleles of GLEANR_897. This chromosomal region, furthermore, is associated with a recent selective sweep. Similarly, the mainland Sonora and Baja Peninsula populations exhibit intermediate reaction mass sizes (Knowles and Markow 2001), and evidence of an old polymorphism between converted and unconverted GLEANR_897 haplogroups. Finally, the Catalina Island population exhibits the smallest reaction mass (Knowles and Markow 2001), and evidence for neither an old polymorphism nor a novel paralog. Future genetic studies of these proteins will shed light on their potential role in intersexual dynamics and determination of reaction mass size.
Extensive research in a broad range of taxa has demonstrated that proteins involved in sexual reproduction evolve rapidly (Swanson and Vacquier 2002; Clark et al. 2006; Panhuis et al. 2006). The complex history exhibited by the protease gene family examined here, however, includes pseudogenation, duplication, gene conversion, and positive selection. Although many of these processes previously have been observed in reproductive proteins (Aguadé 1998; Cirera and Aguadé 1998; Swanson and Vacquier 1998), their integration in a single gene family represents a novel and intriguing observation in the study of reproductive protein evolution. The divergence of these genes between four well-structured populations of D. mojavensis with evidence of ejaculate–female coadaptation (Knowles and Markow 2001; Pitnick et al. 2003; Knowles et al. 2005; Kelleher and Markow 2007), furthermore, suggests an exciting role for gene family evolution in the mediation of intersexual dynamics. Documenting this unique evolutionary history in a female reproductive protein highlights the underexplored “female side” of reproductive tract interactions.
Gene duplication recently has emerged as an integral aspect of reproductive protein evolution in Drosophila. Lineage-specific duplicates are common among Drosophila reproductive proteins (Cirera and Aguadé 1998; Loppin et al. 2005; Dorus et al. 2008; Findlay et al. 2008), particularly within the repleta species group (Kelleher et al. 2007; Wagstaff and Begun 2007; Almeida and Desalle 2008, 2009). Genomewide patterns of gene gain and loss across twelve Drosophila genomes, furthermore, indicates proteins involved in sexual reproduction turn over more rapidly than other functional classes (Hahn et al. 2007). Finally, D. melanogaster genes with copy-number polymorphism are enriched for proteins expressed in the male accessory gland (Dopman and Hartl 2007), the primary site for production of seminal fluid protein in Drosophila (reviewed in Wolfner 2002). Elucidating the role of gene family evolution in determining reproductive success, mediating intersexual dynamics, or both, presents an exciting avenue for future research.
Acknowledgments
The authors acknowledge Michael Nachman and Giovanni Bosco for use of equipment and reagents, Nathan Clark for generously analyzing structural data, Tom Hartl for technical assistance, and Kevin Thornton and Matthew Dean for helpful discussion. Michael Nachman, Matthew Hahn, Luciano Matzkin, Willie Swanson, and the members of the Nachman lab provided helpful comments that significantly improved the manuscript. This research was funded by a National Science Foundation (NSF) doctoral dissertation improvement grant to E.S.K. and the Center for Insect Science at the University of Arizona. E.S.K. was supported by an NSF Integrative Graduate Education and Research Traineeship in Evolutionary, Functional, and Computational Genomics at the University of Arizona and a dissertation fellowship from the American Association of University Women.
References
- Aguadé, M., 1998. Different forces drive the evolution of ACP26Aa and ACP26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics. 150 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, F. C., and R. Desalle, 2008. Evidence of adaptive evolution of accessory gland proteins in closely related species of the Drosophila repleta group. Mol. Biol. Evol. 25 2043–2053. [DOI] [PubMed] [Google Scholar]
- Almeida, F. C., and R. Desalle, 2009. Orthology, function and evolution of accessory gland proteins in the Drosophila repleta group. Genetics 181 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova, M., R. Nielsen and Z. Yang, 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and H. A. Lindfors, 2005. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol. Biol. Evol. 22 2010–2021. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., H. A. Lindfors, M. E. Thompson and A. K. Holloway, 2006. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics 172 1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera, S., and M. Aguadé, 1998. Molecular evolution of a duplication: the sex-peptide (Acp70A) gene region of Drosophila subobscura and Drosophila madeirensis. Mol. Biol. Evol. 15 988–996. [DOI] [PubMed] [Google Scholar]
- Clark, N. L., and W. J. Swanson, 2005. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 1 e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, N. L., J. E. Aagaard and W. J. Swanson, 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131 11–22. [DOI] [PubMed] [Google Scholar]
- Clark, N. L., G. D. Findlay, X. Yi, M. J. MacCoss and W. J. Swanson, 2007. Duplication and selection on abalone sperm lysin in an allopatric population. Mol. Biol. Evol. 24 2081–2090. [DOI] [PubMed] [Google Scholar]
- Dopman, E. B., and D. L. Hartl, 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104 19920–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus, S., Z. N. Freeman, E. R. Parker, B. D. Heath and T. L. Karr, 2008. Recent origins of sperm genes in Drosophila. Mol. Biol. Evol. 25 2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, W. G., 1996. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press, Princeton, NJ.
- Fisher, R. A., 1915. The evolution of sexual preference. Eugen. Rev. 7 115–123. [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Findlay, G. D., X. Yi, M. J. Maccoss and W. J. Swanson, 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6: e178. [DOI] [PMC free article] [PubMed]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraczkiewicz, R., and W. Braun, 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 19 319. [Google Scholar]
- Galindo, B. E, V. D. Vacquier and W. J. Swanson, 2003. Positive selection in the egg receptor for abalone sperm lysin. Proc. Natl. Acad. Sci. USA 100 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper, J., and W. J. Swanson, 2006. Molecular population genetics of the gene encoding the human fertilization protein zonadhesin reveals rapid adaptive evolution. Am. J. Hum. Genet. 79 820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets, S., 2000. Rapid evolution of reproductive barriers driven by sexual conflict. Nature 403 886–889. [DOI] [PubMed] [Google Scholar]
- Gavrilets, S., and D. Waxman, 2002. Sympatric speciation by sexual conflict. Proc. Natl. Acad. Sci. USA 99 10533–10538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W., M. V. Han and S. G. Han, 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm, D., B. S. Mautz, M. F. Wolfner, C. F. Aquadro and W. J. Swanson, 2007. Evidence of amino acid diversity-enhancing selection within humans and among primates at the candidate sperm-receptor gene PKDREJ. Am. J. Hum. Genet. 81 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, T. I., M. Vose and S. Gavrilets, 2007. Genetic differentiation by sexual conflict. Evolution 61 516–529. [DOI] [PubMed] [Google Scholar]
- Hexter, A., 1968. Selective advantage of the sickle-cell trait. Science 160 436–437. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A. L., 1994. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256 119–124. [DOI] [PubMed] [Google Scholar]
- Innan, H., 2003. The coalescent and infinite-site model of a small multigene family. Genetics 163 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, E. S., and T. A. Markow, 2007. Reproductive tract interactions contribute to isolation in Drosophila. Fly 1 33–37. [DOI] [PubMed] [Google Scholar]
- Kelleher, E. S., W. J. Swanson and T. A. Markow, 2007. Gene duplication and adaptive evolution of digestive proteases in Drosophila female reproductive tracts. PLoS Genet. 3 1541–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Knowles, L. L., and T. A. Markow, 2001. Sexually antagonistic coevolution of a postmating prezygotic reproductive character in desert Drosophila. Proc. Natl. Acad. Sci. USA 98 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, L. L., B. B. Hernandez and T. A. Markow, 2005. Non-antagonistic interactions between the sexes revealed by the ecological consequences of reproductive traits. J. Evol. Biol. 18 156–161. [DOI] [PubMed] [Google Scholar]
- Lawniczak, M. K., and D. J. Begun, 2007. Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol. Biol. Evol. 24 1944–1951. [DOI] [PubMed] [Google Scholar]
- Loppin, B., D. Lepetit, S. Dorus, P. Couble and T. L. Karr, 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr. Biol. 15 87–93. [DOI] [PubMed] [Google Scholar]
- Machado, C. A., L. M. Matzkin, L. K. Reed and T. A. Markow, 2007. Multilocus nuclear sequences reveal intra- and interspecific relationships among chromosomally polymorphic species of cactophilic Drosophila. Mol. Ecol. 16 3009–3024. [DOI] [PubMed] [Google Scholar]
- Matzkin, L. M., 2008. The molecular basis of host adaptation in cactophilic Drosophila: molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics 178 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T. A., 1996. Evolution of Drosophila mating systems. Evol. Biol. 29 73–106. [Google Scholar]
- Martinsohn, J. T., A. B. Sousa, L. A. Guethlein and J. C. Howard, 1999. The gene conversion hypothesis of MHC evolution: a review. Immunogenetics 50 168–200. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351 652–654. [DOI] [PubMed] [Google Scholar]
- Moy, G. W., S. A. Springer, S. L. Adams, W. J. Swanson and V. D. Vacquier, 2008. Extraordinary intraspecific diversity in oyster sperm bindin. Proc. Natl. Acad. Sci. USA 105 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. D. Siggia et al., 2005. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics 171 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., 1998. Deleterious mutations in animal mitochondrial DNA. Genetica 102/103 61–69. [PubMed] [Google Scholar]
- Nei, M., and A. P. Rooney, 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum, D. M., and M. F. Wolfner, 1999. Wise, winsome, or weird? Mechanisms of sperm storage in female animals. Curr. Top. Dev. Biol. 41 67–97. [DOI] [PubMed] [Google Scholar]
- Ohno, S, 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Otto, S. P., and P. Yong, 2002. The evolution of gene duplicates. Adv. Genet. 46 451–483. [DOI] [PubMed] [Google Scholar]
- Panhuis, T. M., and W. J. Swanson, 2006. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics 173 2039–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis, T. M., N. L. Clark and W. J. Swanson, 2006. Rapid evolution of reproductive proteins in abalone and Drosophila. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, G. A., 1979. Sexual selection and sexual conflict, pp. 123–166 in Sexual Selection and Reproductive Competition in Insects, edited by M. S. Blum and N. A. Blum. Academic Press, London.
- Patterson, J. T., 1946. A new type of isolating mechanism in Drosophila. Proc. Natl. Acad. Sci. USA 32 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick, S., G. T. Miller, K. Schneider and T. A. Markow, 2003. Ejaculate-female coevolution in Drosophila mojavensis. Proc. Natl. Acad. Sci. USA 270 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar, L., 2005. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 62 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx, S. R., and R. C. Phillips, 2006. Allelic divergence precedes and promotes gene duplication. Evolution 60 881–892. [PubMed] [Google Scholar]
- Prokupek, A, F. Hoffmann, S. I. Eyun, E. Moriyama, M. Zhou et al., 2008. An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution. 62 2936–2947. [DOI] [PubMed] [Google Scholar]
- Reed, L. K., M. Nyboer and T. A. Markow, 2007. Evolutionary relationships of Drosophila mojavensis geographic host races and their sister species Drosophila arizonae. Mol. Ecol. 16 1007–1022. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381 232–234. [DOI] [PubMed] [Google Scholar]
- Robertson, S. A., 2007. Seminal fluid signaling in the female reproductive tract: lessons from rodents and pigs. J. Anim. Sci. 85 E36–E44. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1995. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. 11 621–625. [DOI] [PubMed] [Google Scholar]
- Saitou, N, and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sawyer, S. A., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6 526–538. [DOI] [PubMed] [Google Scholar]
- Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch, 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford, J. B., 1969. Heterosis and evolution of duplications. Am. Nat. 103 407–432. [Google Scholar]
- Sprang, S. R., R. J. Fletterick, L. Graf, W. J. Rutter and C. S. Craik, 1988. Studies of specificity and catalysis in trypsin by structural analysis of site-directed mutants. Crit. Rev. Biotechnol. 8 225–236. [DOI] [PubMed] [Google Scholar]
- Srinivasan, A., A. P. Giri and V. S. Gupta, 2006. Structural and functional diversities in lepidopteran serine proteases. Cell. Mol. Biol. Lett. 11 132–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman, H. H., B. W. Kozyak, A. Nelson, D. M. Thesier, L. T. Su et al., 2004. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428 415–418. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 1998. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. 281 710–712. [DOI] [PubMed]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., C. F. Aquadro and V. D. Vacquier, 2001. Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol. Biol. Evol. 18 376–383. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. Wong, M. F. Wolfner and C. F. Aquadro, 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics 168 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2000. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., 1993. Relaxed natural selection in human populations during the Pleistocene. Jpn. J. Genet. 68 539–547. [DOI] [PubMed] [Google Scholar]
- Thornton, K. R., 2007. The neutral coalescent process for recent gene duplications and copy-number variants. Genetics 177 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, L. M., and H. E. Hoekstra, 2006. Adaptive evolution of fertilization proteins within a genus: variation in ZP2 and ZP3 in deer mice (Peromyscus). Mol. Biol. Evol. 23 1656–1669. [DOI] [PubMed] [Google Scholar]
- Turner, L. M., and H. E. Hoekstra, 2008. Reproductive protein evolution within and between species: maintenance of divergent ZP3 alleles in Peromyscus. Mol. Ecol. 17 2616–2628. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2007. Adaptive evolution of recently duplicated accessory gland protein genes in desert Drosophila. Genetics 177 1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118 279–294. [PubMed] [Google Scholar]
- Wang, X., W. E. Grus and J. Zhang, 2006. Gene losses during human origins. PLoS Biol. 4 e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner, M. F., 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88 85–93. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., 2007. “S.P.E.R.M.” (seminal proteins (are) essential reproductive modulators): the view from Drosophila. Soc. Reprod. Fertil. Suppl. 65 183–199. [PubMed] [Google Scholar]
- Wong, A., M. C. Turchin, M. F. Wolfner and C. F. Aquadro, 2008. Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol. Biol. Evol. 25 497–506. [DOI] [PubMed] [Google Scholar]
- Wiesenfeld, S. L., 1968. Selective advantage of the sickle-cell trait. Science 160 437. [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15 568–573. [DOI] [PubMed] [Google Scholar]
- Yang, Z., W. S. Wong and R. Nielsen, 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22 1107–1118. [DOI] [PubMed] [Google Scholar]