Abstract
DNA lesions that block replication can be bypassed by error-prone or error-free mechanisms. Error-prone mechanisms rely on specialized translesion synthesis (TLS) DNA polymerases that directly replicate over the lesion, whereas error-free pathways use an undamaged duplex as a template for lesion bypass. In the yeast Saccharomyces cerevisiae, most mutagenic TLS of spontaneous and induced DNA damage relies on DNA polymerase ζ (Polζ) activity. Here, we use a distinct mutational signature produced by Polζ in a frameshift-reversion assay to examine the role of the yeast mismatch repair (MMR) system in regulating Polζ-dependent mutagenesis. Whereas MMR normally reduces mutagenesis by removing errors introduced by replicative DNA polymerases, we find that the MMR system is required for Polζ-dependent mutagenesis. In the absence of homologous recombination, however, the error-prone Polζ pathway is not affected by MMR status. These results demonstrate that MMR promotes Polζ-dependent mutagenesis by inhibiting an alternative, error-free pathway that depends on homologous recombination. Finally, in contrast to its ability to remove mistakes made by replicative DNA polymerases, we show that MMR fails to efficiently correct errors introduced by Polζ.
Keywords: DNA damage, mutagenesis, recombination, replication
Mutations generally impair fitness and are important contributors to genome instability and disease, but a low level of mutagenesis is required to provide raw material for evolution. Spontaneous mutations result from endogenous metabolic processes and can be attributed either to errors made when copying an undamaged DNA template or to errors introduced when replicating over a DNA lesion. Mistakes of the first type are corrected by the proofreading activity of the replicative DNA polymerases or by the postreplicative mismatch repair (MMR) machinery (1, 2). In addition to monitoring replication fidelity (spellchecker function), the MMR system also monitors the fidelity of homologous recombination (3). By reducing interactions between sequences that are not identical, MMR-associated antirecombination activity limits genomic rearrangements between dispersed repeats. Finally, because of its ability to recognize helical distortions, the MMR machinery also binds to damage-containing DNA, specifically triggering checkpoint signaling in higher eukaryotes (4).
In terms of the contribution of DNA damage to mutagenesis, some lesions are miscoding and promote the insertion of an incorrect nucleotide (5). Other lesions such as abasic sites and bulky covalent attachments, however, completely block the progress of replicative DNA polymerases in vitro. Such polymerase-blocking lesions must be bypassed in vivo to avoid cell cycle arrest and possible apoptosis. One type of bypass involves insertion of a nucleotide opposite the blocking lesion by a specialized translesion synthesis (TLS) DNA polymerase. Such bypass can be error-free or error-prone depending on the specific TLS polymerase recruited and/or the nature of the lesion (6, 7). The second type of tolerance mechanism is strictly error-free and involves the use of an undamaged DNA strand, typically from the sister chromatid, as a template to extend the blocked 3′ end past the lesion. Such bypass can occur through homologous recombination, which involves the invasion of an intact duplex, or through a template-switching mechanism that likely involves replication fork regression (8).
The yeast Saccharomyces cerevisiae possesses three TLS polymerases: Polζ (zeta), Polη (eta), and Rev1. Polζ is generally considered to be a highly error-prone polymerase; it is required not only for most induced mutagenesis but also for a substantial proportion of spontaneous mutations (9). Although Polζ alone can perform lesion bypass in vitro, its unusual ability to extend mispaired 3′ ends suggests that it cooperates with other DNA polymerases to complete lesion bypass in vivo (10). Most Polζ-dependent mutagenesis requires the Rev1 protein, which acts either as a deoxycytidyltransferase to specifically insert cytosine opposite lesions (11, 12) or as a structural protein to aid in Polζ-dependent bypass (13). In contrast to the mutagenesis associated with Polζ and Rev1, Polη is best known for its ability to bypass thymine-thymine dimers (14) and 7,8-dihydro-8-oxoguanine (15) in an error-free manner. In addition to lesion bypass activity, TLS polymerases share 2 additional properties: (i) they are much more error-prone than the replicative DNA polymerases on undamaged DNA templates and (ii) they lack the 3′-to-5′ exonucleolytic proofreading activity of replicative DNA polymerases. Whether errors introduced by TLS polymerases can be removed by the MMR machinery has not been previously examined.
The studies presented here were designed to determine whether Polζ-dependent mutational intermediates are edited by the MMR machinery in yeast. For these analyses, we took advantage of the distinct mutational signature produced by Polζ in the lys2ΔA746 frameshift-reversion assay. Our previous studies demonstrated that Polζ is specifically required for the production of “complex” mutations in which the selected frameshift is accompanied by 1 or more nearby base substitutions (16). Here we find that TLS by Polζ is greatly reduced upon loss of functional MMR. The requirement of MMR for TLS depends on homologous recombination, indicating a previously unrecognized role for MMR in determining the mechanism of lesion tolerance in yeast.
Results
The lys2ΔA746 allele reverts to lysine prototrophy via compensatory net +1 frameshifts that occur within an ≈150-bp reversion window defined by stop codons in alternative reading frames. In a wild-type (WT) background, 83% of reversion events are simple +1 insertions and of these, 90% are associated with homopolymer runs >3N. In an MMR-deficient background, the reversion rate is elevated several hundredfold and the distribution of revertants is even more skewed, with 99% being +1 events associated with runs >3N (17). In contrast to the strong mutator phenotype of MMR-defective strains, the reversion rate of the lys2ΔA746 allele is elevated only 2-fold in a nucleotide excision repair (NER)-deficient background. There is a very striking increase in complex insertions (from 6% to ≈25% of total mutations) upon NER loss, however, with all of the increase occurring at 2 discrete hotspots referred to as HS1 and HS2 (16, 18). Because the HS1/HS2 events are completely dependent on the presence of Polζ and are specifically enhanced when NER is defective (16, 19), they can be attributed to the error-prone bypass of unrepaired DNA damage. The HS1/HS2 events are the focus of the studies reported here.
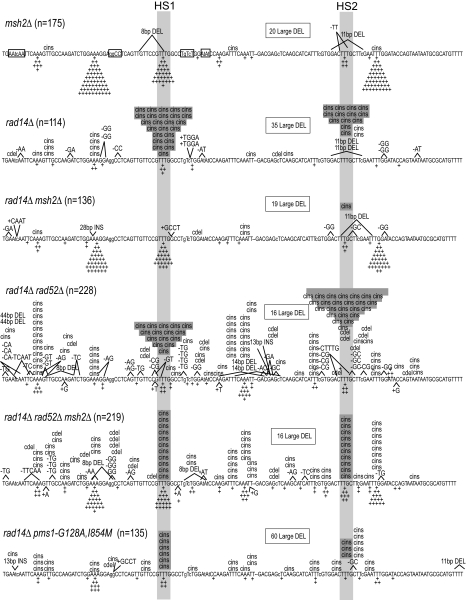
The specific question we wanted to ask is whether the HS1/HS2 events in an NER-defective rad14Δ mutant are subject to correction by the MMR machinery. If so, then their rate should increase further in a rad14Δ msh2Δ double mutant that is devoid of mismatch-recognition activity. The problem with this general approach is that the strong mutator phenotype associated with Msh2 loss would be expected to completely obscure complex mutations at HS1/HS2. To reduce the msh2Δ mutator phenotype, we constructed the “No Run” lys2ΔA746-NR allele, which is missing homopolymer runs of >3N within the reversion window, and hence the positions where most reversion events occur in MMR-deficient strains (see previous discussion). As predicted, there was a much smaller increase in Lys+ rate in the lys2ΔA746-NR msh2Δ mutant (7.3-fold relative to WT; Table 1) than in the original lys2ΔA746 msh2Δ mutant (190-fold relative to WT) (17). In the lys2ΔA746-NR msh2Δ mutant, 82% of the revertants had a simple +1 mutation and 95% of these were in 3N runs (Fig. 1). As observed with the lys2ΔA746 allele upon NER elimination, there was a small, 1.8-fold, increase in the rate of Lys+ prototrophs in the lys2ΔA746-NR rad14Δ mutant (Table 1). As expected, there was a strong accumulation of complex insertions at HS1 and HS2, with these constituting 32% of the total revertants (Fig. 1). We chose to use a rad14Δ mutant in these analyses because, in contrast to NER proteins such as Rad1 and Rad10, the only known function of Rad14 is in NER. In the WT parent strain, 6% of the reversion events were complex insertions, and only half of these were at HS1/HS2 (Table 1).
Table 1.
Reversion of the lys2Δ A746-NR allele in repair-deficient strains
| Genotype | Lys+ rate × 10−9 (95% CI) | Lys+ rate relative to WT | HS1/HS2 complex insertions* |
Non-HS1/HS2 complex insertions |
||
|---|---|---|---|---|---|---|
| Number | Rate × 10−10 | Number | Rate × 10−10 | |||
| WT | 4.6 (3.1–6.6) | 1.0 | 4/128 | 1.4 (0.97–2.1) | 4/128 | 1.4 (0.97–2.1) |
| msh2Δ | 34 (27–40) | 7.3 | 0/175 | <1.9† | 3/175 | 5.8 (4.6–6.9) |
| rad14Δ | 8.4 (6.1–11) | 1.8 | 37/114 | 27 (20–36) | 12/114 | 8.8 (6.4–12) |
| rad14Δ msh2Δ | 31 (25–37) | 6.6 | 1/136 | 2.3 (1.8–2.7) | 3/136 | 6.8 (5.5–8.2) |
| rad14Δ rad52Δ | 41 (33–49) | 8.8 | 39/228 | 69 (56–84) | 65/228 | 120 (94–140) |
| rad14Δ rad52Δ msh2Δ | 77 (64–90) | 17 | 21/219 | 73 (61–86) | 48/219 | 170 (140–200) |
| rad14Δ pms1Δ | 29 (23–36) | 6.3 | 1/123 | 2.4 (1.9–2.9) | 7/123 | 17 (13–20) |
| rad14Δ pms1-G128A,I854M | 9.8 (7.3–13) | 2.1 | 9/135 | 6.5 (4.9–8.7) | 29/135 | 21 (16–28) |
| rad52Δ | 15 (9.1–22) | 3.2 | 23/173 | 20 (12–29) | 63/173 | 54 (33–80) |
| rad52Δ msh2Δ | 49 (37–63) | 11 | 7/189 | 18 (14–23) | 38/189 | 99 (74–130) |
CI, confidence interval.
*Compilations of the HS1 and HS2 complex event types identified in each strain are presented in Tables S2 and S3, respectively.
†Rate calculated assuming the presence of one event.
Fig. 1.
lys2ΔA746-NR reversion spectra in repair-defective backgrounds. The sequence of the ≈150-bp reversion window is shown. The site of the 1-nt deletion that defines the allele is indicated by a dash and additional changes from the WT sequence are in lowercase. The original positions of runs >3N are boxed in the msh2Δ sequence. Simple insertions are indicated by “+” below the sequence and other mutation types are indicated above the sequence. The positions of HS1 and HS2 are indicated by light gray shading; complex insertions (“cins”) at these positions are shaded dark gray. “Large DEL” refers to 95-nt deletions with endpoints in 10-nt direct repeats; “cdel” corresponds to −2 events associated with nearby base substitutions. The number (“n”) of independent revertants sequenced from each background is indicated.
Polζ-Dependent Lesion Bypass at HS1/HS2 Depends on MMR.
We predicted that loss of Msh2 in a rad14Δ background would have 1 of 2 effects on the rate of HS1/HS2 complex events. If MMR edits Polζ errors, then the rate of complex insertions at HS1/HS2 should be greatly elevated in the rad14Δ msh2Δ double mutant relative to the rad14Δ single mutant. If, on the other hand, Polζ-dependent mutational intermediates are not subject to MMR, then the rate of HS1/HS2 complex events should not be affected by Msh2 loss. Given the 4-fold increase in Lys+ rate in the rad14Δ msh2Δ double mutant relative to the rad14Δ single mutant, we predicted that at least 8% of revertants should be HS1/HS2 complex events in the double mutant. Unexpectedly, only a single complex mutation at HS1/HS2 was found among 136 revertants sequenced from the double mutant background (Fig. 1), which translates into an ≈10-fold reduction in the rate of HS1/HS2 complex events upon Msh2 loss (Table 1). These data suggest that Msh2, and presumably MMR, either promotes or is required for Polζ-dependent bypass of the relevant lesion(s) at HS1 and HS2.
The Role of Msh2 in Promoting Error-Prone TLS Is Recombination-Dependent.
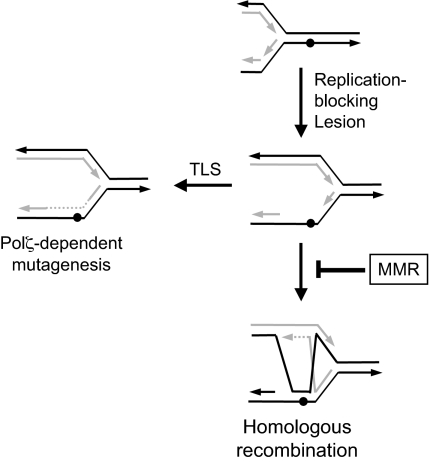
One can imagine a scenario in which it is either the antirecombination or the spellchecker activity of the yeast MMR machinery that is relevant to Polζ-dependent lesion bypass. As shown in Fig. 2, replication blockage and subsequent reinitiation downstream would create a lesion-containing gap that could be filled by either TLS or homologous recombination. Recombination-mediated gap filling using the sister chromatid as a template would displace a D-loop from the invaded duplex, which could then pair with the lesion-containing strand. If the resulting distortion triggers MMR-associated antirecombination activity, the intermediate would be reversed and recombination effectively blocked. Lesion bypass via homologous recombination thus would be very inefficient in the presence of MMR, and the alternative TLS pathway would be favored. In the absence of MMR, recombinational bypass would be favored over mutagenic TLS. In the alternative spellchecker-related scenario, replication past a lesion would trigger MMR-directed removal of the newly-synthesized strand to create a gap behind the fork. Although gap filling by a replicative DNA polymerase would likely initiate another round of MMR (analogous to the “futile repair” proposed to occur with methylated bases) (4), Polζ-mediated gap filling might be refractory to further repair cycles. In the absence of the spellchecker-generated gaps, TLS would thus be greatly reduced.
Fig. 2.
The MMR system regulates TLS through its antirecombination activity. See text for explanation.
The relevance of the antirecombination versus spellchecker activity of MMR to Polζ-dependent mutagenesis can be distinguished by disabling homologous recombination. If MMR promotes TLS by preventing recombination, then the requirement of MMR for TLS should disappear in a rad52Δ background where recombination is no longer possible. If the role of MMR in TLS is simply to generate gaps, then TLS should not be influenced by the presence/absence of the Rad52 recombination protein. We previously demonstrated a Rev3-dependent accumulation of complex events in the lys2ΔA746 system upon loss of the Rad52 recombination protein (16). Similarly, deletion of RAD52 elevated the lys2ΔA746-NR reversion rate 3.2-fold and the proportion of events that were complex insertions increased from 6% to 50% (Table 1 and Fig. S1). In the rad14Δ rad52Δ double mutant, the increase in the Lys+ rate, the total rate of complex events, and the rate of HS1/HS2 complex events were slightly more than additive (Table 1 and Fig. 1). Of particular significance, there was no decrease in HS1/HS2 events in the rad14Δ rad52Δ msh2Δ triple mutant relative to the rad14Δ rad52Δ double mutant. Thus, in contrast to the striking MMR dependence of TLS at these positions when recombination was functional, Msh2 was not important in promoting Polζ-dependent TLS when Rad52 was absent. We conclude that MMR promotes TLS at HS1/HS2 by preventing the efficient use of homologous recombination as an alternative bypass mechanism.
Polζ-Dependent TLS in a Separation-of-Function MMR Mutant.
The MMR protein Pms1 forms a heterodimer with Mlh1 to coordinate the downstream processing steps that occur after Msh2-dependent mismatch recognition (20). We previously described pms1 separation-of-function alleles that retain the spellchecker but eliminate the antirecombination activity of the encoded proteins (21). To confirm the role of MMR-mediated antirecombination in regulating lesion bypass, we introduced either a pms1Δ allele or the pms1-G128A,I854M separation-of-function allele into the lys2ΔA746-NR rad14Δ strain. Consistent with previous results, there was a 27-fold increase in the CAN1 forward mutation rate in the rad14Δ pms1Δ double mutant relative to the rad14Δ single mutant, but only a 2-fold increase in rate in the rad14Δ pms1-G128A,I854M double mutant (3.2 × 10−7, 86 × 10−7, and 6.1 × 10−7, respectively). In terms of lys2ΔA746-NR reversion, the rad14Δ pms1Δ mutant was indistinguishable from the rad14Δ msh2Δ mutant, with the HS1/HS2 complex insertions decreasing ≈10-fold (Table 1 and Fig. S1). Significantly, in the rad14Δ pms1-G128A,I854M mutant, there also was a strong (4-fold) reduction in complex events at HS1/HS2 (Table 1 and Fig. 1). Thus, when the antirecombination activity of Pms1 is specifically impaired, TLS by Polζ becomes less frequent. These data provide additional support for a model in which MMR promotes TLS at HS1/HS2 by specifically inhibiting recombination-mediated lesion bypass.
MMR Does Not Edit Polζ-Dependent Errors.
In a rad52Δ background, where homologous recombination does not occur, the MMR machinery functions only in the removal of DNA synthesis errors. Thus, in the absence of recombination, one can address whether Polζ-dependent mutations are edited by the MMR machinery. In the rad52Δ background, complex insertions composed 50% of the spectrum but less than 30% of these occurred at the HS1/HS2 hotspots that predominate in NER-defective strains (Table 1 and Fig. S1). The HS1/HS2 events are believed to occur by a mechanism of misincorporation slippage that is triggered by a common, discrete lesion (16, 22). Because there is no similar unifying mechanism for the non-HS1/HS2 events, we consider these to be a separate class of event. In contrast to the striking reduction in HS1/HS2 events when MSH2 was deleted in the rad14Δ background, the rate of neither the non-HS1/HS2 nor the HS1/HS2 mutations changed significantly when Msh2 was eliminated in the rad52Δ background (Table 1 and Fig. S1). The lack of an increase in complex insertions indicates that if there is any removal of Polζ-dependent errors by MMR, it is likely to be very inefficient. With regard to errors introduced by the replicative DNA polymerases, base substitution and frameshift intermediates at CAN1 are edited with greater than 90% and 95% efficiency, respectively (23). Finally, the lack of a positive effect of Msh2 on complex mutations in the rad52Δ single mutant provides additional evidence that, at least in the system used here, the MMR machinery is not required to generate lesion-containing gaps that are subsequently filled in by Polζ.
Discussion
Polζ generates distinctive “complex” mutations in the lys2ΔA746 frameshift reversion assay (16), consistent with its propensity to introduce multiple, clustered mutations in vitro (24). We have previously used this signature to examine the genetic requirements for Polζ-dependent mutagenesis (18, 25), and the goal of the current study was to specifically address whether the MMR system affects this process. There are 2 major findings reported here: (i) the antirecombination activity of the MMR system can regulate the mechanism of lesion bypass and (ii) mutational intermediates introduced by Polζ are edited little, if any, by the spellchecker activity of the MMR system.
Spontaneous, Polζ-dependent complex mutations accumulate at 2 discrete hotspots (HS1 and HS2) in an NER-defective (rad14Δ) background, and these events can be attributed to the bypass of endogenous DNA damage. The production of HS1/HS2 complex insertions required the presence of a functional MMR system, with the rate of these events decreasing 10-fold in either a rad14Δ msh2Δ or rad14Δ pms1Δ double mutant. The indistinguishable effects of Msh2 and Pms1 loss is important, as it indicates that it is the MMR system, rather than the Pms1-independent role of Msh2 in processing recombination intermediates (26, 27), that is relevant. The MMR system recognizes base–base mismatches and insertion/deletion loops, as well as some types of DNA damage (2). In the context of DNA replication, the spellchecker activity of the MMR system removes polymerization errors by specifically excising the newly synthesized DNA strand. In the context of recombination, MMR either repairs mismatches/loops to generate gene conversion events or exerts antirecombination activity to reverse/eliminate recombination intermediates (3). Two lines of evidence indicate that it is specifically the antirecombination activity of the MMR system that promotes lesion bypass in the lys2ΔA746-NR system. First, MMR was not required for lesion bypass in a recombination-defective (rad52Δ or rad14Δ rad52Δ) background and second, a pms1 separation-of-function allele that eliminated only the antirecombination activity of the encoded protein had the same effect on complex insertions as the pms1Δ allele.
Homologous recombination and translesion synthesis are alternative modes of lesion bypass, with the former being error free and the latter generally considered to be error prone. A major question concerns the mechanism of pathway choice—what factor(s) determine whether a lesion is bypassed in an error-free versus error-prone manner. The studies reported here demonstrate that the antirecombination activity of an MMR system can regulate this choice. As illustrated in Fig. 2, invasion of the sister chromatid by the lesion-blocked 3′ end would displace a D-loop that pairs with the lesion-containing strand. It is the resulting “mispaired” structure that presumably is detected by the MMR machinery and reverses the recombination process. The feasibility of this model is supported by our earlier work demonstrating that a single potential mismatch is sufficient to block most mitotic recombination between inverted repeat substrates (28). In addition, in vitro work with the bacterial MutS protein has shown that DNA damage, in a manner similar to sequence divergence, impedes the basic RecA-mediated strand exchange reaction (29, 30).
Because the bypass examined here using the lys2ΔA746-NR allele occurs in the absence of exogenous DNA damage, the precise nature of the underlying lesion(s) at HS1/HS2 is not known. We do know, however, that its accumulation requires oxidative metabolism (22), that it is normally a substrate for the NER machinery, and that it likely forms on the lagging-strand template (16). An interesting possibility is that lesions might be bypassed differently when encountered during leading- versus lagging-strand synthesis. At least with regard to recombination and Polζ-dependent TLS in the system used here, however, the interplay between these 2 bypass pathways was not influenced by the direction of DNA replication. Whether Polζ-dependent bypass of lesions at non-HS1/HS2 positions in a rad14Δ mutant is regulated similarly to that at HS1/HS2 could not be determined, as these other complex events occurred too infrequently in the lys2ΔA746-NR assay. The generality of an effect of MMR on TLS, however, is supported by a previous study showing that a functional MMR system enhances sensitivity of yeast to chemotherapeutic drugs in a recombination-dependent manner (31). It was suggested that MMR reduces the efficiency of recombination-mediated bypass when damage is present, and the studies presented here provide a concrete model for how the MMR system modulates pathway choice. Finally, our results may provide an explanation for the very low frequency (1–5%) of Polζ-dependent lesion bypass observed in some plasmid-based yeast transformation studies (32, 33), which contrasts with the high frequency (≈60%) obtained when using lesion-containing single-stranded oligonucleotides (34, 35). Based on the clear MMR dependence of TLS observed in the lys2ΔA746-NR system, we suggest that the specific use of MMR-defective host strains in the plasmid-based studies could account for the low TLS frequency.
In the polymerase-switch model of lesion bypass, bypass is accomplished directly at a stalled fork by sequential proliferating cell nuclear antigen (PCNA)-mediated access of replicative and TLS DNA polymerases to the nascent 3′ end (36). Such polymerase switching might be particularly relevant during leading-strand synthesis, which, in contrast to lagging-strand synthesis, has long been assumed to be continuous. The demonstration in yeast that reinitiation of DNA synthesis can occur during leading- as well as lagging-strand synthesis (37) suggests that TLS may be primarily a gap-filling process to deal with discontinuities that accumulate on both strands behind the fork. The finding that yeast Rev1 is most abundant in G2/M would be consistent with a gap-filling mechanism that functions largely outside of S phase (38). Although the results reported here do not directly address the polymerase-switch versus gap-filling models of TLS, we suggest they are more consistent with the latter model.
The results described here clearly demonstrate that the yeast MMR machinery can play a role in determining whether a lesion is bypassed by error-free recombination or by error-prone TLS. Our experiments cannot address how widespread the involvement of MMR in bypass-pathway choice might be, although it seems unlikely that it would be uniquely limited to the lys2ΔA746 reversion assay. We suggest that the mechanism of lesion bypass will depend on the nature of the underlying lesion, the local sequence context within which the lesion is located (39), and the time during the cell cycle when the bypass occurs. The involvement of MMR in this process reveals a novel way that a system best known for preventing mutagenesis can also promote TLS-dependent genome instability.
Materials and Methods
Plasmids and Strains.
pSR700 contains the lys2ΔA746-NR allele and was constructed by using the Quikchange Site-Directed Mutagenesis Kit (Stratagene) to introduce multiple point mutations into a lys2ΔA746-containing plasmid (pSR585) (17). Primers 5′-gctagctgaaTCaattcaaag, 5′-cgtttggcctGtCtggaTaTccaagatttc, and 5′-ggaaaggaGGcctcagttg (changes are in uppercase and run positions are in italics) were used to interrupt the 6A, 5T and 4A, and 4C runs, respectively.
All strains were derived from SJR195 (MATα ade2–101oc his3Δ200 ura3ΔNco) by lithium acetate transformation. SJR1467 contains the lys2ΔA746-NR allele and was constructed by a 2-step allele replacement using EcoRV-digested pSR700, first selecting His+ transformants and then selecting Lys− segregants. A complete list of the repair-defective derivatives of SJR1467 is provided in Table S1.
Mutation Rates and Spectra.
Yeast strains were grown nonselectively in YEP (1% yeast extract and 2% Bacto-peptone; 2.5% agar for plates) supplemented with 2% dextrose (YEPD) or 2% glycerol and 2% ethanol (YEPGE). Selective growth was on synthetic complete (SC) medium supplemented with 2% dextrose and missing the appropriate nutrient. For determining lys2ΔA746-NR reversion rates and spectra, cells from YEPGE-grown cultures were plated on SC-Lys medium. Data from at least 40 independent cultures were used for each rate determination, and mutation spectra were derived from independent Lys+ revertants as described previously (16). Forward mutation at CAN1 was determined by individually resuspending at least 20 colonies excised from YEPGE plates in H2O and then plating appropriate dilutions on SC − arginine medium supplemented with 60 μg/mL l-canavanine sulfate. Mutation rates and 95% confidence intervals were determined by the maximum likelihood method using Salvador 2.0 software (40). The mutation rate (and corresponding confidence interval) for a given type of mutation was calculated by multiplying the total Lys+ rate by the proportion of the relevant mutation type in the corresponding spectrum.
Supplementary Material
Acknowledgments.
We thank Brenda Minesinger for constructing the original lys2ΔA746-NR allele and especially Nayun Kim for suggesting that the antirecombination activity of MMR might promote TLS by Polζ. This work was supported by National Institutes of Health Grants GM038464 and GM064796 to S.J.-R.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812715106/DCSupplemental.
References
- 1.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: Key regulators of genetic recombination. Cytogenet Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 4.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: Am Soc Microbiol Press; 2006. [Google Scholar]
- 6.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 7.Rattray AJ, Strathern JN. Error-prone DNA polymerases: When making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg EC. Suffering in silence: The tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 10.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 11.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 12.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 13.Haracska L, et al. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Nature. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 15.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 16.Harfe BD, Jinks-Robertson S. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 17.Harfe BD, Jinks-Robertson S. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4766–4773. doi: 10.1128/mcb.19.7.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minesinger BK, Jinks-Robertson S. Roles of RAD6 epistasis group members in spontaneous Polζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2005;169:1939–1955. doi: 10.1534/genetics.104.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdulovic AL, Minesinger BK, Jinks-Robertson S. Identification of a strand-related bias in the PCNA-mediated bypass of spontaneous lesions by yeast Polη. DNA Repair. 2007;6:1307–1318. doi: 10.1016/j.dnarep.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Prolla TA, Christie D-M, Liskay RM. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welz-Voegele C, et al. Alleles of the yeast PMS1 mismatch-repair gene that differentially affect recombination- and replication-related processes. Genetics. 2002;162:1131–1145. doi: 10.1093/genetics/162.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minesinger BK, Abdulovic AL, Ou TM, Jinks-Robertson S. The effect of oxidative metabolism on spontaneous Pol ζ-dependent translesion synthesis in Saccharomyces cerevisiae. DNA Repair. 2006;5:226–234. doi: 10.1016/j.dnarep.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong X, et al. The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabbioneda S, et al. The 9–1-1 checkpoint clamp physically interacts with Polζ and is partially required for spontaneous Polζ-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 26.Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surtees JA, Alani E. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J Mol Biol. 2006;360:523–536. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 28.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calmann MA, Evans JE, Marinus MG. MutS inhibits RecA-mediated strand transfer with methylated DNA substrates. Nucleic Acids Res. 2005;33:3591–3597. doi: 10.1093/nar/gki673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calmann MA, Marinus MG. MutS inhibits RecA-mediated strand exchange with platinated DNA substrates. Proc Natl Acad Sci USA. 2004;101:14174–14179. doi: 10.1073/pnas.0406104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durant ST, et al. Dependence on RAD52 and RAD1 for anticancer drug resistance mediated by inactivation of mismatch repair genes. Curr Biol. 1999;9:51–54. doi: 10.1016/s0960-9822(99)80047-5. [DOI] [PubMed] [Google Scholar]
- 32.Baynton K, Bresson-Roy A, Fuchs RPP. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: A requirement for Rev3 DNA polymerase in translesion synthesis. Mol Cell Biol. 1998;18:960–966. doi: 10.1128/mcb.18.2.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci USA. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsuka C, et al. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kow YW, et al. Mutagenic effects of abasic and oxidized abasic lesions in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:6196–6202. doi: 10.1093/nar/gki926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proc Natl Acad Sci USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdulovic AL, Minesinger BK, Jinks-Robertson S. The effect of sequence context on spontaneous Polζ-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:2082–2093. doi: 10.1093/nar/gkn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Q. New algorithms for Luria-Delbruck fluctuation analysis. Math Biosci. 2005;196:198–214. doi: 10.1016/j.mbs.2005.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.