Abstract
Proteases act in important homeostatic pathways and are tightly regulated. Here, we report an unusual structural mechanism of regulation observed by the 2.5-Å X-ray crystal structure of the serine protease, granzyme C. Although the active-site triad residues adopt canonical conformations, the oxyanion hole is improperly formed, and access to the primary specificity (S1) pocket is blocked through a reversible rearrangement involving Phe-191. Specifically, a register shift in the 190-strand preceding the active-site serine leads to Phe-191 filling the S1 pocket. Mutation of a unique Glu–Glu motif at positions 192–193 unlocks the enzyme, which displays chymase activity, and proteomic analysis confirms that activity of the wild-type protease can be released through interactions with an appropriate substrate. The 2.5-Å structure of the unlocked enzyme reveals unprecedented flexibility in the 190-strand preceding the active-site serine that results in Phe-191 vacating the S1 pocket. Overall, these observations describe a broadly applicable mechanism of protease regulation that cannot be predicted by template-based modeling or bioinformatic approaches alone.
Keywords: protease regulation, register shift, allostery
Proteolysis plays an essential role in many biological pathways, an importance that is emphasized by the wide range and diversity of proteases that account for ≈2–3% of the human proteome (1). Of these, the largest group is the primary specificity (S1) family of serine proteases whose active sites contain three distinct features. The first is the catalytic triad (H57, D102, and S195 in chymotrypsin numbering) that creates a nucleophile to attack the substrate peptide bond. Mutation of any of these residues abolishes activity. Second is the “oxyanion hole” created by the main chain amide groups of S195 and G193. The positive charge in this region neutralizes a negatively charged oxygen atom in the substrate that is transiently formed during catalysis. Last, a conserved salt bridge is formed between the N-terminal amine group and the side chain of D194. This interaction introduces constraints on the active-site architecture that aid in cleavage efficiency.
Given the diversity of irreversible effects that serine proteases can have on a variety of biological pathways, their actions are tightly regulated. During biosynthesis, most serine proteases follow a one-step activation pathway involving precursor processing. They are initially synthesized as inactive precursors (zymogens) in which the active site is unformed. Limited proteolysis at the N terminus removes the inactivating propeptide, thus allowing the protease to fold into a mature, active state. Once activated, serine proteases are controlled by a variety of inhibitors that may be either specific (for example, inhibitors of the serpin family) or nonspecific (such as α2-macroglobulin) to ensure that they exert their effects only within the correct context.
Many proteases incorporate a further level of control and are active only in the presence of small molecule cofactors [typically metal ions (2)] or large ligands such as glycosaminoglycans (3) or other proteins (4). This results in a two-step active-site assembly pathway for activation, from the inactive zymogen to an intermediate with a partially formed active site, and finally the fully active protease. An equilibrium exists between the intermediate and active states that is skewed toward inactivity. Binding of the ligand induces conformational changes in the protease that swing the equilibrium in favor of the fully active state. This conformational control involves transmitting the effects of ligand binding at one site to the distal protease active site, a process termed allostery. Although the method of active-site deformity can differ widely between allosteric proteases, in all cases one of the catalytic residue side chains will be rotated away from the others, which often has the added effect of disrupting the oxyanion hole.
Here, we describe the crystal structure of granzyme C (gzmC), a rodent S1 serine protease expressed by cells of the immune system, which has been proposed to induce apoptosis in target cells (5, 6). However, gzmC cannot cleave a range of peptide substrates (7) and cannot bind diisopropyl fluorophosphate, indicating that the active site is distorted or highly constrained (8). The structure reveals a mode of protease control involving a “gated pathway” in which the oxyanion hole is unformed and the active site is reversibly occluded by the substrate-like side chain of an adjacent residue.
Results
Structure of Wild-Type (WT) GzmC.
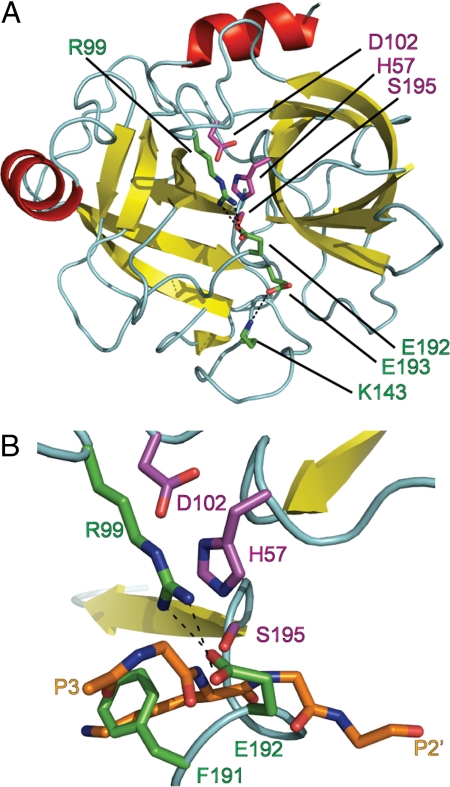
To investigate why gzmC appears catalytically inactive (7, 8), we determined its structure to 2.5 Å (Table 1). Two essentially identical gzmC molecules form a dimer in the asymmetric unit of the crystal, although there is no evidence for dimerization in vivo. Residues Ile-16–His-244 (chymotrypsin numbering of residues is used throughout) are visible in electron density and adopt the expected chymotrypsin-like serine protease fold (Fig. 1A), with the conserved catalytic residues His-57, Asp-102, and Ser-195 adopting canonical and hence potentially functional conformations.
Table 1.
Substrates of wild-type and unlocked granzyme C
| P5–P1↓P1′–P5′ | Signal ratio (wild-type:unlocked) | Protein description | Accession |
|---|---|---|---|
| IIVYL(33)↓YTKKV | 0.086 | 60S ribosomal protein L34 | Q9D1R9 |
| EEGYN(213)↓DGEVD | 0.085 | Acidic leucine-rich nuclear phosphoprotein 32 A | O35381 |
| LVKFM(308)↓CGEKQ | 0.067 | Flap endonuclease 1 | P39749 |
| LEAFM(120)↓AEVED | 0.053 | ATP-dependent RNA helicase DDX42 | Q810A7 |
| NILMF(891)↓TGETE | 0.049 | CTR9 homolog | Q62018 |
| PNTSF(480)↓ASDGS | 0.046 | 5′–3′ exoribonuclease 2 | Q9DBR1 |
| PLEAF(119)↓MAEVE | 0.026 | ATP-dependent RNA helicase DDX42 | Q810A7 |
| ALGFY(225)↓DTSEE | Unlocked only | Cell division cycle 5-related | Q6A068 |
| RLMEL(603)↓SATVE | Unlocked only | 170-kDa Centrosomal protein | Q6A065 |
| PGIFY(11)↓SDSFG | Unlocked only | MCM5 | P49718 |
| RACEM(73)NGEEC | Unlocked only | DNA-binding protein Ikaros | Q03267 |
| KVTFF(296)↓EPSPG | Unlocked only | Staufen homolog 1 | Q9Z108 |
| KFDFM(10)↓ASGED | Unlocked only | GRB2-related adaptor protein 2 | O89100 |
| EKCDY(602)↓MDEVT | Unlocked only | hnRNP U-like protein 2 | Q00PI9 |
| AYIEF(438)↓KSEAD | Unlocked only | Nucleolin | P09405 |
| KPQVM(508)↓AGTVK | Unlocked only | Pescadillo homolog 1 | Q9EQ61 |
| KLDFL(213)↓EGDQK | Unlocked only | EBP2 | Q9D903 |
| PSSVF(113)↓ASEFE | Unlocked only | LTV1 homolog | Q6NSQ7 |
| LVAAY(620)↓SGDSD | Unlocked only | RNA-binding protein 5 | Q91YE7 |
Fig. 1.
Granzyme C active site is occluded. (A) Diagram of granzyme C showing the chymotrypsin-like fold. (B) Enlarged view of the active-site cleft. Granzyme C was superposed with trypsin in complex with BPTI PDB ID code 2PTC). BPTI residues 13–17 indicate likely positioning of substrate P3–P2′. Cyan, loops and turns; red, α-helices; yellow, β-strands. Catalytic residues (magenta) D102, H57, and S195, inactivating residues (green) R99, K143, P191, E192, and E193 and BPTI main chain and P1 side chain residues (orange) are shown as stick models colored by element: red, oxygen; blue, nitrogen. The figures were rendered with PyMOL (32).
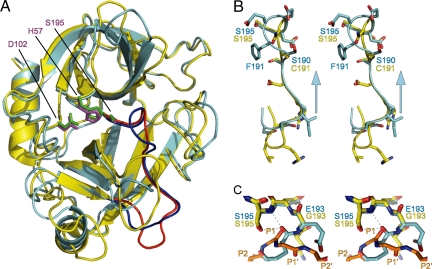
Sequence alignment of gzmC to its closest related homologs, gzmA and gzmB, predicted no insertions or deletions around the active site of gzmC [supporting information (SI) Fig. S1 and ref. 9]. However, a structural comparison of gzmC with granyzme A (61% identity; rmsd. 1.04 Å over 190 Cα atoms; Fig. 2A) revealed an unusual conformation of the active site that could explain gzmC inactivity (Fig. 1B). In particular, the structural data revealed that a register shift (Fig. S1) in the 190-strand that precedes the catalytic Ser-195 results in Phe-191 and Glu-192 bulging into the active site (Fig. 2B). As a result, the S1 pocket is filled by Phe-191, whereas Glu-192 covers the active site by forming a salt bridge with Arg-99 across the catalytic cleft (Fig. 1B). Furthermore, the oxyanion hole is improperly formed as the 192/193 peptide bond is “flipped” so that the amide group points away from the substrate, and the carbonyl oxygen of Glu-192 forms a hydrogen bond with the backbone amide of Ser-195 (thus occupying the space usually filled with a P1 carbonyl; Fig. 2C). Superposition of gzmC with trypsin bound to basic pancreatic trypsin inhibitor (BPTI; rmsd 0.96 Å over 189 Cα atoms) indicates that, separately or collectively, these rearrangements would inactivate the protease and block substrates from interacting with the active site (Fig. 1B).
Fig. 2.
Granzyme C inactivation is caused by a register shift. (A) Superposition of granzyme C (cyan) with human granzyme A (yellow; PDB ID code 1ORF). The active-site triads are shown as stick models: magenta, granzyme C; green, granzyme A. The blue (granzyme C) and red (granzyme A) regions are enlarged in B. (B) Stereoview of the register shift in granzyme C. The arrow indicates the direction of register shift. (C) Stereoview of the oxyanion hole. Trypsin in complex with BPTI (PDB ID code 2PTC) was added to the superposition. BPTI residues 13–17 indicate likely positioning of substrate P3–P2′. Dashed lines indicate hydrogen bonds between granzyme C E193 carbonyl and S195 amide and Oγ. The figures were rendered with PyMOL (32).
Unlocking GzmC Activity.
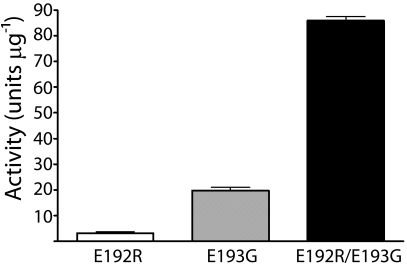
We next investigated the molecular basis for gzmC autoinhibition, by using site-directed mutagenesis to “unlock” activity by removing the constraints identified by the structure. Glu-192 and Glu-193 were mutated, either alone or in combination, to the corresponding residues in mouse granzyme B (Arg-192 and Gly-193) because this is the most closely related protein to gzmC. Activity was assayed on a succinyl-Phe-Leu-Phe thiobenzyl ester substrate because the presence of Phe-191 within the S1 suggested a similar P1 specificity. All three mutants were active, although the relative activity could not be determined in the absence of an active-site titrant (Fig. 3). An R99A mutant did not display activity, suggesting that backbone interactions of Glu-192, rather than steric hindrance by the side-chain salt bridge, are responsible for WT gzmC inactivation.
Fig. 3.
Granzyme C mutants are functional proteases. The indicated mutants of gzmC were assessed for activity on the substrate succinyl-Phe-Leu-Phe thiobenzyl ester. One unit of activity was defined as the amount of protease required to yield a change in A450 nm of 0.001 OD units s−1.
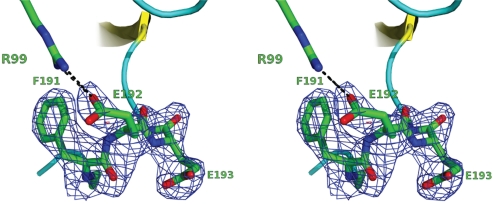
We note that the backbone of Glu-192 adopts stereochemically unfavorable ϕ/ψ geometry (−51.99°/−29.96°), yet is unambiguously modeled in electron density (Fig. 4). This suggests that the H-bond interactions between Glu-192 and Ser-195 provide enough stability to outweigh its unfavorable backbone stereochemistry. Furthermore, the activity of the E192R mutant, which retains glutamate at position 193, demonstrates that the highly conserved glycine at this position is not essential for serine protease function.
Fig. 4.
Stereochemistry of Glu-192. A stereo image of the Fo − Fc omit map, contoured at 3.0 σ, generated after coordinate refinement in the absence of residues Phe-191, Glu-192, and Glu-193. These residues appear as stick models; the salt bridge with Arg-99 is also shown for clarity.
Having demonstrated the catalytic potential of gzmC, we then investigated the effects of common protease cofactors on cleavage of succinyl-Phe-Leu-Phe thiobenzyl ester in an attempt to identify potential activators. However, no increase was detected in the activity of gzmC WT or the E192R/E193G mutant, in the presence of monovalent or divalent cations, common glycosaminoglycans or extracellular matrices, or by varying the pH from 3.1 to 7.4 (Table S1). This suggests that activity is released by binding to a substrate or protein cofactor.
Structure of Unlocked GzmC E192R/E193G.
To assess the changes in the S1 pocket of the unlocked form of gzmC, we determined the crystal structure of the double mutant E192R/E193G protein to 2.5 Å. The structure was essentially identical to that of the WT molecule (rmsd 0.27 Å over 203 Cα atoms) except that residues 145–152 and 186–192 were not visible in the electron density (Fig. S2). This indicates that breaking the H-bond network around Glu-192 results in extreme mobility of the 190-strand. Given that rigidity of the region around the active site of protease is important for function, this suggests that this strand is stabilized by substrate or cofactor binding.
Substrate Specificity and Function of GzmC.
To confirm substrate-induced activation and the preference for phenylalanine at the P1 position, we used proteome-wide N-terminal COFRADIC analysis (10) to identify substrates of WT or E192R/E193G unlocked gzmC in YAC-1 cell lysates. Eighteen substrates of unlocked gzmC were catalogued among the 982 proteins unambiguously identified by mass spectrometry, of which 6 were also cleaved by WT, although the signal ratios indicate that cleavage by WT gzmC was much less efficient (Table 1). Cleavage by WT and unlocked protease occurred at the same position in each shared substrate, indicating that substrate specificity was unaltered in unlocked gzmC. Importantly, this demonstration of cleavage by WT gzmC confirms that it is an autoregulated protease capable of substrate- or cofactor-induced activation.
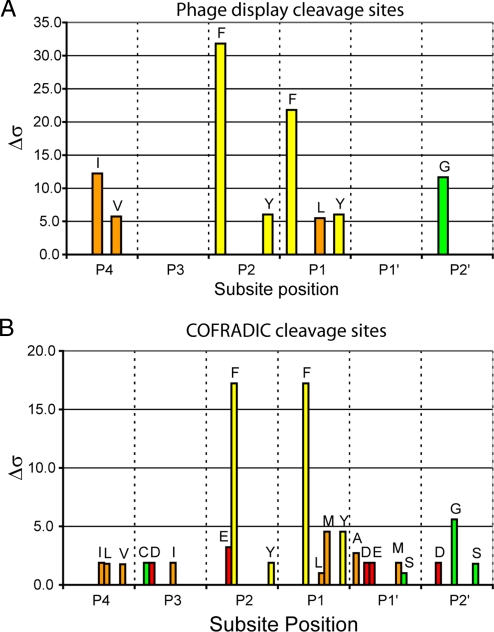
Employing a substrate phage display system, we then examined the extended substrate specificity of gzmC. Alignment of 68 unique sequences from two independent starting libraries treated with unlocked gzmC yielded the consensus recognition sequence [IV]-X-[FY]-[FLY]-X-G. To identify the P1 position, a peptide was synthesized based on this sequence (aminobenzoyl-RIMFFSGATK[dinitrophenyl]S), and cleavage products were identified by mass spectrometry. This showed that cleavage occurs only after the second phenylalanine residue. Importantly, this pattern matches the natural substrate cleavage sites identified by proteomics (Fig. 5).
Fig. 5.
Extended substrate specificity of gzmC. (A) A random nonamer phage display library was subject to 4 rounds of selection with 200 nM gzmC in two separate experiments. Sixty-eight phage sequences randomly selected from both experiments were aligned, and amino acid selectivity at each subsite was analyzed with a Δσ cutoff of 5 applied. (B) The P5–P5′ sequence of 19 gzmC cleavage events identified by proteomics was subject to the same analysis as in A except that the Δσ cutoff was reduced to 1 because of the small sample size.
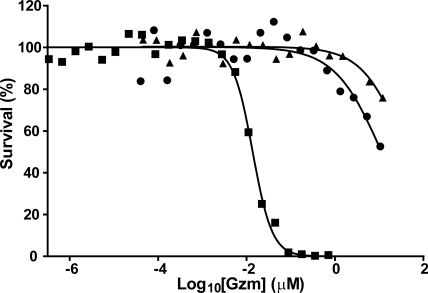
It has been reported that gzmC is cytotoxic when introduced into the cytoplasm of target cells (6). To confirm this, we delivered WT gzmC and gzmC E192R/E193G into P815 mouse mastocytoma cells by using recombinant mouse perforin (Fig. 6). We derived an EC50 for WT gzmC of 39.6 μM, 2,900-fold greater than that of human gzmB, whereas the unlocked mutant showed only a 4-fold improvement (EC50 = 11.2 μM). This suggests that even when catalytically competent, gzmC is an extremely inefficient cytotoxin and is unlikely to be cytotoxic in vivo. Alternative possibilities are that it is cleaves viral proteins as reported for its human homolog gzmH (11), or it is involved in immune signaling as recently shown for gzmA (12).
Fig. 6.
Granzyme C is a poor cytotoxin. P815 cells were treated for 1 h with a sublytic concentration of perforin and varying concentrations of gzmC WT (triangles), gzmC E192R/E193G (circles), or human gzmB (squares). Cells were allowed to recover overnight, and survival was measured by MTT assay. EC50 values were determined by fitting a sigmoidal dose–response curve with variable slope.
Discussion
Our structure reveals a mode of allosteric protease control involving a gated pathway in which the active site of a mature protease is occluded by a neighboring substrate-like side chain, Phe-191. Movement of Phe-191 out of the S1 pocket is required to allow activity and proper formation of the oxyanion hole, and this is most likely regulated by the register shift in the 190-strand. The flexibility required exists because gzmC, along with 10% of S1 family serine proteases, lacks a conserved C191–C220 disulfide bond that would tether the 190-strand and restrict its movement. As with many allosteric proteases, mature gzmC also has features reminiscent of unactivated (immature) zymogens, such as the unformed oxyanion hole (caused by flipping the 192–193 peptide bond) and the distortion of some substrate-binding pockets. Among the mature allosteric proteases, gzmC shows similarity only to Na+-free thrombin, where residue 192 lies over the active-site cleft and obstructs the S2 pocket (13). However, the striking feature of gzmC is the filling of the S1 pocket and the associated register shift.
Register shift in other proteins is controlled by a variety of mechanisms, including cofactor binding (14, 15) and posttranslational modifications such as phosphorylation (16), and we therefore propose that an exosite exists around the surface-exposed 185-loop. Interactions in this region would restore the register of the 190-strand to that observed in other serine proteases, thereby releasing residues 191 and 192 from the S1 pocket, forming the oxyanion hole, and allowing gzmC to accommodate and cleave substrates. In support of this model, a potential cofactor interaction site containing high B factors (and therefore predicted high mobility) exists in the WT gzmC structure around the 185-, 145-, and 220-loops. These regions comprise the so-called activation domain of serine proteases (17), and their involvement in the activation of gzmC would be consistent with the appropriation of zymogen-like features in the inactive state of allosteric proteases. Furthermore, the structure of the unlocked gzmC E192R/E193G mutant shows increased flexibility in both the 190-strand and 185-loop, suggesting that both regions move in concert (Fig. S2). Because these mutations substituted amino acids from granzyme B (which does not display this mechanism of allosteric control despite lacking the C191–C220 disulfide bond) the flexibility observed in gzmC must be inherent to this region of the protease with the strained conformation of residues 192 and 193 providing an energy barrier that locks the molecule in the inactive conformation.
It is important to note that although this mode of regulation is applicable to any class of protease, molecules subject to this mechanism may be difficult to identify a priori. Its conformational nature precludes identification via homology-based approaches, although further understanding of the features of the 190-strand that permit its flexibility may eventually aid in this. Furthermore, cocrystallization of proteases with an inhibitor (as is commonly done) would stabilize the open state of the active site, thereby concealing potential movements of the 190-strand. Despite these issues, there are still candidates in the literature that may be susceptible to this mode of inhibition. For example, mast cell protease 8 is unable to cleave peptide substrates despite a predicted functional fold (18).
In conclusion, our structure of gzmC has unveiled an allosteric mechanism for protease control based on tertiary fold interactions that cannot be easily predicted. This method of control should be considered in cases where proteases are catalytically inert despite containing all of the hallmarks of an active enzyme.
Materials and Methods
Production of Recombinant Granzymes.
Recombinant WT gzmC was amplified with primers 5′-GGCAATTGCACCATCACCATCACCATGAATTCGATGACGACGACAAGATAATCGG (sense) and 5′-GGCAATTGTTAGCTGTGTTTCATCGTTTTCTTTATCC (antisense) from a cDNA kindly provided by R. C. Bleakley (University of Alberta, Edmonton, Canada) and cloned into the pPIC9 vector (Invitrogen). GzmC was mutated by PCR mutagenesis with oligonucleotides 5′-GGAGCTTCCTTTCGCGAGGATTCTGG and its complement (E192R), 5′-GGGAGCTTCCTTCGAAGGGGATTCTGGAGGC and its complement (E193G), and 5′-GATCAAGGGAGCTTCCTTCCGCGGGGATTCTGGAGGCCCGC and its complement (E192R/E193G). Electroporation of Pichia pastoris and purification of the gzmC zymogen from culture supernatant were as described (19, 20). After activation, enterokinase was removed by cation exchange over SP-Sepharose (GE Healthcare). Activated gzmC was analyzed by mass spectrometry and N-terminal sequencing to confirm correct processing by enterokinase.
Functional Assays.
Granzyme C activity was measured by incubating 50 μM succinyl-Phe-Leu-Phe-thiobenzyl ester (Bachem) and 500 μM 5,5′-dithiobis-(2-nitrobenzoic acid) (Sigma) with granzyme in 10 mM Tris, 150 mM NaCl (pH 7.4) and following the increase in absorbance at 405 nm. Phage display and perforin-mediated killing assays were performed as described in ref. 21.
Proteomics.
Culture of YAC-1 cells, treatment with WT or E192R/E193G unlocked gzmC, and subsequent COFRADIC analysis were performed as described in ref. 22.
Crystallization.
A Cartesian Honey bee crystallization robot (Genomic Solutions) was used to establish initial crystallization conditions (100-nL drops). Optimization of crystallization conditions by using the hanging-drop vapor diffusion method led to a reservoir buffer containing 0.2 M ammonium sulfate, 0.25 g/mL PEG 3350, 0.1 M sodium cacodylate (pH 6.6). Crystals were grown by mixing 2 μL of reservoir solution with 2 μL of protein solution [20 mg/mL in 50 mM Hepes (pH 6.8), 200 mM NaCl]. The crystals were flash frozen in liquid nitrogen by using the reservoir solution plus 10% glycerol as a cryoprotectant.
X-Ray Data Collection, Structure Determination, and Refinement.
WT and mutant gzmC crystallized isomorphously in space group P61, diffracted to 2.5-Å resolution, with two molecules in the asymmetric unit. Data for wild-type and mutant crystals were collected at 100 K using an in-house CuKα source, and at the Industrial Macromolecular Crystallography Association Collaborative Access Team beamline 17-ID at the Advanced Photon Source, Chicago, respectively. Raw diffraction images are available at http://arrow.monash.edu.au/hdl/1959.1/61033. The data were merged and processed with MOSFLM (23) and SCALA (24) from the CCP4 suite (25). Five percent of each dataset was flagged for calculation of Rfree (26) with neither a σ nor a low-resolution cutoff applied to the data. A summary of statistics is provided in Table 2.
Table 2.
X-ray diffraction data collection and refinement statistics
| Parameter | Wild-type gzmC | GzmC E192R/E193G |
|---|---|---|
| Data collection | ||
| Space group | P61 | P61 |
| Cell dimensions | ||
| a, b, c, Å | 71.49, 71.49, 206.70 | 71.38, 71.38, 207.17 |
| α, β, γ, ° | 90, 90, 120 | 90, 90, 120 |
| Resolution, Å | 2.5 | 2.5 |
| Rmerge, % | 6.4 (66.0) | 6.7 (71.6) |
| 〈I/σI〉 | 16.5 (2.1) | 15.3 (2.5) |
| Completeness, % | 99.8 (99.6) | 97.6 (98.6) |
| Multiplicity | 3.9 (3.8) | 4.6 (4.5) |
| Structure refinement | ||
| Total no. of observations | 80,056 | 91,680 |
| No. of unique observations | 20,641 | 20,089 |
| Rfree, %* | 25.9 | 23.6 |
| Rcryst, % | 21.5 | 21.2 |
| Nonhydrogen atoms | 3,436 | 3,082 |
| Protein | 3,374 | 3,054 |
| Solvent | 27 | 28 |
| B factors, Å2 | ||
| Mean main chain | 47.2 | 33.2 |
| Mean side chain | 47.6 | 33.5 |
| Mean water molecule | 45.5 | 35.9 |
| rmsd bonded Bs | 1.91 | 1.95 |
| Rmsd from ideality | ||
| Bond lengths, Å | 0.01 | 0.01 |
| Bond angles, ° | 1.6 | 1.3 |
Values in parentheses refer to the highest resolution shell.
*The free R factor was calculated with the 5% of data omitted from the refinement.
The structure of WT gzmC was determined by molecular replacement using PHASER (27) and the structure of granzyme B (PDB identifier 1A1U) as a search model, the closest structural homolog identified by using the FFAS server (28). A “mixed” model consisting of conserved side chains (all other non-alanine/glycine residues truncated at Cγ atom) was then created by using the SCRWL server (28). Two clear peaks in both the rotation and translation functions were evident, and these packed well within the unit cell. Together with the unbiased features in the initial electron density maps, the correctness of the molecular replacement solution was confirmed.
Because the mutant protein crystallized isomorphously with WT, structure refinement of the mutant proceeded by using refined WT coordinates. Structure refinement and building were performed by using the CCP4 suite, REFMAC (29) [incorporating translation, libration, and screw-rotation displacement (TLS) refinement], and COOT (30). Throughout refinement of both structures, tight NCS restraints were imposed on the two molecules. A bulk solvent correction (Babinet model with mask) was used within REFMAC. Water molecules were added to the model by using ARP/wARP (31) when Rfree reached 30%. Solvent molecules were retained only if they had acceptable hydrogen bonding geometry contacts of 2.5–3.5 Å with protein atoms or with existing solvent and were in good 2Fo − Fc and Fo − Fc electron density.
Supplementary Material
Acknowledgments.
We thank the staff at the Industrial Macromolecular Crystallography Association Collaborative Access Team beam line (Advanced Photon Source, Chicago) for technical support. We thank J. Rossjohn and J. Sun (Monash University, Australia) for technical assistance in the early stages of this project. This work was supported by National Health and Medical Research Council (NH & MRC) Program Grant 490900, the Australian Research Council, and the Australian Synchrotron Research Program. A.M.B. is a NH & MRC Senior Research Fellow. J.A.I. is an NH & MRC C. J. Martin Fellow. J.C.W. is an Australian Research Council Federation Fellow. P.V.D., K.G., and J.V. are supported by Fund for Scientific Research Flanders Belgium Research Grants G.0156.05 and G.0077.06, Concerted Research Actions Research Grant BOF07/GOA/012 from Ghent University, Inter University Attraction Poles Grant IUAP06, and European Union Interaction Proteome 6th Framework Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in Protein Data Bank, www.pdb.org [PDB ID codes 3FZZ (granzyme C wild type) and 3G01 (granzyme C E192R/E193G)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0811968106/DCSupplemental.
References
- 1.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: A comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 2.Di Cera E. Thrombin: A paradigm for enzymes allosterically activated by monovalent cations. C R Biol. 2004;327:1065–1076. doi: 10.1016/j.crvi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 3.McRae SJ, Stafford AR, Fredenburgh JC, Weitz JI. In the presence of phospholipids, glycosaminoglycans potentiate factor Xa-mediated protein C activation by modulating factor Xa activity. Biochemistry. 2007;46:4195–4203. doi: 10.1021/bi0617299. [DOI] [PubMed] [Google Scholar]
- 4.Eigenbrot C, Kirchhofer D. New insight into how tissue factor allosterically regulates factor VIIa. Trends Cardiovasc Med. 2002;12:19–26. doi: 10.1016/s1050-1738(01)00139-6. [DOI] [PubMed] [Google Scholar]
- 5.Bailey NC, Kelly CJ. Nephritogenic T cells use granzyme C as a cytotoxic mediator. Eur J Immunol. 1997;27:2302–2309. doi: 10.1002/eji.1830270926. [DOI] [PubMed] [Google Scholar]
- 6.Johnson H, Scorrano L, Korsmeyer SJ, Ley TJ. Cell death induced by granzyme C. Blood. 2003;101:3093–3101. doi: 10.1182/blood-2002-08-2485. [DOI] [PubMed] [Google Scholar]
- 7.Odake S, et al. Human and murine cytotoxic T lymphocyte serine proteases: Subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry. 1991;30:2217–2227. doi: 10.1021/bi00222a027. [DOI] [PubMed] [Google Scholar]
- 8.Jenne D, et al. cDNA cloning of granzyme C, a granule-associated serine protease of cytolytic T lymphocytes. J Immunol. 1988;140:318–323. [PubMed] [Google Scholar]
- 9.Jenne DE, Tschopp J. Granzymes, a family of serine proteases released from granules of cytolytic T lymphocytes upon T cell receptor stimulation. Immunol Rev. 1988;103:53–71. doi: 10.1111/j.1600-065x.1988.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 10.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 11.Andrade F, Fellows E, Jenne DE, Rosen A, Young CS. Granzyme H destroys the function of critical adenoviral proteins required for viral DNA replication and granzyme B inhibition. EMBO J. 2007;26:2148–2157. doi: 10.1038/sj.emboj.7601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metkar SS, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29:720–733. doi: 10.1016/j.immuni.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DJ, Adams TE, Li W, Huntington JA. Crystal structure of wild-type human thrombin in the Na+-free state. Biochem J. 2005;392:21–28. doi: 10.1042/BJ20051217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arac D, et al. Three-dimensional structure of the rSly1 N-terminal domain reveals a conformational change induced by binding to syntaxin 5. J Mol Biol. 2005;346:589–601. doi: 10.1016/j.jmb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg J. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 16.Kern D, et al. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 17.Fehlhammer H, Bode W, Huber R. Crystal structure of bovine trypsinogen at 1–8 Å resolution. II. Crystallographic refinement, refined crystal structure, and comparison with bovine trypsin. J Mol Biol. 1977;111:415–438. doi: 10.1016/s0022-2836(77)80062-4. [DOI] [PubMed] [Google Scholar]
- 18.Gallwitz M, Enoksson M, Hellman L. Expression profile of novel members of the rat mast cell protease (rMCP)-2 and (rMCP)-8 families, and functional analyses of mouse mast cell protease (mMCP)-8. Immunogenetics. 2007;59:391–405. doi: 10.1007/s00251-007-0202-1. [DOI] [PubMed] [Google Scholar]
- 19.Sun J, et al. Expression and purification of recombinant human granzyme B from Pichia pastoris. Biochem Biophys Res Commun. 1999;261:251–255. doi: 10.1006/bbrc.1999.0989. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, et al. Granzyme B encoded by the commonly occurring human RAH allele retains proapoptotic activity. J Biol Chem. 2004;279:16907–16911. doi: 10.1074/jbc.M400563200. [DOI] [PubMed] [Google Scholar]
- 21.Kaiserman D, et al. The major human and mouse granzymes are structurally and functionally divergent. J Cell Biol. 2006;175:619–630. doi: 10.1083/jcb.200606073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Damme P, et al. Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol Cell Proteomics. 2009;8:258–272. doi: 10.1074/mcp.M800060-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Leslie AGW. Joint CCP4 + ESF-EAMCB. Newslett Protein Crystallogr. 1992;26:1. [Google Scholar]
- 24.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 25.Project CC. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT. Assessment of phase accuracy by cross validation: The free R value. Methods and applications. Acta Crystallogr D. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Storoni LC, Read RJ. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr D. 2004;60:1220–1228. doi: 10.1107/S0907444904009990. [DOI] [PubMed] [Google Scholar]
- 28.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: A server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Cohen SX, et al. ARP/wARP and molecular replacement: The next generation. Acta Crystallogr D. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delano WL. The PyMOL User's Manual. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.