Abstract
Analysis of the spatial organization of chromosomes reveals complex three-dimensional networks of chromosomal interactions. These interactions impact gene expression at multiple levels, including long-range control by distant enhancers and repressors, coordinated expression of genes and acquisition of epigenetic states. Major challenges now include deciphering the mechanisms by which loci come together and understanding the functional consequences of these often transient associations.
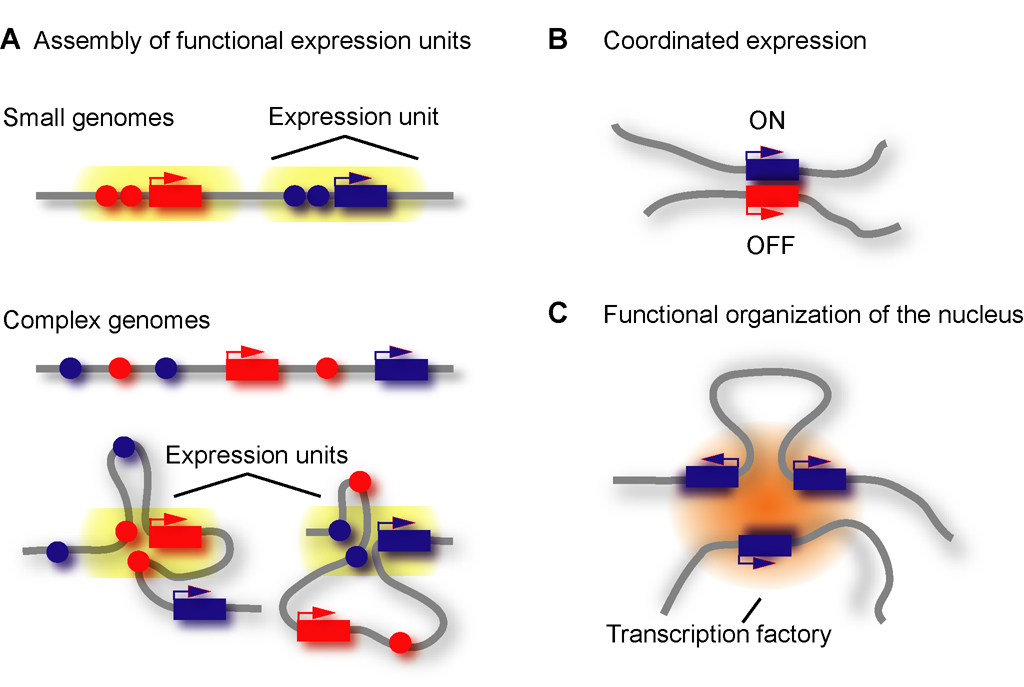
In compact genomes of organisms such as yeast, a gene and its regulatory elements form an uninterrupted genomic segment that constitutes a “regulatory expression unit”. However, in more complex genomes, such as those of human and mouse, genes and their regulatory elements can be dispersed over large genomic distances (1, 2). It has long been hypothesized that communication between widely spaced genomic elements is facilitated by the spatial organization of chromosomes that brings genes and their regulatory elements in close spatial proximity.
The organization of chromosomes has been studied by microscopy and more recently by chromosome conformation capture (3C) (3). 3C is a molecular technique that employs formaldehyde cross-linking and locus-specific PCR to detect physical contacts between genomic loci. These approaches are complementary, with microscopy providing information on single cells, but with relatively low resolution; and 3C allowing much higher resolution analyses, but requiring larger cell populations.
3C and microscopy studies have revealed that long-range chromosomal interactions are widespread, suggesting a surprisingly high level of communication between dispersed genomic elements.
Spatial assembly of expression units
The best-characterized examples of spatial association of genomic elements involve interactions between enhancers and target genes. A well-studied example is that of the beta-globin locus. The locus contains several beta-globin-like genes that are regulated by a single cis-acting element, the Locus Control Region (LCR), which is located ~10–60 kb upstream of the globin gene promoters. The LCR was found to physically associate with the active globin gene (4). Many more examples of long-range looping events have been identified, e.g. in the alpha-globin locus (5, 6), the interleukin gene cluster (7), but also for control of single genes [e.g. (8, 9)].
Highly specific associations between loci located on separate chromosomes have also been described. These trans-interactions can be between enhancers and putative target genes, as in the case of Olfactory Receptor genes (10). However, in other cases they appear to play a role in a higher level of gene control to coordinately regulate multiple loci (Figure 1). One example is the association between the TH2 cytokine locus on mouse chromosome 11 and the Interferon gamma gene on chromosome 10 (11). Expression of these loci is mutually exclusive and their association may provide a window of opportunity to enforce dis-coordinate expression.
Figure 1. Spatial assemblies.
A. Linearly defined expression units in compact genomes and spatially assembled expression units in complex genomes. B. Association between coordinately expressed genes. C. Co-localization of genes at sub-nuclear structures such as transcription factories.
The process of mammalian X-chromosome inactivation involves a specific trans-association between the X-chromosomes. Female cells carry two X-chromosomes, one of which is mostly silenced so that expression levels of X-linked genes are comparable to those in male cells. X-inactivation is initiated at the X-inactivation center. Recently, a transient interaction between the two X-inactivation centers was detected during the developmental stages at which X-inactivation is initiated (12, 13). Analysis of mutations in the inactivation centers showed that their association is intimately involved in the X-inactivation process. X-chromosome pairing provides an elegant mechanism for counting the number of X-chromosomes and for ensuring that their epigenetic fates are linked so that when one chromosome is inactivated the other is not.
These observations suggest an interesting model of what constitutes a “regulatory expression unit” in complex genomes. Whereas in compact genomes genes and their regulatory elements cluster along the linear genome sequence, in more complex genomes expression units are assembled by spatial clustering of genes and distant regulatory elements (Figure 1). This mode of de novo assembly of expression units could provide additional levels of gene regulation by allowing combinatorial association of genes and sets of regulatory elements. For example, for imprinted loci, maternal and paternal alleles associate with different elements to assemble into distinct expression units (14).
Chromosomal interactions are transient
Many of the observations of long-range interactions have been made using 3C and its variations. Performing 3C is relatively simple, but it has proven more complicated to interpret the results, as has been discussed in several reviews (15, 16). In particular, although many of the chromosomal interactions detected with 3C have been confirmed by microscopy, it is difficult to relate 3C signals to actual frequencies of association. In many cases the frequency of co-localization is rather low (less than 10% of cells at a given point in time), in accordance with the fact that chromosome conformation is dynamic and highly variable among individual cells. Therefore the rather rigid looping models that are widely presented, although appealing, can be misleading by ignoring the often highly transient nature of these interactions.
Functions of chromosomal interactions
Observing a specific association between two loci does not by itself reveal a function for that interaction. Additional approaches such as knock-down of proteins that may mediate the interaction (e.g. transcription factors) or deletion of the regulatory element can reveal causal relationships between long-range interactions and gene regulation. Another powerful approach is to analyze co-localization of loci by in situ hybridization combined with simultaneous visualization of RNA production at the gene to determine whether the interaction is correlated in time with gene transcription at the level of single cells. It should be noted that interactions have been observed that correlated with gene transcription but deletion of the interacting regulatory element did not affect expression (10, 17). Although this could indicate that the interaction is not relevant, it could equally reflect our very limited understanding of the role of chromosomal associations in genome regulation.
How do chromosomal associations affect gene expression? Enhancer-promoter interactions could aid in stable recruitment of components of the transcription machinery at the promoter. In addition, enhancer-bound enzymatic activities could be brought in contact with promoter complexes that are then modified e.g. phosphorylated or methylated, leading to modulation of promoter activity. Other types of interactions, such as those between the X-inactivation centers, could allow coordinated assembly of two distinct protein complexes on the interacting partners. Alternatively, given the very transient nature of these associations, the two loci may acquire distinct but stable marks, e.g. DNA methylation, that direct assembly of protein complexes at later time points when the loci no longer interact.
How do loci get together?
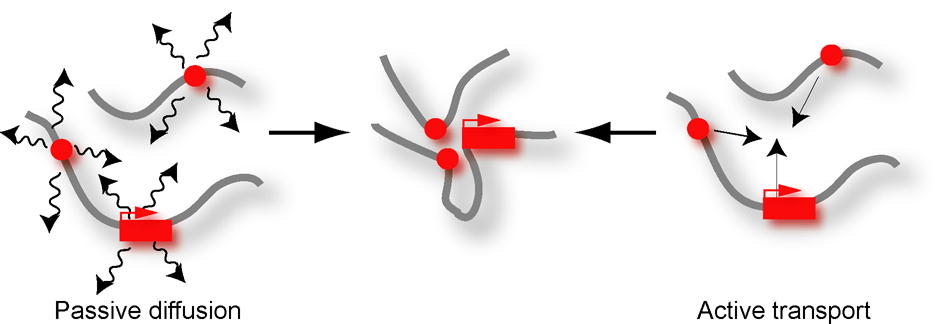
Several models have been proposed (18) by which distant genomic elements can contact each other (Figure 2). Passive models assume that the mobility of loci provides opportunities for random collisions that are then converted into productive interactions dependent upon the affinity and specificity of bound protein complexes. Although these models are appealing, it seems that active processes are required as well to directly guide loci towards each other. For instance, enhancers have been proposed to actively track along chromatin fibers until a receptive promoter is encountered. Recently, it was found that loci can follow rapid and directed trajectories through the nucleus in an actin-dependent fashion (19, 20). The roles of nuclear actin and myosin have been contentious, but these exciting recent results strongly suggest that they play critical roles in facilitating long-range interactions by transporting loci towards each other or to specific sub-nuclear neighborhoods.
Figure 2.
Passive and active models for bringing loci together.
Genomes also contain regulatory elements that modulate interactions between other elements. So-called insulator elements prevent an enhancer from activating a promoter but only when it is positioned in between them. How insulators work is not known in detail, but they too engage in long-range interactions with other elements (21), suggesting they generate looped chromosome structures that somehow facilitate the formation of appropriate assemblies of enhancers and target genes.
Future perspective
Currently significant effort is aimed at comprehensive mapping of chromosomal interactions. Several adaptations of 3C have been developed that allow large-scale detection of genomic interactions using microarrays or deep sequencing. The 4C method (3C-on Chip, or Circular 3C) allows identification of regions throughout the genome that are physically close to a single locus of interest (22, 23). The 5C method (3C-Carbon Copy) is not anchored on a single locus and is used for mapping dense interaction networks throughout large chromosomal regions of interest (24). Although these descriptive mapping studies will yield new insights into the spatial organization of genomes, additional approaches will be essential to unravel the mechanisms by which chromosomal associations affect genome regulation. These approaches include time-resolved imaging of chromosomal loci, molecular and genetic manipulation of the mechanisms that control the sub-nuclear localization and movement of loci, as well as biochemical studies to characterize the complexes that mediate chromosomal associations. Combined these various approaches promise to provide new insights into the three-dimensional aspects of gene regulation.
References and Notes
- 1.Kleinjan DA, van Heyningen V. Am J Hum Genet. 2005 Jan;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West AG, Fraser P. Hum Mol Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 3.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 4.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 5.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou GL, et al. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spilianakis CG, Flavell RA. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 8.Grass JA, et al. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jing H, et al. Mol. Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomvardas S, et al. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 11.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 12.Xu N, Tsai CL, Lee JT. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 13.Bacher CP, et al. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 14.Kurukuti S, et al. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker J. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 16.Simonis M, Kooren J, de Laat W. Nat. Methods. 2007;4:805–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 17.Fuss SH, Omura M, Mombaerts P. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Engel JD, Tanimoto K. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 19.Chuang CH, et al. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 20.Dundr M, et al. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace JA, Felsenfeld G. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonis M, et al. Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Z, et al. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 24.Dostie J, et al. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Dekker is supported by grants from NIH (HG003143), the Keck Foundation and the Cystic Fibrosis Foundation. M. Walhout is acknowledged for suggestions for this article.