Abstract
The Dobzhansky–Muller model posits that intrinsic postzygotic reproductive isolation—the sterility or lethality of species hybrids—results from the evolution of incompatible epistatic interactions between species: favorable or neutral alleles that become fixed in the genetic background of one species can cause sterility or lethality in the genetic background of another species. The kind of hybrid incompatibility that evolves between two species, however, depends on the particular evolutionary history of the causative substitutions. An allele that is functionally derived in one species can be incompatible with an allele that is functionally derived in the other species (a derived-derived hybrid incompatibility). But an allele that is functionally derived in one species can also be incompatible with an allele that has retained the ancestral state in the other species (a derived-ancestral hybrid incompatibility). The relative abundance of such derived-derived vs. derived-ancestral hybrid incompatibilities is unknown. Here, we characterize the genetics and evolutionary history of a lethal hybrid incompatibility between Drosophila mauritiana and its two sibling species, D. sechellia and D. simulans. We show that a hybrid lethality factor(s) in the pericentric heterochromatin of the D. mauritiana X chromosome, hybrid lethal on the X (hlx), is incompatible with a factor(s) in the same small autosomal region from both D. sechellia and D. simulans, Suppressor of hlx [Su(hlx)]. By combining genetic and phylogenetic information, we infer that hlx-Su(hlx) hybrid lethality is likely caused by a derived-ancestral incompatibility, a hypothesis that can be tested directly when the genes are identified.
SPECIATION often involves the evolution of intrinsic postzygotic reproductive barriers—including the sterility and inviability of hybrids—that limit the potential for genetic exchange between populations or species (Dobzhansky 1937; Coyne and Orr 2004). Hybrid sterility and inviability in animals are usually caused by incompatible gene interactions: often functionally divergent genes from one species are incompatible with interacting genes from another species. Many studies have mapped such hybrid incompatibility genes to small chromosomal regions (Naveira and Fontdevila 1986; Pantazidis et al. 1993; Carvajal et al. 1996; Hollocher and Wu 1996; True et al. 1996; Sawamura and Yamamoto 1997; Naisbit et al. 2002; Presgraves 2003; Tao et al. 2003; Slotman et al. 2004; Moyle and Graham 2005; Sweigart et al. 2006; Masly and Presgraves 2007; Good et al. 2008) and, in several cases, identified the causative genes. These studies reveal that hybrid incompatibilities can involve functionally divergent protein-coding genes (Ting et al. 1998; Barbash et al. 2003; Presgraves et al. 2003; Brideau et al. 2006; Mihola et al. 2009; Phadnis and Orr 2009; Tang and Presgraves 2009), chimeric duplicate genes (Wittbrodt et al. 1989), repetitive DNA (Sawamura and Yamamoto 1997), and gene movement (Masly et al. 2006).
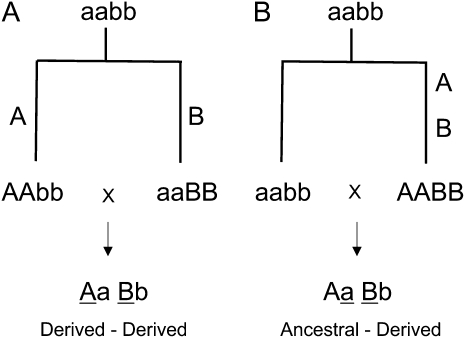
However, none of these individual hybrid incompatibility loci causes sterility or inviability on its own. Rather, as Dobzhansky (1937) and Muller (1940, 1942) first explained, hybrid fitness problems must involve deleterious epistatic interactions that evolve as incidental by-products of divergence (see Orr 1996). In the usual depiction of the so-called Dobzhansky–Muller model, an ancestral population with the two-locus genotype aabb splits into two geographically isolated lineages and each fixes new and different substitutions (yielding AAbb and aaBB lineages, respectively); when brought together in hybrids (AaBb), an incompatibility between these substitutions causes hybrid sterility or hybrid inviability (Figure 1A). Recent theory shows that the evolution of hybrid incompatibilities should follow some simple rules. For example, hybrid incompatibilities should be asymmetric (i.e., A is incompatible with B, but a should be compatible with b), should often be complex (i.e., involve three or more loci; Cabot et al. 1994; Orr 1995), and should snowball with time (i.e., the number of incompatibilities between two populations should increase faster than linearly with divergence; Orr 1995; Orr and Turelli 2001).
Figure 1.—
The Dobzhansky–Muller model for the evolution of postzygotic isolation. An ancestral population aabb splits into two independent populations that then accumulate substitutions. Epistatic interactions between underlined substitutions cause hybrid incompatibilities. (A) A derived-derived incompatibility. (B) A derived-ancestral incompatibility.
In Figure 1A, hybrids suffer from an incompatible epistatic interaction between a derived A allele and a derived B allele (i.e., a derived-derived hybrid incompatibility). But as Muller (1942) pointed out, if both substitutions occur in the same lineage (yielding AABB and aabb lineages; Figure 1B), then hybrids (AaBb) could suffer from an incompatible epistatic interaction between a derived B allele and an ancestral a allele (i.e., a derived-ancestral hybrid incompatibility). Assuming that all substitutions are independent, so that causative substitutions accumulate in both lineages, theory predicts that derived-derived hybrid incompatibilities should be more common (Orr 1995). The reason is that derived alleles can be incompatible with both derived and ancestral alleles, but ancestral alleles can only be incompatible with derived alleles [ancestral alleles must be compatible with one another (Orr 1995)]. If, however, substitutions are not independent, the expected relative frequency of derived-ancestral incompatibilities increases. In the extreme case, in which all substitutions occur in one lineage, only derived-ancestral incompatibilities are possible (Orr 1995). There is good reason to believe that the substitutions involved in hybrid incompatibilities are not independent. Imagine, for instance, that two interacting loci coevolve so that substitution of the A allele favors the subsequent substitution of the B allele at an interacting locus (Presgraves and Stephan 2007; Schlosser and Wagner 2008; Tang and Presgraves 2009). This kind of coevolutionary nonindependence will tend to concentrate substitutions among interacting partner loci in one lineage, enriching for derived-ancestral incompatibilities compared to a scenario of independent substitutions.
Data on the relative abundance of derived-derived vs. derived-ancestral hybrid incompatibilities are lacking as few interacting partners have been mapped and characterized. Incompatible partners causing hybrid lethality have been genetically characterized between Drosophila melanogaster and D. simulans (Hutter et al. 1990; Sawamura et al. 1993; Sawamura and Yamamoto 1993; Brideau et al. 2006), and incompatible partners causing hybrid sterility have been mapped in Drosophila (Pantazidis et al. 1993) and in Mimulus (Sweigart et al. 2006). However, none has established the species lineage in which the functionally derived alleles at the incompatible partner loci evolved. The hybrid incompatibility identified by Masly et al. (2006), who showed that a gene transposition causes male sterility in D. melanogaster–D. simulans hybrids, is a special case: JYalpha, a gene essential for male fertility, is on the fourth chromosome in D. melanogaster but has moved onto the third chromosome in the D. simulans lineage. Thus, hybrid males homozygous for the D. simulans fourth chromosome and the D. melanogaster third are sterile as they lack the JYalpha gene. The transposition of JYalpha caused a derived-ancestral hybrid incompatibility: a derived change (the absence of JYalpha on the fourth in D. simulans) is incompatible with the ancestral state (the absence of JYalpha on the third in D. melanogaster).
Here we characterize the genetics and evolutionary history of a new lethal hybrid incompatibility between D. mauritiana and its sibling species, D. sechellia and D. simulans, three species that diverged nearly simultaneously ∼250,000 years ago (Kliman et al. 2000; McDermott and Kliman 2008). Our analysis builds on an earlier genomewide screen for hybrid incompatibilities between D. mauritiana and D. sechellia in which four hybrid lethal regions were identified, including one near the base of the D. mauritiana X chromosome (Masly and Presgraves 2007). In this article we refine the mapping of this X-linked factor, which we call hybrid lethal on the X (hlx), and we map an incompatible partner, Suppressor of hlx [Su(hlx)], to a small autosomal region. Finally, using comparative mapping, we infer a most-parsimonious history for the evolution of the hlx-Su(hlx) hybrid lethality, which appears to result from a derived-ancestral hybrid incompatibility.
MATERIALS AND METHODS
Fly stocks:
We used stocks of three Drosophila species: D. sechellia w, D. simulans wXD1, and D. simulans wNIG, kindly provided by Jerry Coyne; and a large collection of D. mauritiana w stocks, each bearing single inserts of the P[w+] construct (described in True et al. 1996), kindly provided by Yun Tao. We also used 42 stocks of D. sechellia w that are homozygous for small P[w+]-marked autosomal introgressions of D. mauritiana material. These stocks were produced by selectively introgressing a D. mauritiana P[w+]-marked region into D. sechellia w for 15 generations of repeated backcrossing (for details see Masly and Presgraves 2007). All crosses were done at room temperature (23–24°) on standard cornmeal-agarose medium.
Mapping the X-linked hybrid lethal:
In previous work, Masly and Presgraves (2007) found that D. sechellia lines with small introgressions of cytological region 18DE of the D. mauritiana X chromosome suffer recessive hybrid lethality (see results below). To confirm these original findings, we constructed new introgression lines with both D. sechellia and D. simulans. In particular, following the introgression procedure of Masly and Presgraves (2007), we moved the 18DE region of D. mauritiana, marked with the P[w+]-insert 2E1, into D. sechellia w, D. simulans wXD1, and D. simulans wNIG. Briefly, we crossed, e.g., D. simulans wXD1 females to D. mauritiana 2E1 P[w+]-insert bearing males. In each subsequent generation, we selected P[w+]-bearing hybrid females and backcrossed them to D. simulans wXD1 males (for details see Masly and Presgraves 2007). To score hybrid lethality, we transferred parents to fresh vials three times and pooled progeny numbers from the three vials.
Rather than stop our introgressions at generation 15, we produced 259 sublines over 60 generations of continued introgression. We used the D. simulans wXD1 for these advanced generation sublines as this stock showed the highest fecundity. Of the 259 sublines, 131 ultimately produced viable P[w+]-marked sons and 128 sublines remained hybrid lethal. After 15 generations of introgression, we maintained the sublines by crossing 10 P[w+]-bearing females with 10 w brothers, and scored their progeny for sex and eye color each generation. We collected 5–10 P[w+]-female (and, if available, viable P[w+]-male) progeny from 3–4 lethal and 2–3 viable lines every generation for 45 generations and froze them at −20° for genotyping at molecular markers (see below).
Lethal phase:
To determine if hybrid lethality was embryonic or postembryonic, we scored egg-hatch rates from two kinds of females: those heterozygous for a P[w+]-marked hybrid lethal factor on the X chromosome (for which half of their sons die) and those lacking the hybrid lethal (as controls). We set up individual females in vials containing a small plastic spoon with fly food colored with grape juice and painted with a live yeast suspension (see Hoffmann et al. 1986). All egg collections were done in an incubator at 24°. Every 24 hr, females were transferred to a new vial for three successive transfers, producing four egg-hatch counts per female. After the third transfer, females were transferred to a standard vial with cornmeal-agarose medium and allowed to produce progeny; we then scored the number of w+ and w male and female progeny.
Mapping the autosomal partner:
To map possible autosomal suppressors of hybrid lethality, we crossed lethal-introgression bearing females to males from 42 different viable and fertile lines carrying small D. mauritiana autosomal segments introgressed into D. sechellia. These 42 lines were produced by Masly and Presgraves (2007) and maintained in our lab. For each of the 42 tests, we crossed ∼10 virgin females heterozygous for the lethal X-linked introgression to ∼15 males from each of the 42 autosomal introgressions. Parental adults were transferred every 5–6 days until they ceased to produce progeny. All progeny were scored for sex and eye color.
Molecular markers:
The cytological location for the 2E1 P[w+]-insert was originally inferred from salivary gland squashes to be in cytological subdivisions 18DE (True et al. 1996). The genomic flanking sequences of the P-element (provided by Y. Tao) show that the insert sits in the 5′-UTR of the jog locus in cytological bands 18F2–18F4. To map the X-linked hybrid lethal, we used a combination of microsatellite markers and single-nucleotide differences between lines. We used the D. simulans genome sequence to identify candidate microsatellite markers using the Tandem Repeats Finder software (Benson 1999) and to design primers flanking the microsatellite loci. Each generation we genotyped a subset of w+ females and (when available) recombinant w+ males from selected viable and lethal sublines with three microsatellite and three SNP markers. Microsatellite marker 17.07 is in the Bx locus, in cytological subdivision 17C; microsatellite marker 20.06 is located in cytological band 20D1; and microsatellite marker 20.07 is in the fog locus in cytological band 20D2. SNP markers were surveyed at three genes in cytological region 20EF: CG13865 and CG40485, and su(f).
Genotyping introgression breakpoints:
We isolated genomic DNA following a single fly extraction protocol from Puregene DNA purification kit (Gentra Systems). To genotype microsatellites, we PCR amplified marker regions using standard protocols and visualized species–specific microsatellite array length differences on a 8% polyacrylamide gel stained with ethydium bromide. To genotype SNP differences, we used TILLING (Till et al. 2006) following the protocols of the Transgenomic SURVEYOR mutation detection kit (Transgenomic). This kit uses a mismatch-specific DNA endonuclease to scan for mismatches in heteroduplex DNA. Briefly, to genotype individual flies using TILLING, we PCR amplified a marker region, formed heteroduplex DNA, cut the heteroduplex DNA with SURVEYOR endonuclease, and then visualized the digestion products on a 2% agarose gel. As our loci are X linked, we used different heteroduplex formation steps for the two sexes. To form heteroduplex DNA for P[w+]-introgression males, we mixed equal amounts of PCR product from individual P[w+]-introgression males with PCR product from D. simulans and, separately, from D. mauritiana. For P[w+] introgression females, no DNA mixing was necessary as these females are heterozygous for the introgressed regions. After heteroduplex formation, we treated 30 μl of the sample with SURVEYOR enhancer and SURVEYOR endonuclease (1.5–2 μl of each) and incubated the mixture for 25–30 min at 42°. We stopped the reaction with 1/10 stop solution and froze the samples at −20° until ready for loading in a 2% agarose gel. All SURVEYOR reactions were performed simultaneously with positive controls involving heteroduplexed DNA between two species and negative controls involving pure-species DNA from a single line.
For P[w+]-introgression females, if digestion of PCR products was detected the line was inferred to be heterozygous (D. mauritiana/D. simulans) at the assayed marker. Conversely, if no digestion was detected, the line was inferred to be homozygous for D. simulans material at the assayed marker. For P[w+]-introgression males, if digestion was detected for heteroduplexed DNA between the introgression line and D. simulans but not D. mauritiana PCR products, the line was inferred to be hemizygous D. mauritiana at the assayed marker. Conversely, if digestion was detected for heteroduplexed DNA between the introgression line and D. mauritiana but not D. simulans PCR products, the line was inferred to be hemizygous D. simulans at the assayed marker.
RESULTS
A locus on the D. mauritiana X chromosome causes lethality in D. sechellia and D. simulans genetic backgrounds:
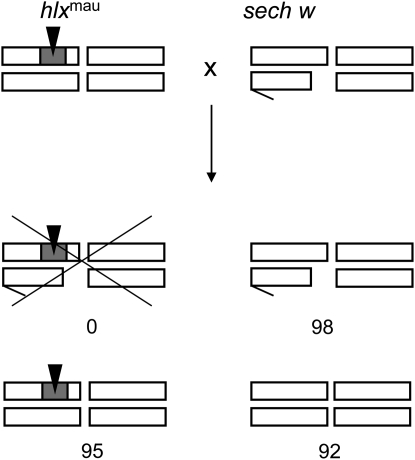
Previous work showed that a genetic factor at the base of the D. mauritiana X chromosome causes lethality when introgressed into an otherwise D. sechellia genetic background (Masly and Presgraves 2007). To confirm this X-linked hybrid lethality, we first generated new introgression lines: for 6–8 generations we backcrossed females carrying the D. mauritiana 2E1 P[w+]-insertion to D. sechellia w males. We then scored progeny from introgression hybrid females heterozygous for the D. mauritiana 2E1 P[w+]-insertion crossed to D. sechellia w males. From this cross we expect four zygotic genotypes: females with and without the 2E1 introgression and males with and without the 2E1 introgression (Figure 2). Flies with the D. mauritiana 2E1 introgression will be red eyed as they carry the P[w+]-insert in an otherwise D. sechellia w background. Figure 2 shows that while both red- and white-eyed females appear in roughly equal numbers, only white-eyed males appear and red-eyed males are absent. These findings show that introgression hybrid males inheriting the 2E1 P[w+]-marked introgression from D. mauritiana are lethal whereas their siblings are viable. We thus named the responsible genetic factor hybrid lethal on the X (hlx). The fact that females heterozygous for the introgression are viable suggests that hlx acts either as a male-specific hybrid lethal or as a recessive hybrid lethal.
Figure 2.—
A locus on the D. mauritiana X chromosome causes lethality in a D. sechellia genetic background. D. sechellia w females heterozygous for hlxmau are crossed to D. sechellia w males. Male offspring that inherit hlxmau are lethal. Open rectangles represent one pair of sex chromosomes and a representative pair of autosomes in D. sechellia; the short, hooked rectangle represents the Y chromosome. The shaded box represents introgressed D. mauritiana genetic material marked with a P[w+]-element, shown as an inverted solid triangle. Numbers below the offspring genotypes are representative for the cross (see Table 1).
To test if the D. mauritiana allele, hlxmau, also causes hybrid lethality in D. simulans, we introgressed 2E1 P[w+]-marked fragments from D. mauritiana into two different D. simulans stocks, wXD1 and wNIG. Table 1 shows that males with mostly D. simulans genetic backgrounds that inherit hlxmau are lethal whereas their siblings are viable. These results show that the D. mauritiana allele of hlx is lethal when introgressed into D. sechellia and into two lines of D. simulans.
TABLE 1.
D. mauritiana hlx causes hybrid lethality in both of its sibling species, D. simulans and D. sechellia
| Females
|
Males
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Species | Line | Generationa | w+ | w | w+/w ratio | w+ | w | w+/w ratio |
| D. simulans | sim wXD1 | 14 | 99 | 105 | 0.943 | 9 | 110 | 0.082 |
| sim wXD1 | 10 | 72 | 71 | 1.014 | 5 | 123 | 0.04 | |
| sim wNIG | 6 | 26 | 25 | 1.04 | 1 | 31 | 0.032 | |
| sim wNIG | 9 | 66 | 62 | 1.065 | 3 | 49 | 0.061 | |
| D. sechellia | sech w | 6 | 95 | 92 | 1.033 | 0 | 98 | 0 |
| sech w | 8 | 101 | 114 | 0.886 | 2 | 105 | 0.019 | |
The number of generations for which the hlx region of D. mauritiana was introgressed into D. simulans or D. sechellia genetic backgrounds.
In some crosses, a small number of P[w+]-bearing introgression males appeared (Table 1). These males could represent rare escapers of hybrid lethality or rare recombinant males that inherit X chromosomes for which the 2E1 P[w+]-insert and hlxmau have become separated by recombination. (Viable P[w+]-bearing introgression males were invariably sterile; not shown.) To determine if most w+ males were escapers or recombinants, we set up their w+ sisters individually in separate vials and scored their progeny for eye color. If P[w+]-bearing males are escapers, then these crosses should produce escapers at a similar rate. If, however, lethality is largely complete and P[w+]-bearing males have inherited recombinant chromosomes, then individual w+ sisters should fall into two distinct classes: those with nonrecombinant hlxmau-bearing chromosomes should produce almost exclusively white-eyed sons; and those with recombinant hlxmau-free chromosomes should produce a 1:1 ratio of red- and white-eyed sons. Table 2 shows that the presence of P[w+]-bearing males is best explained by recombination. There are few, if any, escapers of hlxmau-based hybrid lethality.
TABLE 2.
Occasional viable w+ hybrid males are recombinants, not escapers
| Females
|
Males
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Line | Generationa | w+ | w | w+/w ratio | w+ | w | w+/w ratio | Class | |
| 1. Original line | sim wXD1 | 55 | 20 | 28 | 0.71 | 9 | 23 | 0.39 | |
| 2. Sublines | sim wXD1 | 56 | 9 | 15 | 0.60 | 0 | 13 | 0.00 | Lethal |
| 3. | sim wXD1 | 56 | 15 | 10 | 1.50 | 0 | 15 | 0.00 | Lethal |
| 4. | sim wXD1 | 56 | 28 | 28 | 1.00 | 0 | 36 | 0.00 | Lethal |
| 5. | sim wXD1 | 56 | 13 | 12 | 1.08 | 9 | 11 | 0.82 | Recombinant |
| 6. | sim wXD1 | 56 | 4 | 7 | 0.57 | 2 | 4 | 0.50 | Recombinant |
| 7. | sim wXD1 | 56 | 21 | 17 | 1.24 | 15 | 20 | 0.75 | Recombinant |
The number of generations for which the hlx region of D. mauritiana was introgressed into D. simulans or D. sechellia.
hlxmau causes postembryonic hybrid lethality:
We next tested if lethality was embryonic or postembryonic. For these experiments, we used D. simulans wXD1 introgression females as their fecundity was higher than the D. sechellia w and D. simulans wNIG lines. We set up 15 hlxmau/hlxsim females from each of three viable and three lethal lines, allowed them to lay eggs for four consecutive 24-hour periods, and scored the number of hatched and unhatched eggs for each after 28 hr. We found no significant difference in the percentage of unhatched eggs between the lethal and viable lines (lethal: 40.7% ± 3.8%; viable: 46.2% ± 5.9%; permutation test P = 0.417). Hybrid lethality therefore appears to be postembryonic.
Fine-scale mapping of hlx:
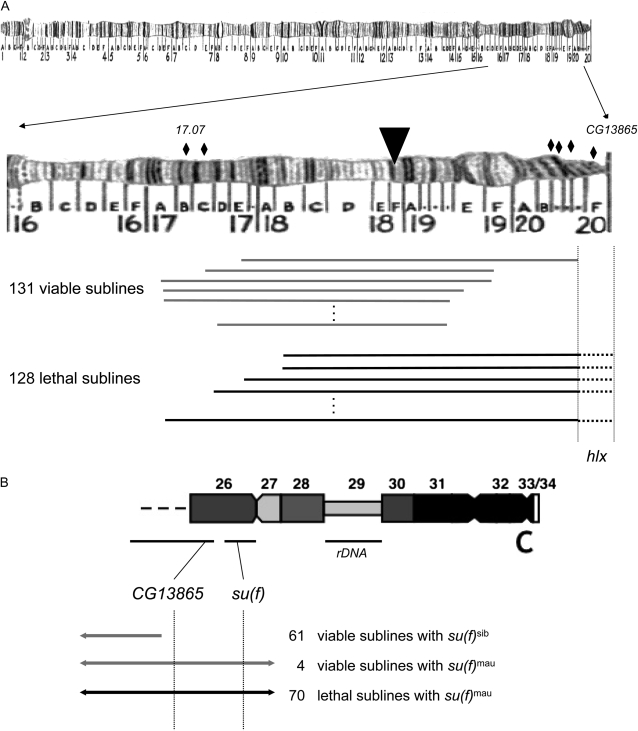
For fine-scale genetic mapping, we focused on the D. simulans wXD1 introgression stocks. As recombination occasionally separates the 2E1 P[w+]-insert from hlxmau, we generated many hybrid lethal-bearing sublines and continued the introgression procedure. Over the course of 20–45 additional generations of introgression, we recovered and genotyped 131 viable and 128 lethal P[w+]-bearing sublines for different combinations of six molecular markers on either side of the 18DE region. We tested for a genetic association between hybrid lethality and the species origin of the markers (Table 3). The distal (leftmost) marker shows no association between species origin and hybrid lethality (microsatellite marker 17.07, Fisher's exact P = 0.365). In contrast, the SNP marker in CG13865 shows a highly significant association (Fisher's exact P = 7.23 × 10−29): introgression males bearing D. simulans material at CG13865 are always viable whereas introgression males bearing D. mauritiana material at CG13865 are nearly always lethal. The association is not, however, perfect. We recovered four viable P[w+]-bearing males that carry D. mauritiana material at CG13865 (we genotyped these exceptional males twice to confirm these results). Assuming that hybrid lethality is complete, these four males suggest that hlx is proximal to CG13865 (20F3–20F4; Figure 3A). CG13865 is the last, most-proximal gene in the contiguous D. melanogaster (v. 5.1) assembly of the X chromosome. The sequence scaffold bearing CG13865 extends to heterochromatin region h26 (heterochromatin regions in the X are designated h26–h34; Figure 3B; Hoskins et al. 2007). The hlx locus thus resides in the highly repetitive, transposon-rich, and gene-poor pericentric heterochromatin of the X chromosome.
TABLE 3.
Genotype-marker association for six molecular markers around 18DE
| Genotype
|
|||||
|---|---|---|---|---|---|
| Marker | Positiona | Class | mau | non-mau | P-valueb |
| 17.07 | 18368602 | Lethal | 14 | 18 | |
| Viable | 12 | 11 | 0.365 | ||
| 20.06 | 21952484 | Lethal | 19 | 0 | |
| Viable | 4 | 24 | 1.27 e−9 | ||
| 20.07 | 22037065 | Lethal | 72 | 0 | |
| Viable | 4 | 41 | 2.37 e−27 | ||
| CG40485 | 22362444 | Lethal | 94 | 0 | |
| Viable | 4 | 83 | 2.27 e−47 | ||
| CGI3865 | 22415346 | Lethal | 42 | 0 | |
| Viable | 4 | 78 | 7.23 e−29 | ||
| su(f) | XHet:69195 | Lethal | 70 | 0 | |
| Viable | 4 | 61 | 4.22 e−34 | ||
D. melanogaster R5.11 sequence starting coordinate.
P-value for Fisher's exact test.
Figure 3.—
Fine-scale genetic mapping of hlx. (A) A collection of 131 viable lines (shaded) and 128 lethal lines (solid) was used to map the location of hlx to the bracketed interval proximal to CG13865. Diamonds represent position of molecular markers, and the inverted solid triangle represents location of P[w+]-insert. (B) Cytogenetic map of the centromeric heterochromatin of the X chromosome (of D. melanogaster) showing heterochromatic regions h26–h34 and the centromere, (C) Solid and shaded lines represent hybrid lethal and viable sublines genotyped at CG13865 and su(f); mau, D. mauritiana and “sib,” D. simulans or D. sechellia. The number of lines genotyped is shown for each class.
To further refine the position of hlx in the heterochromatin, we genotyped 135 introgression chromosomes for SNP differences at the su(f) locus. The su(f) locus lies in scaffold Xhet (GenBank accession no. CP00208), which maps to cytological regions h26–h27 but is not currently contiguous with the euchromatic assembly (Figure 3B; Hoskins et al. 2007). We also found a significant association at this marker (Fisher's exact test P = 4.22 × 10−34). The four viable P[w+]-bearing males carrying D. mauritiana material at CG13865 also carried D. mauritiana material at su(f), placing the location of hlx proximal to su(f) in the centromeric heterochromatin. Only one known protein-coding gene, ATbp, lies ∼5 kb proximal to su(f). However, our repeated attempts to genotype recombinants at ATbp were unsuccessful. Further traditional genetic mapping of the hlx locus is, for the moment, not feasible.
Hybrid lethality is caused by an X–autosome incompatibility:
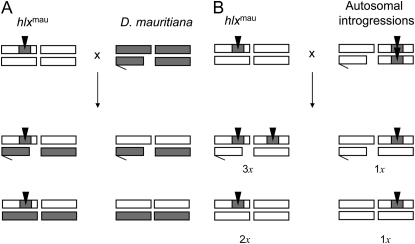
The Dobzhansky–Muller model predicts that hlxmau is incompatible with one or more loci from D. simulans and D. sechellia. In principle, the incompatible partner gene(s) could be located on the autosomes or on the non-D. mauritiana part of the P[w+]-bearing introgression X. To distinguish these possibilities, we tested for an X–autosome interaction. In particular, we crossed D. sechellia females heterozygous for a lethal introgression of hlxmau to D. mauritiana males. All progeny from this cross are heterozygous for the autosomes and, importantly, half of the sons will inherit the hlxmau on an otherwise D. sechellia X chromosome (Figure 4A). If these males are viable, it suggests that some factor(s) in the D. mauritiana autosomal genome can dominantly suppress hlxmau hybrid lethality. We found that, indeed, sons inheriting hlxmau are completely viable when given a set of D. mauritiana autosomes (Table 4, line 1 vs. 2). The same result is obtained when we cross D. simulans females with lethal introgressions of hlxmau to D. mauritiana males (Table 4, line 3 vs. 4). These results show that the D. mauritiana autosomes possess one or more dominant Suppressor of hlx [Su(hlx)] loci. Put differently, hlxmau is incompatible with a recessive autosomal factor(s) from D. sechellia and D. simulans. Importantly, these results also exclude two other explanations. First, hlxmau is not involved in a simple X–X hybrid incompatibility. Second, the lethality of hlxmau introgressions cannot be attributed to linked spontaneous mutations that accumulated during the introgression procedure. If introgression males are lethal because of an X-linked recessive mutation, there is no reason why they ought to be rescued by the D. mauritiana autosomes.
Figure 4.—
(A) Test for an X–autosome interaction. D. sechellia females heterozygous for hlxmau were crossed to D. mauritiana males. If hlxmau-bearing hybrid males are viable, D. mauritiana possesses one or more dominant, autosomal suppressors of hlxmau. (B) A genetic mapping screen for Su(hlx) in D. mauritiana autosomal genome. D. simulans females heterozygous for hlxmau are crossed to D. sechellia males homozygous for small autosomal introgressions from D. mauritiana. If hlxmau-bearing males are viable, the D. mauritiana introgression possesses a dominant Su(hlx).
TABLE 4.
D. mauritiana autosomes suppress hlxmau-based hybrid lethality
| Female progeny
|
Male progeny
|
||||||
|---|---|---|---|---|---|---|---|
| Female parent | Male parent | w+ | w | w+/w ratio | w+ | w | w+/w ratio |
| 1. hlxmau/hlxsech; sech/sech | sech w | 103 | 109 | 0.945 | 0 | 103 | 0 |
| 2. hlxmau/hlxsech; sech/sech | mau w | 105 | 102 | 1.029 | 95 | 103 | 0.922 |
| 3. hlxmau/hlxsim; sim/sim | sim wXD1 | 47 | 57 | 0.825 | 0 | 60 | 0 |
| 4. hlxmau/hlxsim; sim/sim | mau w | 97 | 92 | 1.054 | 87 | 95 | 0.916 |
A genetic screen for Su(hlx) loci on the D. mauritiana autosomes:
To map the autosomal Su(hlx) partner loci, we crossed heterozygous hlxmau-bearing D. simulans wXD1 introgression females to males from 42 different lines of D. sechellia homozygous for small autosomal P[w+]-marked introgressions from D. mauritiana (Figure 4B). [The latter are a subset of the viable and fertile autosomal introgressions (on average ∼2 Mb in size) maintained from the original screen by Masly and Presgraves (2007).] We used hlxmau-bearing D. simulans wXD1 lines for these crosses because their fecundity is substantially higher than that of hlxmau-bearing D. sechellia w lines. For each of the 42 crosses, four zygotic genotypes will be produced: daughters will possess either the autosomal introgression only or both hlxmau and the autosomal introgression; similarly, sons will possess either the autosomal introgression only or both hlxmau and the autosomal introgression (Figure 4B). If the hlxmau-bearing males remain inviable, a Su(hlx) partner locus does not reside in the autosomal introgression. But if hlxmau-bearing males are rescued, we can infer that a Su(hlx) partner resides in the autosomal introgression (Figure 4B). For most crosses, we were able to distinguish the two kinds of male progeny (those inheriting hlxmau-introgression and those not) by P[w+] dosage. Males inheriting only the autosomal introgression will be heterozygous for a P[w+]-insert and thus have light red (or orange) eye color. In contrast, males with both an autosomal introgression and the hlxmau-introgression are heterozygous for one P[w+]-insert and hemizygous for the other and will thus express, in effect, three doses of P[w+]. These males should therefore have a strong red-eye phenotype. In addition to eye color, the sex ratio among progeny is also informative: in crosses lacking Su(hlx) rescue we expect a 1:2 sex ratio as half of the males die; however, if Su(hlx) rescue occurs we expect a 1:1 sex ratio.
Only one of the 42 autosomal introgressions strongly suppresses the hybrid lethality of hlxmau: hybrid males inheriting hlxmau and an autosomal introgression from the 33F–34A region of D. mauritiana are completely viable (Table 5, line 1). None of the 41 other regions suppressed hybrid lethality (not shown). It is important to note, however, that this screen tests a relatively small fraction of the D. mauritiana genome; more Su(hlx) loci could reside in untested regions.
TABLE 5.
Su(hlx) locus maps to D. mauritiana region 33F-34A
| Female progeny
|
Male progeny
|
||||||
|---|---|---|---|---|---|---|---|
| Female parent | Male parent | 2x | 1x | 2x/1x ratio | 3x | 1x | 3x/1x ratio |
| 1. hlxmau/hlxsim; sim/sim | hlxsech/Y; 33F-34Amau/33F-34Amau | 193 | 178 | 1.084 | 180 | 202 | 0.891 |
| 2. hlxmau/hlxsech; sech/sech | hlxsech/Y; 33F-34Amau/33F-34Amau | 112 | 103 | 1.087 | 76 | 98 | 0.776 |
| 3. hlxmau/hlxsim; sim/sim | hlxsim/Y; 33F-34Amau/33F-34Amau | 105 | 98 | 1.071 | 81 | 90 | 0.900 |
The focal males described above are hybrids with genetic material from three species: a hlxmau-bearing X chromosome from D. simulans wXD1, one set of D. simulans wXD1 autosomes, and one set of D. sechellia w autosomes with a small D. mauritiana P[w+]-marked introgression. As these complex genotypes are not ideal, we performed two further crosses. First, we crossed heterozygous hlxmau-bearing D. sechellia females to D. sechellia males with the 33F–34A introgression from D. mauritiana. As expected, hybrid males bearing hlxmau and the D. mauritiana allele, Su(hlx)mau, in an otherwise D. sechellia w genetic background are fully viable (Table 5, line 2). Second, we introgressed the 33F–34A P[w+]-insert from D. mauritiana into a D. simulans wXD1 genetic background via eight generations of repeated backcrossing. We then crossed heterozygous hlxmau-bearing D. simulans wXD1 females to D. simulans wXD1 males homozygous for a D. mauritiana 33F–34A introgression. We found that hybrid males bearing hlxmau and the D. mauritiana allele, Su(hlx)mau, in an otherwise D. simulans wXD1 genetic background are fully viable (Table 5, line 3). These results show that Su(hlx)mau can rescue lethal hlxmau-bearing hybrid males in both D. sechellia and D. simulans genetic backgrounds.
Previous work estimated that the 33F–34A P[w+]-marked D. mauritiana introgression into D. sechellia is ∼1.6 Mb long (Masly and Presgraves 2007). To refine this estimate, we genotyped 15 additional molecular markers in the region. The distal breakpoint falls between CG6405 and Elf (33E4), and the proximal breakpoint falls between CG16848 and CG16956 (34B11). The introgression is thus 0.975–1.03 Mb long, a region comprising 111–120 predicted genes.
DISCUSSION
The genetic analyses presented here yield two main results. First, we have mapped hlx, a locus that causes hybrid lethality, to the pericentric heterochromatin of the X chromosome. The D. mauritiana allele, hlxmau, causes complete postembryonic hybrid lethality when hemizygous in an otherwise D. sechellia or D. simulans genetic background. Second, we have mapped an autosomal partner locus, Su(hlx), that interacts with hlx. In particular, the D. mauritiana allele, Su(hlx)mau, can completely suppress the hybrid lethality of hlxmau in both D. sechellia and D. simulans genetic backgrounds. These findings show that hlxmau is incompatible with at least one recessive autosomal locus whose functional allelic state is shared by D. sechellia and D. simulans. Below, we infer the evolutionary history of the genetic substitutions leading to the hlx-Su(hlx) hybrid incompatibility and then consider the possible genetic basis for hybrid lethality.
Evolutionary history of the hlx-Su(hlx) hybrid incompatibility:
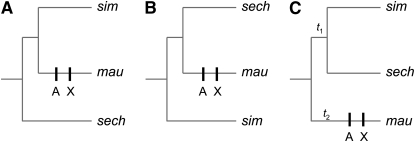
Combining the genetic mapping results with the phylogenetic history of the three D. simulans clade species allows us to make two important inferences about the hlx-Su(hlx) incompatibility (see also Moyle and Nakazato 2008). First, our results show that the D. mauritiana allele of hlx causes hybrid lethality in both D. sechellia and D. simulans genetic backgrounds. The most parsimonious evolutionary history is one in which the hlx substitution(s) causing hybrid lethality is derived in the D. mauritiana lineage. Similarly, we found that the D. mauritiana allele of Su(hlx) suppresses hybrid lethality in both D. sechellia and D. simulans, implying that the substitution(s) causing suppression of hlxmau hybrid lethality is also derived in the D. mauritiana lineage. Taken together, these genetic results imply that the functionally derived hlxmau allele is incompatible with the functionally ancestral Su(hlx) alleles of D. sechellia and D. simulans (Figure 5). The nearly simultaneous splitting of the D. simulans clade species has led to extensive lineage sorting: all three possible genealogical relationships can be detected among different loci (Figure 5; Hey and Kliman 1993; Kliman and Hey 1993; Hilton et al. 1994; Kliman et al. 2000; Ting et al. 2000; Malik and Henikoff 2005; McDermott and Kliman 2008). However, our inference that the causative substitutions at hlx and Su(hlx) occurred in D. mauritiana should be robust to uncertainty in the genealogical relationships at these loci (Figure 5).
Figure 5.—
Inferring the evolutionary history of the substitutions causing the hlx-Su(hlx) hybrid incompatibility. (A) Assuming genealogical histories with either D. sechellia (sech) or (B) D. simulans (sim) as outgroup species, the most parsimonious histories have the causative substitutions at hlx and Su(hlx) derived in the D. mauritiana (mau) lineage. X, X-linked hlx substitution; A, autosomal Su(hlx) substitution. (C) Assuming a genealogical history with D. mauritiana as the outgroup species, the causative substitutions at hlx and Su(hlx) could be derived in D. mauritiana (t1) or in the common ancestor of the D. simulans–D. sechellia (t2); however, given the disparity in branch lengths (t2 ≪ t1), there has been more time for hlx and Su(hlx) to evolve along the external branch leading to D. mauritiana than the very short internal branch of the D. simulans–D. sechellia common ancestor. The functionally derived hlxmau allele therefore appears to be incompatible with functionally ancestral Su(hlx) alleles from D. sechellia and D. simulans.
Second, if hlx and Su(hlx) are derived in the D. mauritiana lineage, we can say something about the order in which the relevant substitutions occurred. Of the two possible orderings—hlx followed by Su(hlx) or Su(hlx) followed by hlx—only one is allowed by natural selection. The derived hlxmau could not evolve first as it causes lethality in an ancestral Su(hlx) genetic background. In contrast, nothing prevents the derived Su(hlx)mau substitution from evolving first, after which the derived hlxmau can evolve in the permissive Su(hlx)mau genetic background. Thus, regardless of which of the three genealogical histories obtains at the hlx and Su(hlx) loci, the relevant substitutions at both most likely occurred in the D. mauritiana lineage. Once the loci have been identified at the molecular level, we will be able to validate this inferred history using molecular population genetics.
Genetic basis of the hlx-Su(hlx) hybrid lethality:
The localization of hlx to the gene-poor pericentric heterochromatin of the X raises the possibility that the hlx-Su(hlx) hybrid lethality is caused by something other than an incompatibility between two protein-coding genes. One possibility is that hlx is a kind of repetitive satellite DNA. If so, then Su(hlx) might be a protein-coding gene that regulates or interacts with heterochromatin. Among the 120 candidate genes in the Su(hlx) region, three have known or predicted chromatin-binding functions: A16, Scm-related gene containing four mbt domains (Sfmbt), and Sir2. A16 and Sfmbt are relatively uncharacterized, but Drosophila Sir2 is of special interest as it has roles in heterochromatin silencing (including suppression of position effect variegation on the X) and sex determination. A loss-of-function mutation at Sir2 in D. melanogaster causes aberrant expression of Sex Lethal in male embryos, disrupting dosage compensation and causing male-specific larval lethality (Rosenberg and Parkhurst 2002). It is therefore possible that Su(hlx) alleles from D. simulans and D. sechellia act as Sir2 loss-of-function mutations in hlxmau introgression males. Although the hlx-Su(hlx) incompatibility causes postembryonic lethality in males, consistent with the Sir2 hypothesis, we cannot be certain that hlxmau causes male-specific lethality as there is no straightforward way to test the viability of hlxmau/hlxmau introgression females.
A second possibility is that an essential gene on the ancestral X chromosome moved to the Su(hlx) autosomal region in the D. mauritiana lineage. In this case, hlxmau introgression males die because they lack an essential gene: the gene is absent from the hlxmau region of the X and from the Su(hlx) autosomal region of D. sechellia and D. simulans. Introgression males with hlxmau can then be rescued when supplied with the gene in the Su(hlx) autosomal region of D. mauritiana. This scenario is similar to the JYalpha-mediated hybrid male sterility described by Masly et al. (2006). Notably, JYAlpha moved from its ancestral position on the heterochromatic dot-fourth chromosome to 3R in the D. simulans lineage. An obvious candidate for gene movement in the hlx-Su(hlx) incompatibility is the viability-essential ribosomal (rDNA) locus. In D. melanogaster, the 18S, 5.8S, and 28S ribosomal RNAs are encoded by a large tandem array of rRNA genes in the pericentric heterochromatin of the X chromosome (h29, Figure 3B). Classical genetic work in D. melanogaster showed that the rDNA locus is the only vital locus in heterochromatin proper (Zhimulev 1998). In species of the D. anannassae complex, the rDNA locus has moved to the fourth chromosome (Roy et al. 2005). We can, however, rule out movement of the rDNA locus in D. mauritiana as in situ hybridization experiments have shown that the rDNA locus is present near the base of the X chromosome in D. simulans, D. sechellia, and D. mauritiana (Lohe and Roberts 2000; Roy et al. 2005). We cannot rule out the possibility that new, viability-essential genes have evolved or moved to the X heterochromatin in the common ancestor of the D. simulans clade species and then subsequently moved off of the X in D. mauritiana.
Relationship of hlx to two other hybrid lethality factors:
In crosses between D. melanogaster and members of the D. simulans species complex, two hybrid lethality factors also map to narrow intervals at the base of the X chromosome. In crosses between D. simulans females and D. melanogaster males, the X-linked Zygotic hybrid rescue (Zhr) factor from D. melanogaster causes dominant embryonic lethality of hybrid daughters (Sawamura et al. 1993; Sawamura and Yamamoto 1993). Zhrmel maps to region h32 of the pericentric heterochromatin of the X (Sawamura et al. 1995; Sawamura and Yamamoto 1997; Zhimulev 1998). Another X-linked hybrid lethal was discovered by chromosomal deletion (deficiency, Df) mapping in F1 hybrid females between D. melanogaster and the D. simulans clade species (Coyne et al. 1998). When D. melanogaster females heterozygous for deficiencies over dominantly marked balancer chromosomes (Df/Bal) are crossed to D. mauritiana males, hybrid daughters inheriting deficiencies in cytological region 20C–20F die whereas their balancer-inheriting sisters are viable (Coyne et al. 1998). Interestingly, Df-bearing hybrid daughters from crosses to D. simulans and D. sechellia are not lethal, consistent with the evolution of a recessive X-linked lethal in region 20C–20F in D. mauritiana. The fact that three hybrid lethals—hlx, Zhr, and the hybrid lethal of Coyne et al. (1998)— map to the same gene-poor pericentric region of the X raises the possibility that the same locus has repeatedly evolved hybrid lethality.
We can exclude the possibility that hlx and the hybrid lethal of Coyne et al. (1998) are the same locus: our mapping results place hlx proximal to 20F3–20F4, whereas new deletion mapping data from our laboratory place the hybrid lethal of Coyne et al. (1998) distal to 20F (M. V. Cattani, unpublished results). We cannot, however, exclude the possibility that hlx and Zhr are the same locus. Zhrmel is thought to be an array of 359-bp repeats belonging to the 1.688 g/cm3 family of satellite DNA specific to D. melanogaster that is incompatible with a maternal factor(s) from its sibling species of the D. simulans clade. Zhrmel causes dominant embryonic lethality in F1 hybrid females from sibling species mothers and D. melanogaster fathers (Sawamura et al. 1993; Sawamura and Yamamoto 1993). Although hybrid males from D. simulans mothers do not normally inherit the X-linked Zhrmel, experimentally introducing Zhrmel kills hybrid males as well (Sawamura and Yamamoto 1997). The hybrid lethality of Zhrmel is thus embryonic, dominant, and independent of sex. These properties contrast with the hybrid lethality of hlxmau, which is postembryonic and either recessive or male specific (see results). The different properties of Zhr and hlx suggest that they are different loci or, at a minimum, functionally distinct alleles. In either case, the mapping of hlx to the pericentric heterochromatin is consistent with an emerging theme: hybrid incompatibilities often involve rapidly evolving heterochromatic elements (Sawamura and Yamamoto 1997; Fishman and Willis 2005) and genes whose products interact with heterochromatin (Barbash et al. 2003; Brideau et al. 2006). If this trend persists as more hybrid incompatibility factors are identified, it could signal that intrinsic postzygotic isolation typically evolves as a byproduct of genomic conflicts rather than ecology (e.g., Henikoff et al. 2001).
Conclusions:
We have identified an apparently simple X–autosome hybrid incompatibility in which the two Dobzhansky–Muller partners appear to be functionally derived in the D. mauritiana lineage. As hlx resides in the unmapped and poorly characterized pericentric heterochromatin of the D. mauritiana X, our immediate efforts will focus on the fine-scale mapping and identification of Su(hlx). Once Su(hlx) is identified, we will determine if hybrid lethality is caused by gene movement, by a protein–DNA incompatibility, or by protein–protein incompatibility. Population genetic analyses of Su(hlx) will then allow us to formally test if the relevant substitutions occurred in the D. mauritiana lineage and to determine the evolutionary forces causing its divergence.
Acknowledgments
We thank Christine Ling and Julienne Ng for technical assistance during the early stages of this project. We are grateful to H. A. Orr, C. Meiklejohn, J. P. Masly, A. Sweigart, P. Gerard, and two anonymous reviewers for helpful discussion and/or comments that improved the manuscript. This work was supported by an Ernst Caspari fellowship from the University of Rochester to M.V.C. and by funds to D.C.P. from the National Institutes of Health (R01-GM079543), the Alexander von Humboldt Foundation, the Radcliffe Institute for Advanced Study at Harvard University, and the University of Rochester.
References
- Barbash, D. A., D. F. Siino, A. M. Tarone and J. Roote, 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau, N. J., H. A. Flores, J. Wang, S. Maheshwari, X. Wang et al., 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 1292–1295. [DOI] [PubMed] [Google Scholar]
- Cabot, E. L., A. W. Davis, N. A. Johnson and C.I. Wu, 1994. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., S. Simeonidis and P. Rooney, 1998. Relative paucity of genes causing inviability in hybrids between Drosophila melanogaster and D. simulans. Genetics 150 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Dobzhansky, T. H., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Fishman, L., and J. H. Willis, 2005. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics 169 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, J. M., M. D. Dean and M. W. Nachman, 2008. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hey, J., and R. M. Kliman, 1993. Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Mol. Biol. Evol. 10 804–822. [DOI] [PubMed] [Google Scholar]
- Hilton, H., R. M. Kliman and J. Hey, 1994. Using hitchhiking genes to study adaptation and divergence during speciation within the Drosophila melanogaster complex. Evolution 48 1900–1913. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A., M. Turelli and G. M. Simmons, 1986. Unidirectional incompatibility between populations of Drosophila simulans. Evolution 40 692–701. [DOI] [PubMed] [Google Scholar]
- Hollocher, H., and C. I. Wu, 1996. The genetics of reproductive isolation in the Drosophila simulans clade: X vs. autosomal effects and male vs. female effects. Genetics 143 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., J. W. Carlson, C. Kennedy, D. Acevedo, M. Evans-Holm et al., 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter, P., J. Roote and M. Ashburner, 1990. A genetic basis for the inviability of hybrids between sibling species of Drosophila. Genetics 124 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 1993. DNA sequence variation at the period locus within and among species of the Drosophila melanogaster complex. Genetics 133 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, R. M., P. Andolfatto, J. A. Coyne, F. Depaulis, M. Kreitman et al., 2000. The population genetics of the origin and divergence of the Drosophila simulans species complex. Genetics 156 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A. R., and P. A. Roberts, 2000. Evolution of DNA in heterochromatin: the Drosophila melanogaster sibling species subgroup as a resource. Genetica 109 125–130. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2005. Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster. PLoS Genet. 1 429–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly, J. P., C. D. Jones, M. A. F. Noor, J. Locke and H. A. Orr, 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313 1448–1450. [DOI] [PubMed] [Google Scholar]
- Masly, J. P., and D. C. Presgraves, 2007. High resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott, S. R., and R. M. Kliman, 2008. Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus DNA sequence data. PLoS One 3 e2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola, O., Z. Trachtulec, C. Vlcek, J. C. Schimenti and J. Forejt, 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323 373–375. [DOI] [PubMed] [Google Scholar]
- Moyle, L. C., and E. B. Graham, 2005. Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics 169 355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle, L. C., and T. Nakazato, 2008. Comparative genetics of hybrid incompatibility: sterility in two Solanum species crosses. Genetics 179 1437–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by J. S. Huxley. Clarendon Press, Oxford.
- Muller, H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Naisbit, R. E, C. D. Jiggins, M. Linares, C. Salazar and J. Mallet, 2002. Hybrid sterility, Haldane's rule and speciation in Heliconius cydno and H. melpomene. Genetics 161 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveira, H., and A. Fontdevila, 1986. The evolutionary history of Drosophila buzzatii. XII. The genetic basis of sterility in hybrids between D. buzzatii and its sibling D. serido from Argentina. Genetics 114 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1996. Dobzhansky, Bateson and the genetics of speciation. Genetics 144 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and M. Turelli, 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55 1085–1094. [DOI] [PubMed] [Google Scholar]
- Pantazidis, A. C., V. K. Galanopoulos and E. Zouros, 1993. An autosomal factor from Drosophila arizonae restores normal spermatogenesis in Drosophila mojavensis males carrying D. arizonae Y chromosome. Genetics 134 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis, N., and H. A. Orr, 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 715–719. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., and W. Stephan, 2007. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol. Biol. Evol. 24 306–314. [DOI] [PubMed] [Google Scholar]
- Rosenberg, M. I., and S. M. Parkhurst, 2002. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109 447–458. [DOI] [PubMed] [Google Scholar]
- Roy, V., L. Monti-Dedieu, N. Chamindale, S. Siljak-Yakovlev, S. Aulard et al., 2005. Evolution of the chromosomal location of rDNA genes in two Drosophila species groups: ananassae and melanogaster. Heredity 94 388–395. [DOI] [PubMed] [Google Scholar]
- Sawamura, K, M.-T. Yamamoto and T. K. Watanabe, 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 133 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura, K, and M.-T. Yamamoto, 1993. Cytogenetical localization of Zygotic hybrid rescue (Zhr), a Drosophila melanogaster gene that rescues interspecific hybrids from embryonic lethality. Mol. Gen. Genet. 239 441–449. [DOI] [PubMed] [Google Scholar]
- Sawamura, K., A. Fujita, R. Yokoyama, T. Taira, Y. H. Inoue et al., 1995. Molecular and genetic dissection of a reproductive isolation gene, zygotic hybrid rescue, of Drosophila melanogaster. Jpn. J. Genet. 70 223–232. [DOI] [PubMed] [Google Scholar]
- Sawamura, K., and M-T. Yamamoto, 1997. Characterization of a reproductive isolation gene, zygotic hybrid rescue, of Drosophila melanogaster by using michrochromosomes. Heredity 79 97–103. [Google Scholar]
- Schlosser, G., and G. P. Wagner, 2008. A simple model of co-evolutionary dynamics caused by epistatic selection. J. Theor. Biol. 250 48–65. [DOI] [PubMed] [Google Scholar]
- Slotman, M., A. della Torre and J. R. Powell, 2004. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics 167 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart, A. L., L. Fishman and J. H. Willis, 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., and D. C. Presgraves, 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Z-B. Zeng, J. Li, D. L. Hartl and C. C. Laurie, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterile loci on the third chromosome. Genetics 164 1399–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B. G., T. Zerr, L. Comai and S. Henikoff, 2006. A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 1 2465–2477. [DOI] [PubMed] [Google Scholar]
- Ting, C.-T., S.-C. Tsaur, M.-L. Wu and C.-I. Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282 1501–1504. [DOI] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur and C.-I. Wu, 2000. The phylogeny of closely related species as revealed by the genealogy of a speciation gene, odysseus. Proc. Natl. Acad. Sci. USA 97 5313–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, J. R., B. S. Weir and C. C. Laurie, 1996. A genomic-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittbrodt, J. D., D. Adam, B. Malitschek, W. Maueler, F. Raulf et al., 1989. Novel putative receptor kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 341 415–421. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., 1998. Polytene chromosomes, heterochromatin and position effect variegation, pp. 1–40 in Advances in Genetics, Vol. 37, edited by J. C. Hall, J. C. Dunlap, T. Friedmann and F. Giannelli. Academic Press, San Diego. [DOI] [PubMed]