Summary
The control of gene expression by enzymes provides a direct pathway for cells to respond to fluctuations in metabolites and nutrients. One example is the proline utilization A (PutA) protein from Escherichia coli. PutA is a membrane-associated enzyme that catalyzes the oxidation of L-proline to glutamate using a flavin containing proline dehydrogenase domain and a NAD+ dependent Δ1-pyrroline-5-carboxylate dehydrogenase domain. In some Gram-negative bacteria such as E. coli, PutA is also endowed with a ribbon-helix-helix DNA-binding domain and acts as a transcriptional repressor of the proline utilization genes. PutA switches between transcriptional repressor and enzymatic functions in response to proline availability. Molecular insights into the redox based mechanism of PutA functional switching from recent studies are reviewed. In addition, new results from cell-based transcription assays are presented which correlate PutA membrane localization with put gene expression levels. General membrane localization of PutA, however, is not sufficient to activate the put genes.
Keywords: PutA, transcriptional regulation, membrane-binding, proline utilization, DNA-binding, multifunctional enzyme
Introduction
We are increasingly discovering that enzymes can be directly involved in gene regulation. In addition to their enzymatic activity, enzymes can have extra functions such as binding to DNA or binding to regulatory protein factors that enable them to control gene expression through a variety of mechanisms (Hall et al., 2004; Jeffery, 2004; Shi and Shi, 2004). The regulation of proline metabolism by the proline utilization A (PutA) protein in Gram-negative bacteria is an excellent example of how enzymes can act as sensors of cellular metabolism. Proline oxidation to glutamate occurs in two steps catalyzed by proline dehydrogenase (PRODH) and Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) (Scarpulla and Soffer, 1978; Menzel and Roth, 1981a; Abrahamson et al., 1983). PutA is a flavoenzyme that links PRODH and P5CDH activities on a single polypeptide. In some Gram-negative bacteria, PutA also encodes a N-terminal DNA-binding domain that enables PutA to function as a transcriptional repressor (Gu et al., 2004). PutA controls transcription of the putA and putP genes in response to proline (Menzel and Roth, 1981b; Wood, 1981; Ostrovsky De Spicer et al., 1991; Brown and Wood, 1992; Muro-Pastor et al., 1997). The putP gene encodes a high-affinity Na+-proline transporter (Chen and Wilson, 1986). Thus, in certain Gram-negative bacteria such as Escherichia coli, PutA is a trifunctional protein that directly links proline metabolism to the control of gene expression.
PutA from E. coli is a polypeptide of 1320 amino acids that purifies mainly as a dimer (Brown and Wood, 1992; Ling et al., 1994). The PRODH domain has been localized to residues 261–612 and X-ray crystal structures of the PRODH domain show that it forms a β8α8 barrel active site which contains a non-covalently bound flavin adenine dinucleotide (FAD) cofactor (Lee et al., 2003). PRODH catalyzes the 2 e- oxidation of proline to Δ1-pyrroline-5-carboxylate (reductive half-reaction). The reduced FAD formed in this step is converted back to oxidized FAD by electron transfer to an acceptor in the cytoplasmic membrane, presumably ubiquinone, to complete the PRODH reaction cycle (oxidative half-reaction) (Wood, 1987). The product of the PRODH reaction, Δ1-pyrroline-5-carboxylate, is subsequently hydrolyzed to γ-glutamic acid semialdehyde which is then oxidized by P5CDH in a NAD+ dependent reaction to generate glutamate and NADH. The P5CDH domain is localized to residues 650–1130 (Ling et al., 1994).
PutA functions as a membrane associated enzyme but is a transcriptional repressor in the cytosol (Wood, 1987; Ostrovsky De Spicer et al., 1991; Brown and Wood, 1993; Muro-Pastor et al., 1997). These disparate functions imply that PutA must change locations in the cell. PutA switches between a DNA-binding transcriptional repressor and a membrane-bound enzyme depending on proline levels. In the absence of proline, PutA accumulates in the cytoplasm and represses transcription of the put genes (Ostrovsky De Spicer et al., 1991; Brown and Wood, 1992). In the presence of proline, PutA binds to the inner cytoplasmic membrane, where it catalyzes the oxidation of L-proline to glutamate (Brown and Wood, 1993; Ostrovsky De Spicer and Maloy, 1993; Zhang et al., 2004). Thus, PutA senses and responds to proline availability in the environment leading to derepression of the put genes and the use of proline as a growth substrate by the cell. Under poor nutrient conditions, proline can be utilized as a source of carbon, nitrogen and energy.
The general mechanism whereby PutA switches its intracellular location and function according to proline levels involves the FAD redox state and conformational changes that favor membrane binding (Wood, 1987; Brown and Wood, 1993; Ostrovsky De Spicer and Maloy, 1993; Surber and Maloy, 1999; Zhang et al., 2004). Here we will review the DNA and membrane binding properties of PutA and discuss mechanistic insights and new findings that support a redox-based mechanism for the functional regulation of PutA as originally proposed by Wood (1987).
DNA binding
Biochemical and structural characterization of the PutA-DNA binding domain have shown that it forms a dimer and is a member of the ribbon-helix-helix (RHH) superfamily of transcriptional regulators (Gu et al., 2004; Larson et al., 2006). The RHH binding domain includes N-terminal residuals 2–43 (Larson et al., 2006). In RHH proteins, the β-sheet recognizes the major groove of the DNA. Lys9 of the RHH anti-parallel β-sheet in PutA has been shown to be critical for PutA-DNA associations as substitution of Lys9 with a Met residue in the PutAK9M mutant eliminates DNA binding (Larson et al., 2006). Transcription repression by PutA occurs in the cytoplasm where it binds to multiple sites in the 419-bp intergenic region of the putP and putA genes thereby repressing the expression of the put genes which are transcribed in opposite directions (Ostrovsky De Spicer et al., 1991; Brown and Wood, 1992; Ostrovsky De Spicer and Maloy, 1993). The dissociation constant (KD) for the overall PutA-DNA complex was estimated to be 45 nM by gel-mobility shift assays (Becker and Thomas, 2001). PutA was reported to induce curvature in the DNA and we have shown that PutA binds to the put control DNA with a Hill coefficient of 1.75 (Ostrovsky De Spicer et al., 1991; Gu et al., 2004).
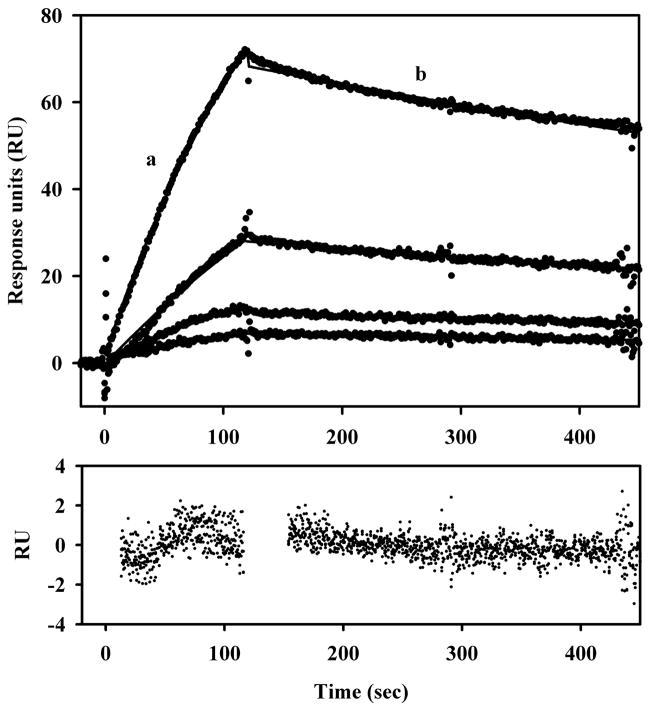
To determine whether PutA-DNA interactions were sensitive to the redox state of the FAD cofactor, PutA-DNA interactions were investigated using spectroelectrochemistry methods. Only a two-fold increase in the dissociation constant (KD) was estimated for the overall PutA-DNA complex upon reduction of the FAD cofactor (KD(ox) ~ 45 nM vs KD(red) ~ 100 nM) (Becker and Thomas, 2001). We have since tested this further using surface plasmon resonance (SPR). Figure 1 shows the kinetic traces of proline reduced PutA binding to the put control DNA immobilized on a streptavidin (SA) sensor chip. The association and dissociation rate constants were determined to be 1.93 × 104 M−1 s−1 and 6.1 × 5 10−4 s−1, respectively. The apparent KD for the PutA-DNA complex in the presence of proline is thus ~ 32 nM. A KD value of ~ 15 nM was calculated for the oxidized PutA-DNA complex from SPR measurements in the absence of proline (data not shown). In agreement with that estimated by spectroelectrochemistry, we observe about a two-fold change in PutA-DNA binding affinity under reducing conditions. The small influence of the FAD redox state on PutA-DNA interactions indicates that PutA functional switching is not significantly mediated by changes in PutA-DNA binding affinity.
Fig. 1.
SPR sensorgrams of the association and dissociation kinetics of PutA-DNA interactionsin the presence of proline. From bottom to top, increasing concentrations of PutA (12.5, 25, 50, and 100 nM) in the presence of 5 mM proline were injected onto a SA sensor chip coated with E. coli put control DNA. The association phase (a) corresponds to the injection of PutA at 60 μl/min for 120 s and the dissociation phase (b) corresponds to the flow of HEPES-N buffer (pH 7.4, 150 mM NaCl) at 60 μl/min for 300 s. For immobilization of the put control DNA, put control DNA was 5′-end labeled by PCR using biotinylated primers. Biotinylated put control DNA was then immobilized onto the surface using a streptavidin derivatized chip (SA chip) as previously described (Buckle, 2001). PutA for these experiments was purified as previously described (Zhang et al., 2004). The data were fit by global analysis to a 1:1 Langmuir binding isotherm as described (Zhang et al., 2004). Signals from the control surface have been subtracted.
Membrane binding
Because only minor changes occur in PutA-DNA interactions, PutA-membrane interactions were then considered to be critical for directing redox-dependent changes in PutA intracellular location and function. Previously, proline was shown to induce PutA-membrane binding (Wood, 1987; Brown and Wood, 1993). To distinguish between substrate binding and FAD reduction, the kinetics of PutA-membrane binding were explored by SPR (Zhang et al., 2004). Kinetic measurements of PutA binding to model lipid bilayers clearly showed that FAD reduction alone regulates PutA-membrane binding. Oxidized PutA showed no binding to immobilized E. coli polar lipid vesicles. Reduction of the FAD cofactor by proline or the chemical reducing agent sodium dithionite induced formation of a tight PutA-membrane complex (KD ≤ 0.01 nM) (Zhang et al., 2004). Clearly, PutA-membrane interactions are highly redox sensitive in contrast to the weak redox influence on PutA-DNA binding. These observations support the hypothesis that PutA functional switching is driven by FAD reduction and induced PutA-membrane binding.
Unlike the well characterized RHH domain of PutA, insights into the membrane binding domain of PutA are just beginning to emerge. Amino acid sequence alignment of various PutA family members reveals that PutA proteins which have the RHH domain along with the two enzymatic domains (i.e., trifunctional PutAs) are generally longer polypeptides (1273–1320 residues) than PutA proteins that lack DNA-binding activity and are strictly bifunctional such as PutA from Bradyrhizobium japonicum (999 residues) (Krishnan and Becker, 2005). Besides the additional N-terminal residues encoding the RHH domain, trifunctional PutAs have 150–190 extra residues at the C-terminus relative to most bifunctional PutAs. This conserved C-terminal extension in trifunctional PutAs implies that the C-terminus may have an important functional role such as membrane binding. Molecular dissection of PutA from E. coli was performed to assess the functional role of the C-terminal region. Sequential C-terminal deletions of PutA eventually identified a region involving residues 1295–1320 that appeared important for membrane binding. Secondary structure analysis of the C-terminal residues in E. coli PutA indicates a potential α-helix from residues 1289–1309 with a hydrophobic face suitable for membrane binding.
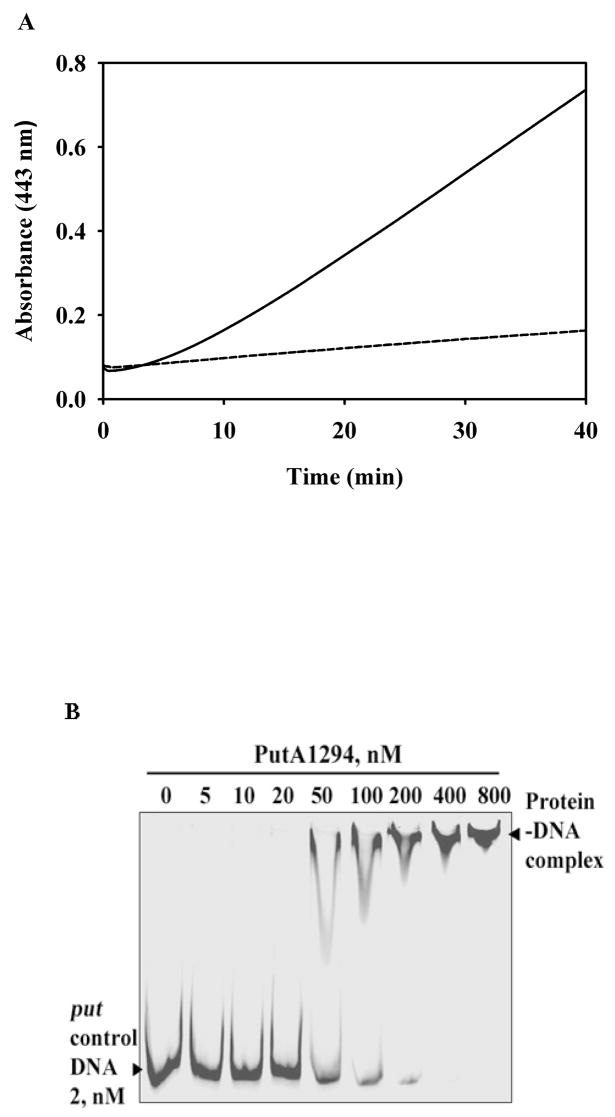
To directly test whether the putative α-helix associates with the membrane we attempted to purify a peptide which contained residues 1289–1309 but so far have been unsuccessful due to insolubility problems with the peptide. Instead, a C-terminal truncation mutant of PutA that is missing the last 26 residues (PutA1294) was characterized and shown to have 10-fold lower functional membrane binding activity (8.0 mU/mg) relative to wild-type PutA (81 mU/mg) (Figure 2A). The lower activity is not due to deficiencies in PRODH activity as PutA1294 exhibits proline: dichlorophenolindophenol (DCPIP) oxidoreductase activity (5.2 U/mg) that is similar to wild-type PutA (5.4 U/mg) (Becker and Thomas, 2001). Furthermore, the UV-visible spectral properties of the FAD cofactor in PutA1294 are the same as wild-type PutA (Becker and Thomas, 2001). Thus, C-terminal residues 1295–1320 appear to be critical for membrane binding and may be part of a membrane binding domain in PutA. We should also note that PutA1294 lacks P5CDH activity suggesting that deleting residues 1294–1320 impacts other functional domains in the C-terminal half of PutA. As a result, we cannot conclude yet whether C-terminal residues 1294–1320 are directly involved in membrane binding.
Fig. 2.
Functional membrane association and DNA-binding assays with PutA1294. A, Full-length PutA (100 μg, solid line) and PutA1294 (100 μg, dashed line) were incubated with 60 mM proline, 4 mM o-aminobenzaldehyde, and inverted membrane vesicles from E. coli strain JT31 putA- (0.1 mg/mL membrane protein) in 20 mM Mops buffer (pH 7.5) at 23 °C. The reactions were monitored at 443 nm as previously described (Becker and Thomas, 2001). The calculated specific activities for full-length PutA and PutA1294 are 81 and 8.0 mU mg−1 of membrane protein, respectively. B, Binding mixtures of TRdyed-700 labeled put control DNA (2 nM) and PutA1294 (0, 5, 10, 20, 50, 100, 200, 400, and 800 nM) were incubated at 20 °C for 20 min in 50 mM Tris buffer (pH 7.5, 50 mM NaCl). The PutA1294-DNA complexes were then separated by native polyacrylamide gel (4 %) electrophoresis at 4 °C as previously described (Gu et al., 2004).
Conformational changes
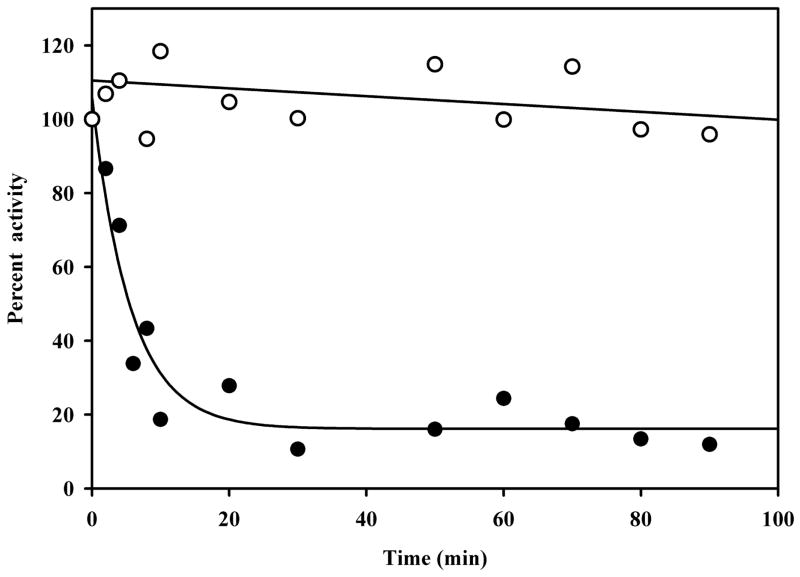
A prerequisite for a protein to be involved in multiple processes would seem to be some degree of flexibility which enables the protein to change its function in response to a signal such as reduction of the FAD cofactor. PutA from E. coli undergoes a structural transition that turns on PutA-membrane binding. In a heat stability test of PutA shown in Figure 3, PRODH activity was observed to decrease at 45 °C with a t1/2 of ~ 6.3 min. A truncated form of PutA that contains only the PRODH domain, PutA86-630, is noticeably more stable with no significant decrease in PRODH activity up to 100 min (see Figure 3) (Zhu and Becker, 2005). Thus, other polypeptide regions such as the RHH, P5CDH and membrane binding domains appear to decrease the stability of the PRODH domain and perhaps create more flexibility in the full-length PutA structure. Conformational changes in PutA most likely involve the membrane-binding domain rather than the RHH domain since reduction of FAD strongly enhances PutA-membrane binding while only weakly influencing PutA-DNA binding.
Fig. 3.
Thermostability of PutA. Full-length PutA (2 mg/ml) and PutA86–630 (2 mg/ml) were incubated in 50 mM potassium phosphate buffer (pH 7.5) containing 50 mM NaCl at 45 °C. At the indicated time points protein samples were removed from the heating mixture and assayed for PRODH activity using the proline:oxidoreductase DCPIP assay as previously described (Becker and Thomas, 2001). Assay results for PutA (closed circles) and PutA86–630 (open circles) are reported as relative percent activity based on the initial PRODH activity at 0 min. A rate constant of 0.18 ± 0.03 min−1 was estimated for the decay in PRODH activity with full-length PutA using best-fit analysis to a single exponential equation. PutA and PutA86–630 were purified as described (Zhu and Becker, 2005; Zhang et al., 2007).
Wood demonstrated that increased PutA-membrane binding affinity coincides with a proline dependent conformational change (Brown and Wood, 1993). Surber and Maloy later reported that proline-dependent flavin reduction increases the relative hydrophobicity of the PutA protein, favoring its interaction with the cytoplasmic membrane (Surber and Maloy, 1999). The increased hydrophobicity of PutA is not due to an oligomeric transition as PutA has been found to exist as a dimer under both oxidizing and reducing conditions (Brown and Wood, 1992). Besides a general increase in hydrophobicity upon reduction of FAD, the only global PutA conformational change that has been identified so far is in a region near the PRODH domain. Controlled potentiometric proteolysis of PutA revealed that a reversible conformational change occurs at a midpoint potential (Em) value of −58 mV (pH 7.5) which is near the Em of the bound FAD cofactor in PutA (−77 mV, pH 7.5) (Zhu and Becker, 2003). The observed redox dependent conformational change was mapped to a region near Arg234, a site which was found to be susceptible to cleavage by chymotrypsin only under reducing conditions (Zhu and Becker, 2003). Intrinsic fluorescence studies of a truncated PutA construct containing residues 86–630 also identified Trp211 as being involved in conformational changes (Zhu and Becker, 2005). An apparent rate constant of 0.59 s−1 was determined for the proline-dependent decrease in Trp fluorescence monitored by stopped-flow fluorescence (Zhu and Becker, 2005). This conformational change is 10-fold slower than the turnover number for PRODH activity in PutA. Therefore, it was proposed that the observed change in Trp fluorescence reports on a structural transition that regulates PutA-membrane binding (Zhu and Becker, 2005). Both the limited proteolysis and fluorescence studies indicate a conformationally labile region near the PRODH domain (residues 263–611) that responds to changes in the redox state of the FAD cofactor. Further studies are required to see whether this flexible domain interacts with the membrane or another domain in PutA that is responsible for membrane binding such as C-terminal residues 1295–1320.
Although the nature of the redox-dependent structural transition that occurs in the PutA polypeptide is not fully understood, molecular details concerning conformational changes in the FAD cofactor have recently become available by X-ray crystallographic studies of the PRODH domain. In crystal structures of oxidized PRODH, the 2′-OH ribityl group of FAD hydrogen bonds to Arg556 (Zhang et al., 2007). Arg556 is an important active site residue that ion pairs with the carboxylate group of proline. Removal of the guanidinium group in the mutant Arg556Met greatly reduces proline binding affinity in PutA (Km > 1 M proline) (Zhang et al., 2007). When the PRODH crystal form is reduced with sodium dithionite, the 2′-OH-Arg556 hydrogen bond is disrupted and a new hydrogen bond is formed between the 2′-OH ribityl group and the N(1) position of the FAD (Zhang et al., 2007). Dithionite reduction also induces a 22° bend in the isoalloxazine ring of the FAD (Zhang et al., 2007). Removal of hydrogen bond interactions with the 2′-OH ribityl group by site-directed mutagenesis of Arg556 or replacement of normal FAD with 2′-deoxy-FAD eliminates reductive activation of PutA-membrane binding (Zhang et al., 2007). Thus, the 2′-OH ribityl group has been proposed to act as a toggle switch between oxidized and reduced FAD to control the oxidized and reduced conformers of PutA that differ so drastically in membrane binding affinity (Zhang et al., 2007). Another important hydrogen bond was identified between Arg431 and the FAD N(5). Disruption of the Arg431-N(5) interaction by site-directed mutagenesis also eliminates PutA-lipid binding in SPR experiments under reducing conditions (Zhang et al., 2007). Therefore, interactions involving PutA residues Arg431 and Arg556, the 2′-OH ribityl group, and the N(1) and N(5) positions of the FAD cofactor are critical for reductive activation of PutA-membrane binding. Studies are underway to determine how redox signals generated at the FAD cofactor are transmitted beyond the PRODH active site to the membrane binding domain.
PutA intracellular localization and activation of the put genes
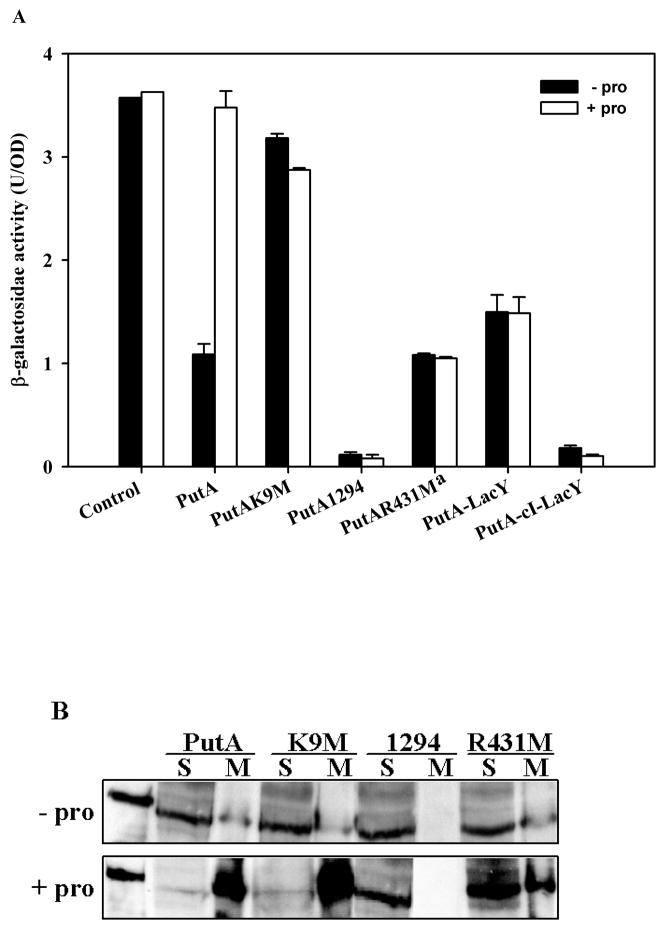
Studies of PutA-DNA binding, PutA-membrane binding, and PutA structural changes have generated strong evidence that proline reduction of the FAD cofactor triggers a conformational change in PutA that induces PutA-membrane binding. Enhanced PutA-membrane binding is consequently predicted to relieve repression of the putA and putP genes (Brown and Wood, 1993; Zhu and Becker, 2003; Zhang et al., 2004; Zhu and Becker, 2005; Zhang et al., 2007). To test this prediction in vivo, cell-based lacZ reporter gene assays were developed to correlate membrane localization with activation of put gene expression. For these assays, the E. coli strain JT31 putA− lacZ− was cotransformed with various PutA-pUC18 constructs (PutA-pUC18, PutAK9M-pUC18, PutA1294-pUC18, and PutAR431M-pUC18) and the reporter construct PputA:lacZ-pACYC184 in which the lacZ gene was positioned under control of the putA promoter as previously described (Gu et al., 2004; Zhang et al., 2007). E. coli strain JT31 putA− lacZ− containing different PutA-pUC18 constructs and the PputA:lacZ reporter construct was grown at 37 °C in M9 minimal medium supplemented with ampicillin (50 μg/ml), kanamycin (40 μg/ml) and chloramphenicol (34 μg/ml) to OD600 ~1.0. PutA expression from the lac promoter on pUC18 was uninduced as no isopropyl-β-D-thiogalactopyranoside (IPTG) was added to the culture medium. Activation of lacZ gene expression by proline was tested by culturing cells at 37 °C in minimal medium supplemented with and without 5 mM proline. Figure 4A shows that repression of the lacZ reporter gene by wild-type PutA is relieved by the addition of proline (5 mM) to the medium with β-galactosidase activity increasing by nearly 3-fold to the same level as the control cells (pUC18 + PputA:lacZ). In Figure 4B, Western blot analysis shows that proline shifts the intracellular location of wild-type PutA from the cytosol to the membrane consistent with membrane binding being critical for activation of put gene expression. Supplementing cells with L-tetrahydro-2-furoic acid, a non-reducing proline analog, does not relieve PutA repression of the lacZ reporter gene (Zhu et al., 2002; Zhang et al., 2007). Thus, proline binding alone to PutA does not activate expression of the put genes, rather FAD reduction is required.
Fig. 4.
Correlation of PutA intracellular location and β-galactosidase activity from the lacZ reporter construct in cells containing various PutA mutant constructs. A, E. coli strain JT31 was grown in minimal medium at 37 °C containing the PputA:lacZ reporter construct and pUC18 constructs expressing wild-type PutA, PutAK9M, PutA1294, PutA-LacY, and PutA-CI-LacY. β-galactosidase activity is reported as mean ± standard errors of the mean of four independent experiments. aData for PutAR431M is from Zhang et al. (2007). Control cells have the PputA:lacZ reporter construct and pUC18 alone. B, Western blot analysis of different PutA proteins. The partitioning of wild-type PutA, PutAK9M, PutA1294 and PutAR431M proteins between the soluble (S) and membrane (M) fractions was analyzed using antibodies generated against purified PutA47 which contains the DNA-binding domain of PutA. PutA proteins were expressed from pUC18 constructs in E. coli strain JT31 grown in the absence and presence of 5 mM proline at 37 °C in minimal medium. Far left-hand lane shows purified wild-type PutA (~ 144 kDa band). Protein bands were visualized by chemiluminescence detection.
We next sought to test residues/regions in PutA that are proposed to have a critical role in DNA binding (PutAK9M), membrane binding (PutA1294), or in the functional switching mechanism (PutAR431M) of PutA. The PutA mutant K9M which is deficient in DNA binding represses lacZ gene expression by only ~ 12 % and 19 % in the absence and presence proline, respectively, relative to control cells (Figure 4A) even though its expression level and localization pattern are similar to wild-type PutA (Figure 4B). These results are consistent with PutAK9M being defective in DNA-binding but not membrane binding (Larson et al., 2006).
To test how defective membrane binding impairs proline-dependent activation of the put genes, we then performed cell-based reported assays with the PutA1294-pUC18 construct. PutA1294 was found by Western analysis to be in the soluble fraction in the absence and presence of proline (Figure 4B). The lack of a proline-induced change in the intracellular location of PutA1294 confirms significantly diminished membrane associations. Figure 2B shows that PutA1294 binds put control DNA similarly to wild-type PutA. Consistent with strong DNA-binding and negligible membrane-binding activity, PutA1294 represses lacZ gene expression (~ 97% repression relative to β-galactosidase activity found in control cells) and is unresponsive to proline (Figure 4A). Thus, PutA1294 appears to behave as super-repressor that exhibits no response to proline. These results demonstrate that PutA-membrane localization is critical to relieve autorepression of the putA gene.
We also examined the PutA mutant R431M which is unresponsive to proline in SPR lipid binding studies (Zhang et al., 2007). Figure 4B shows that proline does not alter the partitioning of the PutA mutant R431M between the soluble and membrane fractions consistent with the lack of PutAR431M-lipid binding in SPR studies (Zhang et al., 2007). Proline also does not relieve PutAR431M repression of the lacZ reporter gene (see Figure 4A) (Zhang et al., 2007). Thus, Arg431 has key role in the redox functional switching mechanism of PutA that directs PutA membrane binding and activation of put gene expression.
Clearly, PutA membrane-binding is necessary to relieve PutA repression of the put genes. To test if membrane localization in general is sufficient to activate put gene expression, PutA was attached to the inner cytoplasmic membrane by fusing the C-terminus of the putA gene to the N-terminal six helices of the E. coli lactose permease lacY gene with and without a flexible linker between the putA and lacY genes (PutA-LacY-pUC18 and PutA-CI-LacY-pUC18, respectively) (Zelazny et al., 1997). The flexible linker (111 bp or 37 residues) was derived from a portion of the λcI repressor gene as previously described (Gorke et al., 2005). The PutA-LacY and PutA-CI-LacY fusion proteins (~ 167 kDa polypeptides) were confirmed to be localized in the membrane by Western blot analysis and were not detected in the soluble fraction (data not shown). The PutA-LacY fusion protein was observed to repress expression of the lacZ reporter gene (~ 58 % relative to control cells) (Figure 4A). Therefore, general localization of PutA on the membrane does not prevent PutA from functioning as a transcriptional repressor. Inserting a flexible linker (PutA-CI-LacY) increased repression of the lacZ reporter gene to ~ 95 % relative to control cells suggesting that varying the proximity of PutA to the membrane does influence the ability of PutA to repress transcription of the put genes. Both fusion proteins were unresponsive to proline indicating that artificially tethering PutA to the membrane disrupts functional PutA-membrane interactions (Figure 4A).
Conclusion
Results from a variety of studies now show conclusively that reduction of the FAD cofactor by proline induces a conformational change in PutA that leads to a shift in the intracellular location of PutA from the cytosol to the membrane thereby relieving repression of the put genes (Brown and Wood, 1993; Surber and Maloy, 1999; Zhang et al., 2004; Zhang et al., 2007). Reduction of the FAD cofactor results in significant conformational changes at the 2′-OH ribityl group and isoalloxazine ring of the FAD (Zhang et al., 2007). These changes are transmitted out of the FAD active site via the FAD N(1) and N(5) positions to drive global conformational transitions involving a flexible region around Trp211 and Arg234. The reduced PutA conformer exhibits enhanced membrane binding that sequesters PutA on the membrane, thus leading to the activation of the put genes. New results by comparing the proline response of wild-type PutA and PutA mutant proteins that are deficient in membrane binding further support this model. Proline-dependent localization of wild-type PutA on the membrane correlates with the activation of lacZ reporter gene expression. In contrast, no change in intracellular location is observed for either PutA431M or PutA1294 which is consistent with the lack of proline activation of lacZ expression in the cell-based reporter assays.
Görke et al. (2005) have shown that sequestration to the membrane in terms of physical separation from the chromosome does not account for the inactivation of a regulatory protein in bacteria. As shown in Figure 4A, artificially attaching PutA to the cytoplasmic membrane (PutA-LacY fusion) does not prevent PutA from acting as a transcriptional repressor of the PputA:lacZ reporter gene. Although the PputA:lacZ reporter gene is encoded on a low copy plasmid, our study still has implications for how PutA-membrane binding relieves repression of the put genes on the chromosome. Plasmids are often thought to be randomly distributed in the cell but studies have shown that plasmids are centrally localized and clustered (Pogliano et al., 2001). Thus, the finding that PutA can still act as a repressor when fused to the membrane supports the model that protein-membrane localization in general is not sufficient to suppress regulatory proteins from binding to chromosomal DNA in bacteria (Görke et al., 2005). Rather, specific conformational changes that are concomitant with membrane localization are needed to prevent protein-DNA binding (Görke et al., 2005).
In the case of PutA, membrane binding is governed by the redox state of the FAD cofactor with PutA binding to the membrane only under reducing conditions. The inability of oxidized PutA to bind to the membrane indicates that a membrane binding domain, perhaps involving residues 1295–1320, is shielded in the oxidized state while the N-terminal DNA binding domain is complexed to DNA (Zhang et al., 2004). Conversely, the PutA RHH domain must be concealed when reduced PutA binds functionally to the cytoplasmic membrane thereby preventing the N-terminal RHH domain from binding DNA (Zhang et al., 2004). Mutually exclusive PutA interactions with the DNA and the membrane were first proposed by Maloy et al. (1997). Therefore, PutA proteins that are defective in communicating FAD reduction (PutAR431M) or in functional membrane binding (PutA1294) are unable to disturb the PutA RHH domain interactions with the put control DNA and relieve repression of the put genes.
Acknowledgments
This work is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. This research was supported by grants from the National Institutes of Health GM061068 and the National Science Foundation MCB0340912. This publication was also made possible by NIH Grant Number P20 RR-017675-02 from the National Center for Research Resources. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- Abrahamson JLA, Baker LG, Stephenson JT, Wood JM. Proline dehydrogenase from Escherichia K12, properties of the membrane-associated enzyme. Eur J Biochem. 1983;134:77–82. doi: 10.1111/j.1432-1033.1983.tb07533.x. [DOI] [PubMed] [Google Scholar]
- Becker DF, Thomas EA. Redox properties of the PutA protein from Escherichia coli and the influence of the flavin redox state on PutA-DNA interactions. Biochemistry. 2001;40:4714–4722. doi: 10.1021/bi0019491. [DOI] [PubMed] [Google Scholar]
- Brown E, Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J Biol Chem. 1992;267:13086–13092. [PubMed] [Google Scholar]
- Brown ED, Wood JM. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J Biol Chem. 1993;268:8972–8979. [PubMed] [Google Scholar]
- Buckle M. Surface plasmon resonance applied to DNA-protein complexes. In: Moss T, editor. DNA-Protein Interactions: Principles and Protocols. Vol 148: Methods in Molecular Biology. Humana Press Inc.; Totowa, NJ: 2001. pp. 535–546. [DOI] [PubMed] [Google Scholar]
- Chen C-C, Wilson TH. Solubilization and functional reconstitution of the proline transport system of Escherichia coli. J Biol Chem. 1986;261:2599–2604. [PubMed] [Google Scholar]
- Görke B, Reinhardt J, Rak B. Activity of Lac repressor anchored to the Escherichia coli inner membrane. Nucleic Acids Res. 2005;33:2504–2511. doi: 10.1093/nar/gki549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D, Zhou Y, Kallhoff V, Baban B, Tanner JJ, Becker DF. Identification and characterization of the DNA-binding domain of the multifunctional PutA flavoenzyme. J Biol Chem. 2004;279:31171–31176. doi: 10.1074/jbc.M403701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr Opin Struct Biol. 2004;14:663–668. doi: 10.1016/j.sbi.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Becker DF. Characterization of a bifunctional PutA homologue from Bradyrhizobium japonicum and identification of an active site residue that modulates proline reduction of the flavin adenine dinucleotide cofactor. Biochemistry. 2005;44:9130–9139. doi: 10.1021/bi050629k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JD, Jenkins JL, Schuermann JP, Zhou Y, Becker DF, Tanner JJ. Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition. Protein Sci. 2006;15:2630–2641. doi: 10.1110/ps.062425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10:109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M, Allen SW, Wood JM. Sequence analysis identifies the proline dehydrogenase and pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coli PutA protein. J Mol Biol. 1994;245:950–956. doi: 10.1006/jmbi.1994.1696. [DOI] [PubMed] [Google Scholar]
- Menzel R, Roth J. Purification of the putA gene product. J Biol Chem. 1981a;256:9755–9761. [PubMed] [Google Scholar]
- Menzel R, Roth J. Regulation of genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J Mol Biol. 1981b;148:21–44. doi: 10.1016/0022-2836(81)90233-3. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor AM, Ostrovsky P, Maloy S. Regulation of gene expression by repressor localization: Biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J Bacteriol. 1997;179:2788–2791. doi: 10.1128/jb.179.8.2788-2791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky De Spicer P, Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc Natl Acad Sci USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky De Spicer P, O’Brian K, Maloy S. Regulation of proline utilization in Salmonella typhimurium: A membrane-associated dehydrogenase binds DNA in vitro. J Bacteriol. 1991;173:211–219. doi: 10.1128/jb.173.1.211-219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Ho TQ, Zhong Z, Helinski DR. Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:4486–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Soffer RL. Membrane-bound proline dehydrogenase from Escherichia coli. J Biol Chem. 1978;253:5997–6001. [PubMed] [Google Scholar]
- Shi Y, Shi Y. Metabolic enzymes and coenzymes in transcription-a direct link between metabolism and transcription? Trends Genet. 2004;20:445–452. doi: 10.1016/j.tig.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Surber MW, Maloy S. Regulation of flavin dehydrogenase compartmentalization: Requirements for PutA-membrane association in Salmonella typhimurium. Biochim Biophys Acta. 1999;1421:5–18. doi: 10.1016/s0005-2736(99)00104-2. [DOI] [PubMed] [Google Scholar]
- Wood J. Membrane association of proline dehydrogenase in Escherichia coli is redox dependent. Proc Natl Acad Sci USA. 1987;84:373–377. doi: 10.1073/pnas.84.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JM. Genetics of L-proline utilization in Eschericia coli. J Bacteriol. 1981;146:895–901. doi: 10.1128/jb.146.3.895-901.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny A, Seluanov A, Cooper A, Bibi E. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc Natl Acad Sci U S A. 1997;94:6025–6029. doi: 10.1073/pnas.94.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang M, Zhu W, Zhou Y, Wanduragala S, Rewinkel D, Tanner JJ, Becker DF. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA-membrane binding. Biochemistry. 2007;46:483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhou Y, Becker DF. Regulation of PutA-membrane associations by flavin adenine dinucleotide reduction. Biochemistry. 2004;43:13165–13174. doi: 10.1021/bi048596g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Becker DF. Flavin redox state triggers conformational changes in the PutA protein from Escherichia coli. Biochemistry. 2003;42:5469–5477. doi: 10.1021/bi0272196. [DOI] [PubMed] [Google Scholar]
- Zhu W, Becker DF. Exploring the proline-dependent conformational change in the multifunctional PutA flavoprotein by tryptophan fluorescence spectroscopy. Biochemistry. 2005;44:12297–12306. doi: 10.1021/bi051026b. [DOI] [PubMed] [Google Scholar]
- Zhu W, Gincherman Y, Docherty P, Spilling CD, Becker DF. Effects of proline analog binding on the spectroscopic and redox properties of PutA. Arch Biochem Biophys. 2002;408:131–136. doi: 10.1016/s0003-9861(02)00535-0. [DOI] [PubMed] [Google Scholar]