Abstract
The C-terminal copper-binding loop in the β-barrel fold of the cupredoxin azurin has been replaced with a range of sequences containing alanine, glycine, and valine residues to assess the importance of amino acid composition and the length of this region. The introduction of 2 and 4 alanines between the coordinating Cys, His, and Met results in loop structures matching those in naturally occurring proteins with the same loop lengths. A loop with 4 alanines between the Cys and His and 3 between the His and Met ligands has a structure identical to that of the WT protein, whose loop is the same length. Loop structure is dictated by length and not sequence allowing the properties of the main surface patch for interactions with partners, to which the loop is a major contributor, to be optimized. Loops with 2 amino acids between the ligands using glycine, alanine, and valine residues have been compared. An empirical relationship is found between copper site protection by the loop and reduction potential. A loop adorned with 4 methyl groups is sufficient to protect the copper ion, enabling most sequences to adequately perform this task. The mutant with 3 alanine residues between the ligands forms a strand-swapped dimer in the crystal structure, an arrangement that has not, to our knowledge, been seen previously for this family of proteins. Cupredoxins function as redox shuttles and are required to be monomeric; therefore, none have evolved with a metal-binding loop of this length.
Keywords: loop modeling, metalloproteins, protein engineering, protein folding, strand-swapped dimerization
Loops link the main secondary structure elements of all globular proteins and are invariably found at the molecular surface. These regions play essential roles by contributing to active sites and facilitating protein interactions. Surface loops can be determinants of protein interactions, and loops that disfavor association may help to maintain a protein as a monomer to enable rapid diffusion in a cell (1). Loop variations are therefore important for altering functionality without influencing a protein's core structure, and can also play a vital role in protein folding (2–4). Despite their importance, de novo loop design is only at a preliminary stage of development, and loop-forming amino acid sequences have not been established. The relationship between loop composition and structure is a feature of protein architecture that is currently poorly understood. Metalloproteins constitute ≈1/3 of structurally characterized proteins and it has been estimated that up to 50% of all proteins bind a metal ion (5). Those metalloproteins whose function necessitates an interaction with a protein partner invariably bind their cofactor via loop residues. The requirements for metal coordination by such loops provide additional constraints to these regions. Altering metal-binding loops can simultaneously tune the properties of a metal site and an important interaction surface.
The β-barrel motif is one of the most widely occurring and the second most abundant in the Protein Data Bank (6, 7). This fold is particularly stable and the β-strand-linking loops are nearly always the location of active sites. In some cases the loops also bind metals, and the cupredoxins are probably the best-studied family of metal-binding β-barrels (8–12). These proteins utilize a type 1 (T1) copper site for electron transfer (ET) in energy transducing processes (10–12). The majority of the ligands; usually a Cys, His and Met, are provided by a single loop, which links the 2 C-terminal β-strands, and makes a sizable contribution to the main interaction surface (10). The His ligand on the loop is in the center of this recognition patch and is the purported conduit for ET. The cupredoxin fold protects the T1 copper site, preventing possible adverse reactivity, and allowing efficient ET. A cupredoxin such as azurin (AZ), which has been characterized in detail and utilized for an array of ET and protein engineering studies (13–23), provides an ideal model system for assessing metal-binding loops.
Studies assessing the role of the T1 copper-binding loop have focused on swapping native sequences between cupredoxins (20–22, 24). In this study, we investigate the importance of metal-binding loop length alone by analyzing AZ variants in which nonnative loops containing only 1 repeating residue, whose side-chains cannot form hydrogen bonds, are introduced [the ligands are not mutated in these studies (see Table 1)]. We have used mainly alanine as it does not impose large backbone alterations and generally does not have significant steric requirements (25). We discover that length and not sequence dictates loop conformation at a T1 copper site. Side-chain variations within the loop can therefore control surface properties and hence facilitate interaction with a particular partner. A loop adorned with methyl groups is sufficient to provide a stable metal site protected from solvent. An empirical relationship is found between solvent accessibility and the Em value of a T1 copper site. A loop of nonnative length results in the formation of a strand-swapped dimer structure.
Table 1.
Loop sequences in the two series of AZ variants prepared in this work compared with those of the WT protein and also AMI
| Protein | Loop sequence | Protein | Loop sequence |
|---|---|---|---|
| AMI* | C-T-P-H-P-F-M | AZ1A1A† | C-A-H-A-M |
| AZ2G2G | C-G-G-H-G-G-M | AZ2A2A | C-A-A-H-A-A-M |
| AZ2A2A | C-A-A-H-A-A-M | AZ3A3A | C-A-A-A-H-A-A-A-M |
| AZ2V2V | C-V-V-H-V-V-M | AZ | C-T-F-P-G-H-S-A-L-M |
| AZ4A3A | C-A-A-A-A-H-A-A-A-M | ||
| AZ4A4A | C-A-A-A-A-H-A-A-A-A-M |
*AMI possesses the shortest naturally occurring T1 copper binding loop.
†This variant did not bind copper.
Results
Protein Isolation.
For all of the mutants prepared (see Table 1) the molecular weights determined by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) are all in agreement with theoretical values (see Table S1). AZ1A1A did not express well, could not be purified (because of the low amounts produced and multiple forms) and did not bind copper (see SI Text). Pure Cu(II) AZ2G2G is not particularly stable in solution (see SI Text), which prevented a detailed characterization of this variant. During the purification of AZ3A3A, blue noncovalently linked dimeric species were observed (see SI Text); possibly a precursor to the arrangement seen in the crystallographic studies (see below).
Spectroscopic Studies.
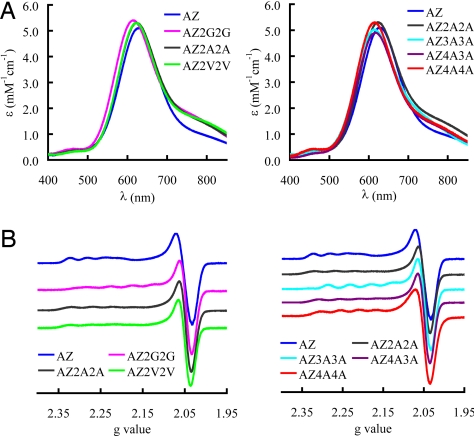
The UV/visible (UV/Vis) spectra of the copper-binding loop variants are compared in Fig. 1A. The positions and molar absorption coefficients of the main S(Cys) → Cu(II) ligand to metal charge-transfer (LMCT) transition, and the weaker LMCT band at higher energy, characteristic of a T1 copper site, are listed in Table S1. The position of the main LMCT band is shifted to slightly higher energy in all cases with the largest effect being 14 nm in AZ2G2G, AZ3A3A, and AZ4A4A.
Fig. 1.
Spectroscopic properties of the loop variants. UV/Vis (A) and EPR (B) spectra of the loop variants of AZ compared with the WT protein. UV/Vis spectra were obtained at 25 °C in 20 mM Tris pH 8.0 for AZ2G2G, AZ2A2A, AZ2V2V, and AZ4A3A; 50 mM Hepes pH 7.6 for AZ3A3A and AZ4A4A; and 10 mM phosphate pH 8.0 for AZ. All EPR spectra were measured at −196 °C and for AZ, AZ2G2G, and AZ3A3A were recorded in 25 mM Hepes pH 7.6 containing 40% glycerol, and those of AZ2V2V, AZ2A2A, AZ4A3A, and AZ4A4A were measured in 50 mM Hepes pH 7.6.
The electron paramagnetic resonance (EPR) spectra of the T1 copper-binding AZ loop variants are shown in Fig. 1B and the parameters are listed in Table S1 (simulations are shown in Fig. S1). All loop mutations have little effect on the EPR spectrum with small changes observed in the separation between gx and gy. In all loop variants gz decreases and Az increases compared with the values found for the WT protein. Certain loop variants exhibited more than 1 T1 copper signal in their EPR spectra, which is most apparent for AZ2G2G (see Fig. S2; for a detailed explanation, see SI Text). This behavior is indicative of metal-site flexibility that is not present in the WT protein, but has been observed in other T1 copper site mutants (15–17, 24).
Active Site Accessibility.
Titrations with azide monitored by UV/Vis spectrophotometry were used to test the accessibility of the copper site in AZ2G2G, AZ2A2A, AZ2V2V, AZAMI [AZ mutant in which the short loop of the cupredoxin amicyanin (AMI) has been introduced (see Table 1)], and the WT protein. The addition of azide to AZ2G2G, AZ2A2A, AZ2V2V, and AZAMI results in a decrease in absorbance for the S(Cys) → Cu(II) LMCT band at ≈600 nm with a band at ≈400 nm (407 nm, 401 nm, 392 nm, and 410 nm for AZ2A2A, AZ2G2G, AZ2V2V, and AZAMI, respectively) appearing, which is due to an azide → Cu(II) LMCT transition. These changes are identical to those seen before in experiments with T1 copper site variants in which the axial ligand has been removed (17, 26) and indicate azide binding to the T1 copper site. Dissociation constants for azide of 0.3, 2.4, 6.6, and 6.9 M for AZ2G2G, AZ2A2A, AZ2V2V, and AZAMI respectively are obtained. In the case of AZ there is no indication of azide binding (no change in the UV/Vis spectrum), even at very high concentrations (1.5 M).
Effect of Loop Mutations on Reduction Potentials.
AZ2G2G, AZ2A2A, AZ2V2V, and AZ4A3A yield quasi-reversible responses on modified gold electrodes giving Em values of 336 mV (pH 7.4), 410 mV (pH 7.6), 426 mV (pH 6.5), and 323 mV (pH 7.5), respectively. For AZ3A3A and AZ4A4A, the electrochemical responses were poorer. AZ3A3A gave a decent response at pH 7.5 and an Em of 332 mV could be determined, in good agreement with a value of 344 mV measured from a redox titration at pH 7.0. In the case of AZ4A4A, an acceptable electrochemical response was obtained at pH 6.5 giving an Em of 397 mV, consistent with a value of 395 mV from a redox titration at pH 7.0 (further details are available in SI Text).
Electron Self-Exchange Reactivity.
The electron self-exchange (ESE) reaction provides a measure of ET capability, which is not influenced by alterations in Em (no driving force). The ESE rate constants (kESE values) for AZ2A2A and AZ4A4A, the variants with the shortest and longest poly-alanine loops, measured using 1H NMR (NMR) at 40 °C, are 5.0 × 104 M−1·s−1 and 1.7 × 104 M−1·s−1 respectively (a more detailed explanation is available in SI Text; see also Fig. S3).
Crystal Structures.
Structures have been determined for AZ2A2A, AZ3A3A, AZ4A3A, and AZ4A4A. In all cases, overall folds are remarkably similar to the β-barrel arrangement of AZ. This is demonstrated by the Cα overlays with the WT protein, which give rmsds of 1.20, 0.44 and 0.84 Å for AZ2A2A, AZ4A3A, and AZ4A4A, respectively [loop residues are excluded from these comparisons except in the case of AZ4A3A, and chain A of 4AZU is used (14)]. The close similarity to AZ is also apparent at the copper sites, which only undergo minor modifications upon making the AZ2A2A, AZ4A3A, and AZ4A4A mutations (see Fig. S4 and Table S2). Comparison of the Cu(II) and Cu(I) (crystals reduced with ascorbate before data collection) structures of AZ2A2A and AZ4A4A reveal little change at the metal sites (see Tables S2 and S3).
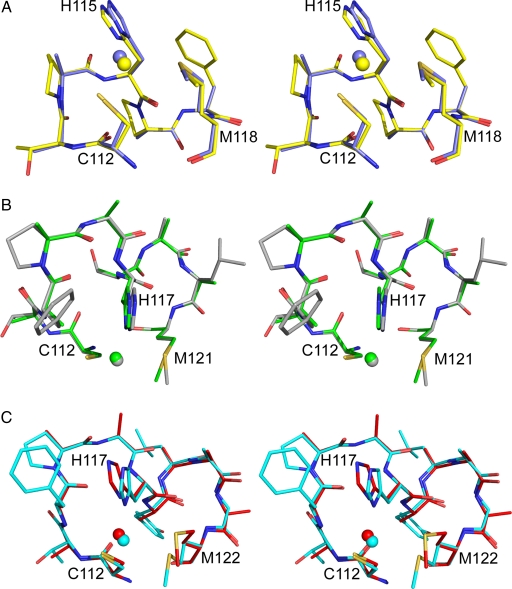
The most remarkable aspect of the structures of the loop mutants is the arrangement that the mutated loop adopts. In the case of AZ2A2A, the introduced loop has a conformation analogous (rmsd of 0.22 Å) to that found in AMI (27), a cupredoxin that naturally possesses the same spacing between the ligands (see Fig. 2A and Table 1) and AZAMI (21) (rmsd = 0.23 Å). For AZ4A3A the ligand spacings on the loop are the same as those in the WT protein (see Table 1) and the structural similarity is even greater (rmsd = 0.15 Å, see Fig. 2B). Overlays of the ligand-containing loops of rusticyanin (28) and auracyanin B (29), which have 4 intervening residues between the Cys and His and the His and Met ligands, with AZ4A4A give rmsds of 0.39 and 0.22 Å, respectively (see Fig. 2C).
Fig. 2.
Stereoviews of the copper-binding loops. Overlays of AZ2A2A (slate) and AMI (yellow) (A), AZ4A3A (green) and AZ (gray) (B), and AZ4A4A (red) and auracyanin B (cyan) (C). The loop residues are shown as stick models, and the copper ions as spheres. Ligands are numbered as in AZ2A2A (A) and AZ4A4A (C).
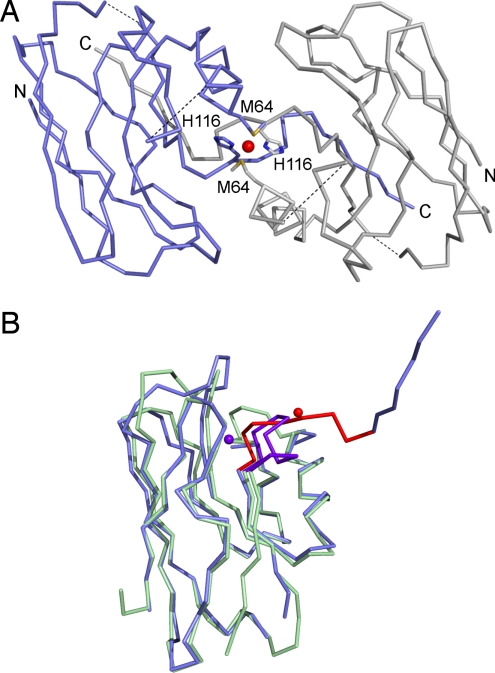
A strand-swapped dimer is found in the crystal structure of AZ3A3A (see Fig. 3A). This involves the C-terminal β-strand of 1 monomer completing the β-barrel of an adjacent molecule in the crystal lattice. The structure of the monomer overlays well with that of the WT protein until residue 114 (rmsd of 1.67 Å). After this point loop structure is lost and the arrangement is completely different to that in the WT protein (see Fig. 3B). The final β-strand from Met-120 to Leu-126 (the C-terminal residue is not observed) forms in AZ3A3A as in AZ, making the same hydrogen bonding contacts with strand 2 and the penultimate strand in the barrel, but these interactions are all with the adjacent monomer (see Fig. 3A). A possible distorted tetrahedral copper site is formed at the dimer interface via the Sδ of Met-64 and the Nδ1 of His-116 (distances of ≈2.0 and ≈2.2 Å, respectively).
Fig. 3.
Cα traces showing the structure of AZ3A3A. (A) The strand-swapped dimer arrangement of AZ3A3A. One monomer is slate and the other is gray; the purported copper ion is a red sphere; and the N- and C-termini are labeled. Because of breaks in the electron density Ser-66 to Asp-71 and Lys-103 to Glu-106 are not modeled and dashed lines link these regions. (B) Overlay of AZ3A3A (slate) with AZ (pale green) with the copper-binding loop and the metal red in the former and purple in the latter.
Discussion
A range of AZ variants have been characterized in which the native copper-binding loop has been replaced with sequences made up from, Ala, Gly and Val residues (Table 1). Certain loop lengths studied match those found naturally in single domain cupredoxins, and some loops of nonnative length have been incorporated. In all cases (except AZ1A1A, which did not bind copper), the introduction of these loops, containing residues whose side chains have no capacity to form hydrogen bonds, can have remarkably little effect on the properties of the T1 copper site. The Cu(II) variants have only subtly altered spectroscopic properties compared with AZ. Therefore, the geometry of the Cu(II) center is largely unaffected by these dramatic variations in loop length and composition. The active site structures, determined crystallographically, of AZ2A2A, AZ4A3A, and AZ4A4A are almost identical to that of the WT protein, with main-chain hydrogen bonding patterns comparable to those of cupredoxins that possess loops of the same length (see below). The Em values are all typical for this family of ET proteins and those of AZ4A3A and AZ are very similar (same spacings between the ligands). It therefore appears that the side chains in the AZ loop have little influence on this key physiological feature, providing that the copper is sufficiently protected from solvent (see below).
The most impressive feature of the AZ2A2A, AZ4A3A, and AZ4A4A variants is the almost identical structure of the C-terminal loop when compared with members of the family that have the same length metal-binding loops, but with very different sequences. The AZ2A2A loop conformation is identical to those of AMI (27) and AZAMI (21), the AZ4A3A loop to that of the WT protein (14, 19) and the AZ4A4A loop to those of rusticyanin (28) and auracyanin B (29). In all cases, this results in the Ala Cβ atoms being in almost identical positions as the Cβ atoms of the corresponding residues in the native loops (see Fig. 2). It is interesting to note that multiple alanine substitutions have been made in a range of proteins, most notably in bovine pancreatic trypsin inhibitor where >1/3 of the residues have been mutated to Ala, with retention of native structure and function (30). Analysis of conserved structural and functional features of cupredoxins has identified a common core that forms a column extending to the copper site (9). Residues in the ligand-containing loop were not highlighted in this study, but conservation of loops that surround the copper ligands was suggested to be important for maintaining the orientation of coordinating amino acids. Our studies with AZ demonstrate that the structure of the metal-binding loop is dictated by length and is essentially independent of sequence.
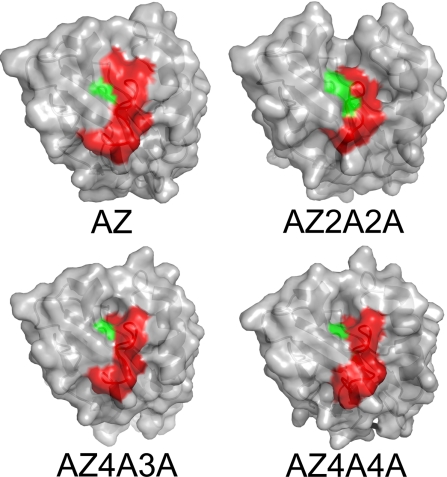
Why would a widely used biological ET system, based on a common structural motif, have evolved to have a surface exposed metal-binding loop whose structure is independent of sequence? To answer this question, the contribution that the metal binding loop residues make to the key surface feature for interactions needs to be considered (see Fig. 4). This typically nonpolar region surrounds the His ligand on the loop and is the only interaction surface identified in this family of proteins (10). The AZ2A2A and AZ4A4A mutations both decrease homo-dimer formation, which occurs via this hydrophobic patch, as indicated by significantly lower kESE values (structures indicate that this is the most likely cause of the kESE decreases, although changes in reorganization energy and electronic coupling cannot be discounted). Physiological partners for AZ in Pseudomonas aeruginosa have not been identified (31), but the effect on kESE indicates that the metal-binding loop is important for protein interactions. The observation that the conformation of this segment is not influenced by sequence means that loops have been able to evolve to provide a surface patch optimized for interacting with a particular partner without compromising a copper site capable of supporting rapid ET.
Fig. 4.
The surface exposed ligand-containing loop. Surface representations showing the contributions that the C-terminal ligand-containing loop (red) and the central His ligand (green) make to the important hydrophobic interaction patch.
Many metalloproteins whose function involves interacting with a protein partner have metal sites bound by surface loops, and we suggest that sequence variability is also present in these regions. One family where this seems to be relevant are the metallochaperones, which protect the metal ion and collect it from, and deliver it to, the correct location. Uptake of metal seems to stabilize the structure of the metal binding loop (32, 33), and may lead to the formation of a structure conducive to interaction with the correct partner. In these proteins the metal needs to be accessible, and the strong preference for a particular oxidation state can provide protection against adverse reactivity. In the cupredoxins, the metal binding loop is not influenced by the presence of the copper ion, because the loop structure is very similar in apo-AZ (34) (loop rmsd = 0.34 Å). This is an important attribute for a site where metal dissociation is not part of the protein's function.
A comparison of the AZ2G2G, AZ2A2A, and AZ2V2V variants allows the influence of solvent accessibility of the copper site to be uncovered. The AZ2G2G mutant exhibits Cu(II) site flexibility (as demonstrated by EPR) and is not stable. Both features are probably a consequence of a more exposed copper as identified by the relatively high affinity of the site for azide [the Kd value is similar to those for the Met-121 variants of AZ missing an axial ligand (17)]. The copper site of AZ2A2A is much less accessible, as demonstrated by the 8-fold lower affinity for azide, consistent with the buried nature of the copper ion in the crystal structure, and the AZ2V2V site is even more protected (to a similar extent as in AZAMI). The Em value of AZ2G2G is the lowest, with that of AZ2V2V considerably higher, just above the value for AZ2A2A. A number of features, including solvent accessibility, copper site geometry, hydrogen bonding, and the dipoles of residues in the vicinity of a T1 copper site (10, 11, 22, 35–39) have been proposed as determinants of Em. The Cu(II) sites in this series of AZ variants are similar (as judged spectroscopically) and the hydrogen bonding pattern around the active site is not expected to vary. We therefore assume that as the active site becomes more protected (molecular models of AZ2G2G and AZ2V2V based on the AZ2A2A structure are consistent with this observation), and the copper environment less polar, the Cu(I) form is stabilized resulting in an increased Em. Adequate protection from solvent is achieved by a short loop adorned with 4 methyl groups, and therefore most sequences will perform this task. The loop length is the same in AZAMI, yet this variant has a much lower Em, which is the same as that of AMI. The loop structure, including side-chain rotamers, of AZAMI matches that in AMI, and in this case it is thought that the dipoles of residues in the loop are a significant factor controlling Em (22, 37).
The AZ3A3A variant has a tendency to form stable dimers in solution and a strand swapped dimer in the crystal structure. Domain swapping involves a β-strand breaking its interactions within 1 barrel and reforming them with an identical partner. All of the noncovalent bonds that stabilize the monomer are still present except in the hinge region, which is the mutated loop. A strand-swapped dimer arrangement has not been seen for a cupredoxin, but in nitrosocyanin an extended β-hairpin between the first 2 strands is involved in trimer formation (40). It thus appears that the loop in AZ3A3A, which only differs in length by 1 residue to that of the WT protein and AZ4A3A, alters stability and favors the formation of a strand-swapped dimer, rather than the desired monomeric β-barrel. Strand-swapped dimerization has been investigated and shortening the hinge region commonly leads to domain swapping (41–43). In AZ, loop contraction is not the only determinant, because AZ2A2A, and other AZ variants with a shortened active site loop (including AZAMI), do not form such structures (21, 22). Loop design experiments have highlighted the importance of Pro residues for this feature, and studies with another β-barrel protein suggest Pro-containing loops prevent domain swapping by not allowing this region to open (44). However, studies on a domain consisting of a 4-stranded β-sheet and a single α-helix have found that a designed Pro in a loop can favor domain swapping (45). In AZ, the native loop has a Pro at position 115, but this is removed in all of the variants studied herein. Only the AZ3A3A mutant forms a strand-swapped dimer, indicating that loop length is the key determinant, and that 3-residue spacings between the ligands on the loop favors domain swapping. For a single domain ET shuttle, this should be avoided and therefore a cupredoxin (or any T1 copper site) has not evolved with this length metal-binding loop.
Materials and Methods
Loop-Mutagenesis.
Loop mutagenesis was carried out using the QuikChange (Stratagene) site-directed mutagenesis kit. pTrcAZ, a pTrc99a derivative harboring the azruin (AZ) gene from Pseudomonas aeruginosa including the signal peptide, was used as the template (20). The Cys112 to Met121 loop of AZ was mutated from C112TFPGH117SALM121 to C112AH114AM116, C112AAH115AAM118, C112GGH115GGM118, C112VVH115VVM118, C112AAAH116AAAM120, C112AAAAH117AAAM121, and C112AAAAH117AAAAM122 (Table 1), using the pairs of primers listed in Table S4. Both strands of the mutated plasmids (pTrcAZ1A1A, pTrcAZ2V2V, pTrcAZ2A2A, pTrcAZ2G2G, pTrcAZ3A3A, pTrcAZ4A3A and pTrcAZ4A4A) were sequenced to verify the mutations.
Cell Growth, Protein Isolation, and Purification.
Procedures used for growing cells and isolating and purifying proteins are described in detail in the SI Text. Pure oxidized AZ2G2G and AZ2V2V have A280/A614 and A280/A622 ratios of ≤4.1 and 2.4, respectively, whereas AZ2A2A, AZ3A3A, AZ4A3A, and AZ4A4A, respectively, have A280/A624, A280/A614, A280/A617, and A280/A614 ratios all in the 1.8 to 2.0 range. All of these variants gave single bands on 15% SDS/PAGE gels.
UV/Vis Spectrophotometry.
UV/Vis spectra were measured at 25 °C on a Perkin-Elmer λ 35 spectrophotometer in either 20 mM Tris pH 8.0 or 50 mM Hepes pH 7.6.
Azide Titrations.
Azide titrations were carried out in 20 mM Tris pH 8.0 at 25 °C. The influence of adding various amounts of azide from a ≈5 M stock solution in 20 mM Tris pH 8.0 (this stock solution had a pH of ≈8.5) on the UV/Vis spectrum was analyzed. The concentration ranges of azide used for the titrations were as follows: 0.05 to 239 mM for AZ2G2G (20 μM); 0.11 to 894 mM for AZ2A2A (22 μM); 0.06 to 1,400 mM for AZ2V2V (32 μM); 5.41 to 1,537 mM for AZ (25 μM) and 5.3 to 1763 mM for AZAMI (29 μM) (pH changes in these experiments were all <0.3).
Determination of Metal Concentrations and Molecular Weights.
The concentration of copper and zinc were determined by atomic absorption spectrophotometry, on a Thermo Electron Corporation (Cambridge) M Series spectrophotometer as described in ref. 21. Molecular weights of all proteins were determined by MALDI-TOF-MS.
EPR Spectroscopy.
X-band EPR spectra were recorded as described in ref. 20. A sample of diphenylpicrylhydrazyl was used as an external reference, and spectral simulations were generated with the program SIMFONIA (Bruker).
Measurement of Reduction Potentials.
The direct measurements of reduction potentials (Em values) were carried out at ambient temperature (22 ± 1 °C), using an electrochemical setup described in ref. 20. Redox titrations with AZ3A3A and AZ4A4A, using the [Fe(CN)6]3−/4− couple were performed at 25 °C in 50 mM phosphate pH 7.0 (46). Further details are provided in the SI Text.
Samples and 1H NMR Spectroscopy.
Samples for 1H NMR experiments on Cu(II) and Cu(I) proteins and also mixtures for kESE determinations were prepared as described in ref. 47. Samples were usually in 10 or 20 mM phosphate pH* 8.0 (kESE of AZ2A2A was determined in 10 mM phosphate pH* 8.0 while that of AZ4A4A was measured in 20 mM phosphate pH * 8.0) and most spectra were acquired at 40 °C. Spectra were obtained using standard 1D and WEFT pulse sequences on a Jeol Lambda 500 spectrometer and were processed as described in ref. 47. Spin-spin (T2) relaxation times were derived from peak widths at half-height, using the relation υ½ = (πT2)−1.
Crystallization and Structure Determination.
Crystallization conditions are described in the SI Text, along with data collection and processing statistics (Table S5). Briefly, X-ray data were collected either in-house or at synchrotron radiation sources, processed with MOSFLM/iMOSFLM (48) and scaled with SCALA [as implemented in the CCP4 suite (49)]. Structures were solved, where necessary, using molecular replacement programs MOLREP and PHASER [as implemented in the CCP4 suite (49)]. Final structures were completed using repeated cycles of interactive model building in COOT (50) and refinement with REFMAC5 [as implemented in the CCP4 suite (49)]. Final structures were validated using COOT and MOLPROBITY (51). LSQMAN (52) was used to superimpose structures and determine rmsds for Cα atoms and PYMOL (see http://pymol.org) was used for preparing all protein structure figures. The coordinates and structure factors were deposited in the Protein Data Bank {PDB ID codes 3FS9 [Cu(II) AZ2A2A, pH 7], 3FSA [Cu(I) AZ2A2A, pH 7], 3FSV (AZ3A3A), 3FSW [Cu(II) AZ4A3A, pH 8], 3FSZ [Cu(II) AZ4A4A, pH 7.6], and 3FT0 [Cu(I) AZ4A4A, pH 7.6]}.
Supplementary Material
Acknowledgments.
We thank Andrew McCarthy (EMBL-Grenoble/ESRF), Adriana Badarau (Newcastle University) and staff of the Diamond Light Source (Liz Duke, Katherine McAuley and David Hall) for assistance with data collection. This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/C504519/1 (to C.D.) and a Royal Society (United Kingdom) University Research Fellowship (to M.J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org {PDB ID codes 3FS9 [Cu(II) AZ2A2A], 3FSA [Cu(I) AZ2A2A], 3FSV (AZ3A3A), 3FSW [Cu(II) AZ4A3A], 3FSZ [Cu(II) AZ4A4A], and 3FT0 [Cu(I) AZ4A4A]}.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811324106/DCSupplemental.
References
- 1.Akiva E, Itzhaki Z, Margalit H. Built-in loops allow versatility in domain-domain interactions: Lessons from self-interacting domains. Proc Natl Acad Sci USA. 2008;105:13292–13297. doi: 10.1073/pnas.0801207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferguson N, et al. Using flexible loop mimetics to extend Φ-value analysis to secondary structure interactions. Proc Natl Acad Sci USA. 2001;98:13008–13013. doi: 10.1073/pnas.221467398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang IJ, Lee JC, Winkler JR, Gray HB. The protein-folding speed limit: Intrachain diffusion times set by electron-transfer rates in denatured Ru(NH3)5(His-33)-Zn-cytochrome c. Proc Natl Acad Sci USA. 2003;100:3838–3840. doi: 10.1073/pnas.0637283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krieger F, Möglich A, Kiefhaber T. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J Am Chem Soc. 2005;127:3346–3352. doi: 10.1021/ja042798i. [DOI] [PubMed] [Google Scholar]
- 5.Thomson AJ, Gray HB. Bio-inorganic chemistry. Curr Opin Chem Biol. 1998;2:155–158. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y. Biosynthetic inorganic chemistry. Angew Chem Int Ed. 2006;45:5588–5601. doi: 10.1002/anie.200600168. [DOI] [PubMed] [Google Scholar]
- 7.Stevens FJ. Homology versus analogy: Possible evolutionary relationship of immunoglobulins, cupredoxins and Cu,Zn-superoxide dismutase. J Mol Recognit. 2007;21:20–29. doi: 10.1002/jmr.861. [DOI] [PubMed] [Google Scholar]
- 8.Adman ET. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 9.Gough J, Chothia C. The linked conservation of structure and function in a family of high diversity: The monomeric cupredoxins. Structure. 2004;12:917–925. doi: 10.1016/j.str.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Dennison C. Investigating the structure and function of cupredoxins. Coord Chem Rev. 2005;249:3025–3054. [Google Scholar]
- 11.Gray HB, Malmström BG, Williams RJP. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 12.Savelieff MG, et al. Experimental evidence for a link among cupredoxins: Red, blue, and purple copper transformations in nitrous oxide reductase. Proc Natl Acad Sci USA. 2008;105:7919–7924. doi: 10.1073/pnas.0711316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker EN. Structure of azurin from Alcaligenes denitrificans. J Mol Biol. 1988;203:1071–1095. doi: 10.1016/0022-2836(88)90129-5. [DOI] [PubMed] [Google Scholar]
- 14.Nar H, Messerschmidt A, Huber R, van de Kamp M, Canters GW. Crystal structure analysis of oxidized Pseudomonas aeruginosa azurin at pH 5.5 and pH 9.0. J Mol Biol. 1991;221:765–772. doi: 10.1016/0022-2836(91)80173-r. [DOI] [PubMed] [Google Scholar]
- 15.Germanas JP, Di Bilio AJ, Gray HB, Richards JH. Site saturation of the histidine-46 position in Pseudomonas aeruginosa azurin: Characterization of the His46Asp copper and cobalt proteins. Biochemistry. 1993;32:7698–7702. doi: 10.1021/bi00081a014. [DOI] [PubMed] [Google Scholar]
- 16.den Blaauwen T, Canters GW. Creation of type-1 and type-2 copper sites by addition of exogenous ligands to the Pseudomonas aeruginosa azurin His117Gly mutant. J Am Chem Soc. 1993;115:1121–1129. [Google Scholar]
- 17.Bonander N, Karlsson BG, Vänngård T. Environment of copper in Pseudomonas aeruginosa azurin probed by binding of exogenous ligands to Met121X (X = Gly, Ala, Val, Leu, or Asp) mutants. Biochemistry. 1996;35:2429–2436. doi: 10.1021/bi9522110. [DOI] [PubMed] [Google Scholar]
- 18.Di Bilio AJ, et al. Reorganization energy of blue copper: Effects of temperature and driving force on rates of electron transfer in ruthenium- and osmium-modified azurins. J Am Chem Soc. 1997;119:9921–9922. [Google Scholar]
- 19.Crane BR, Di Bilio AJ, Winkler JR, Gray HB. Electron tunnelling in single crystals of Pseudomonas aeruginosa azurins. J Am Chem Soc. 2001;123:11623–11631. doi: 10.1021/ja0115870. [DOI] [PubMed] [Google Scholar]
- 20.Yanagisawa S, Dennison C. Loop-contraction mutagenesis of type 1 copper sites. J Am Chem Soc. 2004;126:15711–15719. doi: 10.1021/ja047295r. [DOI] [PubMed] [Google Scholar]
- 21.Li C, et al. Basic requirements for a metal-binding site in a protein: The influence of loop shortening on the cupredoxin azurin. Proc Natl Acad Sci USA. 2006;103:7258–7263. doi: 10.1073/pnas.0600774103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Banfield MJ, Dennison C. Engineering copper sites in proteins: Loops confer native structures and properties to chimeric cupredoxins. J Am Chem Soc. 2007;129:709–718. doi: 10.1021/ja0661562. [DOI] [PubMed] [Google Scholar]
- 23.Shih C, et al. Tryptophan-accelerated electron flow through proteins. Science. 2008;320:1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 24.Buning C, et al. Loop-directed mutagenesis of the blue copper protein amicyanin from Paracoccus versutus and its effect on the structure and the activity of the type-1 copper site. J Am Chem Soc. 2000;122:204–211. [Google Scholar]
- 25.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 26.Yanagisawa S, Dennison C. Reduction potential tuning at a type 1 copper site does not compromise electron transfer reactivity. J Am Chem Soc. 2005;127:16453–16459. doi: 10.1021/ja054426v. [DOI] [PubMed] [Google Scholar]
- 27.Cunane LM, Chen ZW, Durley RCE, Mathews FS. X-ray structure of the cupredoxin amicyanin, from Paracoccus denitrificans, refined at 1.31 Å resolution. Acta Crystallogr D. 1996;52:676–686. doi: 10.1107/S0907444996001072. [DOI] [PubMed] [Google Scholar]
- 28.Barrett ML, et al. Atomic resolution crystal structures, EXAFS, and quantum chemical studies of rusticyanin and its two mutants provide insight into its unusual properties. Biochemistry. 2006;45:2927–2939. doi: 10.1021/bi052372w. [DOI] [PubMed] [Google Scholar]
- 29.Bond CS, et al. Crystal structure of auracyanin, a “blue” copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus. J Mol Biol. 2001;306:47–67. doi: 10.1006/jmbi.2000.4201. [DOI] [PubMed] [Google Scholar]
- 30.Islam MM, Sohya S, Noguchi K, Yohda M, Kuroda Y. Crystal structure of an extensively simplified variant of bovine pancreatic trypsin inhibitor in which over one-third of the residues are alanines. Proc Natl Acad Sci USA. 2008;105:15334–15339. doi: 10.1073/pnas.0802699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijgenboom E, Busch JE, Canters GW. In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of RpoS and ANR. Microbiology. 1997;143:2853–2863. doi: 10.1099/00221287-143-9-2853. [DOI] [PubMed] [Google Scholar]
- 32.Rosenzweig AC. Copper delivery by metallochaperone proteins. Acc Chem Res. 2001;34:119–128. doi: 10.1021/ar000012p. [DOI] [PubMed] [Google Scholar]
- 33.Banci L, et al. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat Chem Biol. 2006;2:367–368. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 34.Nar H, Messerschmidt A, Huber R, van de Kamp M, Canters GW. Crystal structure of Pseudomonas aeruginosa apo-azurin at 1.85 Å resolution. FEBS Lett. 1992;306:119–124. doi: 10.1016/0014-5793(92)80981-l. [DOI] [PubMed] [Google Scholar]
- 35.Libeu CAP, Kukimoto M, Nishiyama M, Horinouchi S, Adman ET. Site-directed mutants of pseudoazurin: Explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences. Biochemistry. 1997;36:13160–13179. doi: 10.1021/bi9704111. [DOI] [PubMed] [Google Scholar]
- 36.Machczynski MC, Gray HB, Richards JH. An outer-sphere hydrogen-bond network constrains copper coordination in blue proteins. J Inorg Biochem. 2002;88:375–380. doi: 10.1016/s0162-0134(02)00364-1. [DOI] [PubMed] [Google Scholar]
- 37.Olsson MHM, Hong G, Warshel A. Frozen density functional free energy simulations of redox proteins: Computational studies of the reduction potential of plastocyanin and rusticyanin. J Am Chem Soc. 2003;125:5025–5039. doi: 10.1021/ja0212157. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Webb SP, Ivanic J, Jensen JH. Determinants of the relative reduction potentials of type-1 copper sites in proteins. J Am Chem Soc. 2004;126:8010–8019. doi: 10.1021/ja049345y. [DOI] [PubMed] [Google Scholar]
- 39.Yanagisawa S, Banfield MJ, Dennison C. The role of hydrogen bonding at the active site of a cupredoxin: The Phe114Pro azurin variant. Biochemistry. 2006;45:8812–8822. doi: 10.1021/bi0606851. [DOI] [PubMed] [Google Scholar]
- 40.Lieberman RL, Arciero DM, Hooper AB, Rosenzweig AC. Crystal structure of a novel red copper protein from Nitrosomonas europaea. Biochemistry. 2001;40:5674–5681. doi: 10.1021/bi0102611. [DOI] [PubMed] [Google Scholar]
- 41.Green SM, Gittis AG, Meeker AK, Lattman EE. One-step evolution of a dimer from a monomer protein. Nat Struct Biol. 1995;2:746–751. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 42.Dickason RR, Huston DP. Creation of a biologically active interleukin-5 monomer. Nature. 1996;379:652–655. doi: 10.1038/379652a0. [DOI] [PubMed] [Google Scholar]
- 43.Ogihara NL, et al. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc Natl Acad Sci USA. 2001;98:1404–1409. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu X, Wang H, Ke H, Kuhlman B. High-resolution design of a protein loop. Proc Natl Acad Sci USA. 2007;104:17668–17673. doi: 10.1073/pnas.0707977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhlman B, O'Neill JW, Kim DE, Zhang KYJ, Baker D. Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc Natl Acad Sci USA. 2001;98:10687–10691. doi: 10.1073/pnas.181354398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldberg M, Pecht I. Kinetics and equilibria of the electron transfer between azurin and the hexacyanoiron (II/III) couple. Biochemistry. 1976;15:4197–4208. doi: 10.1021/bi00664a011. [DOI] [PubMed] [Google Scholar]
- 47.Sato K, Kohzuma T, Dennison C. Active-site structure and electron-transfer reactivity of plastocyanins. J Am Chem Soc. 2003;125:2101–2112. doi: 10.1021/ja021005u. [DOI] [PubMed] [Google Scholar]
- 48.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 49.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA. In: International Tables for Crystallography, Vol. F. Crystallography of Biological Macromolecules. Rossmann MG, Arnold E, editors. Dordrecht, The Netherlands: Kluwer; 2001. pp. 353–356.pp. 366–367. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.