Abstract
Although somatic cell nuclear transfer (SCNT) cloning is more efficient in cattle than in any other species tested so far, there is a high rate of pregnancy failure that has been linked to structural and functional abnormalities of the placenta. We tested the hypothesis that these changes may originate from disturbed embryo–maternal interactions in the peri-implantation period. Therefore, we evaluated the response of the endometrium to SCNT embryos (produced from 7 different fetal fibroblast cell lines) as compared with embryos derived from in vitro fertilization (IVF). SCNT embryos and IVF embryos were cultured under identical conditions to the blastocyst stage (day 7) and were transferred to corresponding recipients, which were slaughtered at day 18 of pregnancy. The mRNA profiles of endometrium samples were obtained using a custom cDNA microarray enriched for transcripts differentially expressed in the endometrium and/or oviduct epithelium during the estrous cycle and/or early pregnancy. Overall, the variation in mRNA profiles was greater in the SCNT group than in the IVF group. Furthermore, 58 transcripts were differentially abundant in endometria from SCNT and IVF pregnancies. Prominent examples are orphan nuclear receptor COUP-TFII and connexin 43, both known to play important roles in uterine receptivity and conceptus placentation. These findings suggest that placental failure in bovine clone pregnancies may originate from abnormal embryo–maternal communication that develops during the peri-implantation period. Endometrium transcriptome profiles may serve as a tool to evaluate SCNT embryos for their ability to establish pregnancy and develop a functional placenta.
Keywords: Bos taurus, cloning, early pregnancy, gene expression, microarray
Cloning by somatic cell nuclear transfer (SCNT) (1) is an important strategic tool for animal breeding and biotechnology. For example, animals carrying rare desired alleles can be multiplied to introduce the desired allele into the breeding population. The life span and capacity of valuable breeding animals and the physiologically limited number of offspring of female animals can be increased, thus enhancing selection intensity. Moreover, cloning allows the propagation of desired genotypes without the risk of genetic recombination that is inherent to sexual reproduction. In animal biotechnology, SCNT using genetically modified donor cells is a powerful approach for the generation of transgenic animals (2) and so far is the only technique facilitating targeted mutagenesis in livestock species (3). Moreover, cloning can be used to multiply transgenic animals and to propagate multitransgenic individuals without segregation of the individual transgenes.
Despite the plethora of important applications of cloning, the efficiency of this technology still is very low. Although SCNT is more efficient in cattle than in any other species tested so far, a recent survey covering the results of bovine cloning in Brazil, Argentina, and the United States over a 5-year period revealed that only 9% of the SCNT embryos transferred to recipients resulted in the birth of a live calf (317 calves from 3374 embryos transferred to 293 surrogate dams). The proportion of live calves per transferred embryos is only 8% and 7% at 1 and 150 days after birth, respectively (4). In general, the failures of cloning are attributed to problems with the reprogramming of a nucleus derived from a differentiated cell. Reprogramming involves changes in the patterns of epigenetic marks, such as DNA methylation and histone modifications, and abnormal patterns of these modifications have been reported in cloned embryos (5–7). Interestingly, the efficiency of cloning can vary dramatically between different donor cell lines. In cattle, for about 1/3 of the donor cell lines the birth rate of live calves per initiated pregnancy was 40%, whereas 1/4 of the donor cell lines failed completely (4). These differences in the birth rate of live calves occurred even when donor cell cultures were used within the same program and did not display detectable chromosomal abnormalities. Thus, it is essential to involve several different donor cell lines in studies trying to determine the underlying pathologies that occur in clone pregnancies.
Although initial pregnancy rates after transfer of cloned bovine embryos were found to be similar to those after artificial insemination or transfer of flushed embryos (8, 9), continued pregnancy loss was observed in recipients of cloned embryos throughout gestation. The survival rate of cloned embryos to term is only 1/3 that of embryos derived by in vitro fertilization (IVF) (9, 10). The high rate of pregnancy failure in recipients of cloned embryos has been linked to the finding of structural and functional abnormalities of the placenta. Losses of pregnancy in surrogate dams in the second and third trimester are associated with placental abnormalities, hydrops, enlarged umbilical cords with dilated vessels, and abnormally enlarged and fewer placental cotyledons (11–13). This abnormal placental development may be present from the early stages after implantation and can be overcome by some embryos that result in the development and birth of live clones (12, 14, 15). Nevertheless, abnormal placental development and associated consequences for maternal–fetal exchange is a main limiting factor in ruminant SCNT pregnancies (16).
The facts that (i) altered trophoblast differentiation has been found in peri-implantation bovine SCNT embryos (16) and (ii) abnormal placental development may be present from the early stages after implantation suggest that placental abnormalities in bovine clone pregnancies may originate from disturbed embryo–maternal communication during the peri-implantation period. To test this hypothesis, we used microarray analysis to compare the response of the maternal environment (i.e., the endometrium) to SCNT embryos vs. IVF embryos.
Results
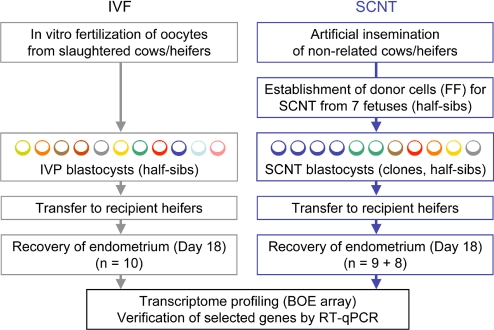
A diagram of the experimental outline is presented in Fig. 1. An IVF protocol was used that has been validated not to produce fetal overgrowth syndrome (17). To exclude specific effects of a particular donor cell culture, SCNT embryos were produced from fibroblast cultures derived from different fetuses (day 150; half-sibs). Thus, the genetic variability in the SCNT embryos can be assumed to be similar to that in the IVF group of embryos, which was produced by IVF of oocytes from different slaughtered cows with semen from a single bull. Results for the embryo transfer experiments are summarized in Table 1. Pregnancy rates (at day 18 of gestation) were 59% for the SCNT group and 77% for the IVF group. In both groups the percentage of pregnancies with 2 embryos at day 18 was ≈40%. Detailed results of the SCNT experiments are presented in Table 2. For 7 of the 9 fetal fibroblast cell lines used for SCNT, pregnancy rates on day 18 were between 25% and 100%.
Fig. 1.
Experimental design used to study the maternal response to SCNT vs. IVF embryos. IVF embryos were produced by fertilization of oocytes from nonrelated cows with semen from a single bull. SCNT embryos were produced from fibroblasts of different fetuses (half-sibs). IVF and SCNT embryos were cultured under identical conditions. Two IVF or SCNT embryos (day 8, grade 1) were transferred per recipient. On day 18, recipients were slaughtered, and pregnancy was verified by the presence of at least 1 normally developed conceptus. Endometrium samples (IVF: n = 10; SCNT: n = 9 from embryos of 4 different genotypes) then were processed for mRNA expression profiling using the BOE array. Differentially abundant transcripts were verified by RT-qPCR analysis in those endometrial samples and in additional samples from 8 other SCNT pregnancies with embryos derived from 3 additional fetal fibroblast donor cell cultures. FF, fetal fibroblasts.
Table 1.
Results of embryo transfer experiments.
| Condition | IVF | SCNT |
|---|---|---|
| Embryo transfers | 13 | 29 |
| Pregnancies | 10 (77%) | 17 (59%) |
| Pregnancies with 1 embryo | 6 | 10 |
| Pregnancies with 2 embryos | 4 | 7 |
Table 2.
Overview of SCNT experiments.
| Cell Line # | # Recipients | # Day 18 Pregnancies | Pregnancy Rate [%] |
|---|---|---|---|
| 1 | 1 | 0 | 0 |
| 3 | 2 | 0 | 0 |
| 6 | 2 | 1 | 50 |
| 7 | 1 | 1 | 100 |
| 8 | 7 | 5 | 71 |
| 11 | 3 | 3 | 100 |
| 13 | 4 | 2 | 50 |
| 17 | 5 | 4 | 80 |
| 20 | 4 | 1 | 25 |
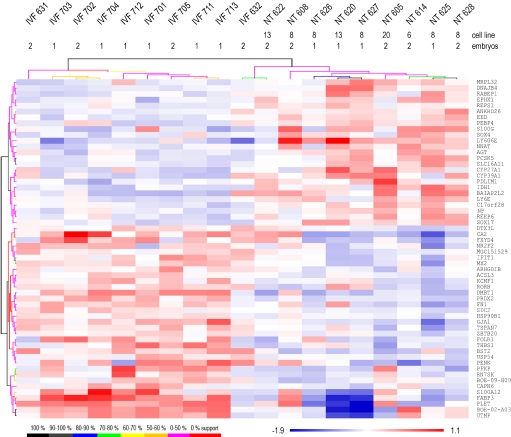
Total RNA was isolated from intercaruncular endometrium samples (10 IVF pregnancies and 9 SCNT pregnancies with embryos from 4 different genotypes) and was used for gene expression profiling with a custom cDNA microarray, the Bovine Oviduct and Endometrium (BOE) array, that contains 950 different transcripts, most of which are differentially expressed in bovine endometrium and/or oviduct epithelium during the estrous cycle or early pregnancy (18). Overall, the variation of gene expression levels in the endometrial samples was greater for SCNT than for IVF pregnancies (P < 0.00001); the mean coefficient of variation of normalized signal values for the differentially expressed genes was 1.87% in the IVF group and 2.74% in the SCNT group. Moreover, 58 transcripts were differently abundant (with a false-discovery rate of 5.24%) between endometria from SCNT and from IVF pregnancies (Table S1). Further, the abundance of 33 transcripts was lower in endometrial samples derived from SCNT pregnancies, with highest mean downregulation (−2.9-fold) for fatty acid binding protein 3 (FABP3) mRNA. The mRNA for lymphocyte antigen 6 complex, locus G6E (LY6G6E) showed highest mean upregulation (2.3-fold) in endometrium of SCNT pregnancies. Cluster analysis for the individual endometrial samples is illustrated in Fig. 2. This analysis detected relatively clear separation of IVF and SCNT samples: only 1 IVF sample (IVF632) was found in the same branch as the SCNT samples, but this sample was adjacent to the IVF samples. Transcript levels were relatively uniform in the IVF samples, but a much larger variation was observed in endometria from SCNT pregnancies. Therefore, the expression differences for individual SCNT samples were much higher than the mean difference for most of the significant transcripts. Strongest deviations from the IVF samples were detected for the SCNT samples #620 and #627.
Fig. 2.
Cluster analysis based on differentially expressed genes. Hierarchical cluster analysis based on mean centered values (log 2 value of a gene minus mean of all log 2 values of this gene) of the significant genes for samples and genes was performed (HCL support tree, Pearson correlation, MultiExperiment Viewer 4.0). The trees were calculated with 100 iterations, and support for the tree is shown by a color code. The fetal fibroblast cell line used for SCNT and the number of concepti present at the time of sample collection are indicated.
The expression profiles of pregnancies with SCNT embryos of the same genotype did not cluster together (cell line number indicated in Fig. 2), suggesting that the transcriptome response of the endometrium is influenced more by the origin of the embryo (SCNT vs. IVF) than by its genetic background. The number of embryos found at the time of slaughter (1 or 2) had no obvious effect on the cluster analysis of the significant genes in the SCNT group and the IVF group (Fig. 2). Significance analyses for IVF pregnancies (2 concepti: n = 4, 1 conceptus: n = 6) and SCNT pregnancies (2 concepti: n = 5, 1 conceptus: n = 4) were performed to test whether there are significant differences between single and twin pregnancies. No significant changes were found between single-embryo and twin pregnancies for IVF even at higher false-discovery rates. mRNA levels for 3 genes (placenta-expressed transcript protein [PLET], fatty acid binding protein 3, and S100 calcium binding protein A12) were higher in endometria of twin SCNT pregnancies than in endometria of single SCNT pregnancies (see Table S2).
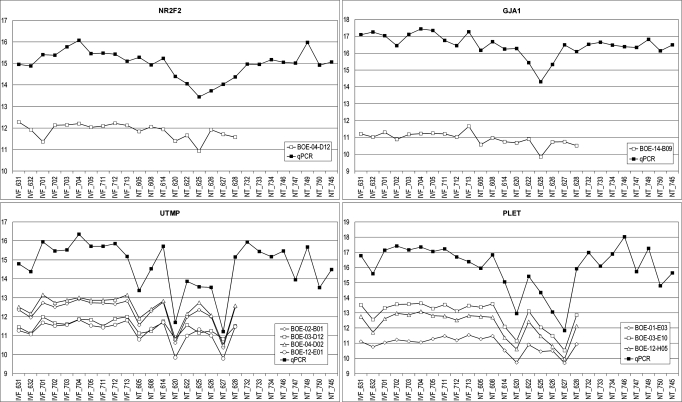
The mRNA levels of 9 selected genes were analyzed by quantitative real-time RT-PCR (qPCR) in the same endometrial samples and in 8 additional samples from clone pregnancies derived from 3 further genetically distinct donor cell lines for a total of 17 SCNT pregnancies (Table 3). Although only relatively small differences in transcript levels were detected for many genes, qPCR analysis mainly confirmed the microarray analysis. Fig. 3 presents a comparison of array and qPCR data for the single endometrial samples for nuclear receptor subfamily 2, group F, member 1 (NR2F2), gap junction protein, alpha 1, 43 kDa (connexin 43) (GJA1), uterine milk protein precursor (UTMP), and PLET. A high correlation between array data and qPCR data and also between different cDNA clones corresponding to the same transcript provides additional data supporting the greater variation in mRNA expression levels in SCNT endometria as compared with IVF pregnancies. No significant differences in the expression of the 9 selected genes were found between single and twin pregnancies for either the IVF pregnancies (2 concepti: n = 4, 1 conceptus: n = 6) or the SCNT pregnancies (2 concepti: n = 7, 1 conceptus: n = 10) pregnancies.
Table 3.
Validation of array data by real-time qRT-PCR analysis
| Gene | Mean ± SEM [CP]a |
FC SCNT/IVFb |
P-value qPCRc | Q-value Arrayd | ||
|---|---|---|---|---|---|---|
| IVF | SCNT | qPCR | Array | |||
| PLET | 18.13 ± 0.18 | 19.55 ± 0.40 | −2.7 | −2.6 | 0.015 | 0.032 |
| UTMP | 19.51 ± 0.18 | 20.75 ± 0.33 | −2.4 | −2.0 | 0.012 | 0.031 |
| GJA1 | 17.98 ± 0.11 | 18.83 ± 0.15 | −1.8 | −1.8 | <0.001 | <0.001 |
| PENK | 16.48 ± 0.20 | 17.16 ± 0.20 | −1.6 | −1.7 | 0.032 | 0.052 |
| NR2F2 | 19.61 ± 0.11 | 20.26 ± 0.16 | −1.6 | −1.5 | 0.007 | 0.052 |
| MX2 | 17.50 ± 0.19 | 18.02 ± 0.13 | −1.4 | −1.5 | 0.032 | 0.034 |
| LY6E | 25.40 ± 0.49 | 23.71 ± 0.18 | 3.2 | 1.4 | <0.001 | 0.027 |
| CYP39A1 | 18.82 ± 0.21 | 18.37 ± 0.18 | 1.4 | 1.5 | 0.136 | 0.027 |
| CYP27A1 | 21.42 ± 0.14 | 21.08 ± 0.11 | 1.3 | 1.5 | 0.070 | 0.052 |
aMean ± SEM CP: mean and standard error of the mean of crossing points of the replicates in the experimental groups (IVF n = 10, SCNT n = 17).
bFC SCNT/IVF: expression fold change of SCNT versus IVF.
cP-value: ANOVA p-value.
dQ-value: statistical value of the SAM procedure.
Fig. 3.
Comparison of expression profiles obtained by array analysis and qPCR. Microarray data are shown as normalized values (generalized logarithm). Data from the qPCR are shown as 35–CP for optimal visualization of array and qPCR data in the same diagram. The IDs of the BOE array cDNA clones are indicated.
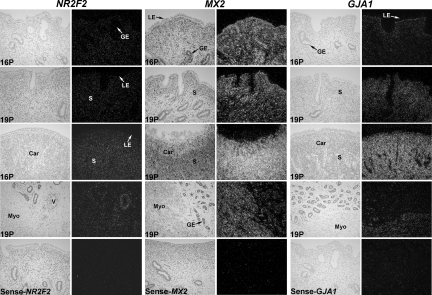
In situ hybridization analyses were conducted and detected NR2F2 mRNA mainly in the stroma and blood vessels in both the intercaruncular and caruncular areas of the endometrium and myometrium of uteri from day 16 and day 19 pregnant heifers (Fig. 4). MX2 mRNA was abundant in the stroma, glands, immune cells, and blood vessels of both the intercaruncular and caruncular areas of the endometrium in day 16 pregnant heifers and also in the myometrium of day 19 pregnant heifers. GJA1 mRNA was present in the stroma in the intercaruncular endometrium and was particularly abundant in the caruncular stroma and myometrium. The overall abundance of GJA1 mRNA increased in the stroma between days 16 and 19 of pregnancy. GJA1 and NR2F2 mRNAs were not detected in the endometrial epithelia.
Fig. 4.
In situ localization of NR2F2, MX2, and GJA1 mRNA in the bovine uterus on days 16 and 19 of pregnancy. Cross-sections of the uterine wall from pregnant (P) heifers were hybridized with radiolabeled antisense or sense bovine NR2F2, MX2, or GJA1 cRNAs and are presented under brightfield and darkfield illumination after counterstaining with hematoxylin. Car, caruncle; GE, glandular epithelium; LE, luminal epithelium; Myo, myometrium; S, stroma; V, blood vessel. All photomicrographs are displayed at the same width of field (450 μm). Original magnification: 25×.
Discussion
The hypothesis that abnormal placental development in SCNT pregnancies may originate from disturbed embryo–maternal communication during the peri-implantation period of early pregnancy was tested by comparing mRNA levels in the endometrium of SCNT and IVF pregnancies. Although in vivo-derived embryos are considered the reference standard for comparing the effects of assisted reproduction techniques on embryonic development, the present study used IVF embryos as a control for the identification of specific responses of the endometrium to SCNT embryos. The IVF protocol used in this study had been shown previously to produce fetuses that are not different from in vivo-derived fetuses in growth and endocrine parameters (17). Our experimental approach allowed for (i) similar levels of genetic variation in both groups, (ii) maintenance of embryos after IVF or SCNT under exactly the same conditions, and (iii) selection of the best blastocysts according to morphological criteria for transfer to recipients. The number of embryos transferred was limited to 2 to maintain normal embryo–maternal interactions. As reported previously, the initial embryo development after transfer to recipients was similar for IVF and SCNT embryos (8, 9). Because all pregnancies from the fetal fibroblast cell lines were terminated on day 18, no data are available on the ability of SCNT embryos from these cell lines to develop further. However, we successfully generated cloned transgenic cows (pregnancy and calving rates, 38% and 15%, respectively) (19) using a similar nuclear transfer protocol and fetal fibroblast cells. Furthermore, we found a similar response to IFNT in the endometrium samples from SCNT pregnancies and in the IVF pregnancies, as shown by the expression level of IFNT–induced genes present on the BOE array. The similarity of response suggests that maternal recognition of pregnancy is not compromised in SCNT pregnancies.
The gene expression profiles in endometrium samples from SCNT pregnancies were more variable than those from IVF pregnancies, as predicted by the concept that stochastic reprogramming of differentiated nuclei after transfer to enucleated oocytes results in variability caused by epigenetic mechanisms (7). This concept is supported by the finding that endometrial gene expression profiles from pregnancies with SCNT embryos of the same genotype did not cluster together. Despite the large variation in endometrium transcriptome profiles in SCNT pregnancies, we detected significant differences in transcript levels between SCNT and IVF pregnancies. Our experimental design using pregnancies with SCNT embryos from 4 different donor cell lines for the initial array experiment excludes the possibility that differences observed between SCNT and IVF pregnancies were caused by the specific effects of a single donor cell line or genotype. This consideration is important, because of the large effect of the donor cell line on the developmental potential of cloned embryos (4). Interestingly, for no genes did the level of expression vary depending on the number of developing embryos in endometrium of IVF pregnancies, but higher levels of 3 genes were found in endometria of twin pregnancies for the SCNT group, based on the array data. However, qPCR expression data for PLET, which were measured for all 17 SCNT pregnancies (2 concepti: n = 7, 1 conceptus: n = 10), revealed no significant difference. These data show that the differences in the response of the endometrium to SCNT and IVF embryos are not caused by the number of embryos. In summary, these data show that significant differences in the response of the endometrium to SCNT embryos and IVF embryos of multiple embryonic genotypes are inherent to SCNT technology.
Interestingly, an important role in implantation and/or placentation already has been shown or suggested for many of the differentially expressed genes. Transcript levels of the uterine milk protein (LOC286871, UTMP) are upregulated in bovine endometrium during the ovulatory phase (20) and during early pregnancy (21). In contrast, UTMP mRNA levels were decreased in endometrium from SCNT pregnancies. Functional studies indicate a role of UTMP in mediating immunosuppressive effects of progesterone on the endometrium (22). Expression of myxovirus resistance 2 (MX2), an IFN-stimulated gene (23), also was reduced in the endometria from cloned pregnancies, and MX2 mRNA was observed primarily in the endometrial stroma, glands, and immune cells (Fig. 4). MX proteins belong to the antiviral proteins that are induced by type I IFN in response to infections caused by a wide range of single-strand RNA viruses (24). Interestingly, in dairy cattle MX2 gene expression also is increased in peripheral blood leukocytes between days 16 and 20 of pregnancy, suggesting that the innate immune system is activated during early pregnancy (25). These results suggest that immune responses of the endometrium to the SCNT-derived conceptus may be dysregulated, and such dysregulation has been suggested as a major cause of inadequate placentome development that leads to immune-mediated abortion in cloned pregnancies (26). Indeed, MHC1 is overexpressed in SCNT-derived blastocysts and placentas (26, 27). Furthermore, differential mRNA expression was found for 2 steroid hydroxylase genes [cytochrome P450, family 27, subfamily A, polypeptide 1 (CYP27A1), cytochrome P450, family 39, subfamily a, polypeptide 1 (CYP39A1)] and a number of transcription factors, such as SRY (sex determining region Y)-box 4 (SOX4) and 17 (SOX17), and RAR-related orphan receptor B (RORB). Regulation by ovarian hormones has been shown for SOX4 (28) and SOX17 (29). The most interesting transcription factor gene is NR2F2, which was downregulated in endometrium from SCNT pregnancies. NR2F2 mRNA was observed primarily in the endometrial stroma and blood vessels (Fig. 4), as previously observed in the mouse. NR2F2 (COUP-TFII) belongs to the orphan nuclear receptor superfamily. Uterine-specific Nr2f2-mutant mice are infertile because of implantation failure in which both embryo attachment and uterine decidualization are impaired (30, 31). Interestingly, decreased fecundity was observed in heterozygous Nr2f2-mutant mice (32), suggesting that the decrease of NR2F2 transcript levels in endometrium from clone pregnancies may be functionally relevant. Another interesting candidate is GJA1 with downregulated transcript levels in SCNT pregnancies. During bovine synepitheliochorial placentation, GJA1 protein was observed in the caruncular stroma (33). In the present study, GJA1 mRNA was detected in the endometrial stroma and was particularly abundant in the caruncular stroma and myometrium. Similar to the present study, a striking increase of stromal GJA1 was observed in the intercaruncular and caruncular endometrial stroma at the onset of implantation between days 18 and 21 of pregnancy in sheep (34), suggesting that reduced GJA1 mRNA expression in bovine clone pregnancies may affect placentation negatively. Indeed, conditional deletion of the Gja1 gene in the stromal cells of the mouse uterus led to impaired production of key angiogenic factors and a striking impairment in the development of new blood vessels within the stromal compartment, resulting in the arrest of embryo growth and early pregnancy loss (35).
Preparation of the endometrium for embryo attachment and implantation in all studied mammals, including ruminants (21, 23, 36), involves carefully orchestrated spatiotemporal alterations in transcriptome profiles. Our results strongly support the hypothesis that placental failure in bovine clone pregnancies, which manifests at later stages, originates from abnormal embryo–maternal communication during the peri-implantation period of early pregnancy. Indeed, endometrial transcriptome profiles may serve as a tool to evaluate and optimize SCNT embryos for their ability to establish pregnancy successfully, develop a functional placenta, and produce viable offspring, outcomes necessary to realize the full potential of SCNT for ruminants.
Materials and Methods
Production of IVF and SCNT Embryos, Embryo Transfer, and Collection of Endometrium Samples.
SCNT and IVF procedures were performed as described by Hiendleder et al. (37). After SCNT or IVF, embryos were cultured under identical conditions (protocol IVF1; ref. 17) to the blastocyst stage (day 7). Briefly, presumptive embryos were cultured in 400-μl droplets of synthetic oviduct fluid culture medium enriched with 5% ECS, 40 μl/ml of 50× BME Amino Acids Solution (#B6766, Sigma Aldrich), and 10 μl/ml of 100× MEM Non-essential Amino Acid Solution (#M7145, Sigma Aldrich) covered with mineral oil. The culture atmosphere was 5% CO2, 5% O2, and 90% N2 at 39 °C and maximum humidity. Two SCNT or IVF blastocysts (grade 1) were transferred per recipient heifer (day 7 of estrous cycle). Preparation of recipient animals and embryo transfer were done as described by Klein et al. (21). The recipients were slaughtered 11 days later, and the uteri were recovered (20). Animals were termed “pregnant” if filamentous trophoblast tubes and at least 1 embryonic disc were observed under a stereomicroscope. A twin pregnancy was diagnosed if 2 embryonic discs were detected. Endometrial tissue samples were collected, preserved, and processed for isolation of RNA as previously described (38).
Array Analysis.
The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE13663. Production of arrays and array hybridization were done as described previously (38). Array evaluation was done using AIDA Image Analyzer software. Background was subtracted with the Lowest Grid Dot function. Raw data were normalized with the BioConductor package vsn (39). For quality control, normalized data were analyzed with a distance matrix and a heatmap based on pair-wise distances (BioConducter package geneplotter). Significance analysis was performed using the Microsoft Excel add-in, significance analysis of microarrays method (SAM, 2-class unpaired) (40).
Quantitative Real-Time RT-PCR.
A 2-step real-time RT-qPCR was conducted as described recently (41). Real-time qPCR reactions using the LightCycler DNA Master SYBR Green I protocol (Roche Diagnostics) were performed. All amplified PCR fragments were sequenced to verify the resulting PCR product (4base lab). The primers listed in Table S3 were used to amplify specific fragments referring to the selected transcripts. The annealing temperature and the appropriate fluorescence acquisition points for quantification within the fourth step of the amplification segment were as indicated. The cycle number required to achieve a definite SYBR Green fluorescence signal (CP) was calculated by the second derivative maximum method (LightCycler software version 4.05, Roche). The CP is correlated inversely with the logarithm of the initial template concentration. The differences between the groups were analyzed using 1-way ANOVA. The normal distribution was tested by the Kolmogorov-Smirnov method, followed by a t test to find the significant differences (Sigma-Stat, version 2.03, Systat Software).
In Situ Hybridization Analysis.
Crossbred nulliparous beef heifers were synchronized to estrus and bred using semen from a single bull in a timed artificial insemination protocol. Bred heifers were slaughtered on either day 16 or 19 postmating, and pregnancy was confirmed by the recovery of a conceptus. Portions of the uterus ipsilateral to the corpus luteum were fixed in 4% paraformaldehyde in PBS (pH 7.2) overnight and then embedded in paraffin. Cell-specific expression of mRNAs in cross-sections of bovine uteri (n = 6 per day) was determined using radioactive in situ hybridization analysis conducted as described previously (42). Partial cDNAs for bovine endometrial NR2F2, MX2, and GJA1 mRNAs were cloned by RT-PCR using specific primers and then sequenced to confirm identity (data not shown). All slides for each respective gene were exposed to photographic emulsion for the same period. Images of representative fields were recorded under brightfield or darkfield illumination using a Nikon Eclipse 1000 photomicroscope (Nikon Instruments Inc.) fitted with a Nikon DXM1200 digital camera.
Supplementary Material
Acknowledgments.
We thank Katy Schulke, Anna Gröbner, and Katrin Mitko for assistance with tissue sampling, Angela Sachsenhauser and Karin Gross for excellent technical assistance, and the McGregor Beef Cattle Research Center of Texas AgriLIFE Research for provision, management, and breeding of heifers. This study was supported by grants from the Deutsche Forschungsgemeinschaft (FOR 478, FOR 1041, GRK 1029) and by the Bundesministerium für Bildung und Forschung (Fertilink, Compendium).
Footnotes
Conflict of interest: no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811841106/DCSupplemental.
References
- 1.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 2.Schnieke AE, et al. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278(5346):2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- 3.McCreath KJ, et al. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405(6790):1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 4.Panarace M, et al. How healthy are clones and their progeny: 5 years of field experience. Theriogenology. 2007;67(1):142–151. doi: 10.1016/j.theriogenology.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 5.Dean W, et al. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98(24):13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos F, et al. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;13(13):1116–1121. doi: 10.1016/s0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- 7.Shi W, Zakhartchenko V, Wolf E. Epigenetic reprogramming in mammalian nuclear transfer. Differentiation. 2003;71(2):91–113. doi: 10.1046/j.1432-0436.2003.710201.x. [DOI] [PubMed] [Google Scholar]
- 8.Heyman Y, et al. Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol Reprod. 2002;66(1):6–13. doi: 10.1095/biolreprod66.1.6. [DOI] [PubMed] [Google Scholar]
- 9.Lee RS, et al. Cloned cattle fetuses with the same nuclear genetics are more variable than contemporary half-siblings resulting from artificial insemination and exhibit fetal and placental growth deregulation even in the first trimester. Biol Reprod. 2004;70(1):1–11. doi: 10.1095/biolreprod.103.020982. [DOI] [PubMed] [Google Scholar]
- 10.Wells DN. Animal cloning: Problems and prospects. Revue Scientifique et Techique. 2005;24(1):251–264. [PubMed] [Google Scholar]
- 11.Wells DN, Misica PM, Tervit HR. Production of cloned calves following nuclear transfer with cultured adult mural granulosa cells. Biol Reprod. 1999;60(4):996–1005. doi: 10.1095/biolreprod60.4.996. [DOI] [PubMed] [Google Scholar]
- 12.Hill JR, et al. Evidence for placental abnormality as the major cause of mortality in first-trimester somatic cell cloned bovine fetuses. Biol Reprod. 2000;63(6):1787–1794. doi: 10.1095/biolreprod63.6.1787. [DOI] [PubMed] [Google Scholar]
- 13.Batchelder CA, et al. Effect of the nuclear-donor cell lineage, type, and cell donor on development of somatic cell nuclear transfer embryos in cattle. Cloning Stem Cells. 2005;7(4):238–254. doi: 10.1089/clo.2005.7.238. [DOI] [PubMed] [Google Scholar]
- 14.Hoffert KA, et al. Measures of maternal-fetal interaction in day-30 bovine pregnancies derived from nuclear transfer. Cloning Stem Cells. 2005;7(4):289–305. doi: 10.1089/clo.2005.7.289. [DOI] [PubMed] [Google Scholar]
- 15.Chavatte-Palmer P, et al. Ultrasound fetal measurements and pregnancy associated glycoprotein secretion in early pregnancy in cattle recipients carrying somatic clones. Theriogenology. 2006;66(4):829–840. doi: 10.1016/j.theriogenology.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 16.Arnold DR, Bordignon V, Lefebvre R, Murphy BD, Smith LC. Somatic cell nuclear transfer alters peri-implantation trophoblast differentiation in bovine embryos. Reproduction. 2006;132(2):279–290. doi: 10.1530/rep.1.01217. [DOI] [PubMed] [Google Scholar]
- 17.Hiendleder S, et al. Tissue-specific effects of in vitro fertilization procedures on genomic cytosine methylation levels in overgrown and normal sized bovine fetuses. Biol Reprod. 2006;75(1):17–23. doi: 10.1095/biolreprod.105.043919. [DOI] [PubMed] [Google Scholar]
- 18.Bauersachs S, Mitko K, Blum H, Wolf E. Technical note: Bovine oviduct and endometrium array version 1: A tailored tool for studying bovine endometrium biology and pathophysiology. Journal of Dairy Science. 2007;90(9):4420–4423. doi: 10.3168/jds.2007-0132. [DOI] [PubMed] [Google Scholar]
- 19.Grosse-Hovest L, et al. Cloned transgenic farm animals produce a bispecific antibody for T cell-mediated tumor cell killing. Proc Natl Acad Sci USA. 2004;101(18):6858–6863. doi: 10.1073/pnas.0308487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauersachs S, et al. Gene expression profiling of bovine endometrium during the oestrous cycle: Detection of molecular pathways involved in functional changes. J Mol Endocrinol. 2005;34(3):889–908. doi: 10.1677/jme.1.01799. [DOI] [PubMed] [Google Scholar]
- 21.Klein C, et al. Monozygotic twin model reveals novel embryo-induced transcriptome changes of bovine endometrium in the preattachment period. Biol Reprod. 2006;74(2):253–264. doi: 10.1095/biolreprod.105.046748. [DOI] [PubMed] [Google Scholar]
- 22.Arck P, Hansen PJ, Mulac Jericevic B, Piccinni MP, Szekeres-Bartho J. Progesterone during pregnancy: Endocrine-immune cross talk in mammalian species and the role of stress. American Journal of Reproductive Immunology. 2007;58(3):268–279. doi: 10.1111/j.1600-0897.2007.00512.x. [DOI] [PubMed] [Google Scholar]
- 23.Spencer TE, Sandra O, Wolf E. Genes involved in conceptus-endometrial interactions in ruminants: Insights from reductionism and thoughts on holistic approaches. Reproduction. 2008;135(2):165–179. doi: 10.1530/REP-07-0327. [DOI] [PubMed] [Google Scholar]
- 24.Pavlovic J, Schroder A, Blank A, Pitossi F, Staeheli P. Mx proteins: GTPases involved in the interferon-induced antiviral state. Ciba Found Symp. 1993;176:233–243. doi: 10.1002/9780470514450.ch15. discussion 243–237. [DOI] [PubMed] [Google Scholar]
- 25.Gifford CA, et al. Regulation of interferon-stimulated genes in peripheral blood leukocytes in pregnant and bred, nonpregnant dairy cows. Journal of Dairy Science. 2007;90(1):274–280. doi: 10.3168/jds.S0022-0302(07)72628-0. [DOI] [PubMed] [Google Scholar]
- 26.Davies CJ, et al. Major histocompatibility antigen expression on the bovine placenta: Its relationship to abnormal pregnancies and retained placenta. Anim Reprod Sci 82- 2004;83:267–280. doi: 10.1016/j.anireprosci.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Aston KI, et al. Global gene expression analysis of bovine somatic cell nuclear transfer blastocysts and cotyledons. Mol Reprod Dev. 2008;76(5):471–482. doi: 10.1002/mrd.20962. [DOI] [PubMed] [Google Scholar]
- 28.Hunt SM, Clarke CL. Expression and hormonal regulation of the Sox4 gene in mouse female reproductive tissues. Biol Reprod. 1999;61(2):476–481. doi: 10.1095/biolreprod61.2.476. [DOI] [PubMed] [Google Scholar]
- 29.Garcia C, Calvo E, Nieto A. The transcription factor SOX17 is involved in the transcriptional control of the uteroglobin gene in rabbit endometrium. J Cell Biochem. 2007;102(3):665–679. doi: 10.1002/jcb.21324. [DOI] [PubMed] [Google Scholar]
- 30.Kurihara I, et al. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genetics. 2007;3(6):e102, 1053–1064. doi: 10.1371/journal.pgen.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petit FG, et al. Deletion of the orphan nuclear receptor COUP-TFII in uterus leads to placental deficiency. Proc Natl Acad Sci USA. 2007;104(15):6293–6298. doi: 10.1073/pnas.0702039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takamoto N, et al. Haploinsufficiency of chicken ovalbumin upstream promoter transcription factor II in female reproduction. Mol Endocrinol. 2005;19(9):2299–2308. doi: 10.1210/me.2005-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfarrer CD, Heeb C, Leiser R. Expression of gap junctional connexins 26, 32 and 43 in bovine placentomes during pregnancy. Placenta. 2006;27(1):79–86. doi: 10.1016/j.placenta.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel S, Winterhager E, Pfarrer C, Traub O, Leiser R. Modulation of connexin expression in sheep endometrium in response to pregnancy. Placenta. 2004;25(4):287–296. doi: 10.1016/j.placenta.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Laws MJ, et al. Gap junction communication between uterine stromal cells plays a critical role in pregnancy-associated neovascularization and embryo survival. Development. 2008;135(15):2659–2668. doi: 10.1242/dev.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauersachs S, et al. Embryo-induced transcriptome changes in bovine endometrium reveal species-specific and common molecular markers of uterine receptivity. Reproduction. 2006;132(2):319–331. doi: 10.1530/rep.1.00996. [DOI] [PubMed] [Google Scholar]
- 37.Hiendleder S, et al. Tissue-specific elevated genomic cytosine methylation levels are associated with an overgrowth phenotype of bovine fetuses derived by in vitro techniques. Biol Reprod. 2004;71(1):217–223. doi: 10.1095/biolreprod.103.026062. [DOI] [PubMed] [Google Scholar]
- 38.Mitko K, et al. Dynamic changes in messenger RNA profiles of bovine endometrium during the oestrous cycle: Focus on mammalian embryogenomics. Reproduction. 2008;135(2):225–240. doi: 10.1530/REP-07-0415. [DOI] [PubMed] [Google Scholar]
- 39.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulbrich SE, Schoenfelder M, Thoene S, Einspanier R. Hyaluronan in the bovine oviduct–modulation of synthases and receptors during the estrous cycle. Mol Cell Endocrinol. 2004;214(1–2):9–18. doi: 10.1016/j.mce.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Spencer TE, et al. Discovery and characterization of endometrial epithelial messenger ribonucleic acids using the ovine uterine gland knockout model. Endocrinology. 1999;140(9):4070–4080. doi: 10.1210/endo.140.9.6981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.