Abstract
To better understand the mechanism of steps in early transcription by RNA polymerase II (pol II), we investigated the molecular determinants of transcript slipping within complexes assembled on promoters containing a pre-melted transcription bubble from –9 to +3. Transcript slippage occurs when an RNA transcript contains a repetitive sequence that allows the transcript to slip back and pair with the template strand of the DNA at a new register before transcription continues. We established the contributions of individual transcription factors, DNA elements, and RNA length to slipping on a heteroduplex template using a highly purified human pol II transcription system. We found that transcripts slip at a very defined point in the transcription reaction, after pol II completes phosphodiester bond synthesis at register +5. This point is set by the position of the polymerase active site on the DNA template, as opposed to the length of the transcript, as well as by a repetitive CUCU sequence that must occur from +2 to +5. Interestingly, slipping at this juncture is induced by TATA-binding protein and transcription factor IIB and requires a TATA box but not a transcription factor IIB recognition sequence. We propose a model in which transcribing complexes, upon completing phosphodiester bond synthesis at register +5, enter one of two branches in which they either complete productive synthesis of the transcript or undergo multiple rounds of transcript slipping.
Transcription of mRNA by pol3 II is a complex process that involves the highly regulated interplay of promoter DNA, many protein factors, and the transcript RNA itself. The promoter DNA influences the level of transcription and the position at which RNA synthesis begins through sequences in the regulatory and core promoter elements (1, 2). In addition, the conformation of promoter DNA can regulate the level of transcription, both in the presence and absence of histones (3–7). The general transcription factors, including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, are required for pol II to transcribe in a specific fashion in most, if not all, genes (8). Regulated gene-specific transcription in cells requires a plethora of additional factors, including activators, repressors, co-regulators, and chromatin-modifying factors (2, 6, 8, 9). In addition to the promoter and protein factors, biochemical studies have shown that the RNA transcript is not simply the product of the reaction, but it also stabilizes elongation complexes through the RNA: DNA hybrid and influences transformations that occur during early transcription (3, 10, 11). Although the core factors required for pol II to synthesize a transcript in vitro are understood, structural and biochemical studies are still uncovering new information about the mechanism by which this process occurs and is regulated.
Biochemical studies have shown that the transcription reaction is composed of a series of steps that minimally include the following: preinitiation complex formation, open complex formation, initiation, escape commitment, promoter escape, elongation, termination, and reinitiation. During preinitiation complex formation, pol II and the general transcription factors assemble at the promoter (8). Before transcription initiates, the DNA around the start site melts from approximately –9 to +3 with the help of the ATP-dependent helicase activity of TFIIH to form open complexes that contain a transcription bubble (4, 12). In vitro, transcription can occur in the absence of TFIIE and TFIIH from negatively supercoiled DNA (13, 14), as well as from heteroduplex templates containing mismatches from –9 to +3 that create a bubble (15). When provided with nucleotides, open complexes initiate transcription and synthesize 2–3-nt RNA transcripts, typically in an abortive fashion (13, 16, 17). Initiation complexes commit to proceed forward through the reaction upon addition of the fourth nucleotide during a step termed escape commitment (11, 18). Complexes then proceed through promoter escape, during which numerous structural re-arrangements occur en route to formation of stable elongation complexes (19).
Biochemical studies have provided insight into many of the transformations that occur during promoter escape, including changes in the transcription bubble, transcript slippage, pausing of ternary complexes, release of TFIIB from the promoter, and formation of a stable RNA:DNA hybrid (4, 19–23). Kinetic studies have identified the rate-limiting step in transcription in vitro as occurring during promoter escape (3, 24). Experiments monitoring the melted region of DNA during promoter escape have shown that the upstream edge of the transcription bubble initially remains anchored while the downstream edge extends, thereby increasing the size of the bubble (4, 22). After the bubble extends to 18 melted base pairs and synthesis of minimally a 7-nt RNA occurs, the upstream edge of the melted region re-anneals in a process termed bubble collapse; when this occurs the stability of transcribing complexes increases (22). During promoter escape, transcribing complexes can undergo transcript slippage (20, 21). Transcript slippage can occur when the RNA transcript contains a repetitive sequence, such as the CUCUCU observed in RNAs transcribed from the adenovirus major late promoter (AdMLP). The repeat sequence allows the transcript to slip back by 2 bases and re-pair with the template strand at a different register before transcription continues, resulting in transcripts that are minimally 2 nt longer than expected. Additional rounds of slipping and extension can occur, resulting in ladders of products that increase in size by increments of 2 nt. Transcript slippage during promoter escape has been shown to be a sensor for ternary complex stability; once complexes complete the structural transformations that occur en route to becoming stable, processive complexes, then the ability of the transcript to slip is greatly reduced (21).
Here we investigated the properties of transcript slippage in complexes assembled on AdMLP DNA containing a pre-melted transcription bubble from –9 to +3. Complexes begin slipping at a very defined point in the transcription reaction, after pol II completes phosphodiester bond synthesis at register +5. The position at which transcript slippage occurs is set by the position of the polymerase active site on the DNA template, as opposed to the length of the RNA transcript. Slipping at this juncture in the reaction is induced by TBP and TFIIB; it does not occur with pol II itself. Slipping also requires a TATA box, but it does not depend on a BRE. We propose that upon completing phosphodiester bond synthesis at register +5, transcribing complexes either complete productive synthesis of the transcript or undergo multiple rounds of transcript slipping.
EXPERIMENTAL PROCEDURES
Preparation of Transcription Factors and Templates—Recombinant human TBP, TFIIB, TFIIF, and native human pol II were prepared as described previously (24, 25). The wild-type bubble template contained the AdMLP core promoter sequence from –40 to +20, with the sequence of the nontemplate strand changed to match that of the template strand from –9 to +3 (see Fig. 1A). The template and non-template strand oligonucleotides (Invitrogen) were annealed in buffer containing 2 mm Tris (pH 7.9), 0.2 mm EDTA, and 250 mm KCl by heating to 95 °C for 3 min and then cooling to 25 °C at a rate of 0.1 °C/s using a thermocycler. The G-oligo (Fig. 3A) was ordered from Invitrogen.
FIGURE 1.
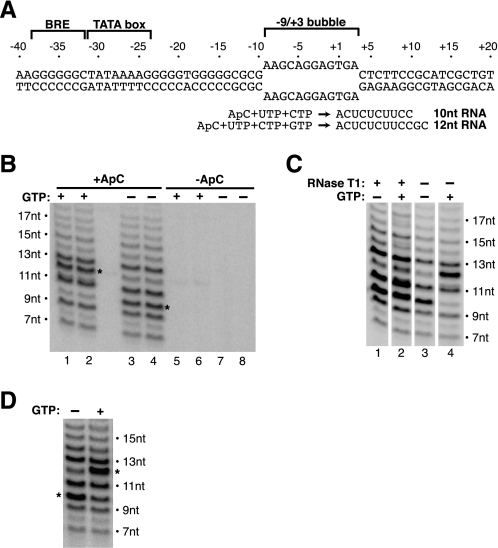
Transcript slippage occurs on a bubble template. A, schematic of the bubble template used in experiments, which contains the AdMLP from –40 to +20 with a heteroduplex from –9 to +3 to create the transcription bubble. The BRE and TATA box are indicated. B, slipped products do not incorporate GTP, and are all initiated with ApC. Transcription reactions were performed in the presence and absence of GTP and ApC as indicated. The expected length products (10 or 12 nt) are denoted with asterisks. C, only the position of the non-slipped transcript changes after treatment with RNase T1. Transcription proceeded for 20 min in the presence or absence of GTP as indicated. Reactions were either stopped or treated with RNase T1 for an additional 10 min. Transcription continued during the RNase treatment, and therefore the bands in lanes 1 and 2 are darker than those in lanes 3 and 4. D, transcripts that slip cannot be elongated by the addition of GTP. Reactions were performed using pulse-chase labeling as described in the text.
FIGURE 3.
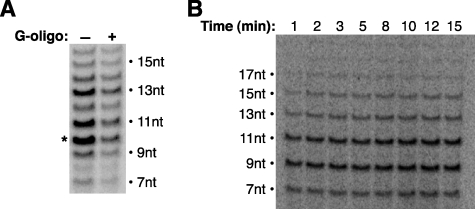
Transcript slippage occurs after register +4 and prior to +9. A, DNA oligonucleotide (G-oligo) that inhibits the step of escape commitment represses both transcript slippage and synthesis of the productive transcript. Reactions were performed on the wild-type template (+11G) in the absence of GTP; the expected 10-nt product is indicated with an asterisk. When included, the G-oligo (1 μm) was added to reactions just prior to addition of the nucleotides. B, transcript slippage occurs prior to the rate-limiting step of productive transcription. Reactions were performed on the +6G template with pulse-chase labeling, and transcription was stopped at the time points indicated.
In Vitro Transcription Assays—Reactions were 20 μl and performed in a buffer containing 10% glycerol, 10 mm Tris (pH 7.9), 10 mm Hepes (pH 7.9), 50 mm KCl, 4 mm MgCl2, 1 mm dithiothreitol, 50 μg/ml bovine serum albumin, and 7 units of RNase Out (Invitrogen). Transcription factors and template DNA were used at the following final concentrations: 2 nm TBP, 10 nm TFIIB, 4 nm TFIIF, 1–3 nm pol II, and 1–2 nm DNA template and were incubated at 30 °C for 20 min. Transcription was initiated by addition of 500 μm ApC (Sigma), 100 μm UTP, and 25 μm [α-32P]CTP (5 μCi). When included GTP was added at 100 μm. For pulse-chase reactions, a pulse NTP solution was added for 15 s, resulting in final concentrations of 500 μm ApC, 100 μm UTP, and 1 μm [α-32P]CTP (5 μCi). A chase NTP solution was then added that increased the CTP concentration from 0.1 to 1 mm, and in some experiments included 100 μm GTP. After 20 min of transcription, reactions were stopped using 3.1 m ammonium acetate, 20 mm EDTA, 10 μg of carrier yeast RNA, and 15 μg of proteinase K. Transcripts were ethanol-precipitated, resolved by 20% denaturing PAGE, and visualized using phosphorimagery.
RESULTS
Transcript Slippage on a Heteroduplex Template Exhibits Unique Characteristics—To investigate the mechanism of early transcription by pol II, we utilized a highly purified human transcription system containing TBP, TFIIB, TFIIF, pol II, and the AdMLP. To observe in vitro transcription using this minimal set of factors that lacked TFIIE and TFIIH, we took advantage of a linear AdMLP template containing mismatched sequence from –9 to +3, thereby creating a heteroduplex that mimicked the transcription bubble (Fig. 1A). Using the initiating dinucleotide ApC, along with CTP and UTP, a 10-nt RNA was synthesized from this template. Addition of GTP extended the product to 12 nt. When we performed transcription reactions with this bubble template, we observed a pattern of several products as opposed to only the expected products of 10 and 12 nt (Fig. 1B). All of the products initiated at the start site because bands virtually disappeared in control reactions performed in the absence of ApC (Fig. 1B, compare lanes 5–8 with lanes 1–4). The pattern of bands we observed was reminiscent of that documented by Pal and Luse (20, 21) when they first reported that pol II can undergo transcript slippage during early transcription. In addition to the expected products of 10 and 12 nt, we observed predominant bands spaced by 2 nt (e.g. 7, 9, 11, and 13 nt), leading us to conclude that these products arose from transcript slippage.
There are, however, differences between the transcript slippage we observed and that described previously. First, we observed products shorter than the expected product (7 and 9 nt, and 11 nt in the presence of GTP). Second, in the presence of GTP, the sizes of the slipped products did not change, but the size of the expected product shifted from 10 to 12 nt (Fig. 1B, compare lanes 1 and 2 with lanes 3 and 4). This result indicated that the slipped transcripts never reached register +11 where the first G could be incorporated. To further probe this, we treated reactions with RNase T1, which cleaves 3′ of single-stranded G residues. The RNase treatment caused no change in the ladder of slipped products, but it did shorten the expected product synthesized in the presence of GTP from 12 to 10 nt, also indicating that the slipped RNAs do not contain G residues (Fig. 1C).
Observing that the slipped products do not incorporate GTP suggested that the complexes undergoing transcript slippage were not capable of proceeding through promoter escape to reach register +11. To test this directly, we performed a pulse-chase experiment and monitored the fate of transcripts produced within the first 15 s after addition of nucleotides to preinitiation complexes. Reactions were pulsed for 15 s with [α-32P]CTP, UTP, and ApC, and then chased with a large excess of unlabeled CTP for 10 min. GTP was then added to determine the extent to which slipped RNAs could be extended by two nucleotides. As shown in Fig. 1D, the pattern and intensity of slipped products did not change upon addition of GTP; however, the expected product shifted from 10 to 12 nt. This indicated that the slipped products were either aborted or were part of ternary complexes not capable of resuming productive transcription once slipping had begun.
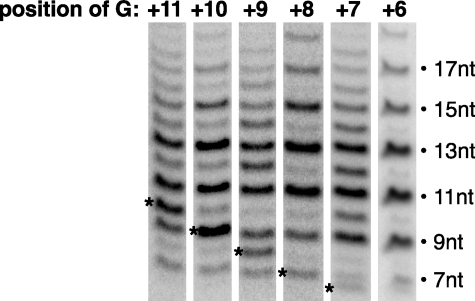
Transcript Slippage on a Bubble Template Occurs after Phosphodiester Bond Synthesis at Register +5—To define the first point in the reaction at which transcript slippage could occur, we created a series of bubble templates in which the first G/C (nontemplate/template strand) was placed at every position from +11 (wild type) back to +6, which is the first position a G can be placed while still maintaining a repetitive CUCU sequence. Omitting GTP from reactions allowed us to pause transcribing complexes at specific positions during early transcription and ask whether or not slipping occurred. In the absence of GTP, we observed slipped products from each template (Fig. 2). The expected length of the non-slipped (productive) products shifted with different templates in accordance with the position of the first G. By contrast, all templates showed the same pattern of slipped products as follows: transcripts spaced 2 nt apart beginning with 7 nt, with the predominant slipped transcripts being 9, 11, and 13 nt. This suggested that on the AdMLP bubble template, slipping first occurred at register +5 after synthesis of ACUCU. At this point the transcript can slip back, with the 3′ end moving from register +5 to register +3, re-pair with the DNA, and be extended to 7 nt, after which the cycle of slipping can occur again. The predominant products of 9, 11, and 13 nt that we observed resulted from 2, 3, and 4 rounds of slipping and extension. Less prominent products of 15, 17, and 19 nt were also observed, indicating that additional rounds of slipping occurred. We were not able to see the 5-nt RNA produced from paused complexes on the +6G template, most likely because all RNA produced on this template underwent transcript slippage.
FIGURE 2.
Transcript slippage on a bubble template occurs after bond synthesis at register +5. The pattern of slipped products does not change when transcription is paused at six different points during early transcription by GTP omission. The position of the G on the nontemplate strand of each promoter is indicated. The expected products are indicated by asterisks.
To further probe the position in the transcription reaction at which slipping on a bubble template occurs, we took advantage of an inhibitor we previously found to block a very specific step in early transcription. The inhibitor, a DNA oligonucleotide consisting of 20 guanosine residues (G-oligo), blocks a step in transcription termed escape commitment that occurs during translocation of the pol II active site after synthesis of a 4-nt RNA (11). We reasoned that if slipping occurs after phosphodiester bond synthesis at register +5, then an inhibitor that blocks transcription prior to this point should inhibit transcript slippage as well as synthesis of the productive transcript. When we added the G-oligo to transcription reactions, both the expected 10-nt product and the slipped products were inhibited (Fig. 3A). This shows that slipping occurs after synthesis of a 4-nt RNA. Moreover, because slipping was inhibited by the G-oligo, we can conclude that the slipped products arise from complexes that proceed through the normal escape commitment step in the transcription reaction.
Our previous kinetic studies found that the rate-limiting step in early transcription occurs after synthesis of an 8-nt RNA as the polymerase active site translocates to the ninth register (3). Our studies here predict that transcribing complexes begin slipping prior to the rate-limiting step. Although slipped RNAs reach lengths beyond 8 nt, it was not known whether complexes undergoing slipping would proceed through the typical slow transformation. To address these issues, we measured the rate at which slipped transcripts accumulate on the +6G template using pulse-chase conditions to limit detectable transcription to a single round. We found that the pattern and intensity of slipped products did not change over time (Fig. 3B). This shows that complexes begin slipping prior to the rate-limiting step during promoter escape, that slipping itself (be it one or multiple rounds) is quite rapid, and that complexes undergoing slipping do not proceed through the slow transition that typically occurs after synthesis of an 8-nt RNA, despite the lengths of many slipped products being longer than 8 nt.
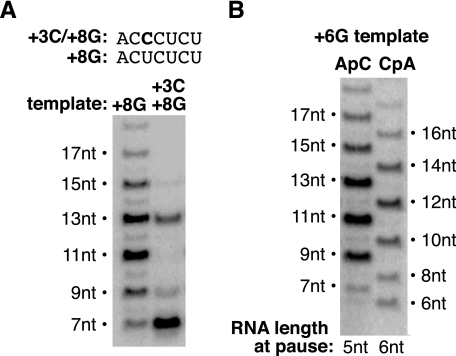
The Point at Which Transcript Slippage Occurs Is Set by the Repeat Sequence from +2 to +5 and the Position of the Polymerase Active Site on the DNA—We next asked whether slipping required that the CUCU repeat sequence that allows slipping to occur be located from +2 to +5. In the context of the +8G template we changed the sequence at +3 from T/T to G/G (nontemplate/template strand); therefore, the RNA transcribed in the absence of GTP would have the sequence ACCCUCU (see the sequences at the top of Fig. 4A). Therefore, the only CUCU repeat sequence is present at +4 to +7. As shown in Fig. 4A, transcript slippage did not occur on this template. This indicates that it is not simply the presence of a CUCU repeat in the RNA that induces transcript slippage on a bubble template. Rather, the CUCU repeat needs to occur at a specific register during early transcription, namely from +2 to +5, for slipping to occur.
FIGURE 4.
Position at which complexes begin to slip is influenced by DNA sequence and the position of the pol II active site on the DNA. A, CUCU repeat from +2 to +5 is required to observe transcript slippage. The sequences of the expected 7-nt products produced from the +3C/+8G and +8G templates are indicated. B, initiating transcription at –1 rather than +1 changes the sizes of slipped products, but it does not alleviate slipping. Transcription on the +6G template was initiated at +1 with ApC or at –1 with CpA.
We previously found that the points at which escape commitment and the rate-limiting step occur are set by the length of the transcript as opposed to the position of the pol II active site on the DNA template (3, 11). We speculated that this might be a general feature of transitions that occur during early transcription; hence we asked whether the point in the reaction at which transcript slippage occurs is set by the length of the RNA (5 nt) or by the position of the pol II active site on the DNA (register +5). To do so, we took advantage of the ability of pol II to initiate transcription at –1 in the presence of the dinucleotide CpA. We monitored transcript slippage from the +6G promoter in the absence of GTP when the start site was changed from +1 to –1 by replacing ApC with CpA. Doing so increased the length of the RNA from 5 to 6 nt at the point at which ternary complexes paused, without changing the position of the pol II active site along the DNA. As shown in Fig. 4B, moving the initiation site from +1 to –1 on the +6G promoter did not change the amount or pattern of slipped products that occurred, rather it changed the size of the products. In other words, a pattern of 6-, 8-, 10-, 12-, and 14-nt products was observed with CpA instead of the 7-, 9-, 11-, 13-, and 15-nt products observed with ApC. Hence, the position at which slipping occurred did not correlate with the length of the RNA that contained the CUCU repeat; slipping occurred at register +5 after synthesis of either a 5- or a 6-nt transcript. Rather, the position at which slipping occurred (register +5) correlated with the position of the pol II active site on the promoter.
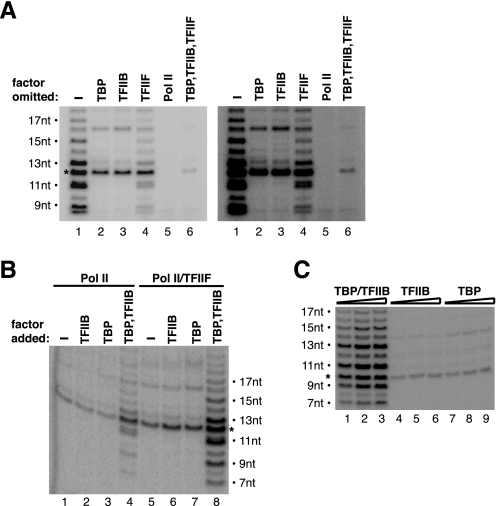
TBP and TFIIB Are Required to Induce Transcript Slippage on a Bubble Template—pol II itself, in the absence of general transcription factors, can specifically initiate transcription from a bubble template when a dinucleotide primer is used (26). This provided the opportunity to probe the contributions that the general transcription factors make to transcript slippage. We first performed experiments in which each factor in the transcription reaction was individually omitted. As shown in Fig. 5A, when either TBP or TFIIB was omitted, slipping no longer occurred (compare lane 1 with lanes 2 and 3). When TFIIF was omitted (Fig. 5A, lane 4), slipping occurred although to a lower extent. Omitting all three general factors resulted in a single 12-nt product produced by pol II alone (Fig. 5A, lane 6), although the amount of product was far less than that observed when any single general factor was omitted. (To better see the product in Fig. 5A, lane 6, a darker exposure of the gel is shown to the right.)
FIGURE 5.
TBP and TFIIB are both required to induce slipping on a bubble template. A, leaving either TBP or TFIIB out of transcription reactions eliminates transcript slippage. Reactions were performed on the wild-type template in the presence of GTP (productive 12-nt product is indicated with an asterisk). The factors were omitted from reactions as indicated. A darker exposure of the same gel is shown to the right. B, TBP and TFIIB function together to cause slipping. Reactions were performed on the wild-type template in the presence of GTP, with the factors added in the combinations indicated. C, higher concentrations of either TBP or TFIIB do not force transcript slippage. TBP, TFIIB, and both factors together were added to reactions at 1×, 3×, and 10× the standard concentrations. Transcription occurred on the wild-type template in the absence of GTP.
We also performed reactions in which each general factor was individually added to pol II alone or to pol II/TFIIF. We again found that both TBP and TFIIB were needed in order for slipping to occur, and that the amount of slipped product was increased by TFIIF (Fig. 5B). Finally, we speculated that perhaps TBP or TFIIB alone could force transcript slippage to occur if present at higher concentrations. When TBP and TFIIB were individually titrated to 10-fold higher concentrations, transcript slippage did not occur (Fig. 5C). We conclude that transcript slippage on a bubble template requires both TBP and TFIIB and is enhanced by TFIIF.
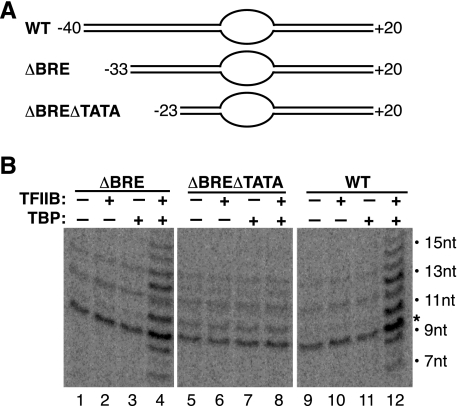
Slipping on a Bubble Template Requires a TATA Box—To determine whether specific protein-DNA contacts involving TBP and TFIIB were important for slipping, we deleted the core promoter from upstream, first removing the BRE and then the TATA box (Fig. 6A). As shown in Fig. 6B (lanes 1–4), transcript slippage did not require the BRE; in its absence TBP and TFIIB induced transcript slippage. This indicated that contacts between TFIIB and DNA were likely not the cause of slipping. Further deleting the DNA to omit the TATA box eliminated slipping (Fig. 6B, lanes 5–8). We conclude that the TATA box is required for slipping to occur, consistent with a model in which DNA-bound TBP facilitates, but is not sufficient for, transcript slippage.
FIGURE 6.
A TATA box, but not a BRE, is required for TBP/TFIIB-induced transcript slippage. A, schematics of the mutant templates that lacked either the BRE or the BRE and TATA box. B, complexes assembled on the ΔBREΔTATA template do not undergo transcript slippage. Transcription in the absence and presence of TBP and TFIIB was monitored from the mutant and wild-type templates in the absence of GTP. The expected 10-nt product is indicated by an asterisk.
DISCUSSION
To further understand the mechanism of the early steps in transcription by human pol II, we investigated the molecular determinants of transcript slippage on a heteroduplex template. We determined the contributions of individual transcription factors, DNA elements, and RNA length to slipping. We found that the point in the transcription reaction at which slipping can first occur is after phosphodiester bond synthesis at register +5. This point is dictated by the position of the polymerase active site on the DNA and a repetitive CUCU sequence occurring from +2 to +5 in the RNA. TBP and TFIIB are required for slipping on a bubble template, as are contacts between TBP and the TATA box. We propose a model in which the transcription reaction branches at register +5 between productive complexes and complexes that undergo transcript slippage. Moreover, slippage is a sensor for contacts in the network of TBP, TFIIB, and pol II interactions that are important during early transcription.
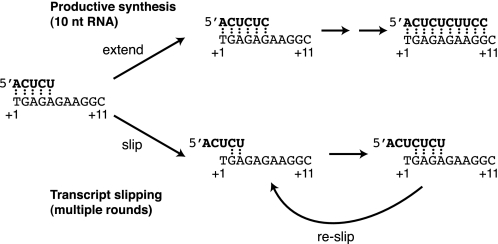
Fig. 7 shows a model in which transcribing complexes, upon completing bond synthesis at register +5, either proceed through promoter escape or undergo one or more rounds of transcript slippage. Complexes that slip give rise to transcripts that are minimally 7 nt in length and increase in size by 2 nt with each additional round of slipping. We found that the pattern of slipped products did not change as transcription was paused at each register beyond +5, indicating that slipping could first occur at +5. Kinetic experiments showed that complexes begin slipping prior to the rate-limiting step, which is complete by register +9. When the template was mutated such that the CUCU repeat first occurred from +4 to +7, slipping did not occur, indicating that on the heteroduplex template the position at which the reaction branches between productive transcription and slipping is uniquely positioned at register +5. Our data also show that once complexes begin slipping, they cannot resume productive transcription. Similarly, once complexes commit to proceeding with productive transcription, they do not slip despite a repetitive sequence that would permit slipping to occur at register +7. This means that in complexes that undergo slipping, the pol II active site never progresses beyond register +5. Prior to register +5, both slipped and productive complexes undergo the same transformations during early transcription because the G-oligo (which blocks translocation of the pol II active site after synthesis of a 4-nt RNA (11)) inhibited both slipping and synthesis of the productive transcript.
FIGURE 7.
A model depicting that after synthesis at register +5, the transcription reaction branches and complexes either proceed through promoter escape or begin rounds of transcript slippage. See text for a more complete discussion of the model.
Some of the characteristics that define transcript slippage on a heteroduplex template are different from other steps we have characterized that occur during early transcription. For example, the point in the reaction at which slipping can occur is set by the position of the pol II active site on the DNA. This is different from the step of escape commitment and the rate-limiting step, the positions of which are both set by the length of the RNA (4 and 8 nt, respectively) (3, 11). This indicates that slipping is more tightly linked to the conformation of the DNA, for example the position and size of the transcription bubble, as opposed to the length of the RNA:DNA hybrid. We also found that transcript slippage depends on the general transcription factors TBP and TFIIB, whereas both escape commitment and the rate-limiting step depends only on pol II; these transformations occur in the absence of general factors (3, 11). These contrasting characteristics demonstrate that the contributions made by transcription factors, DNA elements, and RNA length to the mechanisms by which steps in early transcription occur are not universal.
Transcript slippage was first characterized by Pal and Luse (20, 21) using linear AdMLP templates and HeLa nuclear extracts. They observed products in 2-nt increments longer than expected, found that slipping was significantly reduced at register +8/+9, and found that slipping was greatly reduced when transcription was initiated at –1 with the dinucleotide CpA. In these experiments, the ability of a complex to undergo transcript slippage was a reflection of ternary complex stability; when complexes completed the stabilizing structural transformations that occur during promoter escape, the ability of the transcript to slip was greatly reduced. Slipping on a heteroduplex template is different in that it occurs uniquely at register +5, results in products both shorter and longer than the expected transcript, occurs whether transcription initiates at –1 or +1, and likely reflects the role of protein-protein and protein-DNA contacts involving TBP, TFIIB, and pol II during early transcription. The ability of transcripts to slip may be a general sensor for unique structural transformations that occur during early transcription.
The observation that TBP and TFIIB induce transcript slippage on a heteroduplex template in a TATA box-dependent fashion indicates that slipping is a sensor for contacts that occur between TBP/TATA, TFIIB, and pol II. Previous studies have shown that contacts between TFIIB and pol II influence steps that occur after preinitiation complexes have formed (reviewed in Ref. 27). Therefore, slipping could be triggered by TFIIB-pol II interactions that are critical for the mechanism of early transcription. The requirement for TBP and a TATA box to observe slipping could either indicate that the TBP-DNA complex is required to recruit and position TFIIB in preinitiation complexes or that direct contacts between pol II and TBP also participate in inducing complexes to slip. In this manner, the network of contacts between TBP-TATA, TFIIB, and pol II that are critical for forming preinitiation complexes also have the potential to impede productive transcription.
This work was supported by Grant MCB-0517296 from the National Science Foundation.
Footnotes
The abbreviations used are: pol, polymerase; TBP, TATA-binding protein; AdMLP, adenovirus major late promoter; nt, nucleotide; BRE, transcription factor IIB recognition sequence.
References
- 1.Smale, S. T., and Kadonaga, J. T. (2003) Annu. Rev. Biochem. 72 449–479 [DOI] [PubMed] [Google Scholar]
- 2.Kadonaga, J. T. (2004) Cell 116 247–257 [DOI] [PubMed] [Google Scholar]
- 3.Hieb, A. R., Baran, S., Goodrich, J. A., and Kugel, J. F. (2006) EMBO J. 25 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holstege, F. C. P., Fiedler, U., and Timmers, H. T. M. (1997) EMBO J. 16 7468–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, T.-K., Ebright, R. H., and Reinberg, D. (2000) Science 288 1418–1421 [DOI] [PubMed] [Google Scholar]
- 6.Shilatifard, A. (2006) Annu. Rev. Biochem. 75 243–269 [DOI] [PubMed] [Google Scholar]
- 7.Workman, J. L. (2006) Genes Dev. 20 2009–2017 [DOI] [PubMed] [Google Scholar]
- 8.Thomas, M. C., and Chiang, C. M. (2006) Crit. Rev. Biochem. Mol. Biol. 41 105–178 [DOI] [PubMed] [Google Scholar]
- 9.Naar, A. M., Lemon, B. D., and Tjian, R. (2001) Annu. Rev. Biochem. 70 475–501 [DOI] [PubMed] [Google Scholar]
- 10.Kireeva, M. L., Komissarova, N., Waugh, D. S., and Kashlev, M. (2000) J. Biol. Chem. 275 6530–6536 [DOI] [PubMed] [Google Scholar]
- 11.Kugel, J. F., and Goodrich, J. A. (2002) Mol. Cell. Biol. 22 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holstege, F. C. P., van der Vliet, P. C., and Timmers, H. T. M. (1996) EMBO J. 15 1666–1677 [PMC free article] [PubMed] [Google Scholar]
- 13.Goodrich, J. A., and Tjian, R. (1994) Cell 77 145–156 [DOI] [PubMed] [Google Scholar]
- 14.Parvin, J. D., and Sharp, P. A. (1993) Cell 73 533–540 [DOI] [PubMed] [Google Scholar]
- 15.Pan, G., and Greenblatt, J. (1994) J. Biol. Chem. 269 30101–30104 [PubMed] [Google Scholar]
- 16.Luse, D. S., and Jacob, G. A. (1987) J. Biol. Chem. 262 14990–14997 [PubMed] [Google Scholar]
- 17.Jacob, G. A., Luse, S. W., and Luse, D. S. (1991) J. Biol. Chem. 266 22537–22544 [PubMed] [Google Scholar]
- 18.Kugel, J. F., and Goodrich, J. A. (2000) J. Biol. Chem. 275 40483–40491 [DOI] [PubMed] [Google Scholar]
- 19.Dvir, A. (2002) Biochim. Biophys. Acta 1577 208–223 [DOI] [PubMed] [Google Scholar]
- 20.Pal, M., and Luse, D. S. (2002) Mol. Cell. Biol. 22 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal, M., and Luse, D. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5700–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal, M., Ponticelli, A. S., and Luse, D. S. (2005) Mol. Cell 19 101–110 [DOI] [PubMed] [Google Scholar]
- 23.Zawel, L., Kumar, K. P., and Reinberg, D. (1995) Genes Dev. 9 1479–1490 [DOI] [PubMed] [Google Scholar]
- 24.Kugel, J. F., and Goodrich, J. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver, J. R., Kugel, J. F., and Goodrich, J. A. (2005) J. Biol. Chem. 280 39860–39869 [DOI] [PubMed] [Google Scholar]
- 26.Keene, R. G., and Luse, D. S. (1999) J. Biol. Chem. 274 11526–11534 [DOI] [PubMed] [Google Scholar]
- 27.Deng, W., and Roberts, S. G. (2007) Chromosoma (Berl.) 116 417–429 [DOI] [PubMed] [Google Scholar]