Abstract
Expression of many MHC genes is enhanced at the transcriptional or posttranscriptional level following exposure to the cytokine IFN-γ. However, in this study we found that IFN-γ down-regulated the constitutive expression of the neonatal Fc receptor (FcRn), an MHC class I-related molecule that functions to transport maternal IgG and protect IgG and albumin from degradation. Epithelial cell, macrophage-like THP-1 cell, and freshly isolated human PBMC exposure to IFN-γ resulted in a significant decrease of FcRn expression as assessed by real-time RT-PCR and Western blotting. The down-regulation of FcRn was not caused by apoptosis or the instability of FcRn mRNA. Chromatin immunoprecipitation and gel mobility shift assays showed that STAT-1 bound to an IFN-γ activation site in the human FcRn promoter region. Luciferase expression from an FcRn promoter-luciferase reporter gene construct was not altered in JAK1- and STAT-1-deficient cells following exposure to IFN-γ, whereas expression of JAK1 or STAT-1 protein restored the IFN-γ inhibitory effect on luciferase activity. The repressive effect of IFN-γ on the FcRn promoter was selectively reversed or blocked by mutations of the core nucleotides in the IFN-γ activation site sequence and by over-expression of the STAT-1 inhibitor PIAS1 or the dominant negative phospho-STAT-1 mutations at Tyr-701 and/or Ser-727 residues. Furthermore, STAT-1 might down-regulate FcRn transcription through sequestering the transcriptional coactivator CREB binding protein/p300. Functionally, IFN-γ stimulation dampened bidirectional transport of IgG across a polarized Calu-3 lung epithelial monolayer. Taken together, our results indicate that the JAK/STAT-1 signaling pathway was necessary and sufficient to mediate the down-regulation of FcRn gene expression by IFN-γ.
The neonatal Fc receptor (FcRn)3 for IgG was first characterized in the intestinal epithelial cells of neonatal rodents; however, its expression has recently been identified in a variety of cell types and tissues including epithelial cells, endothelial cells, macrophages, and dendritic cells in rodents and humans of all ages (1–4). The structure of FcRn is similar to that of MHC class I Ags, being composed of a heavy chain (45 kDa in humans and 50 kDa in rodents) that is noncovalently attached to a light chain β2-microglobulin (12 kDa) (5, 6). However, FcRn is not capable of presenting Ags to T cells because its Ag binding groove is too narrow (7). Despite this, FcRn is identified as a transport receptor involved in mediating the transfer of IgG from the maternal to the fetal/newborn blood in placental and/or intestinal tissues (8, 9). FcRn, therefore, plays a major role in the passing on maternal immunity to newborns, possibly in all mammals. FcRn also functions in the maintenance of IgG and albumin homeostasis by salvaging either of them from degradation (8, 10, 11). In the model proposed by Brambell et al., IgG is taken into cells by pinocytosis or endocytosis from the surrounding tissue fluid or blood (12). FcRn in acidic compartments, such as the endosome, binds and recycles IgG out of the cell to avoid IgG degradation in the lysosome (8, 12). In fact, FcRn displays pH-dependent binding of IgG or albumin; specifically, FcRn preferentially binds IgG or albumin at acidic pH (6 – 6.5) and releases IgG or albumin at neutral pH (7–7.4) (8, 11, 12). The transport and protective properties for IgG by FcRn are fully supported by several studies in which mice deficient in either β2-microglobulin or FcRn heavy chain exhibit either failure of transport of maternal IgG or significant reduction in the serum half-life of IgG (3, 10, 13, 14). Recently, FcRn is also shown to play a role in phagocytosis (15).
IFNs are multifunctional cytokines that have antiviral, antiproliferative, antitumor, and immunomodulatory effects (16, 17). In the case of IFN-γ, the cell membrane receptor for IFN-γ is composed of two subunits, IFN-γR1 and IFN-γR2. Upon binding to IFN-γ, the IFN-γ receptor rapidly associates with the Janus tyrosine kinases JAK1 and JAK2. JAK enzymes phosphorylate one another and then subsequently phosphorylate the IFN-γ receptor, which results in the formation of a docking site for the latent cytoplasmic transcription factor named STAT-1, a member of the STAT (signal transducer and activator of transcription) protein family (18). Upon phosphorylation, STAT-1 homodimerizes, translocates to the nucleus, and regulates gene transcription by binding to IFN-γ-activated sequences (GAS) in the IFN-γ-inducible genes. Homodimerization of STAT-1 is mediated by the binding of the phosphorylated tyrosine 701 of one STAT-1 monomer to the Src homology 2 domain of another. However, maximal transcriptional activity by active STAT-1 homodimers also requires STAT-1 phosphorylation at serine 727 (19–21). It has been found that STAT-1 phosphorylation plays a critical role in IFN-mediated innate immunity to microbial infection (22). STAT-1 signaling can also be negatively regulated by the protein inhibitor of activated STAT-1 (PIAS1) and suppressor of cytokine signaling (SOCS) (23). More interestingly, IFN-γ can also regulate expression of its inducible genes in a STAT-1-independent manner (24–27), suggesting that multiple signaling pathways in parallel play important roles in the biological response to IFN-γ.
The pivotal roles in the protection and transport of IgG have led to an increasing interest in the mechanism that regulates FcRn expression regarding both constitutive and stimulated expression. MHC class I and related molecules include HLA-A, HLA-B, HLA-C, HLA-F, HLA-G, HLA-H, MR1, MIC A/B, CD1, and FcRn. Expression of several MHC class I genes significantly increases at the transcriptional or posttranscriptional level following exposure to IFN-γ in a variety of tissues and cells (28–33). Although the transactivating roles of IFN-γ in MHC class I and its related molecules are well established, at present little is known about whether and how IFN-γ regulates FcRn gene expression. In an effort to identify the role of IFN signaling in regulation of the FcRn receptor, we unexpectedly found, for the first time, that IFN-γ down-regulated human FcRn expression and function. Furthermore, our study showed that activation of STAT-1 is required for IFN-γ-induced down-regulation of FcRn expression. STAT-1-repressed FcRn transcription may act through sequestering the transcriptional coactivator CREB binding protein (CBP)/p300, thus reducing the level of CBP/p300 at the human FcRn promoter.
Materials and Methods
Cell lines, Abs, reagents
Human lung-derived Calu-3 adenocarcinoma cells were obtained from American Type Culture Collection (HTB-55) and maintained in a 1:1 mixture of DMEM and Ham’s F-12 medium (Invitrogen). Human 2fTGH cells, a cell line derived from the human fibrosarcoma HT1080 cell line, and the 2fTGH-derived cell lines U3A (STAT-1 deficient) and U4A (JAK1 deficient) were gifts from Dr. G. Stark (Cleveland Clinic Foundation, Cleveland, OH). HeLa-E2A4 (JAK1 deficient) was from Dr. R. A. Flavell (Yale University School of Medicine, New Haven, CT). The human intestinal epithelial cell lines HT-29 and Caco-2 and the macrophage-like THP-1 cells were obtained from Dr. R. S. Blumberg (Harvard Medical School, Boston, MA). The human intestinal epithelial cell line T84 was from Dr. W. Song (University of Maryland, College Park, MD). All epithelial and fibrosarcoma cells were maintained in DMEM complete medium (Invitrogen). The THP-1 cell line or freshly isolated human PBMCs (Institute of Human Virology, Baltimore, MD) were cultured in complete RPMI 1640 medium (Invitrogen). All complete medium was supplemented with 10 mM HEPES, 10% FCS, 2 mM L-glutamine, nonessential amino acids, and penicillin (0.1 μg/ml)/streptomycin (0.292 μg/ml) in a humidified atmosphere of 5% CO2 at 37°C.
HRP-conjugated donkey anti-rabbit or rabbit anti-mouse Ab was purchased from Pierce, and purified human IgG was from Jackson ImmunoResearch Laboratories. Anti-STAT-13, anti-phospho-STAT-1 (tyrosine 701), anti-phospho-STAT-1 (serine 727), and anti-p300 Abs were from Cell Signaling Technology. Human recombinant IFN-γ was from R&D Systems. All DNA-modifying enzymes were purchased from New England Biolab.
Semiquantitative RT-PCR and quantitative real-time RT-PCR
Semiquantitative RT-PCR and real-time RT-PCR were performed as previously described (34). In brief, total RNA was isolated from stimulated and mock-stimulated cells (2 × 106/ml) in TRIzol reagents (Invitrogen) according to the manufacturer’s instructions. Semiquantitative RT-PCR was performed using a one-step RT-PCR kit (Qiagen). Primers for amplification of FcRn and GAPDH have been previously described (34). Thirty cycles of PCR amplification were performed in a 20-μl volume. Each cycle consisted of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. An additional 10 min was applied for the final extension. PCR products were resolved on 1.5% agarose gels and visualized by staining with ethidium bromide. Integrated density values for the FcRn binds were normalized to the GAPDH values to yield a semiquantitative assessment.
The freshly isolated human PBMCs (106 cells/ml) were stimulated with IFN-γ (25 ng/ml) for 24 h. The total RNA samples were extracted. The RNA (400 ng/reaction) was reverse transcribed to yield first-strand cDNA using SuperScript III (Invitrogen). Real-time RT-PCR was performed using FcRn and GAPDH primers (34) and the SYBR Green Supermix kit (Bio-Rad Laboratories) in a Chromo 4 thermocycler (MJ Research). FcRn expression was calculated following normalization to GAPDH levels by the comparative ΔΔ threshold cycle method. All reactions were performed for 40 cycles: 15 s at 94 °C, 15 s at 58 °C, and 20 s at 72 °C. The specificity of the amplification reactions was confirmed by melt curve analysis. The Opticon Monitor 3.1 software package (Bio-Rad Laboratories) was used for real time RT-PCR.
Construction of expression or reporter plasmids and mutagenesis
Construction of the human FcRn promoter-luciferase reporter plasmid phFc-RnLuc containing sequences from −1801 to +863 of the human FcRn promoter has been previously described (34). The mutant derivative plasmids pM1 and pM2 were constructed by overlapping PCR mutagenesis to disable the putative GAS sequence (see Fig. 4B), using phFcRnLuc as a template. The primer pairs for pM1 (5′-GGAAGCCAACTACTCATATGAATCTCTTTCTGTG-3′ and 5′-AGGATTAGTGGACGTTCAGCTGGTTCAGAG-3′) or pM2 (5′-TTATATGATTCAATGGCTTAGACATGTGCAGAATAG-3′ and 5′-TATGAAGTCTTTCCTTCCTTCCTTCCTTGCCTC) were used (the mutations are underlined). The expression plasmid encoding wild-type STAT-1 (pSTAT-1) and the phosphorylation site mutant plasmid pSTAT-1Y701F were kindly provided by Dr. K. Nakajima (Osaka City University Medical School, Osaka, Japan) and Dr. D. Geller (University of Pittsburgh, Pittsburgh, PA). The FLAG-tagged STAT-1 and PIAS1 expression plasmids were kind gifts from Dr. K. Shuai (University of California, Los Angeles, CA). The FLAG-tagged pSTAT-1Y701F, pSTAT-1S727A, or pSTAT-1Y701F/S727A was constructed by the overlapping PCR mutagenesis method. The primer pair (5′-AGGAACTGGATTTATCAAGACTGAGTTGAT-3′ and 5′-TTAGGGCCATCAAGTTCCATTGGCTCTGGT-3′) was used to substitute tyrosine 701 with a phenylalanine residue (underlined). The primer pair (5′-GACAACCTGCTCCCCATGGCTCCTGAGGAG-3′ and 5′-TGTGGTCTGAAGTCTAGAAGGGTGAACTTC-3′) was used to change serine 727 to alanine (underlined). The murine JAK1 expression construct was obtained from Dr. J. Ihle (St Jude Children’s Research Hospital, Memphis, TN). The integrity of the DNA fragments in the plasmids was confirmed by DNA sequence analysis.
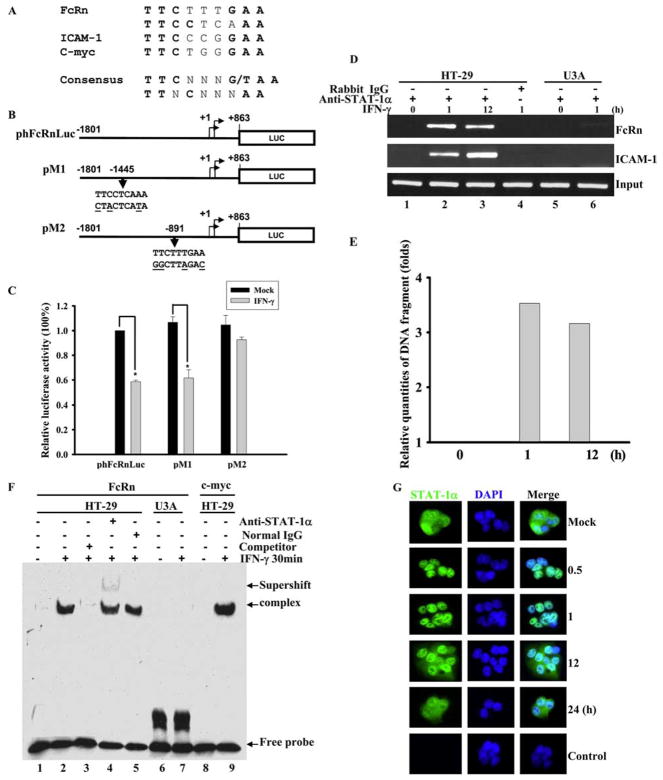
FIGURE 4.
Identification of IFN-γ responsive element in human FcRn promoter. A and B, The putative STAT-1 binding sequences in FcRn gene promoter. A, STAT-1 binding sequences in the promoter of ICAM1 and c-myc were used as a positive control. The consensus STAT-1 sequence is in boldface. N represents any nucleotide. B, A schematic representation of the luciferase reporter constructs. The positions of the base count are shown (GenBank accession no. AC010619). Reporter construct phFcRnLuc contains the FcRn promoter sequence from −1801 to +863 kb. The putative GAS mutations (underlined bases) in constructs pM1 and pM2 are also shown. Arrowheads indicate the position of the STAT-1 binding site in relation to the transcription start site of the FcRn gene. Luc, Luciferase. C, Identification of GAS sequence in response to IFN-γ stimulation. Wild-type 2fTGH cells were transiently transfected with phFcRnLuc, pM1, and pM2 constructs. Twenty-four hours after transfection, cells were either mock-treated (filled bar) or treated with IFN-γ (open bar) for 4 h. Cells were then harvested and protein extracts were prepared for the luciferase assay as described in Materials and Methods. Luciferase activity was measured and normalized to Renilla luciferase content. The results show the mean value from three independent experiments. *, p < 0.05. D and E, Detection of the in vivo binding of STAT-1 protein to the human FcRn promoter in a ChIP assay. D, Formaldehyde-crosslinked chromatin was prepared from both mock-treated and IFN-γ-treated HT-29 (lanes 1– 4) or STAT-1-null U3A (lanes 5 and 6) cells as described in Materials and Methods. ChIP assays were performed using STAT-1-specific Abs (lanes 1–3, 5, and 6) or isotype-matched IgG (lane 4) as a negative control. Immunoprecipitated chromatin was subjected to PCR analysis using FcRn and ICAM-1 specific primers. The equivalent amount of chromatin in the immunoprecipitations was monitored by PCR amplification of input chromatin as an internal control. ChIP assay was performed at least three times. E, Quantitative real-time RT-PCR analysis of chromatin immunoprecipitated PCR products for FcRn at the indicated times. F, EMSA analysis of binding activities of DNA probe with nuclear extracts from HT-29 cells treated with (+) or without (−) IFN-γ. DNA binding was performed using a DNA probe of human FcRn promoter with nuclear extracts from HT-29 or U3A cells treated with or without IFN-γ (25 ng/ml) for 30 min. A 26-bp fragment spanning the putative STAT-1 binding sequence corresponding to the GAS was used as a biotin-labeled probe. Binding specificity of these complexes was examined by competition assays with a 100-fold molar excess of unlabeled STAT-1-specific probe (lane 3). Supershift experiments were performed in the presence of the STAT-1 Ab, resulting in the formation of a slow migrating supershift band (lane 4). Free-labeled probes are also indicated. G, Immunofluorescence images of STAT-1 cellular localization at the indicated times after exposure to IFN-γ (25 ng/ml). HT-29 cells stimulated with IFN-γ were stained with Alexa Fluor 458-labeled-STAT-1-specific Ab, and translocation of STAT-1 into the nucleus was detected by immunofluorescence microscopy as described in Materials and Methods. For correlation of the STAT-1 protein (green) with nuclear morphology, cell nuclei were counterstained with DAPI (blue). The images were merged as indicated. STAT-1 is in green, nucleus is in blue, and colocalization is gray/blue.
Immunoprecipitation, gel electrophoresis, and Western blotting
Immunoprecipitation was done as described previously (36). Protein was precipitated with anti-FLAG Ab. The immunoreactive products were eluted from the protein G complex with gel loading buffer at 95°C. Gel electrophoresis and Western blot were performed as previously described (35, 36). Protein concentrations were determined by the Bradford method. The cell lysates were resolved by electrophoresis on a 12% SDS-polyacrylamide gel under reducing conditions. Proteins were electrotransferred onto a nitrocellulose membrane (Schleicher & Schuell). The membranes were blocked with 5% nonfat milk, probed separately with affinity-purified rabbit anti-FcRn peptide (CLEWKEPPSMRLKARP) Ab for 1 h, followed by incubation with HRP-conjugated donkey anti-rabbit Ab. All blocking, incubation, and washing were performed in PBST solution (PBS and 0.05% Tween 20). Proteins were visualized by an ECL method (Pierce).
Determination of mature FcRn mRNA stability
Stability of the mature FcRn mRNA transcript was determined by using an actinomycin D inhibition assay as described previously (37, 38). Briefly, after 24 h of HT-29 cells being treated with or without IFN-γ, 5 μg/ml actinomycin D (Sigma-Aldrich) was subsequently added to each culture to stop the further production of mature FcRn transcript. Following the addition of actinomycin D, cell viability was analyzed by trypan blue exclusion and did not significantly change over the course of the experiment. HT-29 cells were collected from the cultures at 0, 1, 2, 4, 8, and 10 h following the addition of actinomycin D, and total RNA was isolated. The level of FcRn mRNA was quantified for each time point by semiquantitative RT-PCR or quantitative real-time PCR as described above.
Nuclear run-on assay
The rate of mature FcRn transcription was determined by nuclear run-on as described in detail previously (38, 39). Briefly, 5 × 107 THP-1 cells were collected 24 h following stimulation in the presence or absence of IFN-γ and washed twice with PBS before resuspension in 5 ml of cell lysis buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.5% Nonidet P-40 for 5 min at 4°C. Nuclei were collected by centrifugation at 300 × g for 10 min at 4°C, resuspended in 500 μl of nuclear freezing buffer containing 50 mM Tris-HCl (pH 8.3), 40% glycerol, 5 mM MgCl2, and 0.1 mM EDTA, and stored at −80°C until use for nuclear run-on. Nuclear run-on and RNA isolation were preformed in the presence of biotin-16-UTP (Roche). To control for the possibility of nonbiotin-labeled RNA contamination, replicate sets of nuclei were used in the nuclear run-on that did not contain biotin-16-UTP. Dynabeads M-280 (Invitrogen) were used to capture the biotin-labeled RNA molecules from the purified nuclear RNA, and beads were washed twice with 2× SSC plus 15% formamide and once with 2× SSC and resuspended in 30 μl of RNase-free H2O before the preparation of random hexamer-primed cDNA as described in the paragraph titled Semiquantitative RT-PCR and quantitative real-time RT-PCR above except for the primer pair used for GAPDH (5′-GCCACTAGGCGCTCACTGTTCTCTC-3′ and 5′-CTCCTTGCGGGGAACAGCTACCCTGC-3′) and FcRn (5′-GAGCCTGGGCGCAGGTGAGGGCCGC-3′ and 5′-GCGACAGGTGGTTCCCAGCCTCAGGC-3′). Primers located in the intronic region are underlined. All samples that did not contain biotin-16-UTP were found to be negative for the presence of GAPDH and mature FcRn transcripts.
Immunofluorescence and detection of apoptosis by TUNEL
HT-29 cells were cultivated on coverslips for 24 h. The coverslips were rinsed in PBS and cells were cold-fixed in 4% paraformaldehyde in PBS for 30 min at 4°C. Subsequent procedures were done at room temperature. After two washings with PBS, the coverslips were permeabilized (3% BSA and 0.2% Triton in PBS) for 30 min. Cells were incubated with affinity-purified rabbit anti-STAT-1 in PBST (0.05% Tween 20 and PBS) with 3% BSA for 1 h. Cells were then incubated with Alexa 458 Fluor-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) in PBST with 3% BSA. Cell nuclei were counterstained with 5 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes) in PBS. After each step the cells were washed three times with 0.1% Tween 20 in PBS. To mount coverslips, the ProLong antifade kit was used (Molecular Probes). Images were captured using a ×100 oil-immersion objective on a Zeiss inverted microscope linked to a DeltaVision deconvolution imaging system.
In situ detection of apoptotic cells was performed with the TUNEL kit from Roche. After IFN-γ (50 ng/ml) treatment, HT-29 cells undergoing cell death were identified. Briefly, IFN-γ- or mock-treated cells were fixed with a freshly prepared fixation solution (4% paraformaldehyde in PBS (pH 7.4)) for 1 h at room temperature, and then incubated in permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate) for 2 min on ice, and the TUNEL procedure was conducted according to the manufacturer’s instructions. For the correlation of TUNEL with nuclear morphology, cells were counterstained with DAPI. To confirm the specificity of TUNEL, cells were treated with 3000 U/ml DNase I at room temperature for 10 min to induce DNA strand breaks before labeling procedures. In negative controls, terminal TdT was omitted from the labeling reaction mixture. Samples were viewed by fluorescence microscopy with excitation at 320 –580 nm.
Transient transfection and luciferase assay
Transient transfection and luciferase assay were done as previously described (34). Briefly, cells were transiently transfected with Effectene according to instructions from the manufacturer (Qiagen). In each cotransfection, 2 × 106 cells were transfected with a DNA mix containing 0.95 μg of firefly luciferase reporter plasmid and 0.05 μg of Renilla luciferase pRL-TK control plasmid. Cotransfection experiments with the STAT-1, JAK1, or PIAS1 expression plasmid included an additional 1.0 μg of the plasmid. The following day, the cells were cultured with or without IFN-γ. The cells were harvested 24 h after treatment and assayed for the expression of Renilla and firefly luciferase using the dual luciferase kit (Promega) according to the recommended protocol in a Victor 3 luminometer (PerkinElmer). The values for firefly luciferase were normalized to the Renilla luciferase activity and expressed as fold activation over the vector background.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed according to the manufacturer’s recommendations (Upstate Biotechnology) and as previously described (34). In brief, HT-29 cells (5 × 106 cells) were incubated with or without IFN-γ (25 ng/ml) for 1–12 h. The cells were fixed with 1% formaldehyde. The nuclei were isolated and sonicated 20 times on ice for 10 –20 s with 90-s breaks (Sonifier 350; Branson) between each sonication interval to shear the DNA to 200–1000 bp. A small aliquot (20 μl) was saved as “input DNA” for PCR analysis by reversing histone-DNA crosslinks by heating at 65°C for 4 h. Chromatin was immunoprecipitated from 200-μl aliquots at 4°C by mild agitation overnight with 5 μg of Ab specific for STAT-1, phospho-STAT-1 (tyrosine 701), and phospho-STAT-1 (serine 727) or with 5 μg of normal rabbit IgG as negative control. Immune complexes were collected by incubation with protein A-agarose. To analyze the target region, the immunoprecipitated chromatin DNA samples were amplified by PCR with primer pairs for FcRn (5′-GGAAAGACTTCATATTATATGATTC-3′ and 5′-GCAACTGTCACCTCTATCCGAGTTC) or ICAM-1 (5′-GATTGCTTTAGCTTGGAAATTC-3′ and 5′-GGAGCCATAGCGAGGCTGAG-3′). DNA samples or input DNA fractions were analyzed by 35 cycles of PCR (94°C for 30 s, 58°C for 30 s, and 72°C for 30 s) in 20-μl reaction mixtures. PCR products were subjected to electrophoresis by using 2% agarose gels in TAE (Tris-acetate-EDTA) buffer and visualized by ethidium bromide.
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared using a nuclear and cytoplasmic extraction kit according to the manufacturer’s instructions (Pierce). IFN-γ (25 ng/ml)-treated HT-29 cells (1 × 107) were used. The double-stranded oligonucleotides (5′-TGATTCAATTTCTTTGAAATGTGCAG-3′) containing a putative GAS sequence (underlined) from the FcRn promoter was used. The double-stranded oligonucleotides (5′-CCCTTTCTGGGAAGTCCGGGT-3′) containing the GAS sequence (underlined) from the c-myc promoter (25) were used as a positive control. The DNA was labeled with a biotin 3′-end DNA labeling kit (Pierce). In brief, 4 μg of nuclear extracts were incubated in binding buffer (10 mM Tris (pH 7.9), 50 mM NaCl, 5 mM MgCl2, 50 mM KCl, and 50% glycerol) with 50 ng/ml poly(deoxyinosinic-deoxycytidylic acid) (poly(dI-dC)) and a 20-fmol final concentration of biotin-labeled, double-stranded oligonucleotide for 20 min at room temperature. For competition assays, samples were preincubated with a 100-fold excess of a nonlabeled oligonucleotide. For the supershift assay, 0.8 μg of each Ab specifically directed against STAT-1 was preincubated with the nuclear extracts in the absence of poly(dI-dC) for 30 min at 22°C. Subsequently, poly(dI-dC) was added and incubated for 5 min, followed by the addition of a probe for an additional 20 min. The samples were loaded on a 5% native polyacrylamide gel in 0.5× Tris-borate-EDTA buffer at 80 volts for 2 h. The gels were blotted onto a nylon membrane (Bio-Rad Laboratories), blocked, incubated with HRP-avidin, and developed using the LightShift chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instruction. Visualization of the chemiluminescent signal on the membrane was achieved by exposing to X-ray film (Kodak).
IgG binding assay
IgG binding assays were performed as previously described (34) with the following modifications. Calu-3 cells (1 × 107) were lysed by shaking in PBS (pH 6.0 or 8.0) with 0.5% CHAPS (Sigma) and protease inhibitor mixture on ice for 1 h. Cytoplasmic supernatants containing 0.5 mg of soluble proteins were incubated at 4°C overnight with human IgG-Sepharose (Amersham Pharmacia Biotech). The unbound proteins were removed with PBS (pH 6.0 or 8.0) containing 0.1% CHAPS. The adsorbed proteins were boiled with reducing electrophoresis sample buffer at 95°C for 5 min. The eluted fractions were subjected to Western blot analysis with affinity-purified rabbit anti-FcRn peptide Ab.
IgG transcytosis
IgG transport was performed with a modification of previously described methods (34, 40, 41). Calu-3 cells were grown onto Transwell filter inserts (Corning Costar) to form a monolayer exhibiting transepithelial electrical resistances (700 ohms/cm2). Transepithelial electrical resistance was measured using a tissue-resistance measurement equipped with planar electrodes (World Precision Instruments). Monolayers were equilibrated in HBSS and mock-treated or stimulated with IFN-γ (25 ng/ml) for 24 h. Thereafter, human IgG at a final concentration of 0.5 mg/ml was added to the apical or basolateral medium. Monolayers were incubated for 1 h with IgG or chicken IgY at 37°C. An aliquot of the buffer was collected into which apically and basolaterally directed IgG or IgY transport was conducted. Transported proteins were analyzed by reducing SDS-PAGE and Western blot-ECL. NIH Image software (National Institutes of Health, Bethesda, MD) was used to determine the relative band intensities of a blot.
Statistical analysis
Data from three independent experiments were initially analyzed by ANOVA to detect significant changes between the stimulated and mock-stimulated cells. Additional statistical evaluation of the differences in expression of FcRn genes was measured by Student’s t test with a Bonferroni correction. All results are expressed as mean values. A value of p < 0.05 was considered significant.
Results
Exposure of cells with IFN-γ down-regulates the expression of FcRn
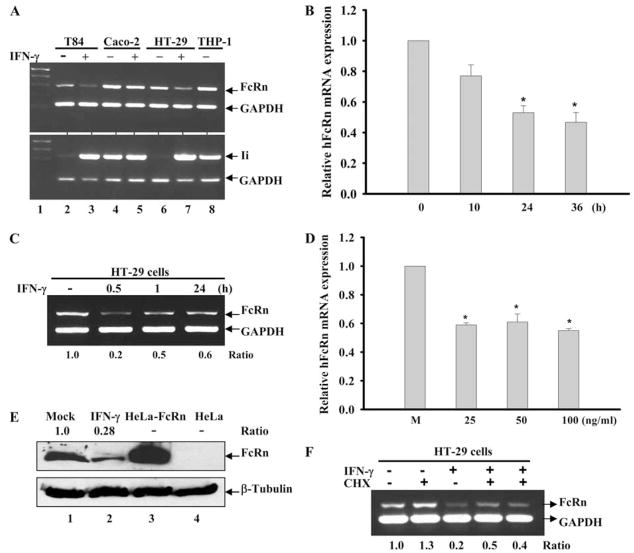
IFN-γ has been shown to enhance the expression of the MHC genes at the transcriptional or posttranscriptional level (28, 29). To determine whether IFN-γ regulates human FcRn gene expression, we treated human intestinal epithelial cell lines that express FcRn (2, 40) with IFN-γ (50 ng/ml). Our data showed that FcRn gene expression in T84 and HT-29 cells was significantly down-regulated in response to IFN-γ treatment as shown by semiquantitative RT-PCR (Fig. 1A). To rule out whether this decrease in FcRn was the result of general transcriptional decreases in the cell, we also measured the transcript for the MHC class II-associated invariant (Ii) chain, a molecule highly up-regulated by IFN-γ. Transcript levels for Ii (Fig. 1A, bottom panel) were significantly increased by IFN-γ, suggesting that the transcriptional down-regulation of FcRn is specific. In the regulation of FcRn mRNA, Caco-2 cells were to some extent refractory to IFN-γ stimulation (Fig. 1A, lanes 4 and 5). In real-time RT-PCR assays, IFN-γ decreased the mRNA levels 40 –50% over the mock-stimulated cells after 24 and 36 h (Fig. 1B). To ascertain whether the IFN-γ needs to be maintained in the medium to induce gene repression, the HT-29 cells were incubated with IFN-γ (25 ng/ml) for 0.5, 1, and 24 h, completely washed at least six times, and then incubated for a further 24 h. As shown in Fig. 1C, the levels of FcRn mRNA were down-regulated at least 50% at 0.5 h of exposure to IFN-γ (second lane from left) in comparison with that of mock-treated cells (far left lane). FcRn expression was down-regulated by IFN-γ in a dosage range between 25 and 100 ng/ml; the lowest dosage could be 5 ng/ml (data not shown). IFN-γ decreased the mRNA level 40% at 25 ng/ml as measured by real-time RT-PCR (Fig. 1D). The decreased expression of FcRn protein in HT-29 cells was shown by Western blotting in IFN-γ-stimulated cells (Fig. 1E, top panel, lanes 2– 4) in comparison with mock-stimulated cells (lane 1). Lysates from HeLa-FcRn and HeLa were used as a positive (Fig. 1E, lane 3) and negative (lane 4) controls. To establish whether this transcriptional repression requires new protein synthesis, we performed additional experiments where the levels of FcRn mRNA were determined following treatment with cycloheximide (CHX), an established inhibitor of protein synthesis. In these experiments we used a concentration of CHX (25 μg/ml) at which >95% of protein synthesis is blocked within 1 h (42). The results showed that the IFN-γ-induced transcriptional repression was totally independent of new protein synthesis. Specifically, by RT-PCR analysis we observed ~60% reduction in FcRn mRNA synthesis following 24 h of exposure to IFN-γ in the presence of CHX, an overall inhibition comparable with that obtained in the absence of CHX (Fig. 1F). These data indicated that preexisting proteins were modified in a ligand-dependent manner to repress the FcRn gene.
FIGURE 1.
Down-regulation of human FcRn expression in epithelial cells by IFN-γ. *, p < 0.05. A, Down-regulation of human FcRn and up-regulation of Ii occur concomitantly in response to IFN-γ treatment. Human intestinal cell lines were treated with (+) IFN-γ (lanes 3, 5, and 7) or without (−) IFN-γ (lanes 2, 4, and 6) (50 ng/ml) for 48 h. Total RNA was isolated by TRIzol reagent and analyzed by semiquantitative RT-PCR for FcRn and Ii mRNA. RNA from THP-1 cells was used as a positive control for Ii amplification (lane 8). GAPDH amplification was used as an internal control. B, Time course effects of IFN-γ on FcRn expression. Quantitative real-time RT-PCR analysis of human FcRn mRNA in HT-29 cells treated with IFN-γ (25 ng/ml) for 10, 24, and 36 h or left untreated. C, IFN-γ incubation time and FcRn expression. Human intestinal HT-29 cells were incubated with or without IFN-γ (25 ng/ml) for 0.5, 1, and 24 h. At the end of the shorter incubation periods, HT-29 cells were washed at least six times and then incubated in fresh medium to reach 24 h. Total RNA was isolated and analyzed by semiquantitative RT-PCR for FcRn and GAPDH. D, Dose-response effects of IFN-γ on FcRn expression. Human intestinal HT-29 cells were treated without (M) or with IFN-γ at the indicated dosages for 24 h. FcRn mRNA was analyzed by quantitative real-time RT-PCR analysis. E, Western blot analysis of FcRn expression. The cell lysates (20 μg) from mock-treated (lane 1) and IFN-γ-stimulated HT-29 (lane 2) were separated by electrophoresis in a 12% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. Cell lysates from HeLa-FcRn (lane 3) and HeLa (lane 4) were used as positive or negative controls, respectively. Proteins were blotted with affinity-purified rabbit anti-FcRn- (top panel) or β-tubulin-specific Ab (bottom panel) and then incubated with HRP-conjugated anti-IgG Ab. The results were visualized with the ECL method. The ratio of the mock group is assigned a value of 1.0, and the values from other groups are normalized to this value. The ratios of FcRn and β-tubulin are shown as indicated. F, Effects of CHX on IFN-γ-mediated repression of FcRn expression. Human intestinal HT-29 cells were incubated with (+) or without (−) the protein synthesis inhibitor CHX (25 μg/ml) for 2 h as indicated. HT-29 cells were subsequently stimulated with (+) or without (−) IFN-γ (25 ng/ml) for 24 h. At the end of the incubation period, total RNA was isolated and analyzed by RT-PCR for FcRn and GAPDH.
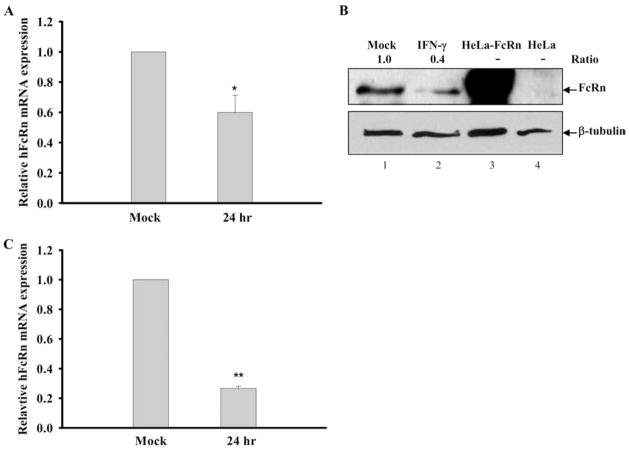
To show FcRn transcription in other cell types in response to IFN-γ repression, human macrophage-like THP-1 cells were treated with IFN-γ (25 ng/ml) and the mRNA level of FcRn was decreased ~40% below that of the mock-stimulated cells (Fig. 2A). As shown in Fig. 2B, the decreased expression of FcRn protein in THP-1 cell lysates was shown by Western blotting (lane 2) in comparison with mock-stimulated cells (lane 1). Cell lysates from HeLa-FcRn and HeLa were used as a positive (Fig. 2B, lane 3) or a negative (lane 4) control. Furthermore, the level of FcRn mRNA from freshly isolated human PBMCs treated with IFN-γ (25 ng/ml) was decreased 75% over the mock-stimulated PBMCs after 24 h as assessed by real time RT-PCR (Fig. 2C). Taken together, these data show that IFN-γ down-regulated the FcRn expression in intestinal epithelial cell lines, human macrophage-like THP-1 cells, and freshly isolated human PBMCs.
FIGURE 2.
Down-regulation of human FcRn expression in THP-1 cells and human PBMCs by IFN-γ. A and C, Effect of IFN-γ treatment on FcRn mRNA expression in THP-1 and PBMCs. The macrophage-like THP-1 cells (A) or freshly isolated human PBMCs (C) were treated with or without IFN-γ (25 ng/ml) for 24 h. The levels of FcRn mRNA were measured by quantitative real-time RT-PCR analysis as described in Materials and Methods. Data are mean ± SD of three independent experiments. *, p < 0.05; **, p < 0.01. B, Western blot analysis of FcRn expression in THP-1. The cell lysates (20 μg) from THP-1 (lane 1), IFN-stimulated THP-1 (lane 2), HeLa-FcRn (lane 3), and HeLa (lane 4) were subjected to 12% SDS-polyacrylamide gel electrophoresis. The proteins were transferred to nitrocellulose membrane and blotted with FcRn- (top panel) or β-tubulin-specific Ab (bottom panel). Blots were then incubated with anti-IgG-HRP and visualized with the ECL method. The ratio of the mock was assigned a value of 1.0, and the values from other groups were normalized to this value. The ratios of FcRn- and β-tubulin are shown above the lanes.
Effect of IFN-γ on FcRn mRNA stability, rate of mRNA transcription, and apoptosis
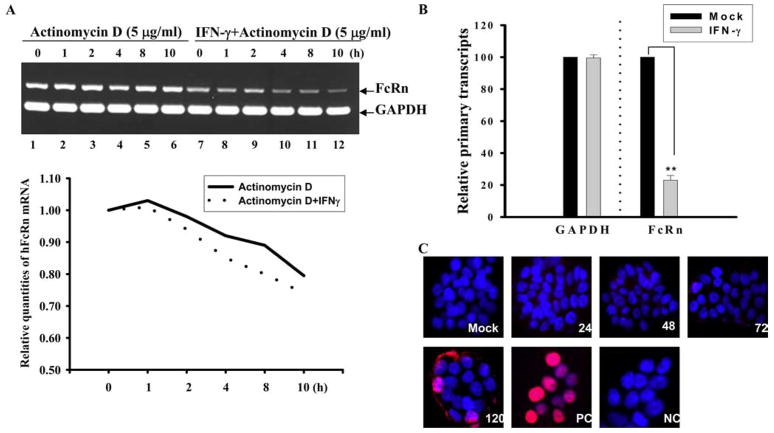
The primary mechanisms that regulate the amount of mRNA produced in mammalian cells are transcript stability and/or the rate of mRNA transcription. As such, we ascertained whether either of these mechanisms was involved in regulating the decrease in mature FcRn mRNA in the absence or presence of IFN-γ. Using an actinomycin D inhibition assay as shown by semiquantitative RT-PCR (Fig. 3A, top panel) and quantitative real-time PCR (Fig. 3A, bottom panel), the half-lives of FcRn mRNA appeared to be similar between mock- and IFN-γ-treated cells for the indicated time period. This suggests that a stability mechanism was not likely responsible for the decrease in FcRn mRNA. In contrast, nuclear run-on analysis indicated that the rate of FcRn mRNA transcription was decreased ~80% in THP-1 cells exposed to IFN-γ (Fig. 3B). Thus, this finding suggests that the decrease in FcRn mRNA induced by IFN-γ-stimulation on a HT-29 or THP-1 cell is due to a decrease in the rate of primary FcRn RNA transcription.
FIGURE 3.
Kinetic studies of FcRn mRNA levels and apoptosis in the absence or presence of IFN-γ. A and B, Kinetic studies of FcRn mRNA levels in the absence or presence of IFN-γ. Human intestinal HT-29 cells were preincubated for 24 h in the absence or presence of IFN-γ (25 ng/ml). Actinomycin D (5 μg/ml) was then added; total cellular RNA was harvested at the indicated time points (1–10 h). Ten nanograms of total RNA were reverse transcribed to cDNA in a final volume of 20 μl. Subsequently, 30 cycles of semiquantitative RT-PCR (A, top panel) or a real-time RT-PCR (A, bottom panel) were performed. Electrophoresis of 10 μl of PCR product was done on 1.5% agarose gel (top panel). FcRn values were normalized for GAPDH with each sample. FcRn product at time 0 before the addition of actinomycin D was plotted as 100% (A, bottom panel). The normalized FcRn mRNA levels are presented in arbitrary units. Solid and dashed lines represent RNA samples isolated from cells cultured in the presence and absence of IFN-γ, respectively. Results are mean of three experiments. B, Nuclear run-on analysis was performed on THP-1 nuclei isolated in the presence of biotin-16-UTP for 30 min. Biotinylated RNA was collected using streptavidin magnetic beads, and the level of FcRn or GAPDH RNA was determined by quantitative real-time RT-PCR. Data are mean ± SD of three independent experiments. **, p < 0.01. C, TUNEL staining of human intestinal epithelial HT-29 cells. After mock treatment or IFN-γ (50 ng/ml) treatment at the indicated times, in situ detection of apoptotic cells was performed on HT-29 cells cultured on coverslips by using an in situ cell death detection kit. Normal human HT-29 cells were stained after treatment with DNase I as a positive control (PC) or stained without terminal deoxynucleotide transferase as a negative control (NC). For the correlation of TUNEL with nuclear morphology, cultures were counterstained with DAPI (5 μg/ml). Red represents apoptosis positive cells. Images were viewed by fluorescence microscopy with excitation at 320 –580 nm.
In addition, activation of the STAT-1 signaling pathway can cause expression of caspase 1 and subsequent apoptosis (43). To further assess the possible role of IFN-γ in inducing apoptosis in our experiment, HT-29 cells were pretreated with or without IFN-γ (50 ng/ml) for the indicated time periods (Fig. 3C). A TUNEL assay demonstrated that IFN-γ induced detectable apoptosis in a small fraction of HT-29 cells only following 120 h of incubation (Fig. 3C). Mock-treated HT-29 cells were stained TUNEL negative at 120 h; cells stained after treatment with DNase I were used as a positive control (Fig. 3C, panel labeled “PC”), and cells without IFN-γ treatment or those stained without TdT were used as a negative control (Fig. 3C, panel labeled “NC”). Collectively, neither instability of FcRn mRNA nor significant apoptosis was induced by IFN-γ when used for this period of time (24 – 48 h) and at these concentrations (≤50 ng/ml) in our experiments.
Identification of STAT-1 binding site in the FcRn promoter
IFN-stimulated response elements (ISRE) and IFN-γ activation site (GAS) motifs are present in a variety of IFN-inducible genes (16, 17). ISRE (consensus sequence AGTTTCNNTTTCNY) and GAS (consensus sequence TTNCNNNAA, TTCNNNG/TAA) binding motifs have been mapped (16, 17, 44). Because FcRn regulation does not require newly synthesized proteins (Fig. 1F), it is possible that transcription factor or factors regulate FcRn expression through a mechanism that involves direct binding to putative regulatory ISRE or GAS elements located within the FcRn gene promoter. To test this, we searched for putative ISRE and GAS sequences along the entire human FcRn promoter (GenBank accession no. AC010619). Computational inspection revealed that the FcRn gene promoter contained no sequence similarity to typical ISRE consensus sequences; however, it had two sequences with a similarity to the STAT-1 consensus target sequence (Fig. 4A). To quickly screen whether these two sequences are functional in the transcriptional repression of FcRn by IFN-γ, we set up a transient cell transfection assay using the FcRn promoter/luciferase reporter gene construct phFcRnLuc (34). We also generated constructs pM1 and pM2, each of which contains mutations of the putative GAS sequence in phFcRnLuc (Fig. 4B). Transient transfection revealed that the phFcRnLuc or pM1 construct had decreased expression of luciferase in response to IFN-γ stimulation in wild-type 2fTGH cells (Fig. 4C). However, transient transfection of the pM2 construct revealed that mutation of this putative GAS sequence significantly increased the luciferase activity in IFN-γ-stimulated cells to a similar level as that in mock-stimulated cells (Fig. 4C). Hence, we conclude that the GAS sequence (TTCTTTGAA) in the human FcRn promoter is functional in response to IFN-γ stimulation (Table I).
Table I.
Comparison of functional GAS elementsa
| Gene | Species | GAS Sequence |
|---|---|---|
| c-myc | Mouse | TTCTGGGAA |
| ICAM-1 | Human | TTCCCGGAA |
| IRF-1 | Mouse | TTCCCCGAA |
| ICSBPb | Human | TTCTCGGAA |
| FcγR1 | Human | TTCCCAGAA |
| IFP 53 | Human | TTCTCAGAA |
| FcRn | Human | TTCTTTGAA |
| Consensus sequence | TTCNNNGAA |
Conserved nucleotides are set in boldface. N represents any nucleotide.
IFN consensus sequence binding protein.
To verify that this putative GAS sequence has the capability to directly bind STAT-1 protein in living cells, a ChIP assay was used to precipitate the STAT-1-DNA complexes with an Ab specific for STAT-1. After cross-linking the DNA with bound STAT-1 proteins in situ in IFN-γ-stimulated vs mock-stimulated HT-29 cells, the DNA fragments containing the STAT-1 sequences in FcRn promoter were precipitated with Ab and measured by PCR amplification. As shown in Fig. 4D, PCR with primers flanking the putative STAT-1 sequences generated a band from DNA coprecipitated with STAT-1 (lanes 2 and 3). In a negative control, immunoprecipitation with normal IgG did not generate any corresponding PCR products (Fig. 4D, lane 4). The STAT-1 binding sequence in the ICAM-1 gene promoter (45) was used as a positive control. As expected, ChIP assays failed to detect DNA bands from U3A cells (Fig. 4D, lanes 5 and 6). A quantitative real-time RT-PCR analysis of chromatin-immunoprecipitated PCR products for FcRn at the indicated time was shown in Fig. 4E. These data suggested that STAT-1 interacts with the putative GAS sequence of the human FcRn promoter after IFN-γ stimulation, at least in HT-29 cells.
To further visualize the capability of STAT-1 protein to directly bind to the putative FcRn GAS site identified from the ChIP assay, EMSAs were conducted using oligonucleotides containing the putative GAS sequence. As shown in Fig. 4F, oligonucleotides formed a complex with extracts from IFN-γ-stimulated cells (lane 2) but not from mock-stimulated cells (lane 1). An oligonucleotide containing the GAS sequence from the c-myc promoter (25) was used as a positive control (Fig. 4F, lane 9). To verify whether the binding was specific, a competition assay was performed. The inducible band could be completely competed away by unlabeled oligonucleotides (lane 3). Supershift analysis revealed that the complex contains a factor that was recognized by Ab specific for the STAT-1 protein (lane 4) but not normal IgG (lane 5). In the above experiments, the dynamics of STAT-1 nuclear transport after exposure to IFN-γ were determined by immunofluorescence staining of STAT-1. In Fig. 4G, STAT-1 appeared in the nucleus 0.5 h following IFN-γ treatment and remained in the nucleus at least 12 h in HT-29 cells. The nucleus was counterstained with DAPI (Fig. 4G, middle column). Taken together, these results identified a GAS site in the FcRn promoter.
Down-regulation of FcRn expression by IFN-γ is dependent on JAK1 and STAT-1 expression
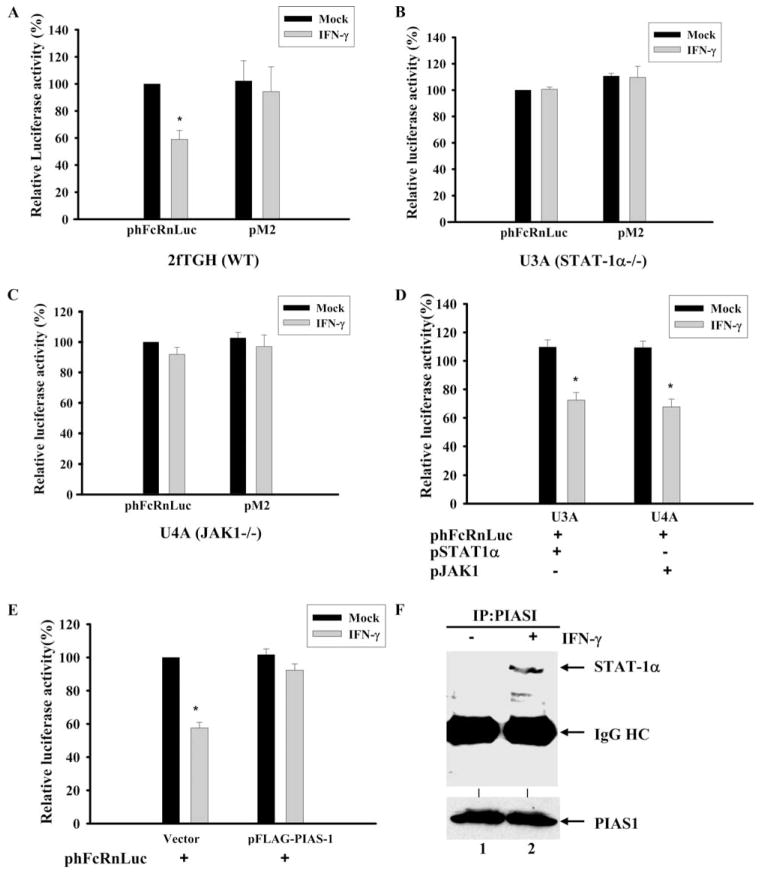
To further investigate the transcriptional repression of FcRn by IFN-γ, we transfected the phFcRnLuc and pM2 plasmids into STAT-1- and JAK1-deficient cells (Fig. 5, B and C). Transient transfection of the phFcRnLuc or pM2 construct into 2fTGH cells yielded similar results upon exposure to IFN-γ (Fig. 5A) as those shown in Fig. 4C. However, when phFcRnLuc and pM2 were transfected into the JAK1- and STAT-1-deficient cell lines U4A and U3A, the luciferase activities were not altered in response to IFN-γ stimulation (Fig. 5, B and C). A similar result was obtained from the JAK1-deficient HeLa-E2A4 cell line (data not shown). When expression of the STAT-1 or JAK1 proteins was rescued by transfection into the U3A and U4A cells, IFN-γ again reduced expression from phFcRnLuc (Fig. 5D). The negative effect of STAT-1 on FcRn transcription was dose dependent, because increased amounts of STAT-1 led to increased suppression of FcRn transcription (data not shown).
FIGURE 5.
Down-regulation of FcRn expression by IFN-γ is dependent on JAK1 and STAT-1 expression. A–C, Wild-type (WT) 2fTGH (A), STAT-1-null U3A (B), and JAK1-null U4A (C) cells were transiently transfected with phFcRnLuc or pM2 construct. D, STAT-1-null U3A or JAK1-null U4A cells were transiently transfected by phFcRnLuc along with pSTAT-1 or pJAK1 constructs. E, The 2fTGH cells were transiently transfected with phFcRnLuc together with vector backbone or pFLAG-PIAS1. Twenty-four hours after transfection, all groups of cells were either mock-treated or treated with IFN-γ for 24 h. Cells were then harvested and protein extracts were prepared for the luciferase assay. Transcriptional activity was measured as firefly luciferase activity and normalized to Renilla luciferase activity. The results show the mean value from three independent experiments. *, p < 0.05. F, Interaction of PIAS1 and STAT-1 proteins. Cell lysates from mock- (lane 1) or IFN-γ-treated (lane 2) 2fTGH cells were immunoprecipitated (IP) with anti-FLAG mAb, and immunoprecipitates were subjected to electrophoresis on a 12% SDS-polyacrylamide gel under reducing conditions and transferred to a nitrocellulose membrane for Western blotting with anti-STAT-1 Ab. Immunoblots were developed with ECL. Experiment was at least performed two times. The cell lysates (bottom row) were blotted to monitor the expression of PIAS1. HC, Heavy chain.
It has been shown that PIAS1 specifically inhibits STAT-1 by directly blocking its DNA binding activity (23). When FLAG-tagged PIAS1 and phFcRnLuc expression plasmids were cotransfected into 2fTGH cells, the luciferase activity was not significantly altered following IFN-γ exposure in comparison with that of mock-treated cells (Fig. 5E). However, the luciferase activity was significantly changed in mock-transfected cells. The interaction of PIAS1 and STAT-1 were verified in our immunoprecipitation-Western blot experiments (Fig. 5F). A STAT-1 protein appeared in the PIASI precipitates from IFN-γ-treated 2fTGH cells (Fig. 5F, lane 2), but not in those from mock-treated cells (lane 1). These results suggested that IFN-γ down-regulated FcRn expression mainly through the activation of STAT-1.
Effect of STAT-1 phosphorylation on IFN-γ-induced FcRn gene repression
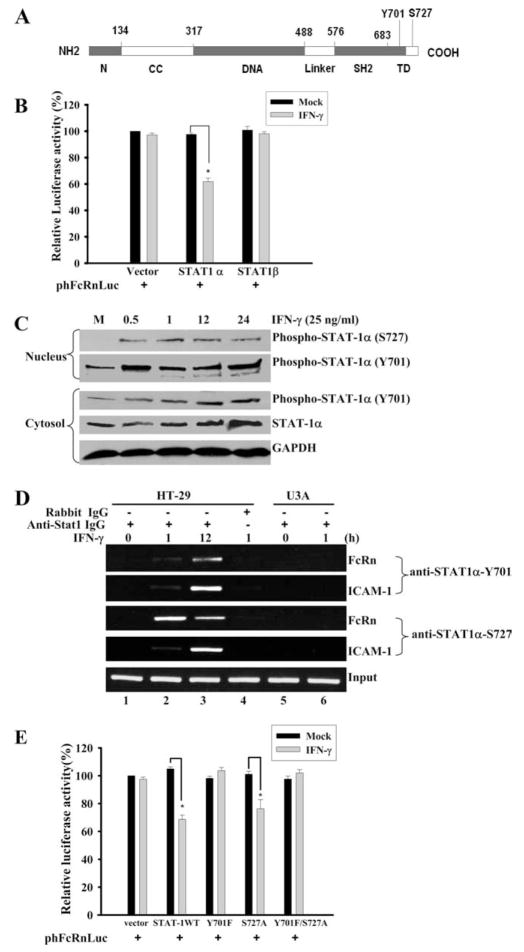
IFN-γ induces phosphorylation of STAT-1 at the tyrosine 701 and serine 727 residues (20, 23) (Fig. 6A). Phosphorylation of STAT-1 at tyrosine 701 is critical for STAT-1 dimerization, nuclear translocation, and DNA binding, whereas phosphorylation at serine 727 is important for optimal transactivational activity of STAT-1 (21). However, the transcriptional suppression of matrix metalloproteinase (MMP)-9 is not dependent on STAT-1 phosphorylation at serine 727 (46). To address this, we first tested the STAT-1β isoform that lacks the transcription activation domain and typically does not activate transcription (16, 17). In a luciferase reporter assay, the STAT-1β isoform failed to restore an IFN-γ-mediated inhibitory effect on the FcRn promoter in STAT-1-deficient U3A cells in comparison with STAT-1α (Fig. 6B). This suggests that the inhibition is dependent on the transcription activation domain of STAT-1. In the dynamic analysis of STAT-1 phosphorylations after IFN-γ stimulation, STAT-1 phosphorylation at tyrosine 701 and serine 727 was enhanced in nucleus (Fig. 6C). To verify that phospho-STAT-1 binds directly to the FcRn promoter, a ChIP assay was used to precipitate the phospho-STAT-1-DNA complexes with Ab specific for STAT-1 phosphorylated at tyrosine 701 or serine 727, respectively. As shown in Fig. 6D, PCR with primers flanking FcRn GAS sequence generated a band from DNA coprecipitated with either anti-phospho-STAT-1 tyrosine 701 or serine 727 in HT-29 cells (lanes 2 and 3), but not in STAT-1-null U3A cells (lanes 5 and 6). In a negative control, immunoprecipitation with normal IgG did not generate detectable PCR products (Fig. 6D, lane 4). The phospho-STAT-1 protein binding to the ICAM-1 gene promoter was used as a positive control in ChIP experiments. To further probe the role of STAT-1 phosphorylation status in regulating FcRn gene expression, phosphorylation sites at tyrosine 701 or serine 727 were mutated alone or in combination. In comparison with pSTAT-1, either pSTAT-1Y701F or pSTAT-1Y701F/S727A resulted in the significant increase of luciferase expression after cotransfection with phFcRnLuc into the U3A cells (Fig. 6E). Mutation of the STAT-1 phosphorylation site at serine 727 did not significantly remove the inhibitory function of IFN-γ (Fig. 6E). Therefore, we conclude that phosphorylation at tyrosine 701 was involved in FcRn repression by IFN-γ.
FIGURE 6.
Differential effects of STAT-1 phosphorylations on IFN-γ-mediated suppression of FcRn gene transcription. A, Schematic diagram of functional domains of STAT-1 protein. N, N-terminal domain; CC, coiled-coil domain; DNA, DNA-binding domain; SH2, Src homology 2 domain; and TD, transactivation domain. Numbers represent the position or length of amino acid in each domain. Y701, Tyrosine 701; S727, serine 727. B, Effects of expression of STAT-1β on FcRn promoter activity. The FcRn promoter construct (0.5 μg) was transiently transfected into U3A cells with empty expression vector pcDNA3 (Vector) and the pSTAT-1α (STAT1 α) and pSTAT-1β (STAT1β) expression vectors (0.5 μg). Twenty-four hours later, transfected cells were treated with or without IFN-γ for 24 h. Cells were then harvested and protein extracts were prepared for the luciferase assay. Transcriptional activity was measured as firefly luciferase activity and normalized to Renilla luciferase activity. *, p < 0.05. C, Dynamic analysis of STAT-1 phosphorylations after IFN-γ stimulation. The 2fTGH cells were treated as indicated above the blot and protein extracts from the nucleus (upper panels) and cytosol (lower panels) were separated by electrophoresis in a 12% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The proteins were blotted with anti-phospho-STAT-1 or anti-STAT-1 Abs, respectively. GAPDH was used as an internal control. Blots are representative of three experiments. M, Mock treated. D, Detection of the in vivo binding of phospho-STAT-1 protein to human FcRn promoter. Formaldehyde-crosslinked chromatin was prepared from both mock-treated and IFN-γ-treated 2fTGH (lanes 1– 4) or STAT-1-null U3A (lanes 5 and 6) cells as described in Materials and Methods. ChIP assays were performed using STAT-1-specific (lanes 1–3, 5, and 6) or isotype-matched (lane 4) Abs as a negative control. Immunoprecipitated chromatin was subjected to PCR analysis using FcRn- or ICAM-1-specific primers. The equivalent amount of chromatin in the immunoprecipitations was monitored by PCR amplification of input chromatin as an internal control. ChIP assay was performed at least three times. E, Effects of over-expression of phospho-mutant STAT-1 on FcRn promoter activity. The FcRn promoter construct (0.5 μg) was transiently transfected into U3A cells with the empty expression vector pcDNA3 and the pSTAT-1, pSTAT-1Y701F, pSTAT-1S727A, and pSTAT-1Y701F/S727A expression vectors (0.5 μg). Twenty-four hours later, transfected cells were treated with or without IFN-γ for 24 h. Cells were then harvested and protein extracts were prepared for the luciferase assay. Transcriptional activity was measured as firefly luciferase activity and normalized to Renilla luciferase activity. *, p < 0.05.
IFN-γ induces the in vivo association of p300 and STAT-1α, and overexpression of p300 reduces IFN-γ-mediated FcRn gene repression
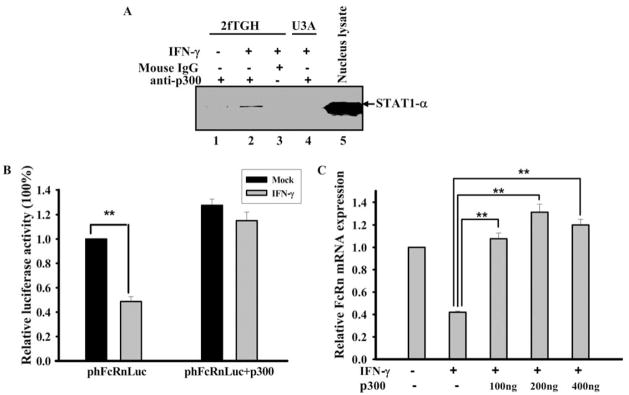
Our data show that the nuclear translocation of STAT-1α correlated with IFN-γ-mediated down-regulation of FcRn gene transcription and that STAT-1α bound directly to the FcRn promoter (Fig. 4). It is possible that nuclear protein(s) interacting with STAT-1α may play a pivotal role in down-regulating FcRn gene expression. It is known that STAT-1α can bind CBP/p300 (47). Therefore, we further examined the possibility that the interaction between STAT-1α and CBP/p300 may lead to down-regulation of FcRn gene expression. Coimmunoprecipitation was used to examine the in vivo association of endogenous p300 and STAT-1α. The 2fTGH and U3A cells were incubated in the absence and presence of IFN-γ and nuclear extracts from these cells were subjected to immunoprecipitation with Ab against p300. The precipitated immune complexes were then blotted for the presence of STAT-1α. In IFN-γ-treated cells, anti-p300 Ab immunoprecipitated a significant amount of STAT-1α (Fig. 7A, lane 2) in comparison with mock-stimulated cells (lane 1). As a negative control, IgG did not immunoprecipitate STAT-1α (lane 3). These results suggest that STAT-1α does not associate with p300 in mock-stimulated cells; however, IFN-γ treatment can induce the in vivo association of STAT-1α and p300.
FIGURE 7.
IFN-γ induces the in vivo association of p300 and STAT-1α, and overexpression of p300 blocks IFN-γ-mediated FcRn gene down-regulation. A, The 2fTGH (lanes 1–3) and U3A (lane 4) cells were treated with IFN-γ (10 ng/ml) or mock treated for 2 h and then nuclear extracts were obtained and subjected to immunoprecipitation. Anti-p300 mAb (lanes 1, 2, and 4) and isotype-matched IgG (lane 3) were used to immunoprecipitate the STAT-1α and p300 complex. The immune complexes were separated by electrophoresis in a 12% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The proteins were blotted with anti-STAT-1α Ab. Immunoblots were developed with ECL. Nuclear extracts (lane 5) were used as a positive control for blotting. B, The 2fTGH cells were transiently transfected with the FcRn promoter construct phFcRnLuc (1 μg), without and with a p300 construct (1 μg), and the total amount of transfected DNA was normalized by pcDNA3. Transfected cells were treated with IFN-γ or mock treated for 12 h. Cells were then harvested 24 h later. Transcriptional activity was determined as firefly luciferase activity and normalized to Renilla luciferase activity. **, p < 0.01. C, HT-29 cells were transiently transfected with increasing amounts (0.1– 0.4 μg) of a p300 construct, and the total amount of transfected DNA was normalized by pcDNA3. Transfected cells were treated with IFN-γ or mock treated for 14 h. FcRn mRNA was analyzed by quantitative real-time RT-PCR analysis. **, p < 0.01.
It is possible that STAT-1α suppresses FcRn gene activation by interfering with the binding of CBP/p300 to the FcRn promoter. Transient transfection assays were first used to examine whether overexpression of p300 could reverse IFN-γ-mediated FcRn suppression. Indeed, overexpression of p300 reversed IFN-γ-induced suppression of luciferase expression driven by the FcRn promoter (Fig. 7B) or FcRn gene expression in a dose-dependent manner (Fig. 7C). Therefore, these data suggest that the IFN-γ-induced interaction between STAT-1α and CBP/p300 is responsible for the down-regulation of FcRn expression by IFN-γ.
IFN-γ reduced bidirectional transport of IgG in polarized lung epithelial monolayers
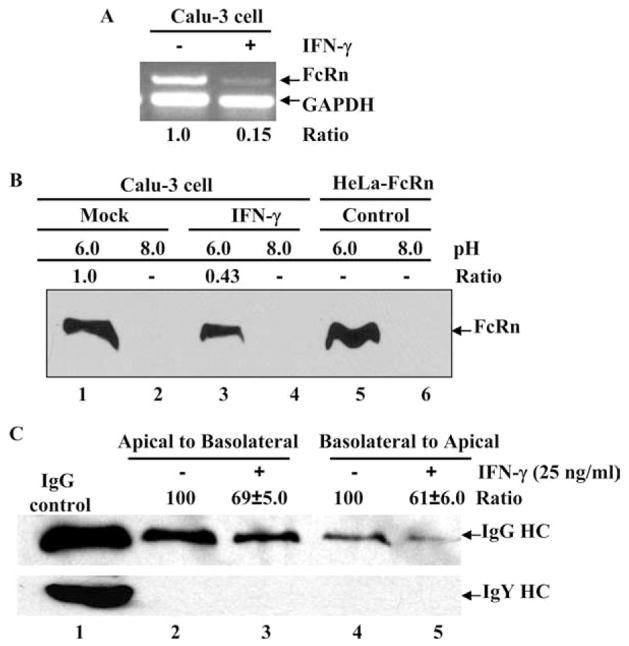
The FcRn protein has been shown to transport IgG bidirectionally in polarized epithelial cells, namely from the apical to the basolateral direction or vice versa (38, 39). We therefore addressed the possibility that IFN-γ-stimulated epithelial cells have altered IgG transcytosis. Calu-3 cells have been previously shown to transcytose dimeric IgA in response to IFN-γ stimulation (48). We established the FcRn expression in Calu-3 cell lines and further verified the FcRn down-regulation by IFN-γ stimulation, as assessed by semiquantitative RT-PCR (Fig. 8A). FcRn binds IgG at acidic pH (6.0) and releases IgG at neutral pH. We further tested whether the decreased expression of FcRn upon IFN-γ exposure affects its ability to bind to its natural ligand, IgG. We incubated cell lysates from Calu-3 cells at either pH 6.0 or pH 8.0 with human IgG-Sepharose. Cell lysates from HeLa cells transfected with FcRn cDNA were used as positive control. As expected, FcRn from HeLa-FcRn cells bound IgG at pH 6.0 but not at pH 8.0 (Fig. 8B, lanes 5 and 6). Our result showed that IFN-γ stimulation decreased cellular FcRn binding to IgG at pH 6.0 (Fig. 8B, lane 3) in comparison with mock-stimulated cells (Fig. 8B, lane 1), suggesting the decreased FcRn expression led to decreased FcRn-IgG complexes. In our transport experiment, after adding human IgG to the apical or basolateral surface of a Calu-3 cell monolayer, we assessed the IgG transported to the opposite basolateral or apical chamber following IFN-γ exposure, respectively. As expected, after 1 h at 37°C intact human IgG applied to the apical or basolateral side was transported across this monolayer. IgG H chain was detected in medium incubated at 37°C (Fig. 8C, upper row). Importantly, IgG transport was decreased ~30% in the apical to basolateral direction (Fig. 8C, lane 3), or 40% in the basolateral to apical direction (Fig. 8C, lane 5) following IFN-γ stimulation, in comparison to the mock-treated monolayer (Fig. 8C, lanes 2 and 4). Treatment of Calu-3 monolayers with IFN-γ for 24 h might result in a leakage of IgG molecules, as shown in human intestinal epithelial cell line T84 (49). Chicken IgY was used as a negative control because it is structurally similar to human IgG but does not bind to human FcRn. As shown in Fig. 8C (bottom panel), chicken IgY was not transported in either direction, suggesting that the transepithelial flux of Abs by passive diffusion through intercellular tight junctions or monolayer leaks does not contribute to the amount of the IgG we detect. Therefore, we concluded that IFN-γ stimulation decreased the IgG transport across the polarized epithelial cells.
FIGURE 8.
Effects of IFN-γ stimulation on the IgG transcytosis. A, Semiquantitative RT-PCR analysis of FcRn mRNA in the human lung epithelial Calu-3 cell line. The Calu-3 cells were treated (+) with IFN-γ (25 ng/ml) (right lane) or left untreated (−) (left lane) for 24 h. Data are representative results for RT-PCR analysis of FcRn expression in Calu-3. Ratios of FcRn-GAPDH are shown as indicated. B, The pH-dependent FcRn binding of IgG. The Calu-3 cells were lysed in sodium phosphate buffer (pH 6.0 or 8.0) with 0.5% CHAPS. Approximately 1 mg of soluble proteins was incubated with human IgG-Sepharose at 4°C. The eluted proteins were subjected to 12% SDS-polyacrylamide electrophoresis and subjected to Western blot analysis. Proteins were probed with affinity-purified rabbit anti-FcRn peptide Ab and HRP-conjugated donkey anti-rabbit Ab. Immunoblots were developed with ECL. The ratio of the mock sample is assigned a value of 1.0, and the values from IFN-γ-treated sample are normalized to this value. C, Calu-3 cells (5 × 105/well) were grown in a 12-well Transwell plate. When the resistance of the monolayer reached 700–1000 ohms/cm2, cells were stimulated with or without IFN-γ (25 ng/ml) for 24 h. Cells were loaded with human IgG (top row) or chicken IgY (bottom row) (0.5 mg/ml) at 4°C in either the apical (lanes 2 and 3) or basolateral (lanes 4 and 5) chamber. Lane 1 represents an IgG or IgY H chain. Cells were warmed to 37°C to stimulate transcytosis, and medium was collected from the nonloading compartment 1 h later and subjected to Western blot-ECL analysis. The results are representative of at least three independent experiments. Band intensities of IgG heavy chain (HC) were compared by densitometry against IgG transported from mock-stimulated cells.
Discussion
Transcriptional regulation of genes hinges on the ordered recruitment of transcriptional polymerase, coactivators, repressors, chromatin modifiers/remodelers, and general transcriptional factors to the promoters of target genes. How the gene transcriptional machinery integrates signals from different biological signaling pathways is a central question for gene regulation. Exposure to IFN-γ can result in the regulation of up to 500 genes in either a positive or a negative way (16, 24). Genes that are negatively regulated by IFN-γ are fewer in number than those positively induced. Among the negatively regulated ones are some of the MMPs, stromelysin, type II collagen, HL-60, neu/HER-2, cell-cycle genes (c-myc, cyclin D, cyclin A), granulocyte chemotactic protein-2, IL-4, prolactin, perlecan, and the scavenger receptor A (SR-A) genes (24, 25, 46, 50–58). In this article we report, for the first time, the effect of IFN-γ on the transcriptional regulation of FcRn.
Activation of the IFN-γ signaling pathway down-regulates the expression of the human FcRn gene, and this down-regulation is dependent on the STAT-1 signaling pathway. This conclusion is supported by several pieces of evidence. First, our results showed that stimulation by IFN-γ decreased the FcRn expression in human intestinal epithelial cells, THP-1 cells, and freshly isolated human PBMC at both the mRNA and protein levels (Figs. 1 and 2). The relative inability of IFN-γ to down-regulate FcRn production in Caco-2 cells may indicate that different control mechanisms regulate transcription of FcRn in this cell type or, more likely, given the relative lack of effect of IFN-γ on Caco-2 and the tight junction integrity of Caco-2 monolayers, that IFN-γ receptors are expressed at a much lower level in this cell type (59). Second, a nuclear run-on assay demonstrated that this down-regulation indeed occurred at transcription initiation (Fig. 3B). Third, we have mapped an IFN-γ-responsive sequence, GAS, to the promoter region of the human FcRn gene by both EMSA and ChIP (Fig. 4). Mutation of this GAS sequence abolished the inhibitory effect of IFN-γ on FcRn promoter (Fig. 4). Fourth, expression of luciferase activity driven by the FcRn promoter following IFN-γ exposure was not affected in STAT-1-null U3A or JAK1-deficient U4A cells in comparison with the wild-type cell 2fTGH (Fig. 5, B and C). However, expression of wild-type STAT-1 or JAK1 proteins in U3A or U4A cells rescued the repressive effect of IFN-γ on the human FcRn promoter (Fig. 5D). Fifth, the inhibitory effect of IFN-γ on the FcRn promoter was abolished by overexpressing PIAS1 protein, a specific inhibitor of STAT-1 protein (Fig. 5, E and F). Sixth, our results indicated that tyrosine 701 phosphorylation of STAT-1 was indispensable for suppression of the FcRn expression, indicating that nuclear translocation and localization of phospho-STAT-1 were required to repress the FcRn gene (Fig. 6). These results provided both biochemical and genetic support for the conclusion that increased phosphorylation of STAT-1 is the mechanism by which IFN-γ treatment leads to FcRn down-regulation. Recent studies have shown that IFN-γ can regulate gene expression by STAT-1-independent pathways (24, 25). Among several genes that are inhibited by IFN-γ, c-myc has been shown to require STAT-1-dependent and STAT-1-independent pathways and, notably, there is a GAS element in the c-myc promoter that is necessary, but not sufficient, to confer the total inhibitory effects of IFN-γ (25). Therefore, our data support the conclusion that the down-regulation of human FcRn expression was mediated via a STAT-1-dependent pathway in response to IFN-γ. However, our data could not exclude the possibility that STAT-1 may bind to sites in other parts, such as introns, of the human FcRn gene. We considered the possibility that IFN-γ induces apoptosis (43) and regulates the expression of the gene at posttranscriptional level (60). However, several facts counter this conjecture. First, down-regulation of human FcRn and up-regulation of Ii occurred concomitantly in response to IFN-γ treatment (Fig. 1A). Second, we failed to detect any noticeable effect of IFN-γ on human FcRn half-life in actinomycin D-treated cells (Fig. 3A), suggesting that the half-life of FcRn mRNA was not affected by IFN-γ. Third, apoptosis was only detected after a 5-day period and then only in a few cells (Fig. 3C). In contrast, a 30-min incubation time for IFN-γ was sufficient to reduce the FcRn gene expression (Fig. 1C). These observations are in agreement with other studies indicating that a high dosage and long time treatment of IFN-γ are necessary to induce apoptosis (61). In addition, the level of IFN-γ repression (40 –50%) on the reporter construct phFcRnLuc was similar to FcRn gene repression in cell lines; this would exclude the possibilities that the down-regulation of FcRn gene expression might be caused by apoptotic effects of IFN-γ or that IFN-γ affects the half-life and stability of FcRn mRNA. Therefore, these complementary experiments eliminate the concerns of apoptotic effects or stability of FcRn mRNA by IFN-γ.
The mechanism of STAT-1-mediated down-regulation of human FcRn expression might be through sequestering of the transcription activator CBP/p300. One potential mechanism by which IFN-γ might normally mediate the repression of FcRn transcription could be via STAT-1 interaction with either constitutive transcription factors or transcription factors that are activated upon exposure to IFN-γ. Although STAT-1 acts as an activator of transcription in numerous genes in response to IFN-γ stimulation, the detailed mechanisms by which STAT-1 switches on and off gene expression are still unclear. As shown in several elegant studies, although STAT-1 is necessary and sufficient to inhibit MMP-9, SR-A, and type II collagen gene transcription by IFN-γ, there are no GAS elements in the promoters of these genes (46, 62). Thus, suppression of the expression of these genes by IFN-γ-activated STAT-1 is probably not dependent on the direct binding of STAT-1 on the gene promoter of these genes. In contrast, the suppression of the MMP-9 or the SR-A gene depends on the ability of activated STAT-1 to interact with other nuclear proteins. Indeed, STAT-1 can interact with a variety of other transcription factors, including STAT-2, CBP, p300, p300/CBP cointegrator protein (pCIP), histone deacetylase 1 (HDAC-1), N-Myc interactor (Nmi), and BRACA1 (25, 47, 63–65). Among these proteins, CBP/p300 serves as a scaffold in transcription complex formation in addition to functioning as histone acetyltransferases. Given the fact that the total amount of CBP/p300 is limited compared with the amount of other transcription regulators, a competition for using CBP/p300 in different signaling pathways has been proposed. In the case of the MMP-9, SR-A, neu/HER-2 genes, activated STAT-1 can competitively bind with CBP/p300, thereby resulting in decreased association of CBP/p300 in the gene promoter and interference with the assembly of functional transcription complexes (47, 53, 62). Our data showed that overexpression of CBP/p300 overcame the inhibitory effect of IFN-γ on the expressions of luciferase in a transfection assay (Fig. 7A) or FcRn mRNA in HT-29 cells (Fig. 7B). However, our data could not exclude the possibility of STAT-1 interacting with other transcription factors. For example, Y-box-binding protein YB-1, RFX5 complex, CIITA, IFN regulatory factor (IRF)-1, and IRF-2 are also involved in the gene repressions by IFN-γ (19, 55, 65, 66). Further work is underway to determine how STAT-1 actually mediates repression of FcRn gene expression.
What might be the biological implications of the down-regulation of FcRn expression by IFN-γ? To date, two biological functions have been attributed for FcRn: transcytosis of IgG across polarized epithelial cells and protection of IgG from degradation. The level of FcRn expression may be critical for the regulation of IgG levels in tissues and blood. First, mucosal Abs are important for mucosal infections (67), and epithelial cells that line mucosal surfaces in vivo express FcRn. Therefore, FcRn transports normal or pathogen-specific neutralizing IgG across polarized cells such as placental or mucosal epithelial cells, potentially “seeding” maternal and mucosal immunity. From our findings, one might speculate whether IFN-γ dampening the expression of the FcRn receptor might lead to the lessening of IgG transport. In an in vitro Transwell model, our results clearly demonstrated that IFN-γ functionally decreased IgG transport in the polarized lung epithelial Calu-3 cell line (Fig. 8). Therefore, IFN-γ may dampen IgG-mediated mucosal immunity by reducing IgG transport in vivo. This result is in contrast to the fact that IFN-γ up-regulates pIgR expression, which is expected to enhance secretory IgA-mediated mucosal immunity (48, 68). Furthermore, our previous finding revealed that TNF-α and IL-1β, via activation of the NF-κB signaling pathway, can up-regulate the functional expression of FcRn (34). Because IFN-γ, TNF-α, and IL-1β are proinflammatory cytokines, FcRn levels may therefore be finely tuned by opposing negative and positive signaling in the maintenance of IgG homeostasis under pathophysiological conditions. Thus, regulation of FcRn expression in vivo likely involves the species, magnitudes, and coordinated actions of proinflammatory cytokines or other functional regulators. Secondly, by mediating the protection of IgG from catabolism, FcRn extends the half-life of pathogenic or autoimmune IgG, potentially promoting the progression of IgG-mediated autoimmune diseases (69, 70). Therefore, by influencing the expression level of FcRn, IFN-γ may be directly coupled to the pathogenesis of IgG-mediated autoimmune diseases. Indeed, IFN-γ has been shown to regulate the intensity or the progression of several autoimmune diseases (71, 72). However, it remains for further investigation whether its regulatory effect in the changing course of an autoimmune disease is, at least in part, through the down-regulation of FcRn expression. This question merits further investigation in a murine model. We also found that IFN-γ down-regulated the expression of mouse FcRn in the macrophage RAW264.7 cell line and in mouse tissues by i.v. injection of IFN-γ (data not shown). Overall, by examining the molecular mechanisms by which IFN-γ regulates FcRn expression, our studies may contribute toward the general understanding of FcRn-mediated mucosal immunity and inflammation. The identification and understanding of IFN-regulated FcRn gene expression may lead to improved therapies for IgG-mediated autoimmune diseases.
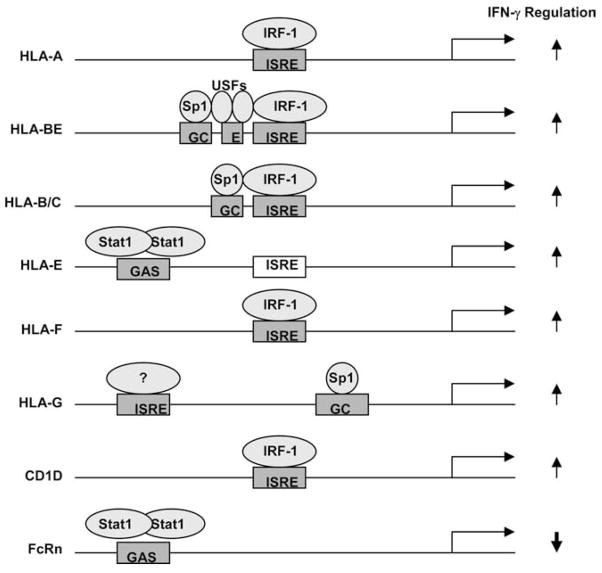
Among MHC class I-related molecules, IFN-γ causes the up-regulations of the MHC genes HLA-A, HLA-B, HLA-C, HLA-F, HLA-G, HLA-H, HLA-E, and CD1 (29). The promoters of HLA-A, HLA-B, HLA-C genes contain a consensus ISRE sequence. IRF-1 is induced by IFN-γ and interacts with the ISRE in HLA gene promoters to stimulate transcription initiation (29). In the special case of HLA-E, although IFN-γ also induces HLA-E expression, the HLA-E gene promoter does not contain a functional ISRE. Instead, two distinct elements in the HLA-E promoter are termed the IFN response region (IRR) and the upstream IFN response region (UIRR). STAT-1 and GATA-1 bind to the IRR and UIRR, respectively, to stimulate transcription from the HLA-E promoter (73). Among the MHC class I-related genes, FcRn is an only molecule that is down-regulated by IFN-γ (Fig. 9). This scenario makes FcRn unique in the response to IFN regulation. Therefore, understanding differences in the mechanisms by which IFN-γ stimulates MHC-I genes and FcRn could be of great interest in the settings of immune responses and autoimmunity. Any differences in the signal transduction pathways leading to differential expression of the FcRn and MHC class I genes would be potential targets for therapeutic intervention aimed at selective activation of one or the other.
FIGURE 9.
Schematic illustration of transcription factors binding to the promoter region of some MHC class I-related genes after IFN-γ treatment. Most information is derived from Gobin et al. (29). The ISREs of HLA-A, HLA-B, HLA-C, and HLA-F bind IRF-1 upon IFN-γ exposure and regulate the IFN-γ-induced transactivation of these genes (29, 32). The putative ISRE of HLA-E did not respond to IFN-γ stimulation, whereas an upstream GAS sequence of HLA-E is responsive to IFN-γ through STAT-1 activation (29, 73). HLA-G is responsive to IFN-γ via an upstream IFN-responsive regulatory sequences (31). Multiple putative ISREs of CD1D are predicted (74), but one is shown here. Human FcRn responds to IFN-γ through STAT-1 activation and binding to an upstream GAS sequence. In addition, several constitutive transcription factors are revealed to bind to the ISRE area. Sp1 binds to the GC-rich sequences in the ISRE areas of HLA-B, HLA-C, and HLA-G. The putative E box 5′ of the ISRE in most HLA-BE alleles is bound by USF-1 and USF-2 (29). Arrows represent the up- and down-regulation of gene expression upon IFN-γ exposure. The schematic structure of the gene promoter is not scaled.
In summary, transcriptional repression of FcRn gene expression by IFN-γ is dependent on activated STAT-1 protein. These findings suggest that the biological consequence of IFN-γ-induced transcription of the FcRn gene is distinct from that of other MHC class I or related genes. Therefore, our observation that FcRn repression by IFN-γ is, to our knowledge, the first demonstration that MHC class I-related genes are regulated negatively by IFN-γ exposure. These results provide proof of principle that IFN-γ differentially modulates expression of the FcRn and of MHC class I or its related genes, whose products often mediate opposing effects on cellular and humoral immunity. Further studies of STAT-1-mediated mechanism of transcriptional repression on FcRn will provide insights into understanding the inhibitory effects of IFN-γ on gene expression in general. Given the important role of FcRn in the maintenance of IgG concentration as well as transport of IgG across placenta and mucosal surfaces, the results from these studies would also provide new information on mucosal protection and vaccine development.
Acknowledgments
We thank Dr. Geoffrey J. Letchworth for providing critiques for the manuscript. We gratefully acknowledge the receipts of cell lines 2fTGH and STAT-deficient cell lines from Dr. George R. Stark, HeLa-E2A-4 cell line from Dr. Richard Flavell, and human intestinal epithelial cells from Dr. Richard Blumberg and Dr. Wenxia Song. We also appreciate the protein expression plasmids encoding STAT-1 from Dr. Koichi Nakajima, JAK from Dr. James N. Ihle, FLAG-tagged STAT-1 and PIAS1 from Dr. Ke Shuai, and CBP/p300 plasmids from Dr. Zhixin Zhang. We thank the technical help of Guozhen Gao and Swati Shah.
Footnotes
This work was supported in part by National Institutes of Health Grants AI67965, AI65892, and AI73139 (to X.Z.), a faculty start-up package (to X.Z.), and MAES competitive grants (to X.Z.) from University of Maryland. Y.B. is in part supported by a fellowship from China Scholarship Council.
Abbreviations used in this paper: FcRn, neonatal Fc receptor; CBP, CREB binding protein; ChIP, chromatin immunoprecipitation; CHX, cycloheximide; GAS, DAPI, 4′,6′-diamidino-2-phenylindole; Ii, invariant chain; IFN-γ activation site; IRF, IFN regulatory factor; ISRE, IFN-stimulated response element; MMP, matrix metalloproteinase; PIAS1, protein inhibitor of activated; SR-A, scavenger receptor A; STAT-1; poly(dI-dC), poly(deoxyinosinic-deoxycytidylic acid).
Disclosures
A patent application related to this work was filed with the U. S. Patent Office by the University of Maryland and the authors Xiaoping Zhu and Xindong Liu on May 18, 2007.
References
- 1.Blumberg RS, Koss T, Story CM, Barisani D, Polischuk J, Lipin A, Pablo L, Green R, Simister NE. A major histocompatibility complex class I-related Fc receptor for IgG on rat hepatocytes. J Clin Invest. 1995;95:2397–2402. doi: 10.1172/JCI117934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Israel EJ, Taylor S, Wu Z, Mizoguchi E, Blumberg RS, Bhan A, Simister NE. Expression of the neonatal Fc receptor, FcRn, on human intestinal epithelial cells. Immunology. 1997;92:69–74. doi: 10.1046/j.1365-2567.1997.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borvak J, Richardson J, Medesan C, Antohe F, Radu C, Simionescu M, Ghetie V, Ward ES. Functional expression of the MHC class I-related receptor, FcRn, in endothelial cells of mice. Int Immunol. 1998;10:1289–1298. doi: 10.1093/intimm/10.9.1289. [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Meng G, Dickinson BL, Li X, Mizoguchi E, Miao L, Wang Y, Robert C, Wu B, Smith PD, et al. MHC class I-related Fc Receptor for IgG is functionally expressed in monocytes, intestinal macrophages, and dendritic cells. J Immunol. 2001;166:3266–3276. doi: 10.4049/jimmunol.166.5.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 6.Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 8.Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I- related receptor FcRn. Annu Rev Immunol. 2000;18:739–766. doi: 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- 9.Firan M, Bawdon R, Radu C, Ober RJ, Eaken D, Antohe F, Ghetie V, Ward ES. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of γ-globulin in humans. Int Immunol. 2001;13:993–1002. doi: 10.1093/intimm/13.8.993. [DOI] [PubMed] [Google Scholar]
- 10.Roopenian DC, Christianson GJ, Sproule TJ, Brown AC, Akilesh S, Jung N, Petkova S, Avanessian L, Choi EY, Shaffer DJ, et al. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J Immunol. 2003;170:3528–3533. doi: 10.4049/jimmunol.170.7.3528. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, Anderson CL. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med. 2003;197:315–322. doi: 10.1084/jem.20021829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brambell FW, Hemmings WA, Morris IG. A theoretical model of γ-globulin catabolism. Nature. 1964;203:1352–1354. doi: 10.1038/2031352a0. [DOI] [PubMed] [Google Scholar]
- 13.Israel EJ, V, Patel K, Taylor SF, Marshak-Rothstein A, Simister NE. Requirement for a β 2-microglobulin-associated Fc receptor for acquisition of maternal IgG by fetal and neonatal mice. J Immunol. 1995;154:6246– 6251. [PubMed] [Google Scholar]
- 14.Ghetie V, Hubbard JG, Kim JK, Tsen MF, Lee Y, Ward ES. Abnormally short serum half-lives of IgG in β 2-microglobulin-deficient mice. Eur J Immunol. 1996;26:690– 696. doi: 10.1002/eji.1830260327. [DOI] [PubMed] [Google Scholar]
- 15.Vidarsson G, Stemerding AM, Stapleton NM, Spliethoff SE, Janssen H, Rebers FE, de Haas M, van de Winkel JG. FcRn: an IgG receptor on phagocytes with a novel role in phagocytosis. Blood. 2006;108:3573–3579. doi: 10.1182/blood-2006-05-024539. [DOI] [PubMed] [Google Scholar]
- 16.Stark GR, I, Kerr M, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 17.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 18.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 19.Eilers A, Georgellis D, Klose B, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Decker T. Differentiation-regulated serine phosphorylation of STAT1 promotes GAF activation in macrophages. Mol Cell Biol. 1995;15:3579–3586. doi: 10.1128/mcb.15.7.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 21.Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy DE, Muller M, Decker T. Specificity of signaling by STAT1 depends on SH2 and C-terminal domains that regulate Ser727 phosphorylation, differentially affecting specific target gene expression. EMBO J. 2001;20:91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M, Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-γ-dependent innate immunity. Immunity. 2003;19:793– 802. doi: 10.1016/s1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- 23.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 24.Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and -independent pathways in IFN-γ-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 25.Ramana CV, Grammatikakis N, Chernov M, Nguyen H, Goh KC, Williams BR, Stark GR. Regulation of c-myc expression by IFN-γ through Stat1-dependent and -independent pathways. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gough DJ, Sabapathy K, Ko EY, Arthur HA, Schreiber RD, Trapani JA, Clarke CJ, Johnstone RW. A novel c-Jun-dependent signal transduction pathway necessary for the transcriptional activation of interferon γ response genes. J Biol Chem. 2007;282:938–946. doi: 10.1074/jbc.M607674200. [DOI] [PubMed] [Google Scholar]
- 27.Klampfer L, Huang J, Kaler P, Sasazuki T, Shirasawa S, Augenlicht L. STAT1-independent inhibition of cyclooxygenase-2 expression by IFNγ; a common pathway of IFNγ-mediated gene repression but not gene activation. Oncogene. 2007;26:2071–2081. doi: 10.1038/sj.onc.1210015. [DOI] [PubMed] [Google Scholar]
- 28.Colgan SP, Morales V, Madara JL, Polischuk JE, Balk SP, Blumberg RS. IFN-γ modulates CD1d surface expression on intestinal epithelia. Am J Physiol. 1996;271:C276–C283. doi: 10.1152/ajpcell.1996.271.1.C276. [DOI] [PubMed] [Google Scholar]
- 29.Gobin SJ, van Zutphen M, Woltman AM, van den Elsen PJ. Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element. J Immunol. 1999;163:1428–1434. [PubMed] [Google Scholar]
- 30.Yang Y, Chu W, Geraghty DE, Hunt JS. Expression of HLA-G in human mononuclear phagocytes and selective induction by IFN-γ. J Immunol. 1996;156:4224– 4231. [PubMed] [Google Scholar]
- 31.Lefebvre S, Moreau P, Guiard V, Ibrahim EC, Adrian-Cabestre F, Menier C, Dausset J, Carosella ED, Paul P. Molecular mechanisms controlling constitutive and IFN-γ-inducible HLA-G expression in various cell types. J Reprod Immunol. 1999;43:213–224. doi: 10.1016/s0165-0378(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 32.Wainwright SD, Biro PA, Holmes CH. HLA-F is a predominantly empty, intracellular, TAP-associated MHC class Ib protein with a restricted expression pattern. J Immunol. 2000;164:319–328. doi: 10.4049/jimmunol.164.1.319. [DOI] [PubMed] [Google Scholar]
- 33.Barrett DM, Gustafson KS, Wang J, Wang SZ, Ginder GD. A GATA factor mediates cell type-restricted induction of HLA-E gene transcription by γ interferon. Mol Cell Biol. 2004;24:6194– 6204. doi: 10.1128/MCB.24.14.6194-6204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Ye L, Christianson G, Yang J, Roopenian DC, Zhu X. NF-κB signaling regulates the functional expression of MHC class I-related neonatal Fc receptor for IgG via intronic binding sequences. J Immunol. 2007;179:2999–3011. doi: 10.4049/jimmunol.179.5.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Peng J, Chen D, Liu X, Ye L, Iijima H, Kadavil K, Lencer WI, Blumberg RS. Calnexin and ERp57 facilitate the assembly of the neonatal Fc receptor for IgG with β2-microglobulin in the endoplasmic reticulum. J Immunol. 2005;175:967–976. doi: 10.4049/jimmunol.175.2.967. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Peng J, Raychowdhury R, Nakajima A, Lencer WI, Blumberg RS. The heavy chain of neonatal Fc receptor for IgG is sequestered in the endoplasmic reticulum by forming oligomers in the absence of β2m association. Biochem J. 2002;367:703–714. doi: 10.1042/BJ20020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen VT, Benveniste EN. IL-4-activated STAT-6 inhibits IFN-γ-induced CD40 gene expression in macrophages/microglia. J Immunol. 2000;165:6235– 6243. doi: 10.4049/jimmunol.165.11.6235. [DOI] [PubMed] [Google Scholar]
- 38.Pongratz G, McAlees JW, Conrad DH, Erbe RS, Haas KM, Sanders VM. The level of IgE produced by a B cell is regulated by norepinephrine in a p38 MAPK- and CD23-dependent manner. J Immunol. 2006;177:2926–2938. doi: 10.4049/jimmunol.177.5.2926. [DOI] [PubMed] [Google Scholar]
- 39.Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Ravazzolo R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. BioTechniques. 2000;29:1012–1017. doi: 10.2144/00295st02. [DOI] [PubMed] [Google Scholar]
- 40.Dickinson BL, Badizadegan K, Wu Z, Ahouse JC, Zhu X, Simister NE, Blumberg RS, Lencer WI. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J Clin Invest. 1999;104:903–911. doi: 10.1172/JCI6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy KM, Yoong Y, Simister NE. Bidirectional transcytosis of IgG by the rat neonatal Fc receptor expressed in a rat kidney cell line: a system to study protein transport across epithelia. J Cell Sci. 2000;113:1277–1285. doi: 10.1242/jcs.113.7.1277. [DOI] [PubMed] [Google Scholar]
- 42.Iozzo RV, Hassell JR. Identification of the precursor protein for the heparan sulfate proteoglycan of human colon carcinoma cells and its post-translational modifications. Arch Biochem Biophys. 1989;269:239–249. doi: 10.1016/0003-9861(89)90105-7. [DOI] [PubMed] [Google Scholar]
- 43.Chin Y, Kitagawa M, Kuida K, Flavell R, Fu X. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol. 1997;17:5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins: comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675– 6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 45.Chang YJ, Holtzman MJ, Chen CC. Interferon-γ-induced epithelial ICAM-1 expression and monocyte adhesion: involvement of protein kinase C-dependent c-Src tyrosine kinase activation pathway. J Biol Chem. 2002;277:7118–7126. doi: 10.1074/jbc.M109924200. [DOI] [PubMed] [Google Scholar]
- 46.Ma Z, Qin H, Benveniste EN. Transcriptional suppression of matrix metalloproteinase-9 gene expression by IFN-γ and IFN-β: critical role of STAT-13. J Immunol. 2001;167:5150–5159. doi: 10.4049/jimmunol.167.9.5150. [DOI] [PubMed] [Google Scholar]
- 47.Ma Z, Chang MJ, Shah RC, Benveniste EN. Interferon-γ-activated STAT-1α suppresses MMP-9 gene transcription by sequestration of the coactivators CBP/p300. J Leukocyte Biol. 2005;78:515–523. doi: 10.1189/jlb.0205112. [DOI] [PubMed] [Google Scholar]
- 48.Loman S, Radl J, Jansen HM, Out TA, Lutter AR. Vectorial transcytosis of dimeric IgA by the Calu-3 human lung epithelial cell line: up-regulation by IFN-γ. Am J Physiol. 1997;272:L951–L958. doi: 10.1152/ajplung.1997.272.5.L951. [DOI] [PubMed] [Google Scholar]
- 49.Madara JL, Stafford J. Interferon-γ directly affects barrier function of cultured intestinal epithelial monolayers. J Clin Invest. 1989;83:724–727. doi: 10.1172/JCI113938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geng YJ, Hansson GK. Interferon-γ inhibits scavenger receptor expression and foam cell formation in human monocyte-derived macrophages. J Clin Invest. 1992;89:1322–1330. doi: 10.1172/JCI115718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkataraman C, Leung S, Salvekar A, Mano H, Schindler U. Repression of IL-4-induced gene expression by IFN-γ requires Stat1 activation. J Immunol. 1999;162:4053– 4061. [PubMed] [Google Scholar]
- 52.Sharma B, Iozzo RV. Transcriptional silencing of perlecan gene expression by interferon-γ. J Biol Chem. 1998;273:4642– 4646. doi: 10.1074/jbc.273.8.4642. [DOI] [PubMed] [Google Scholar]
- 53.Kominsky SL, Hobeika AC, Lake FA, Torres BA, Johnson HM. Down-regulation of neu/HER-2 by interferon-γ in prostate cancer cells. Cancer Res. 2000;60:3904–3908. [PubMed] [Google Scholar]
- 54.Elser B, Lohoff M, Kock S, Giaisi M, Kirchhoff S, Krammer PH, Li-Weber M. IFN-γ represses IL-4 expression via IRF-1 and IRF-2. Immunity. 2002;17:703–712. doi: 10.1016/s1074-7613(02)00471-5. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y, Wang L, Buttice G, Sengupta PK, Smith BD. Interferon γ repression of collagen (COL1A2) transcription is mediated by the RFX5 complex. J Biol Chem. 2003;278:49134– 49144. doi: 10.1074/jbc.M309003200. [DOI] [PubMed] [Google Scholar]
- 56.Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J Immunol. 2006;176:905–913. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- 57.VanDeusen JB, Shah MH, Becknell B, Blaser BW, Ferketich AK, Nuovo GJ, Ahmer BM, Durbin J, Caligiuri MA. STAT-1-mediated repression of monocyte interleukin-10 gene expression in vivo. Eur J Immunol. 2006;36:623– 630. doi: 10.1002/eji.200535241. [DOI] [PubMed] [Google Scholar]
- 58.Kelchtermans H, Struyf S, De Klerck B, Mitera T, Alen M, Geboes L, Van Balen M, Dillen C, Put W, Gysemans C, et al. Protective role of IFN-γ in collagen-induced arthritis conferred by inhibition of mycobacteria-induced granulocyte chemotactic protein-2 production. J Leukocyte Biol. 2007;81:1044–1053. doi: 10.1189/jlb.0806486. [DOI] [PubMed] [Google Scholar]
- 59.Schuerer-Maly CC, Eckmann L, Kagnoff MF, Falco MT, Maly FE. Colonic epithelial cell lines as a source of interleukin-8: stimulation by inflammatory cytokines and bacterial lipopolysaccharide. Immunology. 1994;81:85–91. [PMC free article] [PubMed] [Google Scholar]
- 60.Brand K, Mackman N, Curtiss LK. Interferon-γ inhibits macrophage apolipoprotein E production by posttranslational mechanisms. J Clin Invest. 1993;91:2031–2039. doi: 10.1172/JCI116425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang MC, Liu HP, Demchik LL, Zhai YF, Yang DJ. LIGHT sensitizes IFN-γ-mediated apoptosis of HT-29 human carcinoma cells through both death receptor and mitochondria pathways. Cell Res. 2004;14:117–124. doi: 10.1038/sj.cr.7290210. [DOI] [PubMed] [Google Scholar]
- 62.Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen TM, Rose DW, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatterjee-Kishore M, van den Akker F, Stark GR. Association of STATs with relatives and friends. Trends Cell Biol. 2000;10:106–111. doi: 10.1016/s0962-8924(99)01709-2. [DOI] [PubMed] [Google Scholar]
- 64.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu XS, Ting JP. A 36-amino-acid region of CIITA is an effective inhibitor of CBP: novel mechanism of γ interferon-mediated suppression of collagen a(2)(I) and other promoters. Mol Cell Biol. 2001;21:7078–7088. doi: 10.1128/MCB.21.20.7078-7088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Higashi K, Inagaki Y, Suzuki N, Mitsui S, Mauviel A, Kaneko H, Nakatsuka I. Y-box-binding protein YB-1 mediates transcriptional repression of human α 2(I) collagen gene expression by interferon-γ. J Biol Chem. 2003;278:5156–5162. doi: 10.1074/jbc.M208724200. [DOI] [PubMed] [Google Scholar]
- 67.Kato H, Kato R, Fujihashi K, McGhee JR. Role of mucosal antibodies in viral infections. Curr Top Microbiol Immunol. 2001;260:201–228. doi: 10.1007/978-3-662-05783-4_11. [DOI] [PubMed] [Google Scholar]
- 68.Piskurich JF, Youngman KR, Phillips KM, Hempen PM, Blanchard MH, France JA, Kaetzel CS. Transcriptional regulation of the human polymeric immunoglobulin receptor gene by interferon-γ. Mol Immunol. 1997;34:75–91. doi: 10.1016/s0161-5890(96)00079-x. [DOI] [PubMed] [Google Scholar]
- 69.Akilesh S, Petkova S, Sproule TJ, Shaffer DJ, Christianson GJ, Roopenian DC. The MHC class I-like Fc receptor promotes humorally mediated autoimmune disease. J Clin Invest. 2004;113:1328–1333. doi: 10.1172/JCI18838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petkova SB, Akilesh S, Sproule TJ, Christianson GJ, Al Khabbaz H, Brown AC, Presta LG, Meng YG, Roopenian DC. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: potential application in humorally mediated autoimmune disease. Int Immunol. 2006;18:1759–1769. doi: 10.1093/intimm/dxl110. [DOI] [PubMed] [Google Scholar]
- 71.Park-Min KH, Serbina NV, Yang W, Ma X, Krystal G, Neel BG, Nutt SL, Hu X, Ivashkiv LB. FcγRIII-dependent inhibition of interferon-γ responses mediates suppressive effects of intravenous immune globulin. Immunity. 2007;26:67–78. doi: 10.1016/j.immuni.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 72.Zhang J. Yin and yang interplay of IFN-γ in inflammation and autoimmune disease. J Clin Invest. 2007;117:871– 873. doi: 10.1172/JCI31860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gustafson KS, Ginder GD. Interferon-γ induction of the human leukocyte antigen-E gene is mediated through binding of a complex containing STAT1α to a distinct interferon-γ-responsive element. J Biol Chem. 1996;271:20035–20046. doi: 10.1074/jbc.271.33.20035. [DOI] [PubMed] [Google Scholar]
- 74.Chen QY, Jackson N. Human CD1D gene has TATA boxless dual promoters: an SP1-binding element determines the function of the proximal promoter. J Immunol. 2004;172:5512–5521. doi: 10.4049/jimmunol.172.9.5512. [DOI] [PubMed] [Google Scholar]