Abstract
Factor H-binding protein is a 27-kDa lipoprotein of Neisseria meningitidis discovered while screening the bacterial genome for vaccine candidates. In addition to being an important component of a vaccine against meningococcus in late stage of development, the protein is essential for pathogenesis because it allows the bacterium to survive and grow in human blood by binding the human complement factor H. We recently reported the solution structure of the C-terminal domain of factor H-binding protein, which contains the immunodominant epitopes. In the present study, we report the structure of the full-length molecule, determined by nuclear magnetic resonance spectroscopy. The protein is composed of two independent barrels connected by a short link. Mapping the residues recognized by monoclonal antibodies with bactericidal or factor H binding inhibition properties allowed us to predict the sites involved in the function of the protein. The structure therefore provides the basis for designing improved vaccine molecules.
Neisseria meningitidis, a Gram-negative bacterium that colonizes the upper respiratory tract of 10% of healthy human population, is adapted to grow only in humans. With a frequency of 1 in 100,000 population, the bacterium invades the bloodstream and becomes a severe pathogen, causing sepsis and meningitis. Vaccination with capsular polysaccharides induces serogroup-specific protective antibodies. Meningococcal capsular polysaccharide vaccines are available against serogroups A, C, Y, and W135 (1–3). On the contrary, the development of a vaccine against serogroup B, still responsible for a significant percentage of invasive diseases, has been protracted due to the immunologic cross-reactivity of B polysaccharide with human tissues. Recently, new perspectives to meningococcal B (menB) prevention have been opened by the identification of suitable protein-based vaccine antigens identified by mining the bacterial genome (4). One of the most promising antigens is the factor H-binding protein (fHbp).3 This is a membrane-anchored lipoprotein (3, 5, 6) that binds human factor H, a negative regulator of the alternative complement activation pathway.(7). Coating the bacterial surface with factor H allows the bacterium to mimic a human tissue and avoid complement-mediated lysis. fHbp is expressed by all the pathogenic strains of N. meningitidis and can be classified in three distinct sequence variants (5). This diversity has a remarkable impact on the immunological properties of fHbp given that members of each variant induce a strong protective immunity against meningococcal strains carrying homologous alleles but are ineffective against strains that express distantly related fHbp alleles.
A number of studies using monoclonal antibodies (8–11) have identified residues involved in protective epitopes and factor H binding. Initially, Arg-204 and the cluster Glu-146–Arg-149 were identified as targets of bactericidal monoclonal antibodies elicited by the recombinant fHbp of variant 1 (v.1) (8, 9). The same epitope was later shown to contain also Phe-277, Gly-228, Lys-230, and Glu-233 (10). Recently, Beernink et al. (11) used a panel of monoclonal antibodies obtained by immunizing mice with all three variants of fHbp and identified Gly-121 and Lys-122 as critical for binding by anti-v.1 antibodies. Ser-216 was shown to be important for variant 2 (v.2), whereas amino acid positions 174, 180, and 192 were shown to be key residues to discriminate between variant 2 and variant 3 (v.3).
Although the molecular distribution of residues from 141 to 255 was appreciable onto the structure of C terminus domain, whose solution structure was already solved (12), spatial arrangement of Gly-121 and Lys-122 remained so far undetermined. This incompleteness of information hampered a detailed evaluation of the molecular distribution of variant-specific epitopes, as well as the opportunity to rationalize the reported differences in the ability of monoclonals to induce complement-mediated killing and inhibit the protein binding to human factor H (11). In the present study, we determined the structure of the full-length fHbp by NMR, which improves the knowledge about the distribution of protective epitopes on the protein surface and provides useful indications on the possible localization of the factor H (fH) binding site.
EXPERIMENTAL PROCEDURES
Sample Production and NMR Measurements—Recombinant fHbp v.1 (residues Met-7–Gln-255) was expressed in Escherichia coli as already described. Analytical gel filtration analysis showed that the recombinant protein was eluted in fractions corresponding to a monomeric state of the molecule. Electrospray mass ionization-mass spectrometry spectrum indicated a mass of 27281.43 Da, which corresponded to the cloned construct. NMR spectra were acquired at 298 K on Avance 900, 700, and 500 MHz Bruker spectrometers, all equipped with a triple resonance cryoprobe. The NMR experiments, used for the backbone and the aliphatic side chain resonances assignment recorded on 2H/13C/15N, 13C/15N, and 15N enriched and on unlabeled fHbp samples, are summarized in supplemental Table S1. The 1H, 13C, and 15N resonance assignments of fHbp are reported in supplemental Table S2. All the amide protons of the fHbp protein were assigned, with the only exceptions of Val-8, His-26, Gly-202, and Gly-229. The assignment of the aromatic spin systems was performed with two-dimensional NOESY and total correlation spectroscopy maps acquired on the sample dialyzed against deuterated buffer. Backbone dihedral angle constraints were derived from 15N,13C′,13Cα,13Cβ, and Ha chemical shifts, using TALOS (13). Standard errors of values predicted by TALOS were used as allowed ranges of variations in the dihedral angle constraints.
Distance constraints for structure determination were obtained from 15N-edited and 13C-edited three-dimensional NOESY-HSQC. Residual dipolar couplings have been measured in the presence of an external orienting medium constituted by a binary mixture of C12E5 (penta-ethylene glycol dodecyl ether, > 98% purity, Fluka) and neat n-hexanol (puriss., Fluka), which form a stable liquid crystalline phase made of neutral aggregates (called bicelles) in the temperature range from 298 to 312 K (14). The molar ratio of C12E5 to n-hexanol was 0.96, and the C12E5/water ratio was 7% weight. One-bond 1H-15N coupling constants were measured at 298 K and 800 MHz by using the inphase anaphase (IPAP) method (15). A total of 100 RDC values have been measured from amide signals not overlapped in the HSQC spectrum. Out of them, 73 RDC values, derived from residues not experiencing R1 and/or R2 values outside the average values, were used for structure calculations.
2987 meaningful proton-proton distance restraints (supplemental Fig. S4), together with 158 φ and 158 ψ backbone dihedral angles restraints, were included in structure calculations. The exchangeability of the backbone amide hydrogens with solvent protons was investigated through an 1H-15N HSQC experiment performed on the protein previously extensively dialyzed against deuterated buffer. Hydrogen bond constraints for the slowly deuterium-exchanging amide protons of the β-strands were introduced at later steps of structure calculations.
Structure calculations were performed using the program CYANA-2.1 (16). 900 random conformers were annealed in 13,000 steps. The χ tensor parameters were obtained with FANTAORIENT, and they were optimized through iterative cycles of PSEUDOCYANA until convergence (17).
The family of the best 30 structures in terms of target function was then subjected to restrained energy minimization with the AMBER 10.0 package in explicit water solvent (18). The distance and torsion angle constraints and the RDCs were applied with force constants of 50 kcal mol–1Å–2 and 32 kcal mol–1 radians–2, respectively. The conformational and energetic analysis together with selected quality parameters from PROCHECK-NMR, WHATIF (19, 20) analysis, and QUEEN program (21) of the family of the best 30 structures are reported in supplemental Table S3. The program MOLMOL was subsequently used for structure analysis (22). The root mean square deviation values per residue of the restrained energy minimization (REM) family of 30 conformers for the segment 14–255 are reported in supplemental Fig. S5.
15N R1, R2, and steady-state heteronuclear NOEs were measured with pulse sequences as described by Farrow et al. (23) R2 values were measured using a refocusing time of 450 μs. In all experiments, the water signal was suppressed with the “water flip-back” scheme. Average R1, R2, and 1H-15N NOE values of 0.91 ± 0.04 s–1, 24.2 ± 0.2 s–1, and 0.70 ± 0.03 are found, respectively, at 500 MHz. The experimental relaxation rates were used to map the spectral density function values, J(ωH), J(ωN), J(0) following a procedure available in literature (24, 25).
RESULTS
The far-UV CD spectrum of fHbp had features characteristic of a folded protein with a high content of β-strands, whereas the negative band at 198 nm, characteristic of random coil conformation, was not present (data not shown). Consistently, the 1H-15N HSQC spectra show well dispersed resonances indicative of an overall well folded protein (supplemental Fig. S1).
Heteronuclear relaxation rates, whose values are affected by the dynamic properties of the molecule, were essentially homogeneous along the entire polypeptide sequence, with the exception of residues located at the C and N termini and some loops. The correlation time for the molecule tumbling (τc), as estimated from the R2/R1 ratio, is 20.3 ± 1.5 ns, consistent with the molecular weight of the protein being in the monomeric state.
The solution structure of the full-length fHbp consisted of two domains, fHbpN (residues 8–136) and fHbpC (residues 141–255), composed of 10 and 8 antiparallel β-strands, respectively. fHbpN and fHbpC were connected by a five-residue linker and had a topology of secondary structure elements as shown in Fig. 1A, with an overall three-dimensional structure as shown in Fig, 1B. Both domains are structurally well defined, with only few loops showing some conformational disorder (supplemental Fig. S2). From heteronuclear relaxation measurements (supplemental Fig. S3), it appears that most of these loop regions, and in particular, those including residues 85–90 and 118–123 located in the N-terminal domain, have significantly lower heteronuclear NOE values as a consequence of local motions in the ns–ps time scale. Consistently, they were characterized by a very low number of long range 1H-1H NOEs (supplemental Fig. S4), and consequently, their conformation was less defined (supplemental Fig. S2).
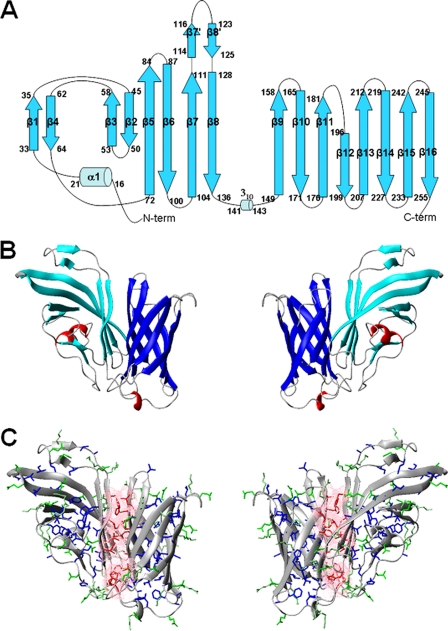
FIGURE 1.
A, topology diagram of the fHbp protein. The α-helices are represented by sky blue cylinders, and the β-strands are cyan arrows. N-term, N terminus; C-term, C terminus. B, ribbon diagram of fHbp. Secondary structure elements are shown. β-strands of the N-terminal domain are shown in cyan and helices are shown in red, whereas β-strands of fHbpC are shown in blue. C, the side chains of the hydrophobic residues involved in interdomains contacts are shown as sticks in red, and pink. Contact surfaces are also reported. The other hydrophobic residues are shown in blue. The charged residues are shown in green. The backbone is shown as a ribbon.
The fHbpN domain has an elongated barrel-like structure with a length of about 49 Å and a width of 22 Å. It is organized in six antiparallel β-strands of different lengths (β2(45–50), β3(53–58), β5(72–84), β6(87–100), β7(104–111), β8(128–136)), forming an extended β-sheet, and two short β-strands (β1(33–35) and β4(62–64)), which face the β-sheet and a short N-terminal α-helix (16–21). In 50% of the family conformers, a short β-hairpin, including residues 114–116 (β7′) and 123–125 (β8′), is observed between β7 and β8 (Fig. 1A), whose presence is supported by the chemical shift index analysis.
The core of fHbpN is characterized by several hydrophobic interactions that involve aromatic and aliphatic residues. In particular, hydrophobic contacts are present between the residues clustered on one side of the β-sheet (Leu-46, Leu-48 (β2), Ala-53 and Tyr-57 (β3), Phe-76, Phe-78 (β5), Phe-96 and Val-98 (β6), Ala-105 and Phe-109 (β7), Ala-135(β8) and those on the two other β-strands β1 (Leu-34 and Leu-36) and β4 (Leu-63), determining the spatial proximity of the two regions and the overall shape of the N-terminal domain. Further long range interactions between residues Leu-34, Thr-35, and Asp-133 and Ala-135 maintainβ1 close toβ8, stabilizing the overall structure. The three-dimensional arrangement of the β-strands defines a hydrophobic core, which is closed on one side by helix α1. Contacts between some residues located in β3, β4, and β5 and those in helix α1 are present that anchor such a helix to the rest of the protein. The latter interactions define the conformation of a long loop (segment 22–32), which is completely solvent-exposed and rich in charged residues. The latter loop, although stabilized by the contacts of two hydrophobic residues (Leu-24 and Leu-31) with the rest of the protein, experiences high conformational disorder. Also, the region between β5 and β6, opposite to the N-terminal helix, is solvent-exposed despite the fact that it is rich in hydrophobic residues (Fig. 1C).
The C-terminal domain fHbpC is constituted by an eight-stranded antiparallel β-barrel (141–255), whose strands are connected by loops of variable lengths. The same arrangement was previously found in the solution structure of the same domain in the isolated fHbpC domain (12). The 310-helix present in the solution structure of isolated fHbpC domainis was still present in the majority of 30 of the full-length conformers, although shorter (141–143) with respect to that observed the isolated fHbpC (138–141). Sizable variations for the backbone NH chemical shifts of fHbpC domain in the two constructs (>0.25 ppm as combined chemical shifts) were observed on β7, β8, β9, β14, β15, and β16 (supplemental Fig. S6). The largest differences were observed for residues located in β7, β8, which formed a long disordered tail in the isolated fHbpC domain. The other four β-strands of the C-terminal domain are located on the one side of the β-barrel and define the domain-domain interface. Interestingly, residues located in β9, β14, β15, and β16 formed a hydrophobic patch in fHbpC molecule. The present structure confirms our previous insights on the role of this region of the β-barrel (12).
The domain-domain interface, formed by about 850 Å2 of buried surface, is stabilized by intermolecular contacts involving exclusively hydrophobic amino acids (Leu-106, Tyr-99, Phe-129, Ile-134, N-terminal domain; Thr-155, Phe-157, Phe-227, Leu-251, C-terminal domain, Fig. 1C). About 70 long range interdomain NOEs were found that defined, together with the RDCs, the reciprocal orientation of the domains.
DISCUSSION
Human complement is the first barrier of innate immunity and kills infectious agents when they try to invade the bloodstream (26). To do this, it attacks anything that is not self. Factor H and C4b are complement components that allow the system to distinguish between invasive agents and self-tissues by coating the latter and avoiding the complement attack. During the evolutionary battle for survival, virtually all pathogens have found multiple and redundant ways to escape the complement-mediated killing (27). One of the most popular strategies among pathogens is to avoid complement by mimicking host surfaces, covering themselves with factor H. fHbp of N. meningitidis is a typical example of this class of molecules. By binding human factor H, this protein allows the bacterium to survive and grow in human blood and cause a terrible disease. This property, together with the ability to induce in humans a strong antibody response, makes this protein an attractive vaccine antigen able to induce antibodies with a double function: killing the bacterium by direct activation of the classical complement cascade and preventing the formation of fH-fHbp complex on the surface of the bacterium.
Based on the knowledge of the complete structure of fHbp, we can identify the location of the variable amino acids (Fig. 2A), the residues involved in binding protective monoclonal antibodies (Fig. 2B), the region bound by antibodies against variant 1 (Fig. 2C), the region bound by antibodies against variant 2 (Fig. 2D), and the residues recognized by the antibodies that inhibit factor H binding (Fig. 2E). The variable residues (Fig. 2A) cluster in the upper part of the molecule. This is consistent with a model where the protein is anchored to the bacterial cell wall through the palmitic acid extension of Cys-1 (5) and exposes the upper part to the outside, where it is under the selective pressure of the immune system. Accordingly, the amino acids known to be part of epitopes (8, 9, 10, 11) are all localized in correspondence of the zone of higher variability (Fig. 2B). Arg-204, Gly-121, and the loop formed by Glu-146 until Arg-149 were identified as involved in the formation of bactericidal v.1 epitopes (8, 9, 11). Arg-204 is located in the loop between β12 and β13, whereas the segment 146–149 corresponds to the region connecting the two fHbp domains and containing the second short helix α2. Gly-121, which was localized in the disordered region of the isolated fHbpC domain, now occupies the loop between β7′ and β8′ strands. NMR-driven epitope mapping carried out on the isolated fHbpC domain identified Phe-141, Lys-199, Arg-204, Glu-146–Arg-149, Phe-227, Lys-230, and Glu-233 as part of the same epitope recognized by the bactericidal monoclonal antibody Mab502 elicited by fFbp v.1 (10). The distribution of these residues on the full-length protein supports the hypothesis that the fHbpC domain contains the major part of the native epitope, which consists of all those residues still accessible in the whole fHbp with the exception of Phe-227 and Glu-233, which are now shielded by the presence of the N-terminal domain. Epitopes of v.2 and v.3 have recently been shown to include residues in positions 174, 216, 180, and 192 (11). Although none of the corresponding monoclonal antibodies were bactericidal individually, nevertheless some pairs of them were able to perform meningococcal killing when combined. The same was true for a pair of monoclonal antibodies raised against v.1 (11). We tried to identify the fHbp regions targeted by these pairs of bactericidal monoclonal antibodies, placing residues recognized by each component at the center of a molecular area of 950 A2, a value chosen as the representative value of the average size for a protein conformational epitope (28–31). The interesting observation that the bactericidal epitopes crucial for the killing of v.1 and v.2 are only partially overlapping suggests that it may be possible to engineer molecules containing epitopes of all variant proteins (Fig. 2, C and D). Finally, Fig. 2E shows that the residues recognized by the monoclonal antibodies that inhibit factor H binding (11) are located In the upper part of both in the N-terminal and in the C-terminal part of the molecule, suggesting that this region is involved in the interaction with factor H.
FIGURE 2.
A, variability profile of the three fHbp variants. Conserved amino acids are colored in blue, conservative substitutions are colored in gray, and variable amino acids are colored in green. B, distribution of amino acids recognized by monoclonal antibodies (Mabs) raised against v.1 (purple) and v.2/v.3 (green). C and D, molecular areas recognized by the bactericidal pairs of monoclonal antibodies against v.1 (orange) and v.2 (gold) (11). E, molecular distribution of fHbp residues recognized by monoclonal antibodies inhibiting (red), partially inhibiting (orange), and not inhibiting (green) the fH binding (11).
In conclusion, the complete structure of fHpb allowed us to map residues involved in important functions, such as binding to factor H and eliciting bactericidal antibodies, and provides the basis to understand the molecular mechanisms behind the function of this important molecule. The structure may be used to design improved vaccine antigens.
Supplementary Material
This work was supported by Ministero dell'Istruzione, Università e della Ricerca Scientifica-Fondo per gli Investimenti della Ricerca di Base (MIURFIRB) Grant RBLA032ZM7.
The atomic coordinates and structure factors (code 2KC0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains seven supplemental figures three supplemental tables, and supplemental references.
Footnotes
The abbreviations used are: fHbp, factor H-binding protein; RDC, residual dipolar coupling; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser effect spectroscopy; HSQC, heteronuclear single quantum correlation; v.1, variant 1; v.2, variant 2; v.3, variant 3.
References
- 1.Borrow, R., Andrews, N., Goldblatt, D., and Miller, E. (2001) Infect. Immun. 691568–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girard, M. P., Preziosi, M. P., Aguado, M. T., and Kieny, M. P. (2006) Vaccine 244692–4700 [DOI] [PubMed] [Google Scholar]
- 3.Snape, M. D., Perrett, K. P., Ford, K. J., John, T. M., Pace, D., Yu, L.-M., Langley, J. M., McNeil, S., Dull, P. M., Ceddia, F., Anemona, A., Halperin, S. A., Dobson, S., and Pollard, A. J. (2008) J. Am. Med. Assoc. 299173–184 [DOI] [PubMed] [Google Scholar]
- 4.Giuliani, M. M., Adu-Bobie, J., Comanducci, M., Aricò, B., Savino, S., Santini, L., Brunelli, B., Bambini, S., Biolchi, A., Capecchi, B., Cartocci, E., Ciucchi, L., Di Marcello, F., Ferlicca, F., Galli, B., Luzzi, E., Masignani, V., Serruto, D., Veggi, D., Contorni, M., Morandi, M., Bartalesi, A., Cinotti, V., Mannucci, D., Titta, F., Ovidi, E., Welsch, J. A., Granoff, D., Rappuoli, R., and Pizza, M. (2006) Proc. Natl. Acad. Sci. 10310834–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masignani, V., Comanducci, M., Giuliani, M. M., Bambini, S., Adu-Bobie, J., Aricò, B., Brunelli, B., Pieri, A., Santini, L., Savino, S., Serruto, D., Litt, D., Kroll, S., Welsch, J. A., Granoff, D. M., Rappuoli, R., and Pizza, M. (2003) J. Exp. Med. 197789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pizza, M., Scarlato, V., Masignani, V., Giuliani, M. M., Aricò, B., Comanducci, M., Jennings, G. T., Baldi, L., Bartolini, E., Capecchi, B., Galeotti, C. L., Luzzi, E., Manetti, R., Marchetti, E., Mora, M., Nuti, S., Ratti, G., Santini, L., Savino, S., Scarselli, M., Storni, E., Zuo, P., Broeker, M., Hundt, E., Knapp, B., Blair, E., Mason, T., Tettelin, H. e., Hood, D. W., Jeffries, A. C., Saunders, N. J., Granoff, D. M., Venter, J. C., Moxon, E. R., Grandi, G., and Rappuoli, R. (2000) Science 2871816–1820 [DOI] [PubMed] [Google Scholar]
- 7.Madico, G., Welsch, J. A., Lewis, L. A., McNaughton, A., Perlman, D. H., Costello, C. E., Ngampasutadol, J., Vogel, U., Granoff, D. M., and Ram, S. (2006) J. Immunol. 177501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsch, J. A., Rossi, R., Comanducci, M., and Granoff, D. M. (2004) J. Immunol. 1725606–5615 [DOI] [PubMed] [Google Scholar]
- 9.Giuliani, M. M., Santini, L., Brunelli, B., Biolchi, A., Arico, B., Di Marcello, F., Cartocci, E., Comanducci, M., Masignani, V., Lozzi, L., Savino, S., Scarselli, M., Rappuoli, R., and Pizza, M. (2005) Infect. Immun. 731151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scarselli, M., Cantini, F., Santini, L., Veggi, D., Dragonetti, S., Donati, C., Savino, S., Giuliani, M. M., Comanducci, M., Di Marcello, F., Romagnoli, G., Pizza, M., Banci, L., and Rappuoli, R. (2008) J. Mol. Biol. 38697–108 [DOI] [PubMed] [Google Scholar]
- 11.Beernink, P. T., Welsch, J. A., Bar-Lev, M., Koeberling, O., Comanducci, M., and Granoff, D. M. (2008) Infect. Immun. 764232–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantini, F., Savino, S., Scarselli, M., Masignani, V., Pizza, M., Romagnoli, G., Swennen, E., Veggi, D., Banci, L., and Rappuoli, R. (2006) J. Biol. Chem. 2817220–7227 [DOI] [PubMed] [Google Scholar]
- 13.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13289–302 [DOI] [PubMed] [Google Scholar]
- 14.Ruckert, M., and Otting, G. (2000) J. Am. Chem. Soc. 1227793–7797 [Google Scholar]
- 15.Ottiger, M., Delaglio, F., and Bax, A. (1998) J. Magn. Reson. 131373–378 [DOI] [PubMed] [Google Scholar]
- 16.Guntert, P. (2000) Methods Mol. Biol. 278353–378 [DOI] [PubMed] [Google Scholar]
- 17.Banci, L., Bertini, I., Huber, J. G., Luchinat, C., and Rosato, A. (1998) J. Am. Chem. Soc. 12012903–12909 [Google Scholar]
- 18.Perlman, D. A., Case, Caldwell, J. W., Ross, W. S., Cheatham, T. C., DeBolt, S. E., Ferguson, D. M., Seibel, G. L., and Kollman, P. A. (1995) Comput. Phys. Commun. 911–41 [Google Scholar]
- 19.Laskowski, R., Rullmann, J., McArthur, M., Kaptein, R., and Thornton, J. (1996) J. Biomol. NMR 8477–486 [DOI] [PubMed] [Google Scholar]
- 20.Vriend, G. (1990) J. Mol. Graph. 852–56 [DOI] [PubMed] [Google Scholar]
- 21.Nabuurs, S., Spronk, C., Krieger, E., Maassen, H., Vriend, G., and Vuister, G. W. (2003) J. Am. Chem. Soc. 12512026–12034 [DOI] [PubMed] [Google Scholar]
- 22.Koradi, R., Billeter, M., and Wüthrich, K. (1996) 1429–32 [DOI] [PubMed]
- 23.Farrow, N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., Forman-Kay, J. D., and Kay, L. E. (1994) Biochemistry 335984–6003 [DOI] [PubMed] [Google Scholar]
- 24.Grzesiek, S., and Bax, A. (1993) J. Biomol. NMR 3185–204 [DOI] [PubMed] [Google Scholar]
- 25.Peng, J. (1992) J. Magn. Reson. 98308–332 [Google Scholar]
- 26.Rittirsch, D., Flierl, M. A., and Ward, P. A. (2008) Nat. Rev. Immunol. 8776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambris, J. D., Ricklin, D., and Geisbrecht, B. V. (2008) Nat. Rev. Microbiol. 6132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies, D. R., Padlan, E. A., and Sheriff, S. (1990) Annu. Rev. Biochem. 59439–473 [DOI] [PubMed] [Google Scholar]
- 29.Farady, C. J., Egea, P. F., Schneider, E. L., Darragh, M. R., and Craik, C. S. (2008) J. Mol. Biol. 380351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lok, S. M., Kostyuchenko, V., Nybakken, G. E., Holdaway, H. A., Battisti, A. J., Sukupolvi-Petty, S., Sedlak, D., Fremont, D. H., Chipman, P. R., Roehrig, J. T., Diamond, M. S., Kuhn, R. J., and Rossmann, M. G. (2008) Nat. Struct. Mol. Biol. 15312–317 [DOI] [PubMed] [Google Scholar]
- 31.Ménez, R., Michel, S., Muller, B. H., Bossus, M., Ducancel, F., Jolivet-Reynaud, C., and Stura, E. A. (2008) J. Mol. Biol. 3761021–1033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.