Abstract
The negative cofactor 2 (NC2) is a protein complex composed of two subunits, NC2α and NC2β, and plays a key role in transcription regulation. Here we investigate whether each subunit contains a nuclear localization signal (NLS) that permits individual crossing of the nuclear membrane or whether nuclear import of NC2α and NC2β depends on heterodimerization. Our results from in vitro binding studies and transfection experiments in cultured cells show that each subunit contains a classical NLS (cNLS) that is recognized by the importin α/β heterodimer. Regardless of the individual cNLSs the two NC2 subunits are translocated as a preassembled complex as co-transfection experiments with wild-type and cNLS-deficient NC2 subunits demonstrate. Ran-dependent binding of the nuclear export receptor Crm1/exportin 1 confirmed the presence of a leucine-rich nuclear export signal (NES) in NC2β. In contrast, NC2α does not exhibit a NES. Our results from interspecies heterokaryon assays suggest that heterodimerization with NC2α masks the NES in NC2β, which prevents nuclear export of the NC2 complex. A mutation in either one of the two cNLSs decreases the extent of importin α/β-mediated nuclear import of the NC2 complex. In addition, the NC2 complex can enter the nucleus via a second pathway, facilitated by importin 13. Because importin 13 binds exclusively to the NC2 complex but not to the individual subunits this alternative import pathway depends on sequence elements distributed among the two subunits.
The negative cofactor 2 (NC2)2 is a protein complex composed of two subunits, NC2α (DRAP1) and NC2β (Dr1). Both subunits are conserved in eukaryotes and essential for Saccharomyces cerevisae viability (1, 2). NC2α and NC2β heterodimerize via histone-fold domains and associate with the promotor-bound TATA-binding protein (3, 4). The resulting NC2-TATA-binding protein-DNA complex sterically hinders the recruitment of transcription factor IIB and in part of transcription factor IIA (5), and thus inhibits transcription initiation (6, 7). The NC2 complex is present on a substantial fraction of human genes (8). Besides mediating TATA-binding protein binding to TATA-containing and TATA-less promoters (9) the NC2 complex can also mobilize TATA-binding protein on the DNA (10). In addition to the well established function as transcriptional repressor (11) several studies have shown that NC2 activates transcription, in vitro and in vivo (12–16). The mechanism underlying the positive effects of NC2 on gene expression is not understood. Although the two NC2 subunits mostly function together, recent studies in S. cerevisae (17), Drosophila (18), and human provided evidence that NC2α and NC2β can associate with different proteins (19–21). In this study, we have analyzed whether human NC2 subunits contain localization signals that permit individual crossing of the nuclear membrane or whether nuclear import of NC2α and NC2β depends on heterodimerization.
During interphase the exclusive site of nucleocytoplasmic exchange is the nuclear pore complex (NPC) (22). Although small molecules can traverse the NPC via diffusion the passage of molecules larger than 40 kDa is restricted by the permeability barrier of the NPC (23). Recent work by Frey and Görlich (24, 25) provides evidence that the permeability barrier of the NPC consists of a hydrophobic meshwork with hydrogel-like properties. The passage of this physical barrier requires soluble nuclear transport receptors (also referred to as karyopherins) that recognize intrinsic signal elements displayed on transport cargoes (26). Import signals named nuclear localization signals (NLSs) can be categorized in classical (cNLS) and non-classical (ncNLS) types. Non-classical NLSs are directly recognized by import receptors, whereas the binding of cNLSs requires an additional importin α adapter protein (27). The most abundant nuclear export signals (NESs) are leucine-rich and interact with the export receptor chromosome region maintenance 1 (Crm1), also known as exportin 1 (28, 29). A steep RanGTP gradient across the nuclear membrane controls the binding and release of transport cargoes (30).
Here we demonstrate that the NC2 subunits are imported into the nucleus as a preassembled complex. Nuclear accumulation of the NC2 complex occurs via two alternative pathways, facilitated by the importin α/β heterodimer and importin 13. The cNLSs present in each subunit have a cumulative effect on the nuclear targeting efficiency of the NC2 complex. In addition to its cNLS the NC2β subunit exhibits also a leucine-rich NES, which is masked upon heterodimerization with NC2α.
EXPERIMENTAL PROCEDURES
Cell Culture—NIH-3T3 (mouse embryonic fibroblast) cells obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ number ACC59) and HeLa P4 cells (31) were cultured in Dulbeccos modified Eagle's medium (Invitrogen). Medium was supplemented with 10% (v/v) fetal bovine serum (Biochrom), antibiotics, and 2 mm glutamine. Cells were maintained in a humidified incubator with 5% CO2 atmosphere at 37 °C.
Expression Constructs—The coding regions of the respective genes were amplified from plasmid DNA using specific primer pairs with appropriate restriction sites.
The bacterial expression constructs were cloned as follows: the coding regions of human NC2α and NC2β as SpeI/HindIII fragments into the respective sites of pET-41a(+) (Novagen); the coding regions of human NC2α and NC2β as NcoI/HindIII fragments into the respective sites of pETM-30 (EMBL, Heidelberg); the coding regions of human NC2α and NC2β as NdeI/BamHI fragments together with six histidine residues as NcoI/NdeI fragments into the NcoI/BamHI sites of pET-11d (Novagen); and the coding region of murine Ubc9 as an EcoRI/XhoI fragment into the respective sites of pGEX-4T-1 (GE Healthcare).
The eukaryotic expression constructs were cloned as follows: the coding regions or gene fragments of human NC2α and NC2β as SalI/BamHI fragments into the respective sites of pEGFP-C1 (Clontech) and pPW1 (modified pEGFP-C1 in which EGFP was replaced by RFP using the restriction sites NheI/BglII); the coding regions of human NC2α and NC2β as BglII/PstI fragments into the respective sites of pEGFP-EGFP-N1 (modified pEGFP-N1 (Clontech) in which a second EGFP was inserted C-terminal of the MCS as SalI/BamHI fragment); the coding regions of human NC2α and NC2β as BglII/SalI fragments from the pEGFP-EGFP-N1 expression constructs into the BglII/SalI sites of pmRFP-N1 (modified pEGFP-N1 in which EGFP was replaced by mono-RFP using restriction sites AgeI/NotI); the gene fragments of NC2α (1–10 amino acids) and NC2β (97–106 amino acids) as SalI/BamHI fragments into the respective sites of pEGFP-EGFP-GST-C1 (modified pEGFP-C1 in which GST was inserted N-terminal of the MCS as the BglII/XhoI fragment and in which a second EGFP was inserted at the N terminus as NheI fragment); the coding region and gene fragments of human importin 13 as EcoRI/XhoI fragments were inserted into the respective sites of pCS2flag (modified pCS2plus (32, 33) in which a FLAG tag was inserted N-terminal of the MCS as a NcoI/EcoRI fragment); the coding regions of human importin β, Xenopus importin 7, and murine importin 9 as NruI fragments were inserted into the StuI sites of pCS2flag; and the coding region of human importin 5 as the StuI/XhoI fragment was inserted into the respective sites of pCS2flag. All constructs were verified by DNA sequencing (Andreas Nolte, Abteilung Entwicklungsbiochemie, Universität Göttingen, Germany).
Site-directed Mutagenesis—Nucleotide exchanges in RFP-NC2α-(K4A) and RFP-NC2α-(K5A) were inserted using sense amplification primers: 5′-TGCAGTCGACATGCCCTCCGCAAAGAAAAAGTACAATGCC-3′ for K4A, 5′-TGCAGTCGACATGCCCTCCAAGGCAAAAAAGTACAATGCC-3′ for K5A. To generate EGFP-NC2β-(K100A), EGFP-NC2β-(R101A), EGFP-NC2β-(L78A/F80A), NC2β-(L78A/F80A)-RFP, and GST-NC2β-(L78A/F80A) site-directed mutagenesis was performed according to the QuikChange site-directed mutagenesis kit protocol (Stratagene). The following oligonucleotides were used: 5′-TGTAAAACAGTAGCATTAGCAAGAAGAAAGGCCAGTTCT-3′ (sense) and 5′-AGAACTGGCCTTTCTTCTTGCTAATGCTACTGTTTTACA-3′ (antisense) for K100A, 5′-AAAACAGTAGCATTAAAAGCAAGAAAGGCCAGTTCTCGT-3′ (sense) and 5′-ACGAGAACTGGCCTTTCTTGCTTTTAATGCTACTGTTTT-3′ (antisense) for R101A, and 5′-GTCATACAAGCACTAGAAAGTGCTGGAGCAGGCTCTTACATCAGTGAAGTA-3′ (sense) and 5′-TACTTCACTGATGTAAGAGCCTGCTCCAGCACTTTCTAGTGCTTGTATGAC-3′ (antisense) for L78A/F80A.
Transfection Experiments—Transfection into HeLa P4 cells was performed with the Effectene™ Transfection Reagent (Qiagen) according to the manufacturer's instructions. The cells were fixed 24 h after transfection with 3% paraformaldehyde in phosphate-buffered saline for 15 min and either analyzed directly by fluorescence microscopy or subjected to indirect immunostaining first. For that purpose, fixed cells were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 10 min, blocked with 3% bovine serum albumin in phosphate-buffered saline, and an anti-FLAG polyclonal (rabbit) antibody (Sigma) and anti-rabbit Alexa 488 antibody or anti-rabbit Alexa 555 antibody (Molecular Probes) used to detect FLAG-importin 13. After washing, cells were stained with 10 μg/ml Hoechst 33258 (Molecular Probes) and mounted in Histogel (Linaris Histogel).
Heterokaryon Assays—Interspecies heterokaryons of human HeLa P4 cells and mouse NIH-3T3 cells were formed as described previously (34). Briefly, HeLa P4 cells were transiently (co-)transfected with plasmid DNA encoding either fluorescently labeled NC2α and NC2β or GFP-QKI-5. Thirty hours post-transfection, non-transfected mouse NIH-3T3 cells were co-plated with the HeLa P4 cells and co-cultured for 18 h. The co-culture was then incubated for 2 h in the presence of 50 μg/ml cycloheximide and another 30 min in 100 μg/ml cycloheximide. The co-cultured cells were washed with phosphate-buffered saline and fused with Sigma HybriMax® warmed to 23 °C for 2 min. Fused cells were washed with phosphate-buffered saline and incubated at 37 °C in medium containing 100 μg/ml cycloheximide for an additional 6 h. The cells were fixed with 3% paraformaldehyde in phosphate-buffered saline for 15 min and counterstained with Hoechst 33258 to distinguish between human and mouse cell nuclei.
In Vitro Transcription and Translation—Transcription and translation of human importin 13 fragments (see also Fig. 6A) were performed from the corresponding SP6 promoter constructs (pCS2flag) in vitro. Using the TnT coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions the proteins were labeled with [35S]methionine (Amersham Biosciences). Reactions were performed at 30 °C for 2 h in a 12.5-μl volume and the samples were then directly used for GST pulldown assays.
FIGURE 6.
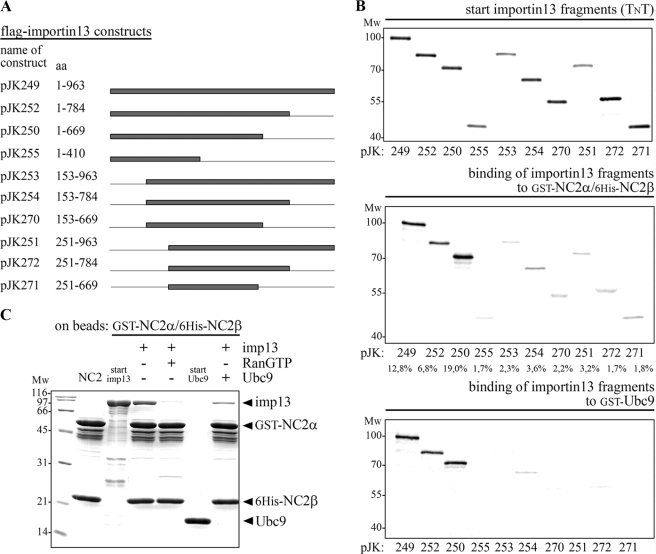
The C terminus of importin 13 is dispensable for recognition of the NC2 complex. A, the names of the importin 13 expression constructs used and the amino acids contained in the constructs are listed. Lines indicate the deleted regions and gray bars represent the different importin 13 fragments. B, the minimal importin 13 fragment that binds to the NC2 complex and UBC9 consists of amino acids 1–669. The two NC2 subunits, GST-NC2α and His6-NC2β, as well as GST-UBC9 were (co-)expressed in E. coli and used as bait after immobilization on glutathione-Sepharose. The immobilized GST-NC2α/His6-NC2β complex and GST-UBC9 were incubated with in vitro transcribed and translated 35S-labeled importin 13 fragments; all from the TnT coupled reticulocyte lysate. Starting material (20% of the importin 13 fragments that were used) and bound fractions were analyzed by SDS-PAGE followed by phosphorimaging (Amersham Biosciences). Among the importin 13 fragments, wild-type importin 13, and amino acids 1–669 of importin 13 showed the highest binding competence for the NC2 complex (see relative binding in percent below the gels, quantified with the program ImageQuant 5.2). A similar binding pattern was observed for UBC9; also here importin 13 fragments lacking the N terminus did not significantly bind to the immobilized substrate. C, the binding sites in importin 13 for the NC2 complex and UBC9 overlap. Importin 13 interacts with the NC2 complex in an UBC9 sensitive manner. Importin 13 was bound specifically to the immobilized GST-NC2α/His6-NC2β complex and this binding was reduced in the presence of equal amounts of UBC9. MW, molecular mass in kilodalton; imp, importin; aa, amino acids.
Recombinant Protein Expression and Purification—Epitope-tagged NC2 complexes were generated as follows: NC2α and NC2β were co-expressed in Escherichia coli BL21(DE3). The cultures were grown at 37 °C to an optical density of 1.0 at 600 nm. After shifting the temperature to 25 °C bacterial protein expression was induced with 0.4 mm isopropyl β-d-thiogalactopyranoside and the cultures were grown for 4 h. The collected bacteria were resuspended in buffer A (50 mm Tris-HCl, pH 7.5, 400 mm NaCl, 5 mm β-mercaptoethanol), lysed by sonication, and the recombinant NC2 complexes were purified on nickel nitrilotriacetic acid-agarose (Qiagen) followed by either gel filtration on Superdex 200 (GE Healthcare) or a second purification step on glutathione-Sepharose 4B (GE Healthcare).
The following proteins were expressed in E. coli BL21(DE3) as indicated and subsequently purified on glutathione-Sepharose 4B or nickel nitrilotriacetic acid-agarose according to the manufacturer's instructions: His6-GST-NC2α, His6-GST-NC2β, and His6-GST-NC2β-(L78A/F80A) at 25 °C for 4 h with 0.2 mm isopropylβ-d-thiogalactopyranoside; and GST-UBC9 at 30 °C for 3 h with 0.2 mm isopropyl β-d-thiogalactopyranoside.
The following transport receptors were expressed in E. coli JM109 or TG1 as described in the literature indicated and were purified on nickel nitrilotriacetic acid-agarose (Qiagen): Xenopus importin α1 (35), human importin β (36), transportin 1 (37), Xenopus importin 7, human importin 5 (38), murine importin 9 (39), human importin 13 (40), and human exportin 1/Crm1 (41). Expression and purification of RanQ69L were performed as described (42).
GST Pulldown Assays—GST fusion proteins (or complexes containing GST fusions) immobilized on glutathione-Sepharose 4B were used as affinity matrix for binding experiments. Appropriate amounts of affinity matrix were incubated for 3 h at 4 °C with bacterial lysates containing expressed import receptors in buffer B (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 5 mm MgCl2, and 5 mm β-mercaptoethanol). The binding experiments were performed in the absence or presence of 2 μm RanQ69L(GTP). After washing three times with ice-cold buffer B, the affinity matrix was boiled in SDS-PAGE sample buffer and the matrix-bound proteins were analyzed by SDS-PAGE followed by Coomassie staining.
For binding to importin 13 fragments, purified GST-NC2α/His6-NC2β and GST-UBC9 were immobilized on glutathione-Sepharose 4B that had been preincubated with 10% bovine serum albumin in buffer C (50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5 mm MgCl2, and 5 mm β-mercaptoethanol). The resulting affinity matrix was washed and incubated with 5 μl of the TnT coupled reticulocyte lysate containing in vitro transcribed and translated 35S-labeled importin 13 fragments in 300 μl of buffer C supplemented with 3% bovine serum albumin. After 3 h at 4 °C, the matrix was washed three times with buffer C, boiled in SDS-PAGE sample buffer, and matrix-bound proteins were analyzed by SDS-PAGE followed by phosphorimaging (Amersham Biosciences).
For interaction studies with exportin 1, purified His6-GST-NC2β (0.4 μm), His6-GST-NC2β-(L78A/F80A) (0.4 μm), and epitope-tagged NC2 complexes (supplemental Fig. S3) were incubated for 3 h at 4 °C with purified recombinant exportin 1 (0.2 μm) in buffer D (50 mm Tris-HCl, pH 7.5, 130 mm NaCl, 2% glycerol, 5 mm MgCl2, and 5 mm β-mercaptoethanol). Binding of exportin 1 was performed in the absence or presence of 2 μm RanQ69L(GTP). Subsequent binding to glutathione-Sepharose 4B (20 μl of matrix) was carried out at 4 °C for 12 h. After washing three times with ice-cold buffer D, the matrix was boiled in SDS-PAGE sample buffer and matrix-bound proteins were analyzed by SDS-PAGE followed by Coomassie staining.
RESULTS
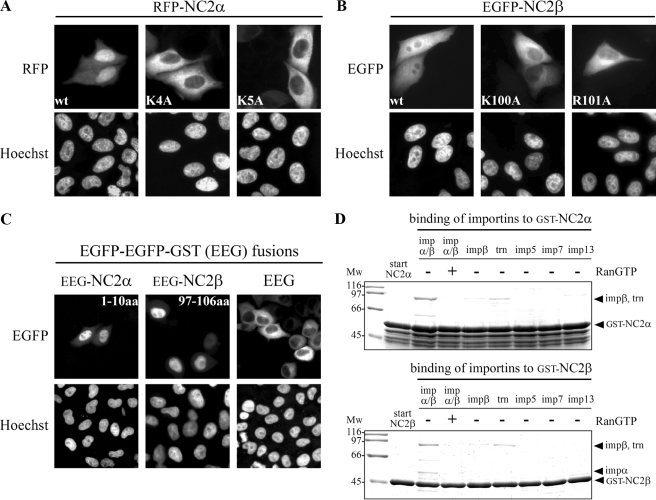
Both NC2 Subunits Exhibit a Monopartite cNLS—Classical NLSs are characterized as short stretches enriched in basic amino acids (43). Based on the consensus motifs for cNLSs (44) each NC2 subunit contains one putative monopartite cNLS. This type of signal follows the four-residue consensus K-(K/R)-X-(K/R) (45) with additional sequence requirements up- and downstream (46). The predicted basic stretch in NC2α (4KKKK7) is located in the unstructured N-terminal region, whereas the relevant sequence in NC2β (100KRRK103) is found in the predicted random coiled C-terminal half of the fourth α helix (5). To functionally characterize the putative signals we first compared the subcellular localization of wild-type and mutated NC2 subunits heterologously expressed in HeLa P4 cells (Fig. 1). To visualize the subcellular distribution, both NC2 subunits were fused to green (EGFP) and red (RFP) fluorescent proteins. Surprisingly, the subcellular distribution of the NC2 subunits showed a strong dependence on the position of the fluorescent fusion protein. Tandem EGFP or RFP fused to the C terminus caused a dominant cytoplasmic distribution of the individual NC2 subunits (supplemental Fig. S1A). In contrast, fusion of EGFP or RFP to the N terminus caused a nuclear localization pattern of NC2α and a rather homogeneous distribution of NC2β (Fig. 1 and supplemental Fig. S1B). Because the subcellular distribution of N-terminal fusion proteins was similar to the localization of endogenous NC2 subunits (data not shown) they were used for further analysis. We observed that wild-type RFP-NC2α no longer accumulated in the nucleus when either lysine residue 4 (K4A) or lysine residue 5 (K5A) was mutated (Fig. 1A). Similar results were found for NC2β, the homogeneous distribution of wild-type EGFP-NC2β was blocked when either lysine residue 100 (K100A) or arginine residue 101 (R101A) of the putative cNLS was substituted for alanine (Fig. 1B). Because the absence of neither critical lysine residues at the second position nor basic amino acid residues at the third position of the four-residue consensus sequence were tolerated, both sequences appeared to represent cNLSs. To analyze whether the predicted sequences are sufficient to mediate nuclear transport of a reporter protein, amino acids 1–10 of NC2α (1MPSKKKKYNA10) and 97–106 of NC2β (97VALKRRKASS106) containing the putative cNLSs (underlined) were fused to EGFP-EGFP-GST (EEG). EEG alone resides exclusively in the cytoplasm when expressed in mammalian cells, because it is too large to enter the nucleus by passive diffusion and does not contain a NLS (Fig. 1C). Fusion of amino acids 1–10 of NC2α or amino acids 97–106 of NC2β to EEG led to efficient nuclear import of the resulting proteins (Fig. 1C). To identify nuclear transport receptors that interact with the individual NC2 subunits, in vitro binding studies were performed. GST-tagged NC2α and NC2β were expressed in E. coli and immobilized on glutathione-Sepharose. The immobilized fusion proteins were incubated with the importin α/β heterodimer, importin β, transportin 1, importin 5, importin 7, and importin 13, all from bacterial lysates (Fig. 1D). After washing, the bound proteins were analyzed by SDS-PAGE followed by Coomassie staining. The importin α/β heterodimer was bound to both NC2 subunits. In the nucleus direct binding of RanGTP to β-family import receptors disintegrates the receptor-substrate interaction. The binding of importin α/β to the NC2 subunits was abolished in the presence of RanGTP, demonstrating its specificity (Fig. 1D). Additionally, weak binding of transportin 1 was observed. However, this interaction was not RanGTP-dependent (data not shown). None of the other import receptors, including importin β alone, bound significantly to the individual NC2 subunits. Together, these results demonstrate that each subunit exhibits a monopartite cNLS, 4KKKK7 in NC2α and 100KRRK103 in NC2β, which are necessary and sufficient for nuclear uptake of the respective NC2 subunit.
FIGURE 1.
Both NC2 subunits exhibit a monopartite cNLS. HeLa P4 cells were transiently transfected with plasmid DNA encoding wild-type (wt) and mutated RFP-NC2α (A), EGFP-NC2β (B), and EGFP-EGFP-GST (EEG), amino acids 1–10 of NC2α or amino acids 97–106 of NC2β fused to EEG (C). The subcellular distribution was examined 24 h after transfection by direct fluorescence. The DNA was counterstained with Hoechst. A, the nuclear accumulation of wild-type RFP-NC2α was blocked when either lysine residue 4 (K4A) or lysine residue 5 (K5A) of NC2α was mutated. B, the homogeneous distribution of wild-type EGFP-NC2β was strongly affected by alanine substitution of either lysine residue 100 (K100A) or arginine residue 101 (R101A) of NC2β. C, the dominant cytoplasmic localization of EEG changed upon fusion to amino acids 1–10 of NC2α (EEG-NC2α-(1–10)) or amino acids 97–106 of NC2β (EEG-NC2β-(97–106)), the localization now becoming nuclear. D, both NC2 subunits bind to importin α/β and the binding is abolished in the presence of RanGTP. Immobilized GST-NC2α and GST-NC2β were incubated with bacterial lysates containing the indicated import receptors. Bound fractions were analyzed by SDS-PAGE and Coomassie stained. Mw, molecular weight; imp, importin; aa, amino acids; trn, transportin 1.
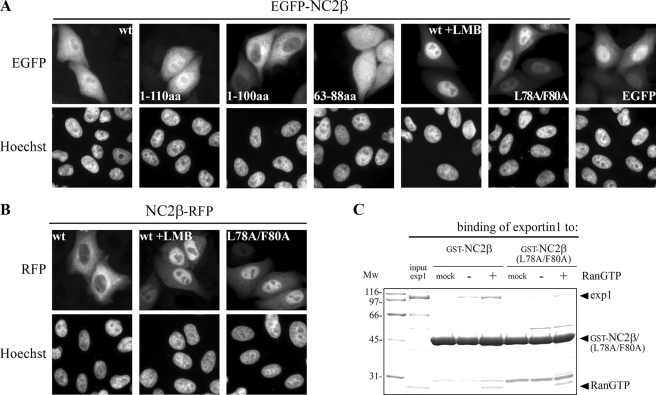
NC2β Contains Also a Leptomycin B-sensitive NES That Is Recognized by Exportin 1—Next we examined whether the NC2 subunits exhibit NESs in addition to the characterized cNLSs. The first hint toward the existence of an NES was provided by the subcellular localization of N-terminal-tagged NC2β. Despite the presence of a cNLS, wild-type EGFP-NC2β showed a homogeneous localization in transfected cells (Fig. 2A). Furthermore, the first 110 amino acids of NC2β fused to EGFP (EGFP-NC2β-(1–110)) distributed homogeneously in transfected cells similar to the pattern of wild-type NC2β. In contrast, the first 100 amino acids of NC2β fused to EGFP (EGFP-NC2β-(1–100)) showed a cytoplasmic localization at steady state. This loss of nuclear uptake can be explained by the missing cNLS (100KRRK103) of NC2β. This fusion protein, however, is not homogeneously distributed like EGFP alone but strictly cytoplasmically, which can only be explained by the presence of an NES within the first 100 amino acids of NC2β. The most abundant and also the best-characterized NESs are leucine-rich (enriched in hydrophobic amino acids) and interact with the export receptor Crm 1/exportin 1 (28, 29). Thus, we examined whether the cytoplasmic localization of fluorescently labeled NC2β was lost on the application of leptomycin B (LMB), a specific inhibitor of exportin 1 (28). We observed that wild-type EGFP-NC2β and NC2β-RFP largely accumulated in the nucleus on LMB treatment for 2 h (Fig. 2, A and B). In contrast to NC2β, the strong cytoplasmic distribution of C-terminal-labeled NC2α was not affected by LMB treatment (data not shown) and N-terminal-labeled NC2α showed a largely nuclear localization in transfected HeLa P4 cells (see Fig. 1A and supplemental Fig. S1B). Hence, these results do not provide evidence for the existence of an NES in NC2α. As LMB impairs various cellular pathways the effects of LMB were additionally verified. The sequence analysis of the first 100 amino acids of NC2β revealed one sequence element (71VIQALESLGF80) that fulfills the loosely defined consensus motif for leucine-rich NES: ΦX2–3ΦX2–3ΦXΦ (Φ= Leu, Ile, Val, Phe, Met) (47, 48). This putative NES was initially confirmed by the fact that amino acids 63–88 of NC2β fused to EGFP (EGFP-NC2β-(63–88)) localized more cytoplasmically than EGFP alone. Furthermore, nuclear export of NC2β was strongly reduced in cultured cells when the last two hydrophobic amino acids of the putative NES were mutated (71VIQALESLGF80-71VIQALESAGA80; mutated residues are underlined), as shown for the EGFP-NC2β-(L78A/F80A) and NC2β-RFP-(L78A/F80A) fusion proteins (Fig. 2, A and B). The results of the mutational analysis resemble the effect of LMB treatment and indicate that the mutated amino acids are part of a hydrophobic NES recognized by exportin 1. The direct interaction of exportin 1 and NC2β was verified by in vitro binding studies. In contrast to import receptors RanGTP is required for the binding of cargoes to export receptors. Recombinant GST-NC2β bound specifically to exportin 1 in the presence of RanGTP, whereas no efficient binding was observed in the absence of RanGTP (Fig. 2C). The RanGTP-dependent binding of exportin 1 to GST-NC2β-(L78A/F80A), however, was reduced. Thus, hydrophobic amino acids leucine (Leu78) and phenylalanine (Phe80) are necessary for the interaction of exportin 1 and NC2β. Together, these results demonstrate that NC2β contains
FIGURE 2.
NC2β exposes a leptomycin B-sensitive NES. HeLa P4 cells were transiently transfected with plasmid DNA coding for wild-type (wt), truncated or mutated NC2β, N-terminal fused to EGFP (A) or C-terminal fused to RFP (B). The subcellular distribution of the gene products was examined 24 h post-transfection by direct fluorescence. The DNA was counterstained with Hoechst. A, wild-type EGFP-NC2β and the first 110 amino acids of NC2β fused to EGFP (EGFP-NC2β-(1–110)) show a homogeneous subcellular distribution that becomes nuclear after addition of 10 ng/ml LMB. Additional deletion of amino acids 101–110 (EGFP-NC2β-(1–100)) results in an exclusively cytoplasmic localization of NC2β. Amino acids 63–88 of NC2β fused to EGFP (EGFP-NC2β-(63–88)) are more cytoplasmically localized than EGFP alone. The homogeneous subcellular distribution of wild-type EGFP-NC2β was reduced when the leucine (L) and phenylalanine (F) residues at positions 78 and 80 were mutated (EGFP-NC2β-(L78A/F80A)). B, wild-type NC2β-RFP shows a dominant cytoplasmic localization that becomes nuclear after addition of LMB. Substitution of the two hydrophobic amino acids at positions 78 and 80 (NC2β-RFP-(L78A/F80A) prevented nuclear export of NC2β-RFP leading to a largely nuclear localization. C, NC2β is recognized by exportin 1 in a RanGTP-dependent fashion. GST-NC2β and GST-NC2β-(L78A/F80A) were incubated with exportin 1 (exp1) in the absence or presence of RanGTP (for details see “Experimental Procedures”). For a negative control exportin 1 was omitted (mock). After binding, glutathione-Sepharose bound fractions were analyzed by SDS-PAGE and Coomassie stained. Input of exportin 1 corresponds to 10% of the protein that was used. The hydrophobic amino acids leucine (Leu78) and phenylalanine (Phe80) are necessary for the binding of exportin 1 to NC2β. Mw, molecular weight; aa, amino acids.
a leucine-rich NES (71VIQALESLGF80), which confers nuclear export via exportin 1. The location of the mapped cNLSs and NES are summarized in supplemental Fig. S2.
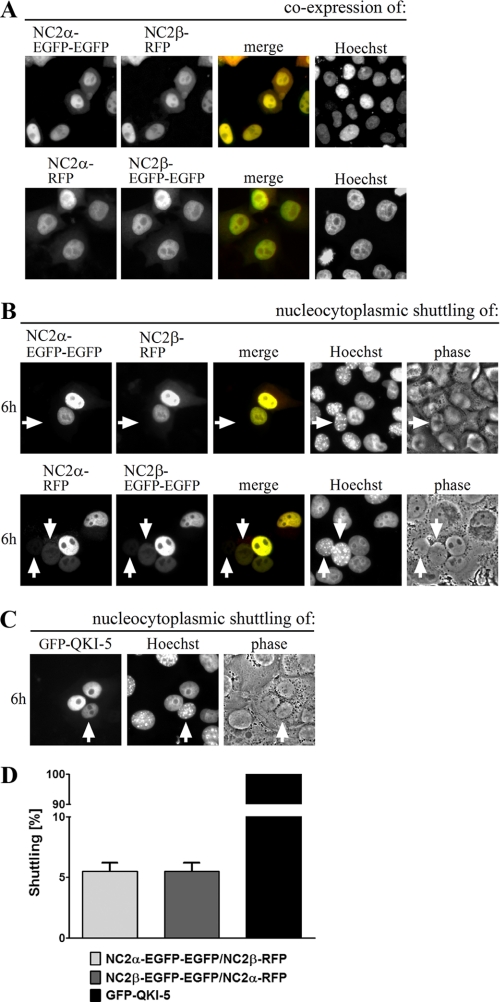
Dimerization with NC2α Masks the NES in NC2β—Many transcription factors contain both types of signals, NLS and NES, which allows them to shuttle between the nucleus and the cytoplasm (49). The identification of the leucine-rich NES in NC2β raised the question whether also the NC2 complex, which is nuclear at steady state (Fig. 3A), can be exported. To answer this question, we performed interspecies heterokaryon assays (34, 50). HeLa P4 cells were transiently co-transfected with plasmid DNA encoding wild-type NC2α and NC2β fused to EGFP-EGFP or RFP (Fig. 3, A and B). The fusion of fluorescent proteins to the C terminus of NC2 subunits seems to favor nuclear export as indicated by the cytoplasmic localization of heterologously expressed NC2 subunits (supplemental Fig. S1A). However, the C-terminal fusions do not block nuclear import in general as shown by the nuclear accumulation of NC2β-RFP in cells treated with LMB (Fig. 2B), and the predominantly nuclear distribution of co-expressed NC2α and NC2β subunits (Fig. 3A). Twenty-four hours post-transfection, an equal amount of untransfected mouse NIH-3T3 cells were seeded to the transfected HeLa P4 cells, the cells were co-cultured, and ultimately fused to form heterokaryons. To inhibit new protein synthesis cycloheximide was added and to distinguish between human HeLa P4 and mouse NIH-3T3 nuclei the cells were stained with Hoechst. Six hours after cell fusion, co-localized NC2α and NC2β were detected in only 5% of the mouse nuclei (white arrows) of heterokaryotic cells (Fig. 3, B and D). This indicates that the NES in NC2β does not allow rapid shuttling of the NC2 complex. RanGTP-dependent binding of exportin 1 to GST-NC2β could no longer be detected when the subunit was dimerized with His6-NC2α. Instead, unspecific binding of exportin 1 to the GST-NC2β/His6-NC2α complex in the absence of RanGTP was observed (supplemental Fig. S3). The shuttling protein QUAKING-5 fused to GFP (GFP-QKI-5) was used as a positive control. In all heterokaryons, GFP-QKI-5 accumulated in the nucleus of non-transfected mouse NIH-3T3 cells (Fig. 3, C and D) demonstrating that QKI-5 shuttles between the nucleus and cytoplasm as previously described (51).
FIGURE 3.
One-way transport of the NC2 complex. HeLa P4 cells were transiently co-transfected with plasmid DNA encoding wild-type NC2α and NC2β fused to EGFP-EGFP or RFP (A and B). The shuttling protein QUAKING-5 fused to GFP (GFP-QKI-5) was overexpressed in HeLa P4 cells (C). A, the subcellular distribution of the gene products was examined 24 h post-transfection by direct fluorescence. Co-expression of NC2α and NC2β leads to a nuclear co-localization shown in yellow (merge). The DNA was counterstained with Hoechst. B, 30 h post-transfection, untransfected mouse NIH-3T3 cells were co-seeded onto the HeLa P4 cells, co-cultured for 18 h, and fused with Sigma HybriMax®. The subcellular distribution of the fusion proteins was examined after an additional 6-h incubation in the presence of cycloheximide. To distinguish between HeLa P4 and NIH-3T3 cell nuclei the heterokaryotic cells were counterstained with Hoechst. Co-localized NC2α and NC2β (shown in yellow, merge) could hardly be detected in the nucleus of mouse NIH-3T3 cells (white arrows). Thus, the NES in NC2β does not allow nucleo-cytoplasmic shuttling of the NC2 complex. C, GFP-QKI-5 accumulates in the nucleus of non-transfected mouse NIH-3T3 cells (white arrow) demonstrating that QKI-5 shuttles between the nucleus and cytoplasm. D, quantification of two independent experiments. 100 heterokaryotic cells were analyzed per condition. Only co-transfected HeLa P4 cells were included in the analysis of the NC2 complex. Bars indicate the mean ± S.D.
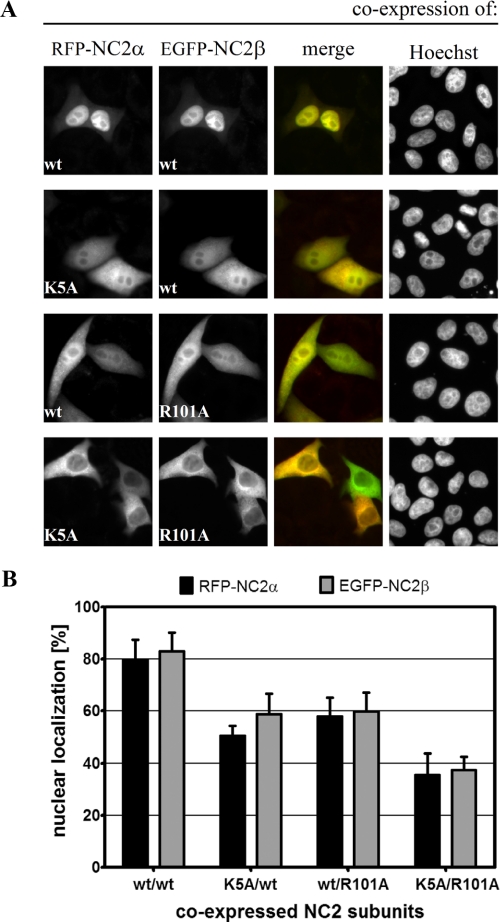
Both cNLSs Are Necessary for Nuclear Import of the NC2 Complex—Next we examined whether nuclear accumulation of the NC2 complex relies on just one or both of the cNLSs of its subunits. For that purpose, we co-expressed the wild-type and cNLS-mutated NC2 subunits described above (see again Fig. 1, A and B) in HeLa P4 cells. Because the heterologously expressed NC2 complex is mainly comprised of fluorescently tagged components, the individual contribution of cNLSs of NC2α and NC2β could be elucidated. The results of these transfection studies are summarized in Fig. 4. Quantitative analysis (Fig. 4B) revealed an 80% nuclear localization of co-expressed wild-type RFP-NC2α and EGFP-NC2β (Fig. 4A, top panel). Mutations in either the cNLS of NC2α (K5A) or the cNLS of NC2β (R101A) did not only reduce the nuclear accumulation of the mutant proteins to 60% or less but also of the corresponding wild-type partner subunit, leading to a homogeneous distribution of the NC2 complexes (Fig. 4A, middle panels). The mutual localization dependence of the NC2 subunits observed in these experiments suggests that the two subunits assemble essentially in the cytoplasm and are transported into the nucleus as heterodimers. In contrast to the mutation of a single cNLS, cytoplasmic accumulation of the NC2 complex was substantially more pronounced when both cNLSs were absent (Fig. 4A, bottom panel). Nuclear import of the cNLS-deficient RFP-NC2α/EGFP-NC2β complex dropped to 35% (Fig. 4B). Similar results were obtained with EGFP-NC2α/RFP-NC2β complexes (supplemental Fig. S4). Thus, these results demonstrate that the two cNLSs have a cumulative effect on the nuclear targeting efficiency of the NC2 complex.
FIGURE 4.
Nuclear accumulation of the NC2 complex via importin α/β requires the cNLSs of both subunits. A, HeLa P4 cells were transiently co-transfected with plasmid DNA encoding wild-type (wt) and mutated RFP-NC2α and EGFP-NC2β, respectively. The subcellular distribution of the gene products was examined 24 h post-transfection by direct fluorescence. The DNA was counterstained with Hoechst. Co-expression of wild-type RFP-NC2α and EGFP-NC2β results in a nuclear co-localization shown in yellow (merge; top panel). Mutation of either the cNLS of NC2α (K5A) or the cNLS of NC2β (R101A) reduced the nuclear accumulation leading to a homogeneous localization of both subunits (middle panels). Nuclear import of the RFP-NC2α/EGFP-NC2β complex was strongly blocked when the cNLSs of both subunits were mutated (bottom panel). B, for quantitative analysis, the mean red (RFP) and green (EGFP) fluorescence value in the nucleus and cytoplasm of 25 cells that co-expressed RFP-NC2α and EGFP-NC2β was measured using the ImageJ Software (NIH). After subtraction of the background value the percentage of nuclear localization of the different NC2 complexes was calculated. Bars indicate the mean ± S.D.
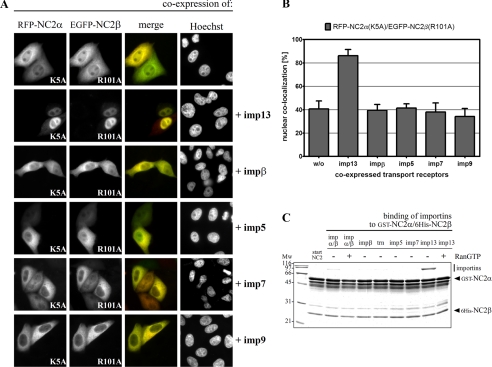
The NC2 Complex Also Enters the Nucleus via Importin 13—To identify nuclear transport receptors that directly bind to the NC2 complex, in vitro interaction studies were performed. For that purpose, GST-tagged NC2α and His6-tagged NC2β were co-expressed in E. coli, affinity-purified on nickel-nitrilotriacetic acid, and immobilized on glutathione-Sepharose. The immobilized GST-NC2α/His6-NC2β complex was incubated with the importin α/β heterodimer, importin β, transportin 1, importin 5, importin 7, and importin 13, all from bacterial lysates (Fig. 5C). After washing, the bound proteins were analyzed by SDS-PAGE followed by Coomassie staining. Surprisingly, importin 13 was much more efficiently bound to the immobilized NC2 complex than the importin α/β heterodimer. Both NC2-receptor interactions were reduced in the presence of RanGTP suggesting functional significance. None of the other transport receptors bound significantly to the NC2 complex. Because of the strong binding of importin 13 to the NC2 complex in vitro, we tested its possible role as NC2 carrier also in cultured cells. For that purpose, FLAG-tagged importin 13 was additionally co-expressed with mutated RFP-NC2α (K5A) and EGFP-NC2β (R101A). The co-expression of exogenous importin 13 led to an efficient nuclear accumulation of the cNLS-deficient RFP-NC2α/EGFP-NC2β complex (Fig. 5A). In contrast to importin 13, the additional co-expression of FLAG-tagged importin β, importin 5, importin 7, and importin 9 did not result in enhanced nuclear uptake of the NC2 complex devoid of cNLSs. Fig. 5B shows a quantitative analysis of the data in Fig. 5A. These results demonstrate that the loss of nuclear import via the classical importin α/β pathway could only be rescued by importin 13. Comparable results were obtained with the cNLS-deficient EGFP-NC2α/RFP-NC2β complex (supplemental Fig. S5). Hence, a second, alternative, transport pathway via importin 13 does exist for the NC2 complex. However, the subcellular distribution of wild-type and cNLS-mutated RFP-NC2α (K5A) was not influenced by co-expression of importin 13 (supplemental Fig. S6A), and the subcellular localization of NC2β-RFP was only slightly changed upon co-expression of importin 13 (supplemental Fig. S6B). These results are in line with previous in vitro binding studies showing that importin 13 does not interact with the monomeric NC2 subunits (see Fig. 1D). In conclusion, importin 13 mediates nuclear import of the NC2 complex but not of its individual subunits. In that way, the NC2 complex behaves like its closest relative, the NF-YB/NF-YC histone-fold heterodimer. Only the heterodimerized NF-YB/NF-YC subunits were specifically recognized by importin 13, as shown previously (52). Because heterodimerization of both NC2α with NC2β and NF-YB with NF-YC is a prerequisite for importin 13 binding, the minimal sequence elements recognized by importin 13 comprise the histone-fold domains of both NC2 subunits. Moreover, basic amino acid residues distributed among the two subunits and conserved between related histone-fold pairs (Lys18, Arg19, Arg40, and Lys63 in NC2α and Lys64, Lys88, Lys95, and Arg102 in NC2β) are crucial for the recognition of importin 13.3
FIGURE 5.
Importin 13 also mediates nuclear transport of the NC2 complex. A, HeLa P4 cells were transiently co-transfected with plasmid DNA encoding mutated RFP-NC2α (K5A), EGFP-NC2β (R101A), and FLAG-tagged import receptors. The subcellular distribution of the gene products was examined 24 h post-transfection by direct fluorescence of the RFP and EGFP fusion proteins. The overlap between the green EGFP fusion protein and the red RFP fusion protein is shown in yellow (merge). The DNA was counterstained with Hoechst. RFP-NC2α-(K5A) and EGFP-NC2β-(R101A) both predominantly remained in the cytoplasm of co-transfected cells. Although co-expression of importin 13 led to a strong nuclear accumulation of the NC2 complex, co-expressed importin β, importin 5, importin 7, and importin 9 did not affect the subcellular distribution of the NC2 complex. B, quantification of nuclear import of the cNLS-deficient NC2 complex. The mean fluorescence value of co-localized RFP-NC2α-(K5A) and EGFP-NC2β-(R101A) was measured in 15 cells using ImageJ software (NIH). The percentage of nuclear localization in the presence of co-expressed import receptors was calculated. Bars indicate the mean ± S.D. C, immobilized GST-NC2α/His6-NC2β complex was incubated with bacterial lysates containing the indicated import receptors. Bound fractions were analyzed by SDS-PAGE and Coomassie stained. The NC2 complex binds strongly to importin 13 and less pronounced to the importin α/β heterodimer. Both cargo-receptor interactions are reduced in the presence of RanGTP, which was used to simulate nuclear conditions. Mw, molecular weight; imp, importin; trn, transportin 1.
The C Terminus of Importin 13 Is Dispensable for Recognition of the NC2 Complex—After confirming that importin 13 indeed represents an alternative transport receptor for nuclear import of the NC2 complex, we wanted to characterize the binding sites in importin 13 for the NC2 complex. Additionally, we compared the binding of NC2 with that of another importin 13 substrate, the sumo-conjugating enzyme UBC9 (40). For that purpose, we generated different importin 13 deletion constructs and tested their NC2 binding potentials (Fig. 6). Independent of the HEAT repeat prediction for importin 13 in Swiss-Prot, the secondary structure prediction programs PHD (Heidelberg) and PSIPRED (53, 54) were used to identify putative α helices within importin 13. The programs predicted 38 and 37 α helices, respectively, which is in agreement with the assumption that all transport receptors of the importin β family share 19 HEAT repeats (55). The deletion constructs were created by cutting between the predicted α helices (putative HEAT repeats) of importin 13, and the importin 13 fragments were in vitro transcribed and translated (Fig. 6, A and B). The immobilized GST-NC2α/His6-NC2β complex and GST-UBC9 were incubated with the 35S-labeled importin 13 fragments and bound fractions were analyzed by SDS-PAGE followed by phosphorimaging. As shown in Fig. 6B, among the importin 13 fragments only amino acids 1–669 and 1–784 of importin 13 were bound to the NC2 complex (middle panel) and UBC9 (lower panel). Importin 13 fragments lacking the N terminus did not significantly bind to the immobilized substrates. Accordingly, only C-terminal truncations of importin 13 (amino acids 1–784 and 1–669, respectively) were able to efficiently accumulate the cNLS-deficient RFP-NC2α/EGFP-NC2β complex in the nucleus of co-transfected HeLa cells (supplemental Fig. S7). Due to a similar binding pattern of the importin 13 fragments to the NC2 complex and UBC9, we also asked whether the binding sites in importin 13 for the NC2 complex and UBC9 overlap. To address this question binding of recombinant purified importin 13 to an immobilized GST-NC2α/His6-NC2β complex was performed in the absence and presence of equal amounts of recombinant, untagged UBC9 (Fig. 6C). The importin 13 binding was reduced in the presence of UBC9, which demonstrates that the binding sites in importin 13 for the NC2 complex and UBC9 overlap.
In summary, our results suggest that the two NC2 subunits preassemble in the cytoplasm prior to nuclear import. The NC2 complex enters the nucleus via two alternative pathways, facilitated either by the importin α/β heterodimer or importin 13. Furthermore, each import pathway depends on sequence elements distributed on both NC2 subunits.
DISCUSSION
In this study, we have analyzed the nuclear import pathways of the human NC2 complex. We identified a cNLS in each NC2 subunit. Because cNLSs bind in an extended conformation to importin α they are most likely located in unstructured regions to fit into the importin α binding pockets (44, 56). The cNLS of NC2α (4KKKK7) is located in the N terminus, whereas the cNLS of NC2β (100KRRK103) is found in the center of the protein. In the native proteins, these regions are predicted to be unstructured or random coiled, respectively (5). In addition, both cNLSs fulfill the consensus motif for a monopartite cNLS as proposed by Chelsky et al. (45). However, none of the additional sequence requirements for monopartite cNLSs summarized by Conti (46) are satisfied in the two NC2 subunits. Differences from preferred amino acids for monopartite cNLSs may account for the weak interaction of the importin α/β heterodimer with the NC2 subunits in vitro. Interestingly, the cNLS of NC2α with four contiguous lysines resembles the downstream cluster of the bipartite cNLS of nucleoplasmin. Perhaps puzzling, whereas the downstream cluster of nucleoplasmin is not sufficient to target a protein into the nucleus (57, 58), amino acids 1–10 of NC2α containing its cNLS (4KKKK7) functioned as an effective monopartite signal when fused to an EGFP-EGFP-GST reporter protein. Both NC2 genes are highly conserved in eukaryotes and are required for viability in yeast (1). However, the monopartite cNLS of neither NC2α nor NC2β is conserved in S. cerevisiae. Therefore, nuclear import of NC2 in yeast must occur via a different transport mechanism.
Despite exposing cNLSs the two human NC2 subunits are not imported individually but assemble essentially in the cytoplasm and are transported into the nucleus as heterodimer. This was demonstrated by transfection experiments in which the loss of the cNLS in one subunit strongly affected the nuclear accumulation of the partner subunit. Nuclear import of the NC2 complex was significantly blocked when the cNLSs of both subunits were mutated. These results point to an additive effect of both cNLSs on the nuclear targeting efficiency of the NC2 complex.
Besides the cNLS also a leucine-rich NES (71VIQALESLGF80) was identified in the NC2β subunit. In S. cerevisiae the last of the four essential hydrophobic positions (underlined) contains tyrosine instead of phenylalanine. However, in contrast to the human subunit three alternative putative sequence elements fulfill the consensus motif for a leucine-rich NES in yeast. The presence of NLS and NES allow NC2β to shuttle between the cytoplasm and nucleus. Phosphorylation near an NLS can either have a stimulatory or inhibitory function for protein transport (59). For example, it has been shown that phosphorylation of residues in close proximity to the NLS of the transcriptional activator NF-AT (60) and Lamin B2 (61) lead to the inhibition of the NLS function. The NC2β subunit is phosphorylated by casein kinase II in vitro and has two serine residues (Ser) in close distance to the cNLS (100KRRKASS106) (3). Substitution of the two serines for glutamate (S105E/S106E) to mimic the effect of phosphorylation did not significantly influence nuclear accumulation of the NC2 complex (supplemental Fig. S8A). In contrast, nuclear import of NC2β-(S105E/S106E) fused to EGFP-EGFP-GST was reduced in cells treated with LMB (supplemental Fig. S8B) indicating that the cNLS function can be inhibited by phosphorylation. Hence, the modulation of nuclear transport rates by cytoplasmic phosphorylation may represent a mechanism for regulating nuclear activity of NC2β.
The results of interspecies heterokaryon assays with fluorescently labeled NC2 subunits demonstrated that the NC2 complex does not shuttle between the nucleus and cytoplasm. Hence, heterodimerization with NC2α masks the NES of NC2β and ensures that only complexed NC2β remains in the nucleus. These findings point toward a scenario in which NC2α also functions independently of NC2β. More detailed, the activity of the NC2 complex could be modulated by displacement of NC2β. Due to the immediate nuclear export of removed NC2β interactions of NC2α with additional nuclear partners such as BTAF1 (19) would not be disrupted.
The NES in NC2β is located at the C terminus of the third α helix (5). This α helix is part of the histone-fold motif that facilitates dimerization of the two NC2 subunits. Based on the structural information provided by Kamada et al. (5), only the valine residue at position 71 is involved in subunit dimerization, whereas none of the other essential hydrophobic residues of the NES are predicted to be involved. Therefore, we assume that binding of exportin 1 to the NC2 complex is blocked via steric hindrance. Likewise, Stommel et al. (62) showed that tetramerization masks the NES of p53 and prevents its export.
In addition to the importin α/β-mediated nuclear transport, a second import pathway via importin 13 does exist for the NC2 complex. This feature is shared by two other recently identified importin 13 cargoes: glucocorticoid receptor (63) and actin-binding protein myopodin (64). These two importin 13 substrates also contain additional cNLSs (65–67).
Surprisingly, endogenous importin 13 alone was not sufficient to facilitate effective nuclear import of the heterologously expressed NC2 complex. One would expect that strong overexpression of importin 13 cargoes can be compensated by constant recycling of the transport receptor. However, very low cellular concentrations of endogenous importin 13 could explain the cytoplasmic retention of the overexpressed NC2 complex. Additionally, the affinity of importin 13 to the NC2 complex might be low in vivo due to phosphorylation of the NC2 subunits (3). As recently described for its closest relative, the NF-YB/NF-YC histone-fold heterodimer, only the dimerized NC2 subunits were recognized by importin 13 (52). Hence, despite their functional divergence these two histone-fold heterodimers rely on the same import mechanism to translocate through the NPC. Interestingly, for importin 13 the most similar protein from S. cerevisiae is Pdr6p/karyopherin 122, which translocates the NC2 antagonist transcription factor IIA into the nucleus (68). The identity of Pdr6p with importin 13 is too low to regard it as a clear orthologue (40). However, Pdr6p might be the functional homologue of importin 13 in yeast. Hence, it would be interesting to investigate whether the yeast NC2 complex (Ydr1-Bur6) is transported by Pdr6p and whether human transcription factor IIA can be imported by importin 13.
To characterize the binding sites in importin 13 for the NC2 complex we compared the binding of several importin 13 fragments. The results of these in vitro interaction studies demonstrated that the C terminus of importin 13 is dispensable for recognition of the NC2 complex. These results were further confirmed by co-transfection experiments with the importin 13 fragments. In contrast to the observations made by Tao and colleagues (69) all FLAG-tagged importin 13 fragments including the N-terminal truncated fragments distributed equally between the cytoplasm and nucleus.4 The loss of the N terminus of importin 13 did therefore not result in a dominant negative regulator. Despite the homogenous distribution, none of the N-terminal-truncated importin 13 fragment facilitated efficient nuclear uptake of the NC2 complex. Hence, the N terminus of importin 13 is essential for recognition and nuclear import of the NC2 complex. Furthermore, we showed that importin 13 cannot bind the NC2 complex and UBC9 at the same time due to overlapping importin 13 binding sites.
For importin β, two major types of cargo-receptor interactions are found. These interactions are either hydrophobic as in the case of the SREBP-2 NLS where minimal contacts are made between importin β and the substrate (70) or electrostatic as observed for the importin β binding domain of importin α (71). Because dimerization of the two NC2 subunits buries the hydrophobic residues of the second α helix and creates a positively charged surface (5) it is reasonable to assume that importin 13 and NC2 interact via electrostatic interactions. The requirement of basic amino acid residues for importin 13-mediated nuclear import of the NC2 complex3 provides first support for this hypothesis.
Supplementary Material
Acknowledgments
We thank Dirk Görlich, José-Manuel Mingot, and Stefan Jäkel (Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany) for providing the expression plasmids for the import factors; Ulrike Kutay (Institute for Biochemistry, Eidgenösische Technische Hochschule Zürich, Switzerland) for providing the expression plasmid for importin 9; Ralph Kehlenbach and Saskia Hutten (Institut für Biochemie und Molekulare Zellbiologie, Universität Göttingen, Germany) for providing the plasmid DNA of the pEGFP-EGFP-GST vector; Frauke Melchior (Institut für Biochemie und Molekulare Zellbiologie, Universität Göttingen, Germany) for providing recombinant, purified UBC9; Stéphane Richard (Departments of Oncology and Medicine, McGill University, Montreal, Canada) for providing the expression plasmid for GFP-QKI-5; Iain Mattaj (EMBL, Heidelberg, Germany) for providing the expression plasmid for Crm1; Volker Haucke and Nadja Jung (Abteilung Membranbiochemie, Freie Universität Berlin, Germany) for providing the plasmid DNA of the pmRFP-N1 vector. We acknowledge Erik Meulmeester and Patrick Walker for critical reading of the manuscript. Christa Bode is acknowledged for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 523: Protein und Membrantransport zwischen zellulären Kompartimenten.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
Footnotes
The abbreviations used are: NC2, negative cofactor 2; NLS, nuclear localization signal; NES, nuclear export signal; Crm1, chromosome region maintenance 1; HEAT, huntingtin, elongation factor 3, protein phosphatase 2A, TOR1; EGFP, enhanced green fluorescent protein; RFP, red fluorescent protein; GST, glutathione S-transferase; LMB, leptomycin B; cNLS, classical nuclear localiztion signal; NPC, nuclear pore complex.
Walker, P., Doenecke, D., and Kahle, J. (February 14, 2009) J. Biol. Chem. 10.1074/jbc.M806820200.
P. Walker and J. Kahle, unpublished observations.
References
- 1.Kim, S., Na, J. G., Hampsey, M., and Reinberg, D. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 820-825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gadbois, E. L., Chao, D. M., Reese, J. C., Green, M. R., and Young, R. A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3145-3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goppelt, A., Stelzer, G., Lottspeich, F., and Meisterernst, M. (1996) EMBO J. 15 3105-3116 [PMC free article] [PubMed] [Google Scholar]
- 4.Mermelstein, F., Yeung, K., Cao, J., Inostroza, J. A., Erdjument-Bromage, H., Eagelson, K., Landsman, D., Levitt, P., Tempst, P., and Reinberg, D. (1996) Genes Dev. 10 1033-1048 [DOI] [PubMed] [Google Scholar]
- 5.Kamada, K., Shu, F., Chen, H., Malik, S., Stelzer, G., Roeder, R. G., Meisterernst, M., and Burley, S. K. (2001) Cell 106 71-81 [DOI] [PubMed] [Google Scholar]
- 6.Inostroza, J. A., Mermelstein, F. H., Ha, I., Lane, W. S., and Reinberg, D. (1992) Cell 70 477-489 [DOI] [PubMed] [Google Scholar]
- 7.Meisterernst, M., and Roeder, R. G. (1991) Cell 67 557-567 [DOI] [PubMed] [Google Scholar]
- 8.Albert, T. K., Grote, K., Boeing, S., Stelzer, G., Schepers, A., and Meisterernst, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10000-10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilfillan, S., Stelzer, G., Piaia, E., Hofmann, M. G., and Meisterernst, M. (2005) J. Biol. Chem. 280 6222-6230 [DOI] [PubMed] [Google Scholar]
- 10.Schluesche, P., Stelzer, G., Piaia, E., Lamb, D. C., and Meisterernst, M. (2007) Nat. Struct. Mol. Biol. 14 1196-1201 [DOI] [PubMed] [Google Scholar]
- 11.White, R. J., Khoo, B. C., Inostroza, J. A., Reinberg, D., and Jackson, S. P. (1994) Science 266 448-450 [DOI] [PubMed] [Google Scholar]
- 12.Cang, Y., and Prelich, G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12727-12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castano, E., Gross, P., Wang, Z., Roeder, R. G., and Oelgeschlager, T. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7184-7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willy, P. J., Kobayashi, R., and Kadonaga, J. T. (2000) Science 290 982-985 [DOI] [PubMed] [Google Scholar]
- 15.Geisberg, J. V., Holstege, F. C., Young, R. A., and Struhl, K. (2001) Mol. Cell. Biol. 21 2736-2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaire, M., Xie, J., Meisterernst, M., and Collart, M. A. (2000) Mol. Microbiol. 36 163-173 [DOI] [PubMed] [Google Scholar]
- 17.Creton, S., Svejstrup, J. Q., and Collart, M. A. (2002) Genes Dev. 16 3265-3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., Vijayadamodar, G., Pochart, P., Machineni, H., Welsh, M., Kong, Y., Zerhusen, B., Malcolm, R., Varrone, Z., Collis, A., Minto, M., Burgess, S., McDaniel, L., Stimpson, E., Spriggs, F., Williams, J., Neurath, K., Ioime, N., Agee, M., Voss, E., Furtak, K., Renzulli, R., Aanensen, N., Carrolla, S., Bickelhaupt, E., Lazovatsky, Y., DaSilva, A., Zhong, J., Stanyon, C. A., Finley, R. L., Jr., White, K. P., Braverman, M., Jarvie, T., Gold, S., Leach, M., Knight, J., Shimkets, R. A., McKenna, M. P., Chant, J., and Rothberg, J. M. (2003) Science 302 1727-1736 [DOI] [PubMed] [Google Scholar]
- 19.Klejman, M. P., Pereira, L. A., van Zeeburg, H. J., Gilfillan, S., Meisterernst, M., and Timmers, H. T. (2004) Mol. Cell. Biol. 24 10072-10082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assmann, E. M., Alborghetti, M. R., Camargo, M. E., and Kobarg, J. (2006) J. Biol. Chem. 281 9869-9881 [DOI] [PubMed] [Google Scholar]
- 21.Iratni, R., Yan, Y. T., Chen, C., Ding, J., Zhang, Y., Price, S. M., Reinberg, D., and Shen, M. M. (2002) Science 298 1996-1999 [DOI] [PubMed] [Google Scholar]
- 22.Fahrenkrog, B., and Aebi, U. (2003) Nat. Rev. Mol. Cell Biol. 4 757-766 [DOI] [PubMed] [Google Scholar]
- 23.Gorlich, D., and Kutay, U. (1999) Annu. Rev. Cell Dev. Biol. 15 607-660 [DOI] [PubMed] [Google Scholar]
- 24.Frey, S., Richter, R. P., and Gorlich, D. (2006) Science 314 815-817 [DOI] [PubMed] [Google Scholar]
- 25.Frey, S., and Gorlich, D. (2007) Cell 130 512-523 [DOI] [PubMed] [Google Scholar]
- 26.Pemberton, L. F., and Paschal, B. M. (2005) Traffic 6 187-198 [DOI] [PubMed] [Google Scholar]
- 27.Gorlich, D., Kostka, S., Kraft, R., Dingwall, C., Laskey, R. A., Hartmann, E., and Prehn, S. (1995) Curr. Biol. 5 383-392 [DOI] [PubMed] [Google Scholar]
- 28.Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997) Cell 90 1051-1060 [DOI] [PubMed] [Google Scholar]
- 29.Stade, K., Ford, C. S., Guthrie, C., and Weis, K. (1997) Cell 90 1041-1050 [DOI] [PubMed] [Google Scholar]
- 30.Mattaj, I. W., and Englmeier, L. (1998) Annu. Rev. Biochem. 67 265-306 [DOI] [PubMed] [Google Scholar]
- 31.Charneau, P., Mirambeau, G., Roux, P., Paulous, S., Buc, H., and Clavel, F. (1994) J. Mol. Biol. 241 651-662 [DOI] [PubMed] [Google Scholar]
- 32.Rupp, R. A., Snider, L., and Weintraub, H. (1994) Genes Dev. 8 1311-1323 [DOI] [PubMed] [Google Scholar]
- 33.Turner, D. L., and Weintraub, H. (1994) Genes Dev. 8 1434-1447 [DOI] [PubMed] [Google Scholar]
- 34.Michael, W. M., Choi, M., and Dreyfuss, G. (1995) Cell 83 415-422 [DOI] [PubMed] [Google Scholar]
- 35.Gorlich, D., Prehn, S., Laskey, R. A., and Hartmann, E. (1994) Cell 79 767-778 [DOI] [PubMed] [Google Scholar]
- 36.Kutay, U., Izaurralde, E., Bischoff, F. R., Mattaj, I. W., and Gorlich, D. (1997) EMBO J. 16 1153-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Gorlich, D. (1997) EMBO J. 16 6535-6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakel, S., and Gorlich, D. (1998) EMBO J. 17 4491-4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muhlhausser, P., Muller, E. C., Otto, A., and Kutay, U. (2001) EMBO Rep. 2 690-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mingot, J. M., Kostka, S., Kraft, R., Hartmann, E., and Gorlich, D. (2001) EMBO J. 20 3685-3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan, T., Kehlenbach, R. H., Schirmer, E. C., Kehlenbach, A., Fan, F., Clurman, B. E., Arnheim, N., and Gerace, L. (2000) Mol. Cell. Biol. 20 5619-5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribbeck, K., Lipowsky, G., Kent, H. M., Stewart, M., and Gorlich, D. (1998) EMBO J. 17 6587-6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dingwall, C., and Laskey, R. A. (1991) Trends Biochem. Sci. 16 478-481 [DOI] [PubMed] [Google Scholar]
- 44.Conti, E., and Kuriyan, J. (2000) Structure Fold Des. 8 329-338 [DOI] [PubMed] [Google Scholar]
- 45.Chelsky, D., Ralph, R., and Jonak, G. (1989) Mol. Cell. Biol. 9 2487-2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conti, E. (2002) Results Probl. Cell Differ. 35 93-113 [DOI] [PubMed] [Google Scholar]
- 47.Bogerd, H. P., Fridell, R. A., Benson, R. E., Hua, J., and Cullen, B. R. (1996) Mol. Cell. Biol. 16 4207-4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, F. J., Beeche, A. A., Hunter, J. J., Chin, D. J., and Hope, T. J. (1996) Mol. Cell. Biol. 16 5147-5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaffman, A., and O'Shea, E. K. (1999) Annu. Rev. Cell Dev. Biol. 15 291-339 [DOI] [PubMed] [Google Scholar]
- 50.Pinol-Roma, S., and Dreyfuss, G. (1992) Nature 355 730-732 [DOI] [PubMed] [Google Scholar]
- 51.Wu, J., Zhou, L., Tonissen, K., Tee, R., and Artzt, K. (1999) J. Biol. Chem. 274 29202-29210 [DOI] [PubMed] [Google Scholar]
- 52.Kahle, J., Baake, M., Doenecke, D., and Albig, W. (2005) Mol. Cell. Biol. 25 5339-5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGuffin, L. J., Bryson, K., and Jones, D. T. (2000) Bioinformatics 16 404-405 [DOI] [PubMed] [Google Scholar]
- 54.Jones, D. T. (1999) J. Mol. Biol. 292 195-202 [DOI] [PubMed] [Google Scholar]
- 55.Petosa, C., Schoehn, G., Askjaer, P., Bauer, U., Moulin, M., Steuerwald, U., Soler-Lopez, M., Baudin, F., Mattaj, I. W., and Muller, C. W. (2004) Mol. Cell 16 761-775 [DOI] [PubMed] [Google Scholar]
- 56.Fontes, M. R., Teh, T., and Kobe, B. (2000) J. Mol. Biol. 297 1183-1194 [DOI] [PubMed] [Google Scholar]
- 57.Dingwall, C., Robbins, J., Dilworth, S. M., Roberts, B., and Richardson, W. D. (1988) J. Cell Biol. 107 841-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robbins, J., Dilworth, S. M., Laskey, R. A., and Dingwall, C. (1991) Cell 64 615-623 [DOI] [PubMed] [Google Scholar]
- 59.Jans, D. A. (1995) Biochem. J. 311, 705-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, J., Shibasaki, F., Price, R., Guillemot, J. C., Yano, T., Dotsch, V., Wagner, G., Ferrara, P., and McKeon, F. (1998) Cell 93 851-861 [DOI] [PubMed] [Google Scholar]
- 61.Hennekes, H., Peter, M., Weber, K., and Nigg, E. A. (1993) J. Cell Biol. 120 1293-1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stommel, J. M., Marchenko, N. D., Jimenez, G. S., Moll, U. M., Hope, T. J., and Wahl, G. M. (1999) EMBO J. 18 1660-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao, T., Lan, J., Lukacs, G. L., Hache, R. J., and Kaplan, F. (2006) Am. J. Respir. Cell Mol. Biol. 35 668-680 [DOI] [PubMed] [Google Scholar]
- 64.Liang, J., Ke, G., You, W., Peng, Z., Lan, J., Kalesse, M., Tartakoff, A. M., Kaplan, F., and Tao, T. (2008) Mol. Cell. Biochem. 307 93-100 [DOI] [PubMed] [Google Scholar]
- 65.Weins, A., Schwarz, K., Faul, C., Barisoni, L., Linke, W. A., and Mundel, P. (2001) J. Cell Biol. 155 393-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faul, C., Huttelmaier, S., Oh, J., Hachet, V., Singer, R. H., and Mundel, P. (2005) J. Cell Biol. 169 415-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Picard, D., and Yamamoto, K. R. (1987) EMBO J. 6 3333-3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Titov, A. A., and Blobel, G. (1999) J. Cell Biol. 147 235-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao, T., Lan, J., Presley, J. F., Sweezey, N. B., and Kaplan, F. (2004) Am. J. Respir. Cell Mol. Biol. 30 350-359 [DOI] [PubMed] [Google Scholar]
- 70.Lee, S. J., Sekimoto, T., Yamashita, E., Nagoshi, E., Nakagawa, A., Imamoto, N., Yoshimura, M., Sakai, H., Chong, K. T., Tsukihara, T., and Yoneda, Y. (2003) Science 302 1571-1575 [DOI] [PubMed] [Google Scholar]
- 71.Cingolani, G., Petosa, C., Weis, K., and Muller, C. W. (1999) Nature 399 221-229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.