Abstract
Dentin matrix protein 1 (DMP1) is present in the extracellular matrix (ECM) of dentin and bone as processed NH2- and COOH-terminal fragments, resulting from proteolytic cleavage at the NH2 termini of 4 aspartic acid residues during rat DMP1 processing. One cleavage site residue, Asp181 (corresponding to Asp197 of mouse DMP1), and its flanking region are highly conserved across species. We speculate that cleavage at the NH2 terminus of Asp197 of mouse DMP1 represents an initial, first-step scission in the whole cascade of proteolytic processing. To test if Asp197 is critical for initiating the proteolytic processing of mouse DMP1, we substituted Asp197 with Ala197 by mutating the corresponding nucleotides of mouse cDNA that encode this amino acid residue. This mutant DMP1 cDNA was cloned into a pcDNA3.1 vector. Data from transfection experiments indicated that this single substitution blocked the proteolytic processing of mouse DMP1 in HEK-293 cells, indicating that cleavage at the NH2 terminus of Asp197 is essential for exposing other cleavage sites for the conversion of DMP1 to its fragments. The NH2-terminal fragment of DMP1 occurs as a proteoglycan form (DMP1-PG) that contains a glycosaminoglycan (GAG) chain. Previously, we showed that a GAG chain is linked to Ser74 in rat DMP1 (Ser89 in mouse DMP1). To confirm that mouse DMP1-PG possesses a single GAG chain attached to Ser89, we substituted Ser89 by Gly89. Data from transfection analysis indicated that this substitution completely prevented formation of the GAG-containing form, confirming that DMP1-PG contains a single GAG chain attached to Ser89 in mouse DMP1.
Key Words: Dentin matrix protein 1, Proteolytic processing, Glycosaminoglycan, Mutation
Introduction
Dentin matrix protein 1 (DMP1) was discovered by cDNA cloning [George et al., 1993]. The distinctive feature of DMP1 is a large number of acidic domains, a property that implicates it is a possible participant in regulating matrix mineralization. This purported biological function is supported by the observations that MC3T3-E1 cells overexpressing DMP1 demonstrate an earlier onset of mineralization and a significantly larger size of the induced mineralized nodules compared to those of the control cells [Narayanan et al., 2001]. Findings from Dmp1-null mice strengthen the conclusion that DMP1 plays an important role in biomineralization [Ye et al., 2004, 2005]. Recent studies also demonstrated the association between mutations in the DMP1 gene and autosomal recessive hypophosphatemic rickets, a novel human disorder [Feng et al., 2006; Lorenz-Depiereux et al., 2006].
DMP1 was originally postulated to be dentin specific, but later its expression was observed in bone and nonmineralized tissues [D'Souza et al., 1997; MacDougall et al., 1998; Ogbureke et al., 2007]. The cDNA of DMP1 has been cloned and sequenced in a number of species including rat [George et al., 1993], mouse [MacDougall et al., 1998], human [Hirst et al., 1997] and crocodilia [Toyosawa et al., 1999]. After translation, rat DMP1 is proteolytically processed into an NH2-terminal (37-kDa) fragment and a COOH-terminal (57-kDa) fragment at 4 bonds, Phe173-Asp174, Ser180-Asp181, Ser217-Asp218 and Gln221-Asp222, in the amino acid sequence [Qin et al., 2003]. Amino acid sequence alignment of DMP1 shows that residues in the region flanking Ser180-Asp181 are highly conserved across a very broad range of species, indicating that the proteolytic cleavage at this site must be related to an important biological function [Qin et al., 2004].
More recently, our group discovered that, in addition to the 37-kDa form, the NH2-terminal fragment of DMP1 also occurs as a proteoglycan, present in the extracellular matrix (ECM) of bone and dentin [Qin et al., 2006]. The proteoglycan variant, referred to as DMP1-PG, possesses a glycosaminoglycan (GAG) chain that is made predominantly of chondroitin-4-sulfate and is linked to the core protein via Ser74 in rat DMP1.
The purpose of this investigation was to determine the functional role of Asp197 of mouse DMP1 (corresponding to Asp181 in rat DMP1) as a key cleavage site residue essential for initiating the proteolytic processing events of DMP1. We also analyzed whether Ser89 of mouse DMP1 (Ser74 in rat DMP1) is the only GAG chain attachment site. In our study, the cleavage site residue Asp197 of mouse DMP1 was substituted by Ala197 (D197A substitution), while the Ser89 residue was substituted by Gly89 (S89G substitution). Results from transfection experiments demonstrated that D197A substitution prevented the proteolytic processing, while S89G substitution completely prevented formation of the glycosaminoglycan form of mouse DMP1.
Materials and Methods
Plasmid Construction
For generation of a DMP1 cDNA construct, we utilized a pBC-KS+ construct carrying full-length mouse DMP1 cDNA and a pcDNA3.1 vector (Invitrogen, San Diego, Calif., USA). Mutagenesis was performed on the pBC-KS+ construct to create 3 types of mutant DMP1 cDNA, using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, Calif., USA) following the manufacturer's protocol. For substitution of residues at cleavage sites, Asp197 and Asp234 in mouse DMP1 were substituted by Ala197 and Ala234, respectively; these 2 substitutions were named D197A and D234A, respectively. For the substitution of GAG chain attachment residue, Ser89 was substituted by Gly89 (designated S89G substitution). These mutations were confirmed by restriction enzyme digestion and DNA sequencing. The mutant DMP1 cDNA fragments were released by EcoRI digestion, and subcloned into pcDNA3.1 vectors.
Cell Culture, Transient Transfection and Western Immunoblotting
The human epithelial kidney 293 (HEK-293) cells were selected for transient transfection, since these cells have a high transfection efficiency for mammalian expression vector, and in our preliminary experiments, DMP1 was clearly detected when these cells were used for transfection. The HEK-293 cells were grown in Dulbecco's modified Eagle's medium (GIBCO-BRL/Invitrogen, Carlsbad, Calif., USA) supplemented with 10% (v/v) fetal bovine serum, 2 mML-glutamine. The pcDNA3.1 constructs containing DNA fragments coding for normal DMP1 as well as those for D197A, D234A and S89G substitutions were transfected into HEK-293 cells using the Lipofectamine 2000 (Invitrogen, Carlsbad, Calif., USA) method. After 16–18 h of transfection, cells were changed to conditioned medium, which was harvested at 72 h. Lyophilized conditioned media from HEK-293 cells or samples containing DMP1 and its processed fragments purified by ion exchange chromatography were separated on 4–20% Tris-glycine SDS-PAGE gels (LifeGels). For isolation of DMP1 and its fragments, the culture medium was loaded onto a Q-Sepharose column (Amersham Biosciences, Piscataway, N.J., USA) connected to a fast protein liquid chromatography system. In this separation, DMP1 and its processed fragments co-elute in early fractions, while DMP1-PG elutes in late fractions.
Proteins separated on SDS-PAGE were visualized by Stains-All staining and/or Western immunoblotting. The following 3 types of antibodies were used for chemiluminescent Western immunoblotting. (1) A polyclonal antibody generated by using peptide AYNHKPIGDQDDNDC (residue 471–485 of mouse DMP1) was used to detect the carboxyl terminus of DMP1; this antibody (designated anti-DMP1-C-785) recognizes the 57-kDa fragment as well as the full-length form of DMP1. (2) A polyclonal antibody raised against peptide GLGPEEGQWGGPSKLDSDEDS (residue 101–121 of mouse DMP1) recognizes the amino terminus of DMP1; this antibody (referred to as anti-DMP1-N-784) is immunoreactive to the 37-kDa fragment and the full-length form of DMP1, but shows a relatively weak reaction to DMP1-PG. (3) A monoclonal antibody (named anti-DMP1-N-clone 9B6.3) generated using the NH2-terminal fragment of DMP1 isolated from rat long bone as the antigen is strongly reactive to DMP1-PG, but shows a relatively weak reaction to the 37-kDa form; previously, this monoclonal antibody was used to successfully isolate DMP1-PG from bone [Qin et al., 2006]. All 3 antibodies were purified by affinity columns.
Results
D197A Substitution Blocked the Proteolytic Processing of Mouse DMP1
Stains-All staining as well as Western immunoblotting showed that the conditioned culture media of HEK-293 cells transfected with the pcDNA3.1 construct carrying either normal mouse DMP1 cDNA or mutant DMP1 contained significant amounts of DMP1 and/or its fragments, whereas the media from cells transfected with empty vector or with vector carrying the DMP1 cDNA insert in the incorrect orientation did not contain any DMP1 or its fragments.
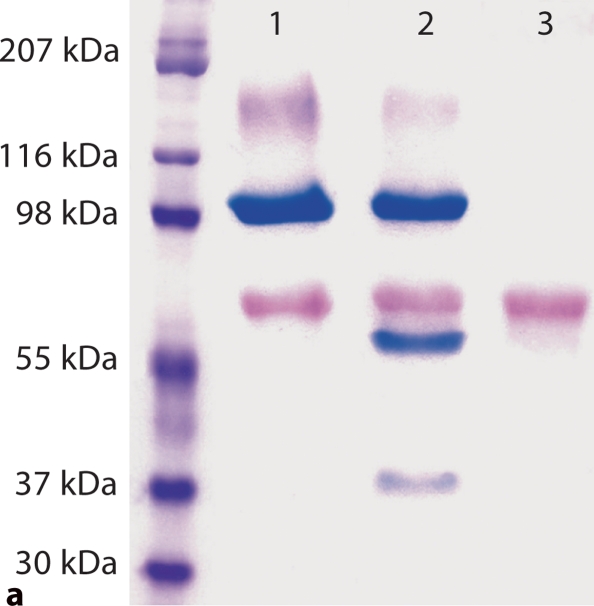
For analysis of the product of the D197A substitution, conditioned culture medium from cells transfected with a construct carrying the D197A substitution as well as that from the normal control were directly loaded onto SDS-PAGE gels for Stains-All staining and Western immunoblotting. As shown in figure 1a, an approximately 100-kDa band (stained blue) representing full-length DMP1 as well as the 57- and 37-kDa fragments were clearly visible in the normal control, whereas in the D197A substitution group, only full-length DMP1 was observed. The findings indicate that the D197A substitution blocked the conversion of DMP1 into N- and C-terminal fragments. The result from Western immunoblotting (fig. 1b) using the anti-DMP1-C-785 and anti-DMP1-N-784 polyclonal antibodies further confirmed the above findings. It should be noted that in the normal control group, a significant amount of uncleaved, full-length DMP1 was observed. This is most likely due to overexpression of the protein driven by the cytomegalovirus promoter in excess amounts that could not be cleaved by endogenous protease.
Fig. 1.
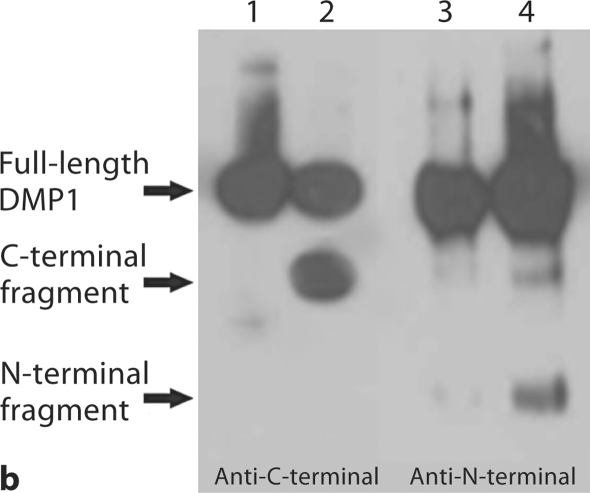
Substitution of a single amino acid residue prevented cleavage of mouse DMP1. a Stains-All staining. Lane 1: conditioned culture medium of cells transfected with pcDNA3.1 construct carrying a mouse DMP1 cDNA that encodes DMP1 protein, in which Asp197, an amino acid residue at a key cleavage site, was substituted by Ala197 (note the absence of the lower molecular weight, blue-stained band). Lane 2: pcDNA3.1 vector carrying normal mouse DMP1 cDNA (note the presence of low molecular weight bands stained blue). Lane 3: empty pcDNA3.1 vector (without DMP1 cDNA). The blue band at approximately 100–110 kDa is full-length DMP1. Blue bands at approximately 57 and 37 kDa are DMP1 fragments. Note that the 37-kDa fragment did not stain as darkly as the 57-kDa fragment, which is most likely due to the less acidic properties of the 37-kDa fragment. The purple band at approximately 65 kDa represents BSA from the culture medium. For each lane, 600 μl of lyophilized cell-conditional culture medium was loaded onto SDS-PAGE gels. b Western immunoblotting using polyclonal antibodies against COOH-terminal region of DMP1 (left 2 lanes, anti-DMP1-C-785) and NH2-terminal region of DMP1 (right 2 lanes, anti-DMP1-N-784). Lanes 1 and 3 are the same sample (D197A) as in lane 1 of a; lanes 2 and 4 are the same as in lane 2 of a (DMP1 without mutation). Note the absence of the DMP1 fragments in the lanes with an amino acid substitution at the proposed cleavage site (lanes 1 and 3). For each lane, 300 μl of lyophilized cell-conditioned culture medium was loaded onto SDS-PAGE gels. Please note that DMP1-PG was hardly recognized by the polyclonal antibody against the NH2-terminal region of DMP1.
D234A Substitution Did Not Block the DMP1 Processing
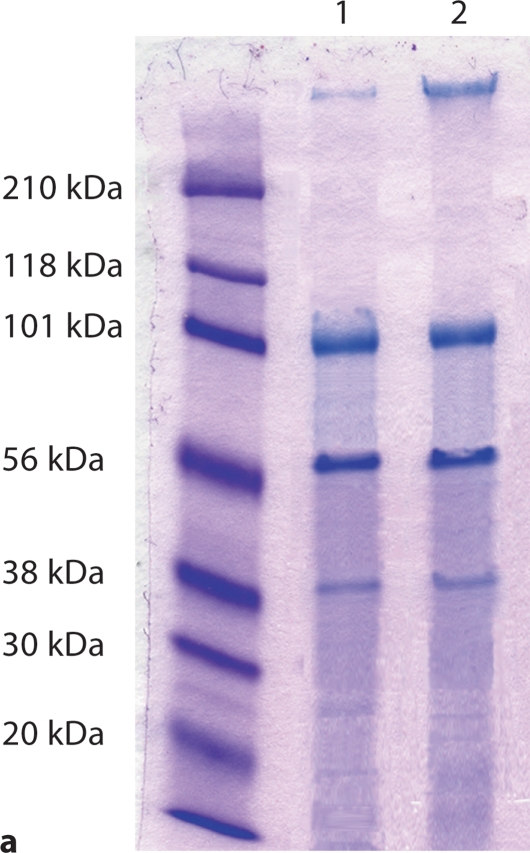
For analysis of the D234A substitution, proteins from the conditioned culture medium of cells transfected with a construct carrying the D234A substitution as well as from the normal control was first purified by ion exchange chromatography, and then fractions containing DMP1 and its fragments, were loaded onto SDS-PAGE gels for Stains-All staining and Western immunoblotting. Both Stains-All staining (fig. 2a) and Western immunoblotting (fig. 2b) of the D234A substitution and normal control groups showed essentially identical protein bands, indicating that the D234A substitution had no effect on the proteolytic processing of DMP1.
Fig. 2.
D234A substitution did not block DMP1 processing. For these experiments, proteins (DMP1 and its processed fragments) purified by ion exchange chromatography were used. Note that DMP1-PG was eluted in different chromatographic fractions and thus was not included in these samples. a About 6 μg of purified protein was loaded onto each lane for Stains-All staining. Note that the pattern of protein bands is similar between the D234A substitution (lane 1) and the normal control group (lane 2). b About 3 μg of purified protein was loaded onto each lane for Western immunoblotting using the antibody against the COOH-terminal region of mouse DMP1 (anti-DMP1-C-785). Lane 1: D234A substitution; lane 2: normal control group. Note the presence of the DMP1 COOH-terminal fragment (approximately 57 kDa) in both groups.
S89G Substitution Prevented GAG Glycosylation of DMP1
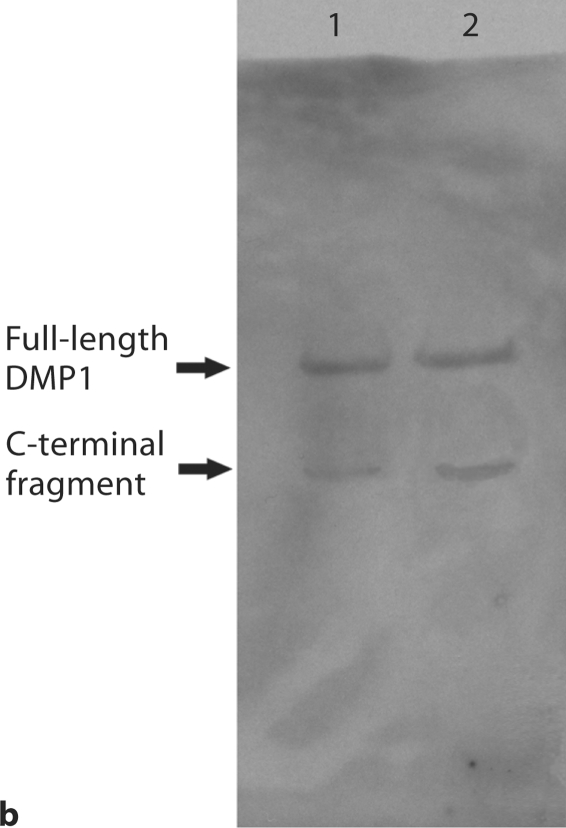
For analysis of the S89G substitution, conditioned culture medium from cells transfected with a construct carrying the S89G substitution as well as from the normal control was directly loaded onto SDS-PAGE gels for Western immunoblotting. As shown in figure 3, Western immunoblotting using the anti-DMP1-N-clone 9B6.3 antibody demonstrated the presence of DMP1-PG, which appeared as broad bands in the normal control, whereas these high molecular weight bands were completely absent in the S89G substitution group. This observation confirmed that Ser89 is the only site linking a single GAG chain to the core protein.
Fig. 3.
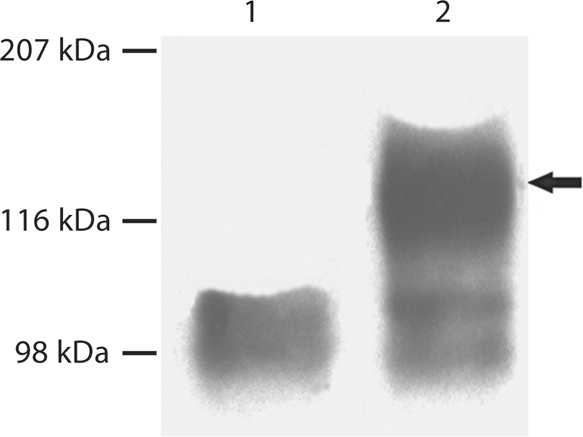
Substitution of Ser89 in mouse DMP1 completely prevented GAG glycosylation of DMP1. 300 μl of cell-conditioned culture medium was loaded onto SDS-PAGE, and the proteins were visualized by Western immunoblotting using a monoclonal antibody against the NH2-terminal region of DMP1 (anti-DMP1-N-clone 9B6.3). This monoclonal antibody [Qin et al., 2006] shows a strong reaction to DMP1-PG, but a weak reaction to the 37-kDa form. Lane 1: conditioned culture medium from cells transfected with the pcDNA3.1 construct carrying a mutant cDNA that encodes mouse DMP1, in which the GAG attachment residue, Ser89, was substituted by Gly89 (S89G). Lane 2: conditioned culture medium from cells transfected with the pcDNA3.1 construct carrying normal mouse DMP1 cDNA. Note the absence of DMP1-PG in lane 1 compared to the intact proteoglycan in lane 2 (arrow).
Discussion
In the ECM of bone and dentin, 3 forms of DMP1 have been identified: (1) the NH2-terminal fragment, (2) the COOH-terminal fragment and (3) DMP1-PG. Based on a number of observations, we have proposed that proteolytic processing of DMP1 is one of the necessary steps in the biomineralization of normal bones and teeth [Qin et al., 2004]. We believe that these variants with distinct biochemical features may play different roles in biomineralization. In vitro mineralization studies have demonstrated that the COOH-terminal fragment promotes mineralization by acting as a nucleator for hydroxyapatite formation [Tartaix et al., 2004, He et al., 2005; Gajjeraman et al., 2007]. Information regarding the function of DMP1-PG is lacking.
Our previous studies showed that all 4 cleavage sites in DMP1 occur at the NH2 termini of aspartyl residues. In this investigation we tested whether Asp197 of mouse DMP1 is a key cleavage site residue, essential for initiating proteolytic processing of DMP1. Our findings showed that substitution of Asp197 by Ala197 blocked the proteolytic cleavage of DMP1, whereas the substitution of another cleavage site residue, Asp234, did not prevent proteolytic processing of DMP1. These findings further strengthened our theory that DMP1 processing is an activation step. Based on these observations, we envision thatcleavage at the NH2 terminus of Asp197 represents an initial, first-step scission in the whole cascade of DMP1 processing. It is likely that conformational changes after the initial cleavage at Asp197 expose other sites for the proteinase(s) to exert its scission; that is, breaking of the bond Ser196-Asp197 starts a chain of proteolytic events, resulting in cleavage of several other peptide bonds. Thus, the cleavage of the Ser196-Asp197 bond would be essential for the release of DMP1 fragments from the precursor, and this step would be critical for the biological functions of DMP1.
We did not detect DMP1 in the culture medium of nontransfected HEK-293 cells. The observation that normal DMP1 is processed in these cells indicated that the proteinase(s) responsible for cleaving DMP1 must exist in a very broad range of tissues or cells. However, the cleavage pattern of DMP1 in HEK-293 cells showed certain differences from that of DMP1 in bone or dentin ECM; in the latter 2 tissue compartments, DMP1 fragments appeared as clusters of bands indicating multiple cleavages, while the protein band pattern in the medium of HEK-293 cells suggested a single cleavage. It is possible that in the ECM of bone and dentin, more than 1 proteinase is involved in DMP1 processing. Further studies are warranted to identify the enzymes responsible for DMP1 processing.
A careful analysis of the amino acid sequence of mouse DMP1 shows that Ser89-Gly90 is 1 of only 2 Ser-Gly dipeptides, an amino acid motif specific for the attachment of GAG chains to serine. The finding that the S89G substitution completely eliminated the proteoglycan form of DMP1 confirmed that DMP1-PG has a single GAG chain linked to Ser89 in mouse DMP1. Future studies involving transgenic mice carrying the S89G mutant transgene may provide valuable information regarding the biological functions of the GAG chain.
Acknowledgment
This work was supported by NIH grant DE005092 (C.Q.).
Abbreviations used in this paper
- DMP1
dentin matrix protein 1
- DMP1-PG
proteoglycan form of DMP1
- ECM
extracellular matrix
- GAG
glycosaminoglycan
- HEK-293
human epithelial kidney 293 cell line
References
- D'Souza R.N., Cavender A., Sunavala G., Alvarez J., Ohshima T., Kulkarni A.B., M. MacDougall. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res. 1997;12:2040–2049. doi: 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- Feng J.Q., Ward L.M., Liu S., Lu Y., Xie Y., Yuan B., Yu X., Rauch F., Davis S.I., Zhang S., Rios H., Drezner M.K., Quarles L.D., Bonewald L.F., White K.E. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38:1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajjeraman S., Narayanan K., Hao J., Qin C., George A. Matrix macromolecules in hard tissues control the nucleation and hierarchical assembly of hydroxyapatite. J Biol Chem. 2007;282:1193–1204. doi: 10.1074/jbc.M604732200. [DOI] [PubMed] [Google Scholar]
- George A., Sabsay B., Simonian P.A., Veis A. Characterization of a novel dentin matrix acidic phosphoprotein: implications for induction of biomineralization. J Biol Chem. 1993;268:12624–12630. [PubMed] [Google Scholar]
- He G., Gajjeraman S., Schultz D., Cookson D., Qin C., Butler W.T., Hao J., George A. Spatially and temporally controlled biomineralization is facilitated by interaction between self-assembled dentin matrix protein 1 and calcium phosphate nuclei in solution. Biochemistry. 2005;44:16140–16148. doi: 10.1021/bi051045l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst K.L., Simmons D., Feng J., Aplin H., Dixon M.J., M. MacDougall. Elucidation of the sequence and the genomic organization of the human dentin matrix acidic phosphoprotein 1 (DMP1) gene: exclusion of the locus from a causative role in the pathogenesis of dentinogenesis imperfecta type II. Genomics. 1997;42:38–45. doi: 10.1006/geno.1997.4700. [DOI] [PubMed] [Google Scholar]
- Lorenz-Depiereux B., Bastepe M., Benet-Pages A., Amyere M., Wagenstaller J., Muller-Barth U., Badenhoop K., S.M. Kaiser, R.S. Rittmaster, A.H. Shlossberg, J.L. Olivares, C. Loris, F.J. Ramos, F. Glorieux, M. Vikkula, H. Juppner, T.M. Strom. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet. 2006;38:1248–1250. doi: 10.1038/ng1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall M., Gu T.T., Luan X., Simmons D., Chen J. Identification of a novel isoform of mouse dentin matrix protein 1: spatial expression in mineralized tissues. J Bone Miner Res. 1998;13:422–431. doi: 10.1359/jbmr.1998.13.3.422. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Srinivas R., Ramachandran A., Hao J., Quinn B., George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci USA. 2001;98:4516–4521. doi: 10.1073/pnas.081075198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbureke K.U., Fisher L.W. SIBLING expression patterns in duct epithelia reflect the degree of metabolic activity. J Histochem Cytochem. 2007;55:403–409. doi: 10.1369/jhc.6A7075.2007. [DOI] [PubMed] [Google Scholar]
- Qin C., Brunn J.C., Cook R.G., Orkiszewski R.S., Malone J.P., Veis A., Butler W.T. Evidence for the proteolytic processing of dentin matrix protein 1: identification and characterization of processed fragments and cleavage sites. J Biol Chem. 2003;278:34700–34708. doi: 10.1074/jbc.M305315200. [DOI] [PubMed] [Google Scholar]
- Qin C., Baba O., Butler W.T. Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med. 2004;15:126–136. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- Qin C., Huang B., Wygant J.N., McIntyre B.W., McDonald C.H., Cook R.G., Butler W.T. A chondroitin sulfate chain attached to the bone dentin matrix protein 1 NH2-terminal fragment. J Biol Chem. 2006;281:8034–8040. doi: 10.1074/jbc.M512964200. [DOI] [PubMed] [Google Scholar]
- Tartaix P.H., Doulaverakis M., George A., Fisher L.W., Butler W.T., Qin C., Salih E., Tan M., Fujimoto Y., Spevak L., Boskey A.L. In vitro effects of dentin matrix protein-1 on hydroxyapatite formation provide insights into in vivo functions. J Biol Chem. 2004;279:18115–18120. doi: 10.1074/jbc.M314114200. [DOI] [PubMed] [Google Scholar]
- Toyosawa S., O'hUigin C., Tichy H., Klein J. Characterization of dentin matrix protein 1 gene in crocodilia. Gene. 1999;234:307–314. doi: 10.1016/s0378-1119(99)00195-x. [DOI] [PubMed] [Google Scholar]
- Ye L., MacDougall M., Zhang S., Xie Y., Zhang J., Li Z., Y. Lu, Y. Mishina, J.Q. Feng. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J Biol Chem. 2004;279:19141–19148. doi: 10.1074/jbc.M400490200. [DOI] [PubMed] [Google Scholar]
- Ye L., Mishina Y., Chen D., Huang H., Dallas S.L., Dallas M.R., Sivakumar P., Kunieda T., Tsutsui T.W., Boskey A., Bonewald L.F., Feng J.Q. Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J Biol Chem. 2005;280:6197–6203. doi: 10.1074/jbc.M412911200. [DOI] [PMC free article] [PubMed] [Google Scholar]